Anatomical Accuracy of Standard-Practice Tractography Algorithms in the Motor System – a Histological Validation in the Squirrel Monkey Brain
Kurt G. Schilling, Yurui Gao, Vaibhav Janve, Iwona Stepniewska, Bennett Landman, Adam Anderson. “Anatomical Accuracy of Standard-Practice Tractography Algorithms in the Motor System – a Histological Validation in the Squirrel Monkey Brain”. Magnetic Resonance Imaging. 2018. doi: 1016/j.mri.2018.09.004
Full text: https://www.ncbi.nlm.nih.gov/pubmed/30213755
Abstract
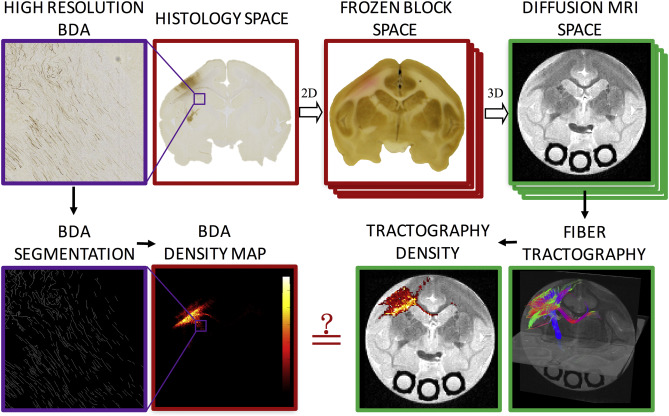
For two decades diffusion fiber tractography has been used to probe both the spatial extent of white matter pathways and the region to region connectivity of the brain. In both cases, anatomical accuracy of tractography is critical for sound scientific conclusions. Here we assess and validate the algorithms and tractography implementations that have been most widely used – often because of ease of use, algorithm simplicity, or availability offered in open source software. Comparing forty tractography results to a ground truth defined by histological tracers in the primary motor cortex on the same squirrel monkey brains, we assess tract fidelity on the scale of voxels as well as over larger spatial domains or regional connectivity. No algorithms are successful in all metrics, and, in fact, some implementations fail to reconstruct large portions of pathways or identify major points of connectivity. The accuracy is most dependent on reconstruction method and tracking algorithm, as well as the seed region and how this region is utilized. We also note a tremendous variability in the results, even though the same MR images act as inputs to all algorithms. In addition, anatomical accuracy is significantly decreased at increased distances from the seed. An analysis of the spatial errors in tractography reveals that many techniques have trouble properly leaving the gray matter, and many only reveal connectivity to adjacent regions of interest. These results show that the most commonly implemented algorithms have several shortcomings and limitations, and choices in implementations lead to very different results. This study should provide guidance for algorithm choices based on study requirements for sensitivity, specificity, or the need to identify particular connections, and should serve as a heuristic for future developments in tractography.
Keywords:
Accuracy; Diffusion magnetic resonance imaging; Dispersion; HARDI; Histology; Tractography; Validation