Shape-Constrained Multi-Atlas Segmentation of Spleen in CT
Z. Xu, B. Li, S. Panda, A. Asman, Kristen. L. Merkle, Peter L. Shanahan, Richard G. Abramson, B. Landman. “Shape-Constrained Multi-Atlas Segmentation of Spleen in CT.” In Proceedings of the SPIE Medical Imaging Conference. San Diego, California, February 2014†
Full Text: https://www.ncbi.nlm.nih.gov/pubmed/?term=Shape-Constrained+Multi-Atlas+Segmentation+of+Spleen+in+CT
Abstract
Spleen segmentation on clinically acquired CT data is a challenging problem given the complicity and variability of abdominal anatomy. Multi-atlas segmentation is a potential method for robust estimation of spleen segmentations, but can be negatively impacted by registration errors. Although labeled atlases explicitly capture information related to feasible organ shapes, multi-atlas methods have largely used this information implicitly through registration. We propose to integrate a level set shape model into the traditional label fusion framework to create a shape-constrained multi-atlas segmentation framework. Briefly, we (1) adapt two alternative atlas-to-target registrations to obtain the loose bounds on the inner and outer boundaries of the spleen shape, (2) project the fusion estimate to registered shape models, and (3) convert the projected shape into shape priors. With the constraint of the shape prior, our proposed method offers a statistically significant improvement in spleen labeling accuracy with an increase in DSC by 0.06, a decrease in symmetric mean surface distance by 4.01 mm, and a decrease in symmetric Hausdorff surface distance by 23.21 mm when compared to a locally weighted vote (LWV) method.
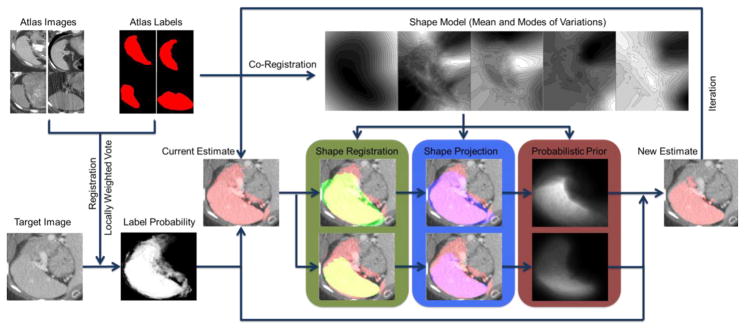
Flowchart of the proposed method. The atlas labels are co-registered to construct a pose-free implicit parametric shape model, including the mean and the modes of variation of the spleen shape. The atlas images are registered to the target image, based on which the atlas labels are propagated to the target space. The locally weighted vote yields the initial fusion result of the registered atlas labels weighted by the local intensity similarity between the registered atlas images and the target image in the form of a fuzzy estimate of label probability and a binary estimate of spleen segmentation. The binary image of the mean shape from the pre-constructed spleen shape model is registered to that of the current estimate with two distinct effective ranges, i.e., (1) the whole volume of both image and (2) the mean shape region, of the similarity metric of registration so that the pose-free shape model is transformed into the target space. The current estimate of the spleen is then projected to the two registered shape models. The shape projections are converted into probabilistic priors to adjust the label probability from locally weighted vote, and then generate a new estimate of the spleen. The estimate can be refined with iterative adjustment.