Research
Lab Publications:
Lab publications can be found at Professor Noble’s NCBI Biblography.
Datasets from lab publications are being made available upon request for NIH-funded studies conducted by the BAGL lab. Please contact Professor Noble by email with such requests. You will be provided with a Box link to download the data spreadsheet. By downloading the data you agree to (1) only use the data for educational or research purposes and not for commercial purposes; (2) allow the BAGL lab to keep record of your download of the data for grant reporting purposes with potential public release of information regarding the institution(s) where you are affiliated; (3) not share the datasets directly with any other individual, but rather direct others to this website to request their own download link; and (4) acknowledge the grants that supported creation of the datasets when the data is used in a publication or presentation. The grant numbers that need to be cited are listed in the downloaded data spreadsheet.
Research Projects:
Digital twins for cochlear implants
Funded by grant R01DC014037 from the NIDCD.
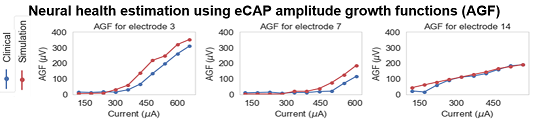
We are developing comprehensive patient-specific cochlear implant electro-anatomical models. The models could permit neural activation simulation, treatment planning, and customized processor programming optimization strategies that have never before been possible.
Our processing pipeline (shown below) involves: (1) localizing the patient cochlear anatomy and electrodes in CT, (2) estimating patient specific tissue resistivity and voltage maps using electric field models, and (3) estimating neural fiber health and activation patterns using neural activation models.
An example of neural activation patterns that we estimate for a CI patient is shown in the following video.
Patient-specific high resolution tissue resistivity maps for cochlear implant electric field modeling
We are currently developing and validating multiple approaches for constructing high resolution electric field models for patient-specific cochlear implant simulation. For example, we are evaluating custom deep learning architectures based on cycle consistent generative adverserial networks (Cycle GAN) to estimate electric fields from tissue label maps. Our network is trained with weak supervision learning with custom, physics-based loss terms. Our proposed method can generate high quality predictions that improve the speed of constructing models.
Ziteng Liu and Jack H. Noble, “Patient-specific electro-anatomical modeling of cochlear implants using deep neural networks,” Proceedings of the SPIE Conference on Medical Imaging, vol. 12034, pp. 12034-14, 2022.
Minh Q. Vu, Erin L. Bratu, Jack H. Noble, “Predicting electrically evoked action potentials in image-based cochlear implants models using ResNet,” Proceedings of the SPIE Conf. on Medical Imaging, vol. 13408-69, 2025.
We are also developing conditional generative adversarial network (cGAN) deep learning techniques to synthesize high resolution μCT from CT images to create high resolution tissue class maps. µCT images offer significantly more detail than CT images, with ~1000x better volume resolution. However, µCT images cannot be acquired in vivo. In this work, we aim to investigate whether a cGAN approach can learn how to produce a very detailed synthetic µCT image using a clinical resolution patient CT scan as input.

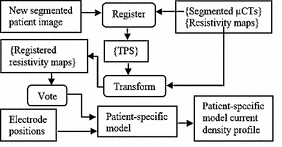 We have also developed traditional atlas-based methods to create patient customized stimulation models. In this approach, we use using high resolution μCTs of cochlear specimens that are non-rigidly registered to patient CT images to facilitate atlas-based tissue resistivity map estimation.
We have also developed traditional atlas-based methods to create patient customized stimulation models. In this approach, we use using high resolution μCTs of cochlear specimens that are non-rigidly registered to patient CT images to facilitate atlas-based tissue resistivity map estimation.
Cakir A., Dawant B.M., Noble J.H., “Development of a microCT-based patient-specific model of the electrically stimulated cochlea,” Lecture Notes in Computer Science – Proceedings of MICCAI, 2017.
Cakir A., Dawant B.M., Noble J.H., “Evaluation of a µCT-based electro-anatomical cochlear implant model,” Proceedings of the 2016 SPIE Conf. on Medical Imaging, vol. 9786, pp. 97860M, 2016.
