Research Interests
Laboratories for Innovation in Global Health Technologies (LIGHT)
The Haselton Laboratory is an active participant in LIGHT. This is a cross-disciplinary initiative between the College of Arts and Science, the School of Engineering, and the School of Medicine. LIGHT aims to accelerate the development of global health technologies in a multidisciplinary research environment addressing the needs of patients and physicians in low-resource settings. LIGHT collaborations between researchers, users, and health practitioners nurture a growing portfolio of technologies that are at different stages of development around the world. These include the development of patient sampling devices, diagnostics for infectious diseases (malaria, TB, HIV, respiratory, and enteric) and maternal health, adherence diagnostics for treatment management, epidemiological surveillance systems, rapid prototyping tools and low-cost training tools. Our model for technology development follows ASSURED: affordable, sensitive, specific, user-friendly, rapid and robust, equipment free and deliverable to end-users, in order to create high quality products that are appropriate and affordable to the 4 billion persons at the base of the global healthcare pyramid.
Adaptive PCR enables point-of-care molecular diagnostic testing using mirror image DNA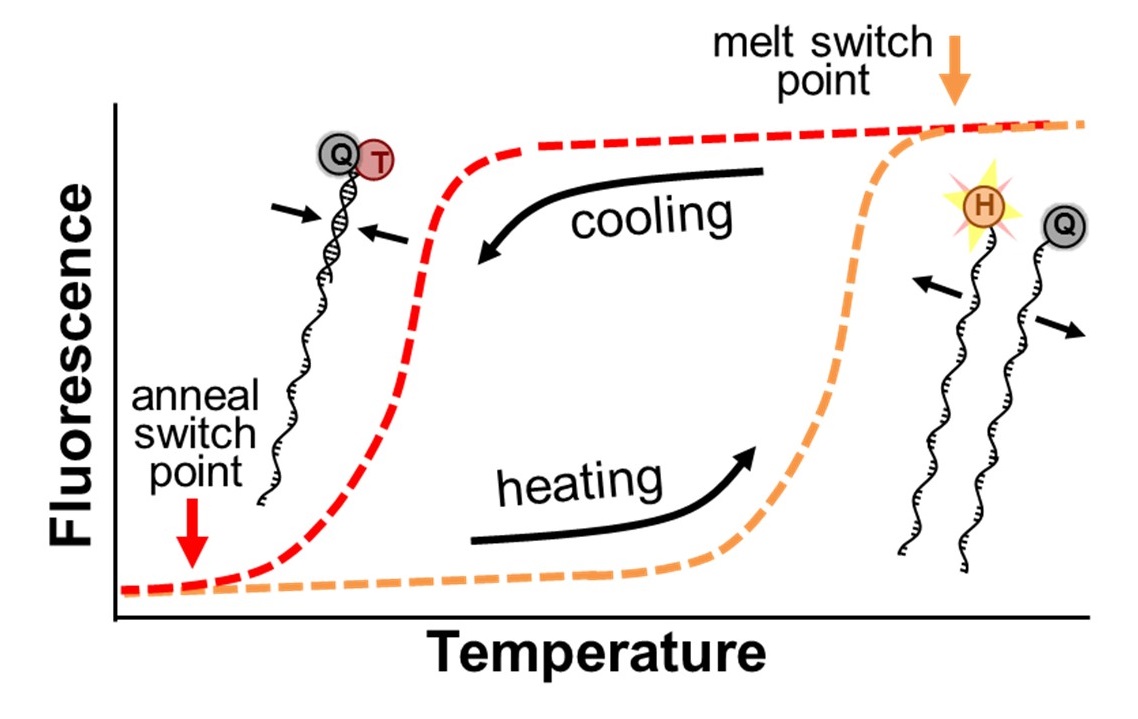
We have invented a new technology that we call Adaptive PCR that lowers the barrier for performing PCR and promises to expand access to this valuable diagnostic tool. The breakthrough is based on the use of synthetic left-handed DNA (L-DNA) molecules to more precisely monitor and control the critical thermal cycling steps. The L-DNAs are designed to represent the DNAs in the PCR reaction that are present in such low concentrations that they cannot be detected. By optically monitoring the interactions of the L-DNAs, Adaptive PCR adjusts cycling conditions to match the ideal state of the reactants, rather than cycling between inflexible thermal set points that are estimated to be ideal. Further details about the technology can be found in our recent publication (Adams, et al. Analytical Chemistry, 2016). The use of L-DNAs simplifies the PCR process in two critical ways. First, because the L-DNAs mimic the actual DNA in the reaction, the approach compensates for the presence of salts or other small molecules that inhibit standard PCR. This enables Adaptive PCR to be performed directly in patient samples without the need for the technical processing to purify DNA. Second, because Adaptive PCR more directly monitors the critical DNA interactions during the reaction rather than relying of indirect temperature measurements, the instrument does not require the upkeep and operation constraints to maintain the thermal calibrations. This enables the development of less expensive, simpler, and more robust thermal cycling instruments. These critical advantages of Adaptive PCR will enable high quality and inexpensive point-of-care diagnostics.
