Research
 |
 |
|
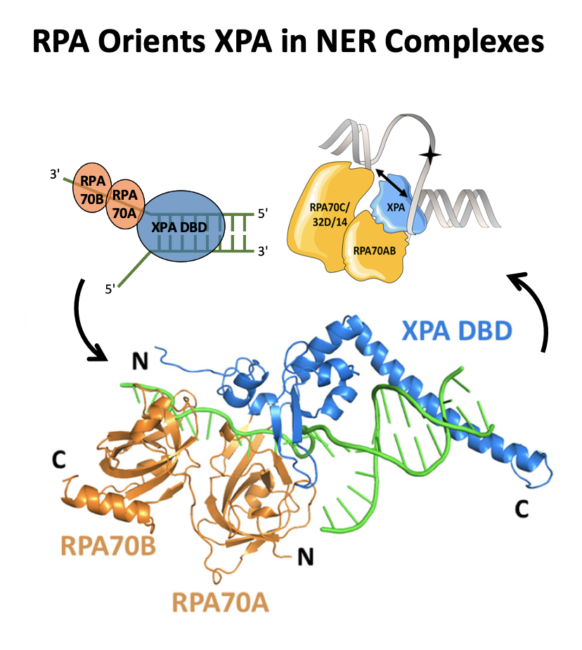
Topolska-Woś AM et al. A key interaction with RPA orients XPA in NER complexes. Nucleic Acids Res. 2020 Feb 28;48(4):2173-2188. doi: 10.1093/nar/gkz1231 |
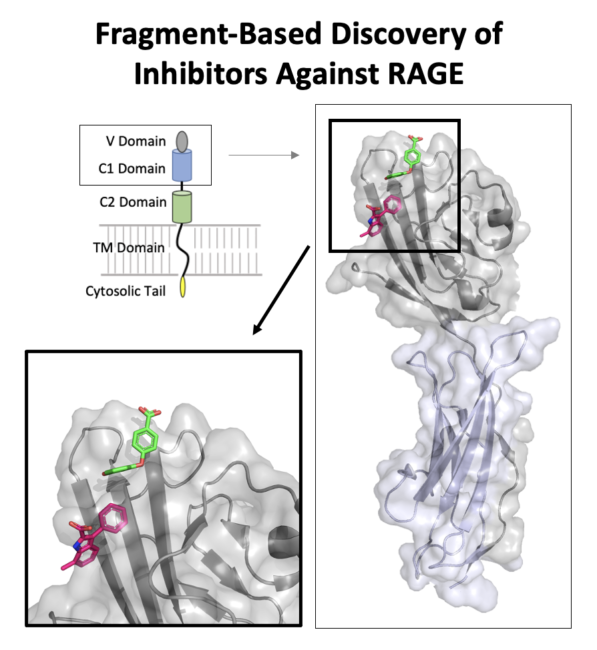
Kozlyuk N et al. A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors. Proteins. 2021 Nov;89(11):1399-1412. doi: 10.1002/prot.26162. |
Overview
The Chazin laboratory investigates the structural basis for recognition, biochemical function and biological specificity of proteins and nucleic acids. Specifically, we study systems involved in bacterial pathogenesis, cell signaling, and cancer at the molecular level. Rather than focusing on specific techniques, we take a problem-oriented approach and use whichever techniques are necessary to answer the question at hand. Often, this requires complementary application of structural approaches including spectroscopy, light scattering, crystallography, and microscopy (see below.) We also prioritize tight coupling of our research to functional analysis. This strategy is by nature very collaborative and collegial, and has stimulated involvement in many multi-investigator projects.
Broadly, the lab is focused on two main areas:
- Genome Maintenance and Propagation Processes
- Protein Interactions in Immunity, Inflammation, and Infection
To investigate these topics, we employ a variety of techniques including (but not limited to) the following:
NMR Spectroscopy, X-ray Crystallography, X-ray and Neutron Scattering, Cryo-Electron Microscopy, Computational Modeling, Calorimetry, Microscale Thermophoresis, Spectroscopy (UV-Vis, Circular Dichroism, Fluorescence) Dynamic and Multi-angle Light Scattering (in collaboration with Wyatt)
Get to know more about the operation, driving force, and interests of this lab in a full interview with Dr. Chazin.
A comprehensive list of the lab’s publications can be found on PubMed.