 Localization and activation simulation of auditory nerve fibers
Localization and activation simulation of auditory nerve fibers
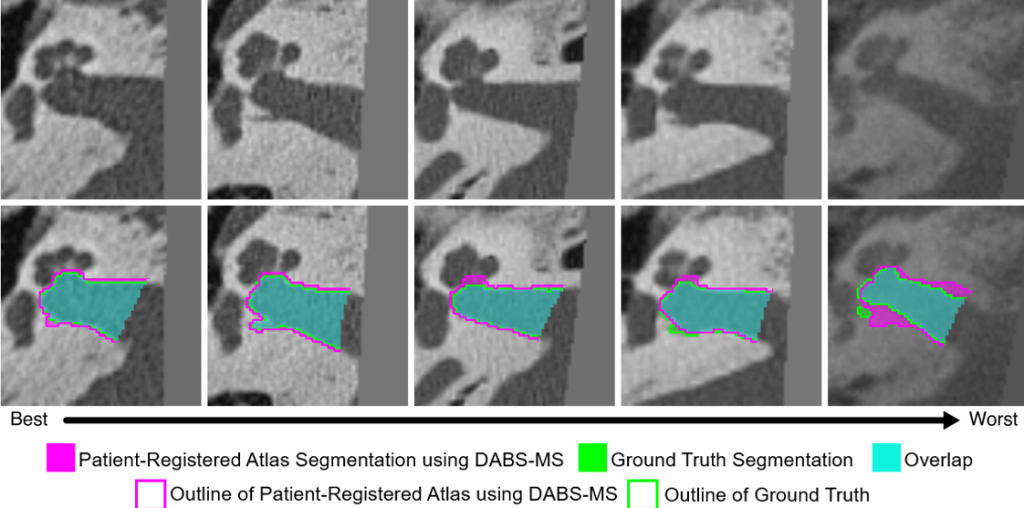
We have developed methods to localize individual auditory nerve fiber bundles within the internal auditory canal and detect nerve fiber health by optimizing health parameters of our neural models so that model simulated physiological measurements match those acquired from the patient’s CI.
These methods provide an unprecedented window into the health of the inner ear, opening the door for studying population variability and intra-subject neural health dynamics.
Hannah G. Mason and Jack H. Noble, “DABS-MS: Deep Atlas-Based Segmentation using Mumford Shah-Functional Self-Supervision,” SPIE Journal of Medical Imaging, vol. 12(4), pp. 055002, 2025.
Z Liu, A Cakir, JH Noble, “Auditory Nerve Fiber Health Estimation Using Patient Specific Cochlear Implant Stimulation Models,” Lecture Notes in Computer Science – Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, vol. 12417, pp 184-194, 2020.
 Image-Guided Cochlear Implant Programming
Image-Guided Cochlear Implant Programming
Cochlear Implants (CIs) restore hearing using an electrode array that is surgically implanted into the cochlea. In recent work, a multidisciplinary team of researchers at Vanderbilt, Drs. Jack Noble, Benoit Dawant, René Gifford, and Robert Labadie, has developed image processing techniques that have permitted accurate detection of the post-operative position of CI electrodes relative to the auditory nerve cells they stimulate (watch video of cochlear implant in CT image). Using these image processing techniques, they have developed the first Image-Guided CI Programming (IGCIP) strategy.The IGCIP strategy is to deactivate CI electrodes that the imaging information suggests create overlapping stimulation patterns (steps of IGCIP system shown in this video). Current studies show that IGCIP leads to significantly better hearing outcomes with CIs. The current goals of this project are to continue IGCIP validation studies and to develop new patient-customized, IGCIP strategies that could provide more objective information to the CI programming process and lead to CI programs that better approximate natural hearing performance.
Noble JH, Labadie RF, Gifford RH, Dawant BM, “Image-guidance enables new methods for customizing cochlear implant stimulation strategies,” IEEE Trans Neural Syst Rehabil Eng. vol. 21(5):820-9, 2013.