Applying Adaptive PCR to develop a point-of-care RT-PCR system to inform COVID-19 and respiratory illness decisions
This project develops a point-of-care device capable of distinguishing between a variety of respiratory illnesses. In light of the current COVID-19 pandemic, it is important to distinguish between viruses like SARS-CoV-2 and influenza, which can present with similar symptoms yet require distinct management strategies, as well as other respiratory viruses and bacterial illness that can be treated with antibiotics. We propose a simplified methodology through Adaptive RT-PCR, creating diagnostics for SARS-CoV-2 (the virus that causes COVID-19) and other respiratory pathogens without RNA extraction. As a collaboration between biomedical engineers and a COVID-19 diagnostic laboratory, we seek to develop a workflow and instrument that are simple-to-use, cost-effective, and suitable for point-of-care settings, tools that can rapidly inform treatment and management strategies. To achieve this goal we will develop multiplexed amplification reagents to create a sensitive and specific respiratory panel that detects SARS-CoV-2, four other viruses, two bacteria, and one control target. Ultimately, we will design and fabricate a self-contained Adaptive RT-PCR instrument suitable for point-of-care settings, while validating this system using characterized human specimens in a CLIA-accredited lab environment. Completion of this project will result in a novel point-of-care tool for both the established and emerging respiratory infections that threaten public health, facilitating rapid treatment, follow-up, infection prevention, and epidemiologic containment.
Applying Adaptive PCR to monitor HIV viral load and provide drug resistance testing in resource constrained settings
This project develops a point-of-care technology for use in antenatal care clinics in resource-constrained settings to identify viremic pregnant women and screen for the presence of genotypic drug resistant mutations. Once identified, their HIV antiretroviral treatment will be adjusted to maintain durable viral suppression to minimize HIV transmission to their unborn infant. This will be tested and compared to the current standard of care which requires sending samples to a reference laboratory and waiting several weeks to obtain results at the point-of-care. This information empowers the treating physicians to more quickly optimize maternal antiretroviral treatment regimens to reduce the risk of mother to child HIV transmission resulting in significant maternal-child health improvements.
“Connected” Diagnostic Tests
The rapid expansion of mobile-phone usage and internet connectivity has the potential to substantially change the way that healthcare is provided, and received, throughout the world. The broad utility of mobile-health (mHealth) tools is reflected in the diversity of the applications presented in literature, including: SMS reminders for treatments, epidemiological modeling, supply-chain management, and point-of-care diagnostic test evaluation. Our efforts have focused on the use of mobile-phones to improve quality control of test analysis (Scherr et al, Scientific Reports 2016) and quality control (Scherr et al, Lab on a Chip 2017). This technology blends point-of-care device fabrication with mobile-phone and server-side software development to improve healthcare around the world.
Low resource extraction and processing of biological samples using surface tension valves.
This Bill & Melinda Gate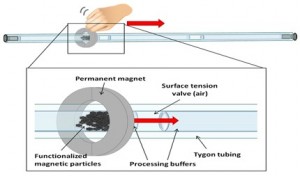 s Foundation funded project (with David Wright in Chemistry and Ray Mernaugh in Biochemistry) is a sample collection, concentration and preparation component for integration with downstream detection components to form a general diagnostic platform suitable for low resource environments. This self-contained processing device captures targets of interest from complex biological matrices on the surface of magnetic carrier beads which convey the biomarker targets through sequential extraction solutions to concentrate them and reduce interferents of downstream biomarker detector performance. The device has advantages over existing extraction technologies, mainly it that it can be implemented to: (1) require little or no power, (2) be suitable for an unskilled user, (3) have rapid time-to-extraction, (4) be low cost, and (5) be adaptable to multiple downstream detection designs. To our knowledge there is no currently available point-of-care extraction device that has the same level of simplicity, convenience, low cost and ease-of-use.
s Foundation funded project (with David Wright in Chemistry and Ray Mernaugh in Biochemistry) is a sample collection, concentration and preparation component for integration with downstream detection components to form a general diagnostic platform suitable for low resource environments. This self-contained processing device captures targets of interest from complex biological matrices on the surface of magnetic carrier beads which convey the biomarker targets through sequential extraction solutions to concentrate them and reduce interferents of downstream biomarker detector performance. The device has advantages over existing extraction technologies, mainly it that it can be implemented to: (1) require little or no power, (2) be suitable for an unskilled user, (3) have rapid time-to-extraction, (4) be low cost, and (5) be adaptable to multiple downstream detection designs. To our knowledge there is no currently available point-of-care extraction device that has the same level of simplicity, convenience, low cost and ease-of-use.
