Projects
Selected Projects in Personalized Structural Biology
Modeling EGFR-KDD, a novel oncogenic driver in lung cancer
In collaboration with Christine Lovly in the Vanderbilt Ingram Cancer Center
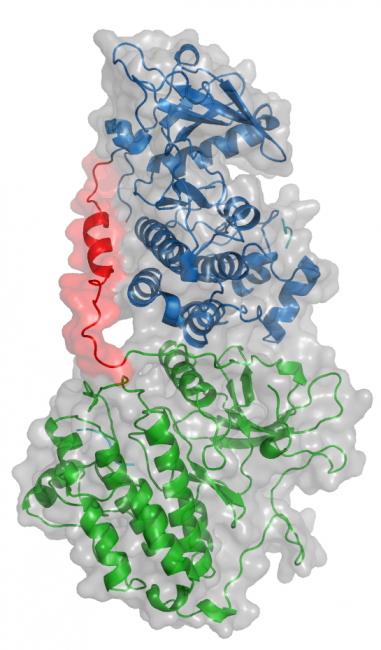 Oncogenic EGFR mutations are found in 10% to 35% of lung adenocarcinomas. Such mutations, which present most commonly as small in-frame deletions in exon 19 or point mutations in exon 21 (L858R), confer sensitivity to EGFR tyrosine kinase inhibitors (TKI). In analyzing the tumor from a 33-year-old male never-smoker, we identified a novel EGFR alteration in lung cancer: EGFR exon 18-25 kinase domain duplication (EGFR-KDD). Through analysis of a larger cohort of tumor samples, we detected additional cases of EGFR-KDD in lung, brain, and other cancers. In vitro, EGFR-KDD is constitutively active, and computational modeling provides potential mechanistic support for its auto-activation. EGFR-KDD-transformed cells are sensitive to EGFR TKIs and, consistent with these in vitro findings, the index patient had a partial response to the EGFR TKI afatinib. The patient eventually progressed, at which time resequencing revealed an EGFR-dependent mechanism of acquired resistance to afatinib, thereby validating EGFR-KDD as a driver alteration and therapeutic target.
Oncogenic EGFR mutations are found in 10% to 35% of lung adenocarcinomas. Such mutations, which present most commonly as small in-frame deletions in exon 19 or point mutations in exon 21 (L858R), confer sensitivity to EGFR tyrosine kinase inhibitors (TKI). In analyzing the tumor from a 33-year-old male never-smoker, we identified a novel EGFR alteration in lung cancer: EGFR exon 18-25 kinase domain duplication (EGFR-KDD). Through analysis of a larger cohort of tumor samples, we detected additional cases of EGFR-KDD in lung, brain, and other cancers. In vitro, EGFR-KDD is constitutively active, and computational modeling provides potential mechanistic support for its auto-activation. EGFR-KDD-transformed cells are sensitive to EGFR TKIs and, consistent with these in vitro findings, the index patient had a partial response to the EGFR TKI afatinib. The patient eventually progressed, at which time resequencing revealed an EGFR-dependent mechanism of acquired resistance to afatinib, thereby validating EGFR-KDD as a driver alteration and therapeutic target.
SIGNIFICANCE:
We identified oncogenic and drug-sensitive EGFR-KDD that is recurrent in lung, brain, and soft-tissue cancers and documented that a patient with metastatic lung adenocarcinoma harboring the EGFR-KDD derived significant antitumor response from treatment with the EGFR inhibitor afatinib. Findings from these studies will be immediately translatable, as there are already several approved EGFR inhibitors in clinical use.
Publications:
Gallant JN, Sheehan JH, Shaver TM, Bailey M, Lipson D, Chandramohan R, Red Brewer M, York SJ, Kris MG, Pietenpol JA, Ladanyi M, Miller VA, Ali SM, Meiler J, Lovly CM. EGFR Kinase Domain Duplication (EGFR-KDD) Is a Novel Oncogenic Driver in Lung Cancer That Is Clinically Responsive to Afatinib. Cancer Discov. 2015 Nov;5(11):1155-63.
Predicting functional impact of missense mutations in KCNQ1
In collaboration with Al George in the Vanderbilt Institute for Integrative Genomics
Long-QT syndrome (LQTS) is an inherited disease associated with mutations in genes encoding cardiac ion channel subunits. This disorder predisposes children and young adults to sudden death due to arrhythmia. An emerging standard-of-care for LQTS patients employs clinical genetic testing to identify LQTS-associated mutations to guide the course of treatment. However, interpreting results from genetic testing is complicated by the presence of variants of unknown significance or VUS for which there is inadequate experimental or clinical evidence to classify whether or not they are pathogenic. We searched extensively in the literature and curated a high-quality set of functionally characterized KCNQ1 mutations.