Noble JH, Gifford RH, Hedley-Williams AJ, Dawant BM, and , Labadie RF, “Clinical evaluation of an image-guided cochlear implant programming strategy,” Audiology & Neurotology, vol. 19, pp. 400-11, 2014.
Noble J.H., Hedley-Williams A.J., Sunderhaus L.W., Dawant B.M., Labadie R.F., Camarata S.M., Gifford R.H., “Initial results with image-guided cochlear implant programming in children,” Otology & Neurotology 37(2), pp. 69-9, 2016
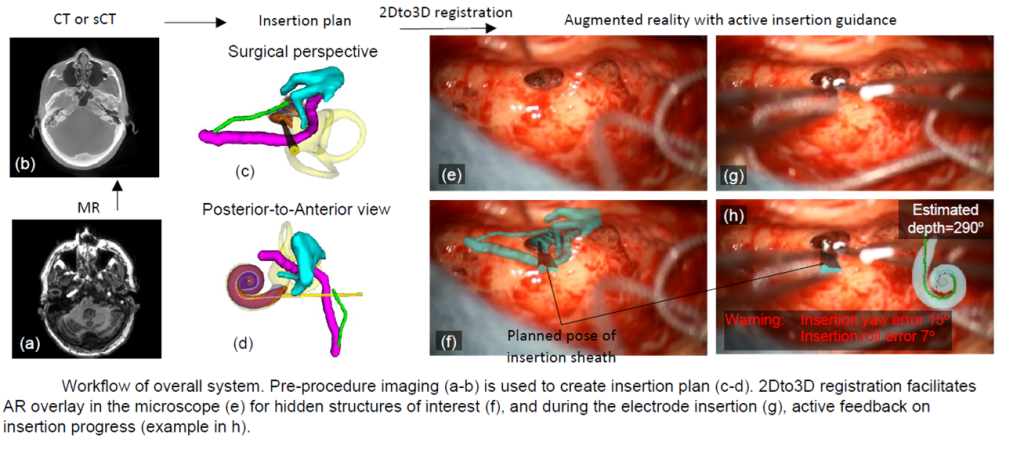
Augmented reality-guided cochlear implant placement
Funded by grant R01DC022099 from the NIDCD.
We are developing methods to aid surgeons in optimizing surgical placement of cochlear implant electrode arrays using intra-operative augmented reality techniques. Optimized array placement leads to less surgical trauma and has been shown to be associated with improved hearing outcomes.
Image-guided Cochlear Implant Insertion Techniques
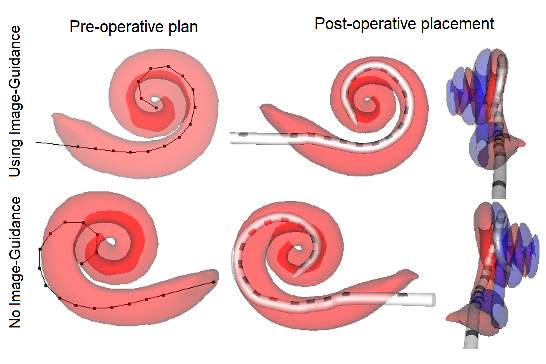
Cochlear implant (CI) electrode arrays are treated as one size fits most, despite the fact that sub-optimal placement of the array is the norm. In this study, we are investigating the use of patient-customized insertion plans, created by analyzing pre-operative CTs. Temporal bone studies show patient customized cochlear implant insertion techniques achieve better positioning of electrode arrays. We are currently evaluating whether these techniques improve electrode positioning in patients.
Jack Noble and Robert Labadie, “Preliminary results with image-guided cochlear implant insertion techniques,” Otology & Neurotology, vol. 39(7), pp. 922-928, 2018.
Kareem O. Tawfik and Mohammad M.R. Khan (co-first author), Ankita Patro, Miriam R. Smetak, David Haynes, Robert F. Labadie, René H. Gifford, and Jack H. Noble, “Cochlear Implantation of Slim Pre-curved Arrays using Automatic Pre-operative Insertion Plans,” Otology & Neurotology, vol. 46(7), pp. 862-70, 2025.