Enhancement of lateral flow assays for molecular detection of biomarkers of infection
While remarkable progress toward WHO recommendations for clinical confirmation of malaria has been made, passive case detection methods relying on self-referred, symptomatic individuals significantly under-estimates infection rates and do not adequately support goals toward eradication. There is an unmet need for new biomarker detection designs that increase sensitivity yet retain simplicity and easy interpretation of the user-centric lateral flow rapid diagnostic tests currently available. The most important performance criterion is sensitivity, otherwise the tool fails to support useful operational scenarios such as mass and focal testing and treatment. Existing laboratory-based designs using PCR and isothermal reactions meet this criterion but their operation is complex.
Our goal is to develop and evaluate an electricity-free nucleic acid diagnostic test. Our approach is based on a single-use cassette the size of current, well-excepted lateral flow immunoassays (LFIAs) for protein biomarker detection. This diagnostic process will require three steps with an optical read-out of the results within a total of 45-50 minutes. Blood from a finger prick is introduced into the interior of the cassette via the absorbent filter where it is mixed with pre-arrayed magnetic beads and reagents for further biomarker concentration and delivery into an isothermal reaction chamber. Biomarker transport and isothermal amplification is achieved by insertion into a reusable housing which couples to the cassette via magnetic and thermal interfaces. Final reaction product is transferred to a lateral flow strip for visual readout.
Coffee Ring Diagnostics for Malaria.
In collaboration with the Wright lab 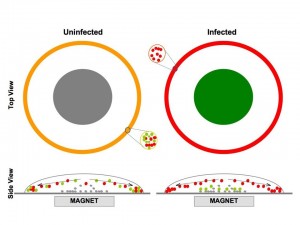 (Chemistry) the goal of this project is to develop a simple, low-cost diagnostic test for malaria infection suitable for locations that lack electricity, refrigeration and highly trained technicians. The idea is based on the phenomenon that causes coffee-ring stains on the kitchen counter: When a liquid like coffee that contains a suspension of fine particles evaporates from a flat surface, the particles tend to accumulate along the outer edge. The proposed malaria test consists of two liquids and specially treated glass slides. Liquid A is mixed with the blood sample. A drop of the mixture is placed on the glass slide and left to dry. Once it has dried, the slide is washed with liquid B. If the washing reveals a purple ring, the person is infected. If it washes clean, then the person is not.
(Chemistry) the goal of this project is to develop a simple, low-cost diagnostic test for malaria infection suitable for locations that lack electricity, refrigeration and highly trained technicians. The idea is based on the phenomenon that causes coffee-ring stains on the kitchen counter: When a liquid like coffee that contains a suspension of fine particles evaporates from a flat surface, the particles tend to accumulate along the outer edge. The proposed malaria test consists of two liquids and specially treated glass slides. Liquid A is mixed with the blood sample. A drop of the mixture is placed on the glass slide and left to dry. Once it has dried, the slide is washed with liquid B. If the washing reveals a purple ring, the person is infected. If it washes clean, then the person is not.
Application of Coffee Ring Phenomenon to Tuberculosis Diagnostics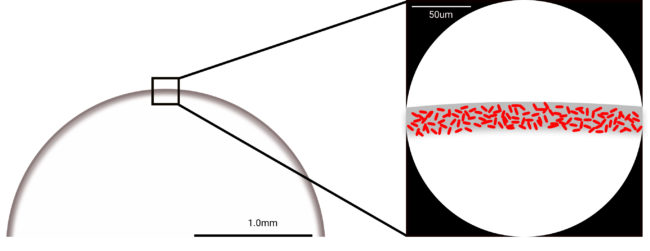
The goal of this NIH funded project (with David Wright in Chemistry, Jonathan Blackburn, and Keertan Dheda at the University of Cape Town, South Africa) is to develop a 100-fold more sensitive Tuberculosis diagnostic based on magnetic concentration and coffee ring formation. Microscopic examination of sputum smear is the most accessible method for detection of Tuberculosis. Unfortunately, sputum smear microscopy has a current limit of detection of 10,000 bacilli per milliliter of sputum which prevents early detection and allows continued spread of infection. This project will develop simple methods for improving the limit of detection of existing bright field microscopy to 100 bacilli/ml. Our approach is based on the magnetic concentration of bacilli followed by high density deposition onto a standard microscopy slide using the coffee ring phenomenon. A variation of this approach will be developed to detect drug resistant tuberculosis. These designs will be packaged into a simple kit format for initial clinical testing of retrospective sputum samples in a laboratory as well as in rural South Africa.
