Molecular determinants of COPI-mediated intracellular trafficking
Sorting of protein and lipids in Golgi is a highly regulated process important for many physiological events. Protein and lipid sorting and transport through Golgi is largely mediated through vesicle trafficking. Coat proteins drive the formation of vesicles by assembling onto the surface of cellular membranes. Different coats are implicated in budding from different compartments and facilitating different routes of transport; for example, (A) clathrin-coated endocytic vesicles form at the plasma membrane constituting the major pathway for internalization of membrane proteins from the cell surface as well as mediate transport of newly synthesized lysosomal hydrolases from trans-Golgi network to the lysosome; (B) COPII-coated vesicles originating from the endoplasmic reticulum(ER) constitute the major route of anterograde transport from ER to the Golgi apparatus; (C) COPI-coated vesicles originate at Golgi, except at the trans-Golgi network, and mediate retrograde transport from the cis end of the Golgi complex to within the Golgi cisternae or back to the rough ER. The selectivity of transport is key to maintaining organelle homeostasis and the functional organization of the cell. Various subprojects within the lab seek to address several key questions related to vesicle-mediated trafficking such as (1)how are cargoes sorted into different vesicles; (2) how are vesicle-mediated trafficking events regulated; (3) what are the intracellular routes serviced by various types of vesicle-mediated processes and (4) how do cargoes and coats regulate to maintain protein and lipid homeostasis in a cell.
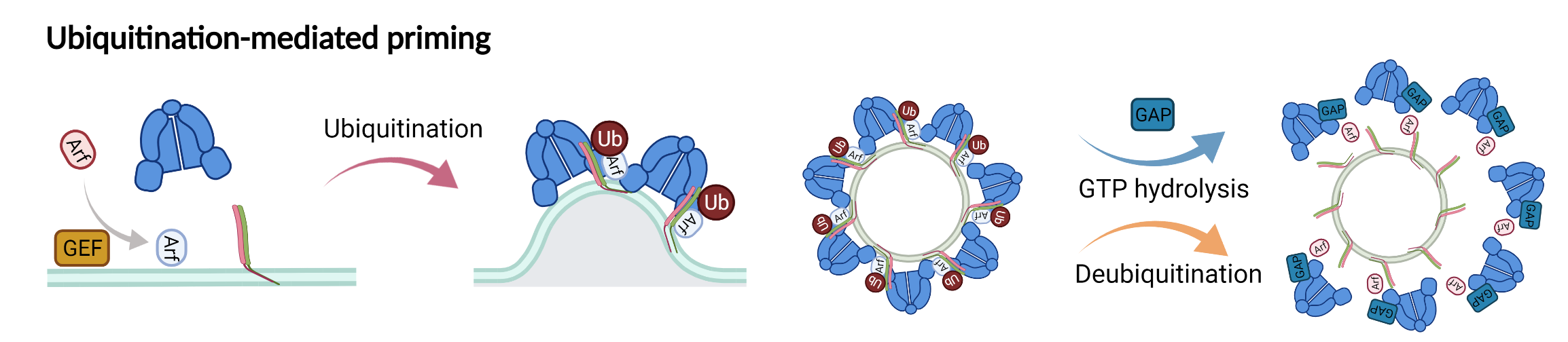
New model showing the role of ubiquitination in driving COPI priming and regulation of SNARE cargoes.