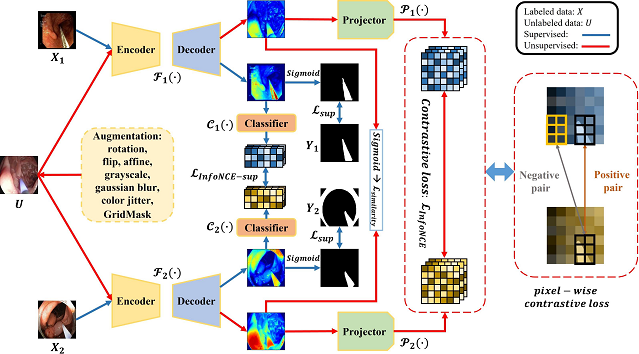
Self-supervised segmentation of cochlear implant insertion tools in surgical videos
Semantic segmentation of tools in surgical microscope video can permit real-time feedback to surgeons regarding adherence to the surgical plan. As manual segmentation of many video frames would be time consuming, we have developed a self-supervised deep learning method to segment cochlear implant insertion tools using the contrastive learning framework shown below.
This contrastive learning framework, which we call “Min-Max Similarity” permits accurately segmenting the CI insertion sheath in real-time and only needs a small number of labelled video frames for training.

A Lou, K Tawfik, X Yao, Z Liu, J Noble, “Min-Max Similarity: A Contrastive Semi-Supervised Deep Learning Network for Surgical Tools Segmentation,” IEEE Trans. on Medical Imaging, vol. 42(10), pp. 2832-2841, 2023
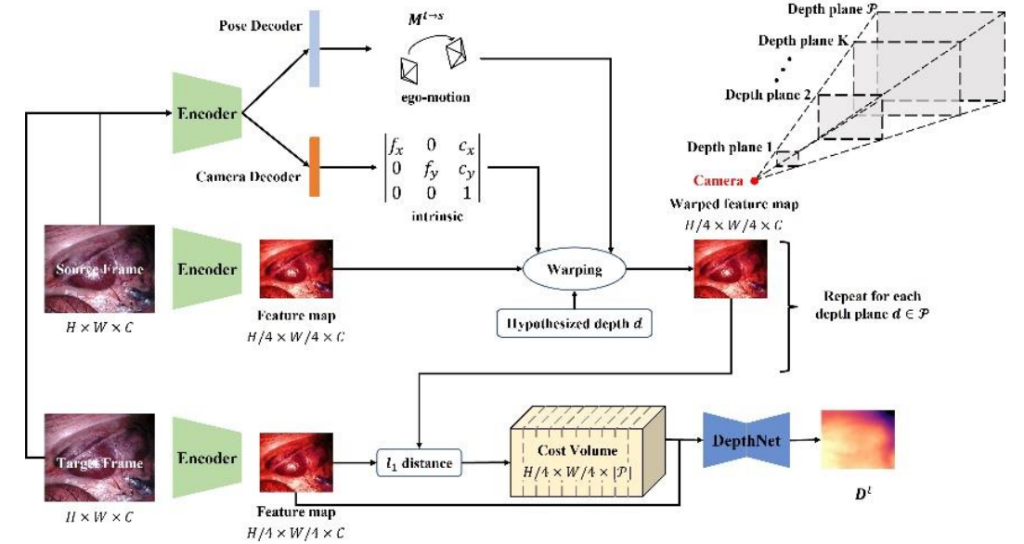
Simultaneous camera self localization and scene depth mapping for surgical microscopes with unknown intrinsic parameters using deep learning
Deep learning techniques have been designed to estimate the microscope position in the surgical scene, the intrinsic parameters of the microscope, and a depth map of the surgical environment simultaneously. The technology relies on self-supervised training strategies and outperforms many state-of-the-art methods.
Ange Lou and Jack H Noble, “WS-SfMLearner: Self-supervised Monocular Depth and Ego-motion Estimation on Surgical Videos with Unknown Camera Parameters,” Journal of Medical Imaging, vol. 12(2), pp. 025003, 2025.