Based on this data set, we compared the functional impact of mutations with their clinical classification to estimate the error rate of clinical classification of KCNQ1 mutations, thus illustrating the necessity of training the predictor on functionally verified variants. We trained a neural network, Q1MutPred, specifically for predicting the functional impact of KCNQ1 mutations on this experimentally validated data set. We showed that Q1MutPred has superior performance over five commonly used genome-wide predictors. Q1MutPred is deployed in a web-based format to allow researchers and medical professional to evaluated novel KCNQ1 mutations.
Publications:
Kroncke BM, Van Horn WD, Smith J, Kang C, Welch RC, Song Y, Nannemann DP, Taylor KC, Sisco NJ, George AL Jr, Meiler J, Vanoye CG, Sanders CR. Structural basis for KCNE3 modulation of potassium recycling in epithelia. Sci Adv. 2016 Sep 9;2(9):e1501228. doi: 10.1126/sciadv.1501228. eCollection 2016 Sep. PubMed PMID: 27626070; PubMed Central PMCID: PMC5017827.
Rationalize the effect of kinase inhibitors in patient with EGFR-RAD51 fusion, a novel therapeutic target in lung cancer
In collaboration with Christine Lovly in the Vanderbilt Ingram Cancer Center
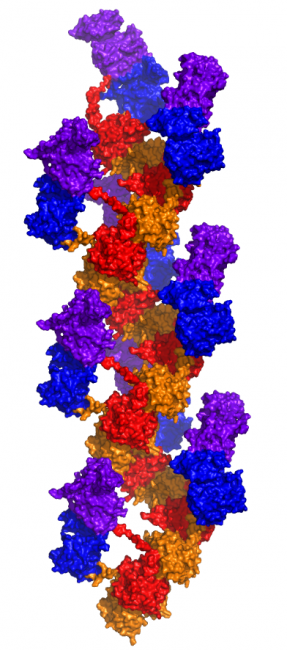 Given the presence of RAD51, a self-assembling filamentous protein, we hypothesized that the EGFR-RAD51 fusion protein can form such partner-driven dimers. To validate this hypothesis, we modeled EGFR-RAD51 based on available experimental structures of RAD51 and the active asymmetric EGFR dimer. Conformational loop sampling with Rosetta demonstrates that it is geometrically feasible for EGFR kinase subunits to adopt the asymmetric (active) dimeric conformation when fused to RAD51. Further, the concatenation of RAD51 subunits could bring tethered EGFR kinase domains close together, increasing their local concentration and leading to further EGFR activation.
Given the presence of RAD51, a self-assembling filamentous protein, we hypothesized that the EGFR-RAD51 fusion protein can form such partner-driven dimers. To validate this hypothesis, we modeled EGFR-RAD51 based on available experimental structures of RAD51 and the active asymmetric EGFR dimer. Conformational loop sampling with Rosetta demonstrates that it is geometrically feasible for EGFR kinase subunits to adopt the asymmetric (active) dimeric conformation when fused to RAD51. Further, the concatenation of RAD51 subunits could bring tethered EGFR kinase domains close together, increasing their local concentration and leading to further EGFR activation.
Publications:
Konduri K, Gallant JN, Chae YK, Giles FJ, Gitlitz BJ, Gowen K, Ichihara E, Owonikoko TK, Peddareddigari V, Ramalingam SS, Reddy SK, Eaby-Sandy B, Vavalà T, Whiteley A, Chen H, Yan Y, Sheehan JH, Meiler J, Morosini D, Ross JS, Stephens PJ, Miller VA, Ali SM, Lovly CM. EGFR Fusions as Novel Therapeutic Targets in Lung Cancer. Cancer Discov. 2016 Apr 21.
Structural analysis of RTEL1 variants in Idiopathic Pulmonary Fibrosis
In collaboration with John Phillips, John Newman, Joy Cogan, Tim Blackwell, Jonathan Kropski, and other members of Vanderbilt’s Undiagnosed Disease Network.
Publications:
Manuscript in preparation:
Spatial Clustering of Familial Interstitial Pneumonia Variants in RTEL1 Enables Prioritization of Novel Patient Mutations
Understand de-repression mechanism of novel deletion 447-455 in p85alpha from PI3K
In collaboration with Carlos Arteaga in the Vanderbilt Ingram Cancer Center
Support the use of drug AZD9291 for a breast cancer patient with acquired T798I mutation in HER2
In collaboration with Carlos Arteaga in the Vanderbilt Ingram Cancer Center
Generate activation hypotheses about heterodimer of HER3 E928G and HER2 L869R
In collaboration with Carlos Arteaga in the Vanderbilt Ingram Cancer Center

©2026 Vanderbilt University ·
Site Development: University Web Communications