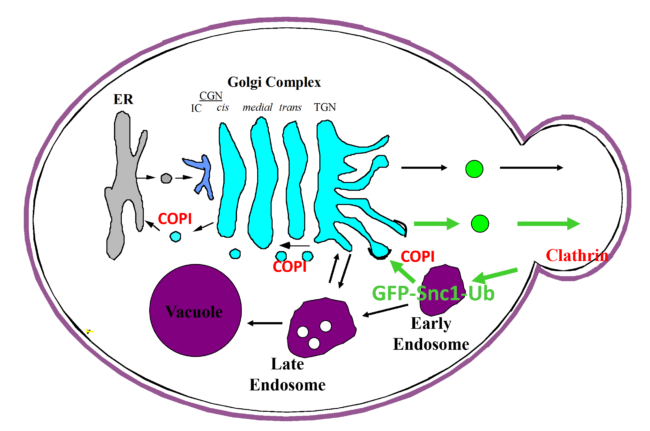
A carton representation for the recognition of ubiquitin by COPI as a sorting signal for retrieval of SNARE Snc1
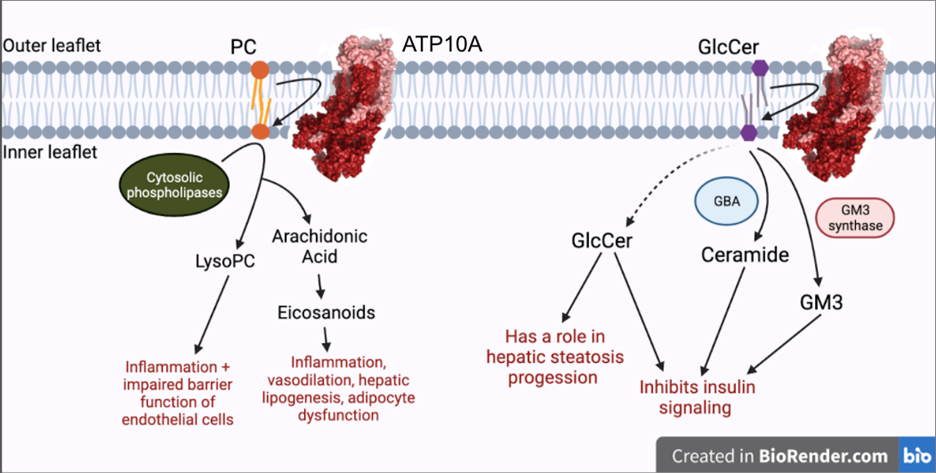
Exploring the role of flippase ATP10A in metabolism
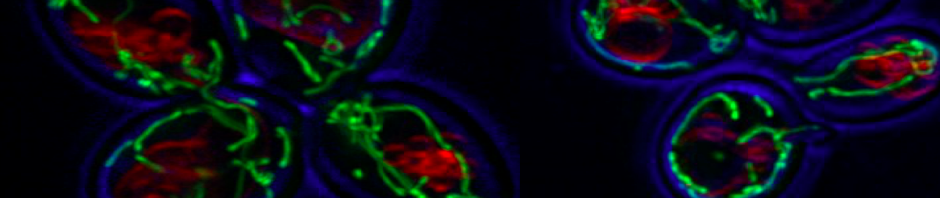
Genetic predisposition and environment play substantial roles in diet-induced obesity, type 2 diabetes, and cardiovascular disease (CVD). Genetic association studies have linked ATP10A, encoding a type IV P-type ATPase (P4-ATPase), to human metabolic disease. ATP10A is a lipid flippase that catalyzes the translocation of phosphatidylcholine and glucosylceramide across the plasma membrane, and these lipids or their metabolites have been independently implicated in metabolic dysfunction. To explore the role of this flippase in metabolism, we created a novel Atp10a knockout (KO) mouse model. Atp10a KO mice display a female-specific excess weight gain during high- fat diet feeding and this is attributable to increased adiposity. We have recently found that ATP10A is specifically expressed in the endothelial cells of multiple tissues. We are now exploring these metabolic phenotypes through the lens of endothelial cell dysfunction. These studies suggest mechanisms by which this flippase contributes to the development of CVD with obesity.