Ange Lou, Yamin Li, Yike Zhang, Jack H. Noble, “Surgical depth anything: Depth estimation for surgical scenes using foundation models,” Proceedings of the SPIE Conf. on Medical Imaging, vol. 13408-10, 2025.
Computer Vision-based Cochlear Implant Insertion Tool Localization for Augmented Reality Surgery
We are developing computer vision-based techniques for automatically localizing cochlear implant insertion tools in real-time. This will permit providing active feedback to the surgeon using an augmented reality interface to assist with following the optimal electrode array insertion plan. If successful, this project will permit improving the electro-neural interface and hearing outcomes with cochlear implants.
Serena Abraham, Jason E. Mitchell, Robert F. Labadie, Jack H. Noble, “Tool pose estimation in cochlear implant surgery using monocular microscopy,” Proceedings of the SPIE Conf. on Medical Imaging, vol. 13408-3, 2025.
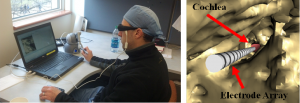 Cochlear Implant Electrode Array Placement Simulator
Cochlear Implant Electrode Array Placement Simulator
The primary goals when placing the CI electrode array are to fully insert the array into the cochlea while minimizing trauma to the cochlea. Studying the relationship between surgical outcome and various surgical techniques has been difficult since trauma and electrode placement are generally unknown without histology. Our group has created a CI placement simulator that combines an interactive 3D visualization environment with a haptic-feedback-enabled controller. Surgical techniques and patient anatomy can be varied between simulations so that outcomes can be studied under varied conditions. With this system, we envision that through numerous trials we will be able to statistically analyze how outcomes relate to surgical techniques.
Turok RL, Labadie RF, Wanna GB, Dawant BM, Noble JH, “Cochlear implant simulator for surgical technique analysis,” Proceedings of the SPIE Conf. on Medical Imaging, 9036, 903619, 2014.
Image segmentation for ear anatomy
Self-supervised Registration and Segmentation of the Ossicles using only one labelled dataset
Recently published image segmentation methods that leverage deep learning usually rely on a large number of manually definedvground truth labels for training. However, it is a laborious and time-consuming task to prepare the dataset. We propose a novel technique using a self-supervised 3D-UNet that produces a dense deformation field between an atlas and a target image that can be used for atlas-based segmentation of the ossicles. Our results show that our method outperforms traditional image segmentation methods and generates a more accurate boundary around the ossicles compared to traditional methods.
Yike Zhang, Jack H. Noble, “Self-supervised registration and segmentation on ossicles with a single ground truth label,” Proceedings of the SPIE Conference on Medical Imaging, vol. 12466, pp. 12466-80, 2023 (in press)
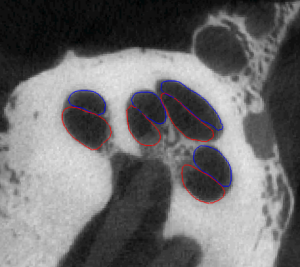 Statistical Shape Model-Based Automatic Segmentation of Intra-Cochlear Anatomy
Statistical Shape Model-Based Automatic Segmentation of Intra-Cochlear Anatomy
Segmentation of intracochlear structures (Scala Tympani, Scala Vestibuli and Media, and Modiolus) would aid surgical guidance and post-surgery analysis of electrode positioning, which could help improve placement outcomes and lead to better hearing outcomes. However, intracochlear structures are too small to be seen in conventional in vivo imaging, thus traditional segmentation techniques are inadequate. In this work, we circumvent this problem by creating a weighted active shape model with micro CT (μCT) scans of the cochlea acquired ex-vivo (see video of Scala Tympani=Red, Scala Vestibuli=Blue, and Modiolus=green manually segmented in μCT). We then use this model to segment conventional CT scans. The model is fit to the partial information available in the conventional scans and used to estimate the position of structures not visible in these images. Quantitative evaluation of our method, made possible by the set of μCTs, results in Dice similarity coefficients averaging 0.77 and surface errors of 0.15 mm. A video of the shapes in the model is shown here (Red=ST, Blue=SV/SM, Green=Promontory).