Development of DNA Logic Operations for Viral Diagnostics.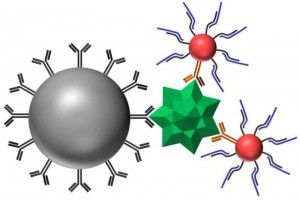
The goal of this NIH funded project (with David Wright in Chemistry, Jim Crowe in the Medical Center) is to develop and evaluate a new paradigm in virus detection. The approach is based on a combination of nanoparticle surface chemistry, tag-specific DNA sequences, and DNA ligation logical AND operators. Components of this project have been used to amplify the impact of DNA biomarkers on detection signal.
Multispectral quantum dot-based retinal imaging.
The unique optical properties of quantum dots offer the potential for new optical methods to study molecular events. This NIH funded project seeks to develop a platform for the real-time, in vivo analysis of multiple cellular and molecular events in the in vivo retina using quantum dot nanocrystals and retinal fluorescence microscopy.
Furthermore, we also seek to use novel retinal imaging agents based on quantum dots as a tool to predict the progression of atherosclerotic lesions in coronary arteries.
Collaborators
The Vanderbilt research environment is defined by collaboration. Only in a select few institutions across the nation can you actually toss a rock from a window in the BME department and literally break a window in the medical center (and even in a number of other science and engineering departments!). No rocks are involved, but below is a sampling of the people who we either currently collaborate with or have collaborated with especially closely.
- Nicholas M. Adams, Department of Biomedical Engineering, Vanderbilt University
- Muktar H. Aliyu, Departments of Medicine and Health Policy, Institute for Global Health, Vanderbilt University Medical Center [link]
- David Aronoff, Division of Infectious Diseases, Department of Medicine, Department of Pathology, Microbiology, & Immunology, Vanderbilt University Medical Center [link]
- Jonathan Blackburn, Institute of Infectious Diseases and Molecular Medicine, University of Cape Town, South Africa [link]
- David Cliffel, Department of Chemistry, Vanderbilt University [link]
- James Crowe, Jr., Departments of Pediatric Infectious Disease and Pathology, Microbiology & Immunology, Vanderbilt University Medical Center [link]
- Keertan Dheda, Division of Pulmonology, University of Cape Town, South Africa [link]
- Suman Das, Division of Adult Infectious Diseases, Department of Medicine, Vanderbilt University Medical Center [link]
- Douglas C. Heimburger, Department of Medicine, Institute for Global Health, Vanderbilt University Medical Center [link]
- Mindy Leelawong, Department of Biomedical Engineering, Vanderbilt University
- Ray L. Mernaugh, Department of Biochemistry, Vanderbilt University Medical Center [link]
- William Pao, Departments of Medicine, Cancer Biology, Pathology, Microbiology & Immunology, Vanderbilt University Medical Center [link]
- John S. Penn, Department of Opthalmology, Vanderbilt Eye Institute [link]
- Sandy J. Rosenthal, Department of Chemistry, Vanderbilt University [link]
- Patricia K. Russ, Department of Biomedical Engineering, Vanderbilt University
- Thomas F. Scherr, Department of Chemistry, Vanderbilt University
- Douglas C. Schmidt, Computer Science & Engineering, Vanderbilt University [link]
- Jonathan E. Schmitz, Department of Pathology, Microbiology, and Immunology, Vanderbilt University [link]
- Phil Thuma, Senior Scientific Advisor, Macha Research Trust, Macha, Zambia [link]
- Sharon M. Weiss, Departments of Electrical Engineering and Physics, Vanderbilt University [link]
- C. William Wester, Division of Adult Infectious Diseases, Department of Medicine, Institute for Global Health, Vanderbilt University School of Medicine [link]
- David W. Wright, Department of Chemistry, Vanderbilt University [link]
Connect with Vanderbilt
©2024 Vanderbilt University ·
Site Development: University Web Communications