Improved Stability of Whole Brain Surface Parcellation with Multi-Atlas Segmentation
Yuankai Huo, Shunxing Bao, Prasanna Parvathaneni, Bennett A. Landman. “Improved Stability of Whole Brain Surface Parcellation with Multi-atlas Segmentation.” SPIE 2018
Full text: https://arxiv.org/abs/1712.00543
Abstract
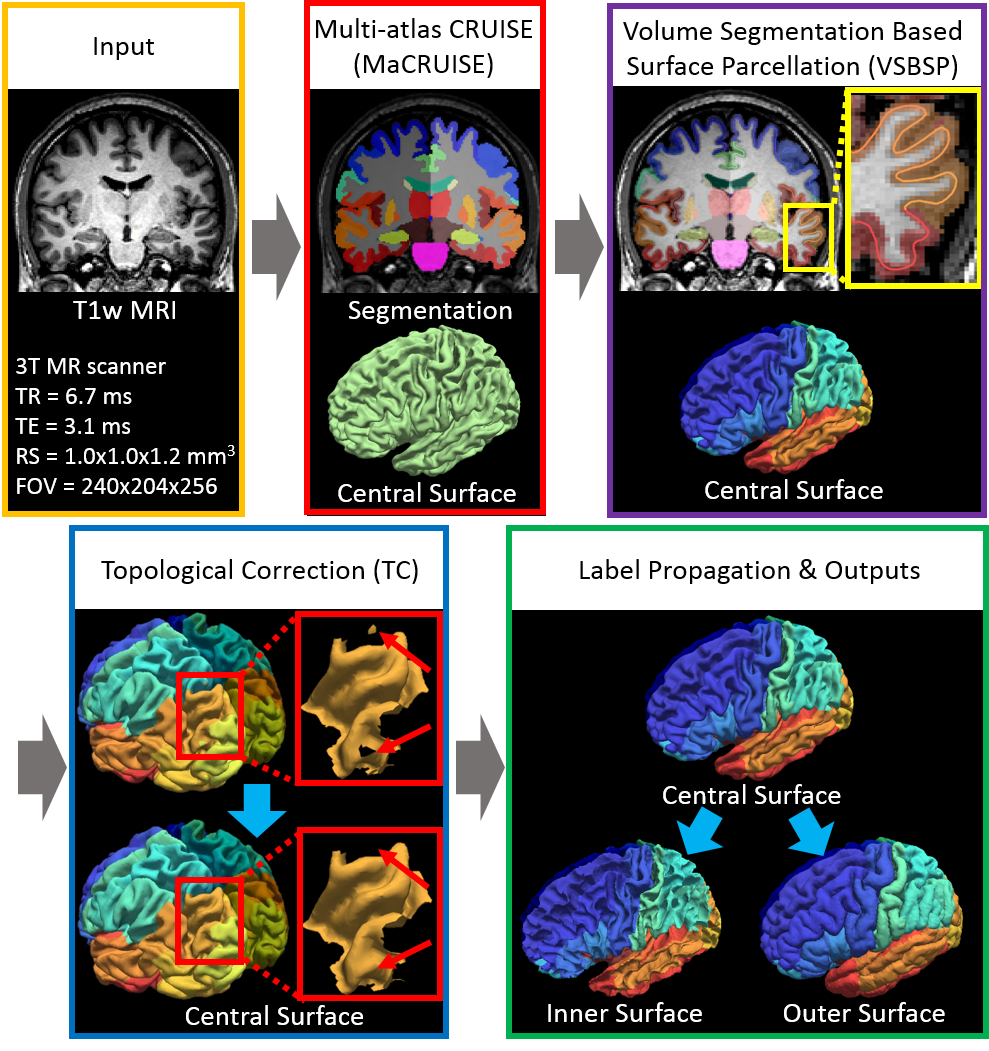
Whole brain segmentation and cortical surface parcellation are essential in understanding the brain’s anatomical-functional relationships. Multi-atlas segmentation has been regarded as one of the leading segmentation methods for the whole brain segmentation. In our recent work, the multi-atlas technique has been adapted to surface reconstruction using a method called Multi-atlas CRUISE (MaCRUISE). The MaCRUISE method not only performed consistent volume-surface analyses but also showed advantages on robustness compared with the FreeSurfer method. However, a detailed surface parcellation was not provided by MaCRUISE, which hindered the region of interest (ROI) based analyses on surfaces. Herein, the MaCRUISE surface parcellation (MaCRUISEsp) method is proposed to perform the surface parcellation upon the inner, central and outer surfaces that are reconstructed from MaCRUISE. MaCRUISEsp parcellates inner, central and outer surfaces with 98 cortical labels respectively using a volume segmentation based surface parcellation (VSBSP), following a topological correction step. To validate the performance of MaCRUISEsp, 21 scan-rescan magnetic resonance imaging (MRI) T1 volume pairs from the Kirby21 dataset were used to perform a reproducibility analyses. MaCRUISEsp achieved 0.948 on median Dice Similarity Coefficient (DSC) for central surfaces. Meanwhile, FreeSurfer achieved 0.905 DSC for inner surfaces and 0.881 DSC for outer surfaces, while the proposed method achieved 0.929 DSC for inner surfaces and 0.835 DSC for outer surfaces. Qualitatively, the results are encouraging, but are not directly comparable as the two approaches use different definitions of cortical labels.