Noble, J.H., Labadie, R.F., Majdani, O., Dawant, B.M., “Automatic segmentation of intra-cochlear anatomy in conventional CT”, IEEE Trans. on Biomedical. Eng., Vol. 58, No. 9, pp. 2625-32, 2011.
Noble, J.H., Gifford, R.H., Labadie, R.F., Dawant, B.M., 2012, “Statistical Shape Model Segmentation and Frequency Mapping of Cochlear Implant Stimulation Targets in CT,” N. Ayache et al. (Eds.): MICCAI 2012, Part II, LNCS 7511, pp. 421-428. 2012.
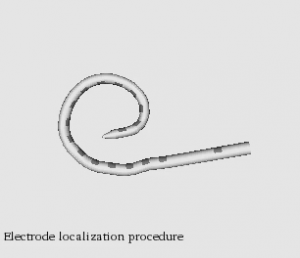 Automatic graph-based localization of cochlear implant electrodes in CT
Automatic graph-based localization of cochlear implant electrodes in CT
To facilitate clinical translation of IGCIP, we are developing fully automated image analysis algorithms that permit accurately locating CI electrodes in post-implantation CT images (video of CI electrodes in CT).We have developed a novel graph-based method for localizing electrode arrays in CTs that is effective for various implant models. It relies on a novel algorithm for finding a path of fixed length in a graph that optimizes an intensity and shape-based cost function and achieves maximum localization errors that are sub-voxel (video of electrode localization procedure). These results indicate that our methods could be used in a clinical IGCIP system.
Y Zhao, S Chakravorti, RF Labadie, BM Dawant, JH Noble, “Automatic graph-based method for localization of cochlear implant electrode arrays in clinical CT with sub-voxel accuracy,” Medical image analysis, vol. 52, pp. 1-12, 2019.
Yiyuan Zhao, Robert Labadie, Benoit Dawant, Jack Noble, “Validation of cochlear implant electrode localization techniques using µCTs,” Journal of Medical Imaging, vol. 5(3), pp. 035001, 2018.
Yiyuan Zhao, Benoit Dawant, and Jack Noble., “Automatic localization of closely-spaced cochlear implant electrode arrays in clinical CTs,” Med. Phys., vol 45 (11), pp. 5030-5040, 2018.
Automatic segmentation of brain tumor resections in intraoperative ultrasound images using U-Net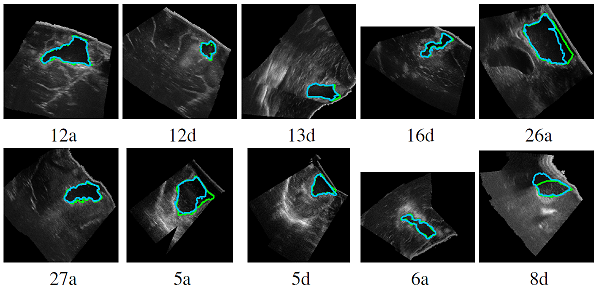
To compensate for the intraoperative brain tissue deformation, computer-assisted intervention methods have been used to register preoperative Magnetic Resonance (MR) images with intraoperative images. In order to model the deformation due to tissue resection, the resection cavity needs to be segmented in intraoperative images. In a collaborative study with Prof. Matthieu Chabanas from the Univ. of Grenoble, we are developing an automatic method to segment the resection cavity in intraoperative ultrasound (iUS) images using U-Net-based deep neural networks.
François-Xavier Carton, Matthieu Chabanas, Florian Le Lann, Jack H. Noble “Automatic segmentation of brain tumor resections in intraoperative ultrasound images,” Journal of Medical Imaging, vol. 7(3), pp. 031503, 2020.
François-Xavier Carton, Jack H. Noble, Florian Le Lann, Bodil K. R. Munkvold, Ingerid Reinertsen, Matthieu Chabanas, “Multiclass segmentation of brain intraoperative ultrasound images with limited data,” Proceedings of the SPIE Conf. on Medical Imaging, 11598-19, 2021.



Connect with Vanderbilt
©2026 Vanderbilt University ·
Site Development: University Web Communications