Evaluation of Atlas-Based White Matter Segmentation with Eve
Andrew J. Plassard, Kendra E. Hinton, Christopher Gonzalez, Vijay Venkatraman, Susan M. Resnick, Bennett A. Landman. “Evaluation of Atlas-Based White Matter Segmentation with Eve” In Proceedings of the SPIE Medical Imaging Conference. Orlando, Florida, February 2015. †
Full text: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4405655/
Abstract
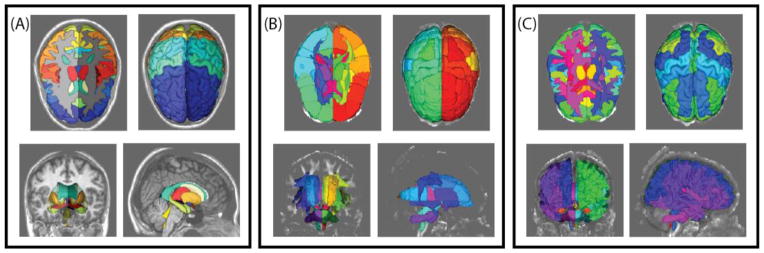
Multi-atlas labeling has come in wide spread use for whole brain labeling on magnetic resonance imaging. Recent challenges have shown that leading techniques are near (or at) human expert reproducibility for cortical gray matter labels. However, these approaches tend to treat white matter as essentially homogeneous (as white matter exhibits isointense signal on structural MRI). The state-of-the-art for white matter atlas is the single-subject Johns Hopkins Eve atlas. Numerous approaches have attempted to use tractography and/or orientation information to identify homologous white matter structures across subjects. Despite success with large tracts, these approaches have been plagued by difficulties in with subtle differences in course, low signal to noise, and complex structural relationships for smaller tracts. Here, we investigate use of atlas-based labeling to propagate the Eve atlas to unlabeled datasets. We evaluate single atlas labeling and multi-atlas labeling using synthetic atlases derived from the single manually labeled atlas. On 5 representative tracts for 10 subjects, we demonstrate that (1) single atlas labeling generally provides segmentations within 2mm mean surface distance, (2) morphologically constraining DTI labels within structural MRI white matter reduces variability, and (3) multi-atlas labeling did not improve accuracy. These efforts present a preliminary indication that single atlas labels with correction is reasonable, but caution should be applied. To purse multi-atlas labeling and more fully characterize overall performance, more labeled datasets would be necessary.