Phospholipid recognition and translocation by P4-ATPases
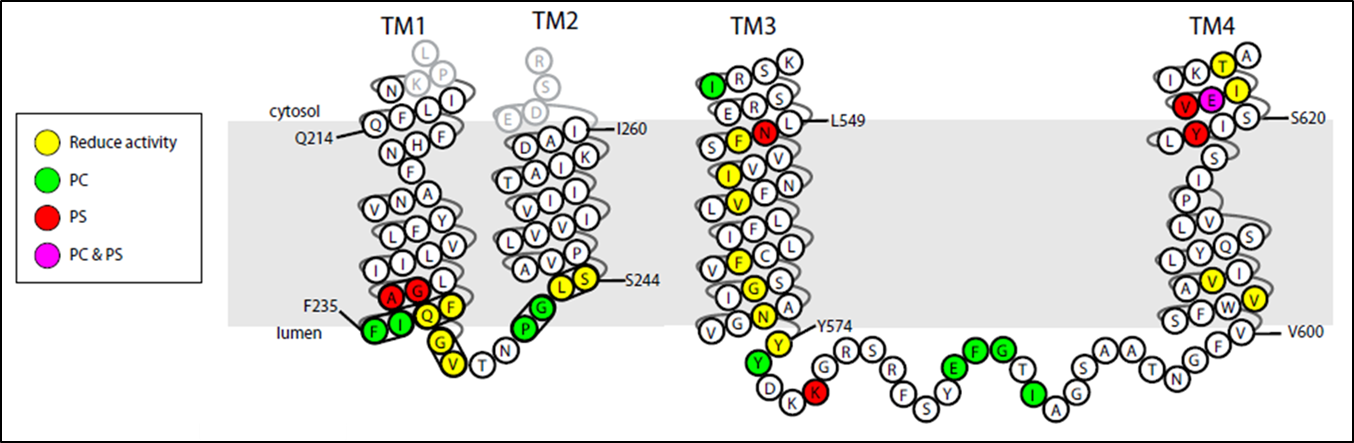
The P4-ATPases, or flippases, flip specific phospholipid species, such as phosphatidylserine, from the extracellular leaflet of the plasma membrane to the cytosolic leaflet, thus producing an asymmetric membrane structure that is conserved among most eukaryotic cells. This phospholipid asymmetry has a major influence on the localization and activity of many different plasma membrane proteins. Moreover, regulated disruption of membrane asymmetry is a signaling device in blood clotting and for recognition of apoptotic cells. Humans have 14 P4-ATPases and members of this protein family are implicated in severe liver and neurological diseases.
A current project in the laboratory is to define the mechanism of substrate recognition and translocation by P4-ATPases using genetic and biochemical approaches. The best characterized P-type ATPases transport small cations across membranes to establish ion gradients so phospholipid molecules are an unusual substrate for this protein family. Our work is suggesting a novel transport mechanism for the P4-ATPases and is providing insight into how these transporters evolved the ability to transport their “giant substrate”. We have so far identified a number of key residues in budding yeast P4-ATPases that are critical for phospholipid headgroup specificity and have found two clusters of residues which act as entry and exit gates for phospholipid movement across the membrane. Current work in the lab is focused on identifying residues involved in phospholipid backbone selectivity as well as identifying the phospholipid substrates of the P4-ATPase Neo1.
Function of P4-ATPases in vesicular trafficking
In addition to establishing membrane asymmetry, we discovered that P4-ATPases play a crucial role in budding protein transport vesicles from Golgi and endosomal membranes. For example, a P4-ATPase called Drs2 translocates phosphatidylserine across the membrane of the trans-Golgi network and this flippase activity is required to bud AP-1/clathrin-coated vesicles that transport proteins from the Golgi to endosomes. Our work has uncovered both positive and negative regulators of Drs2 activity representing proteins and lipids known to have critical roles in vesicular transport. The unidirectional translocation of phosphatidylserine to the cytosolic leaflet should have a dramatic influence on the biophysical properties of the membrane by enhancing the anionic membrane potential of the cytosolic surface as well as by inducing curvature through a bilayer couple mechanism. The protein trafficking events dependent on P4-ATPase activity have a major influence on the protein composition of the plasma membrane and organelles of the secretory and endocytic pathways. A current project in the lab is to determine the mechanism by which P4-ATPases function in protein transport with emphasis on the effects of PS flip on Golgi and endosomal membranes. Another current project is determining the role of PS flippase activity in establishing and maintaining lateral membrane domains (similar to lipid rafts) in budding yeast.