Congrats to Kirsten Diggins, Jocelyn Gandelman, and Caroline Roe on their publication! This protocol aggregates code, scripts, and examples for MEM that we use regularly in courses and research:
Diggins KE, Gandelman JS, Roe CE, Irish JM. Generating Quantitative Cell Identity Labels with Marker Enrichment Modeling (MEM). Current Protocols in Cytometry 2017. NIHMS912131. PMC in progress. Pubmed. DOI.
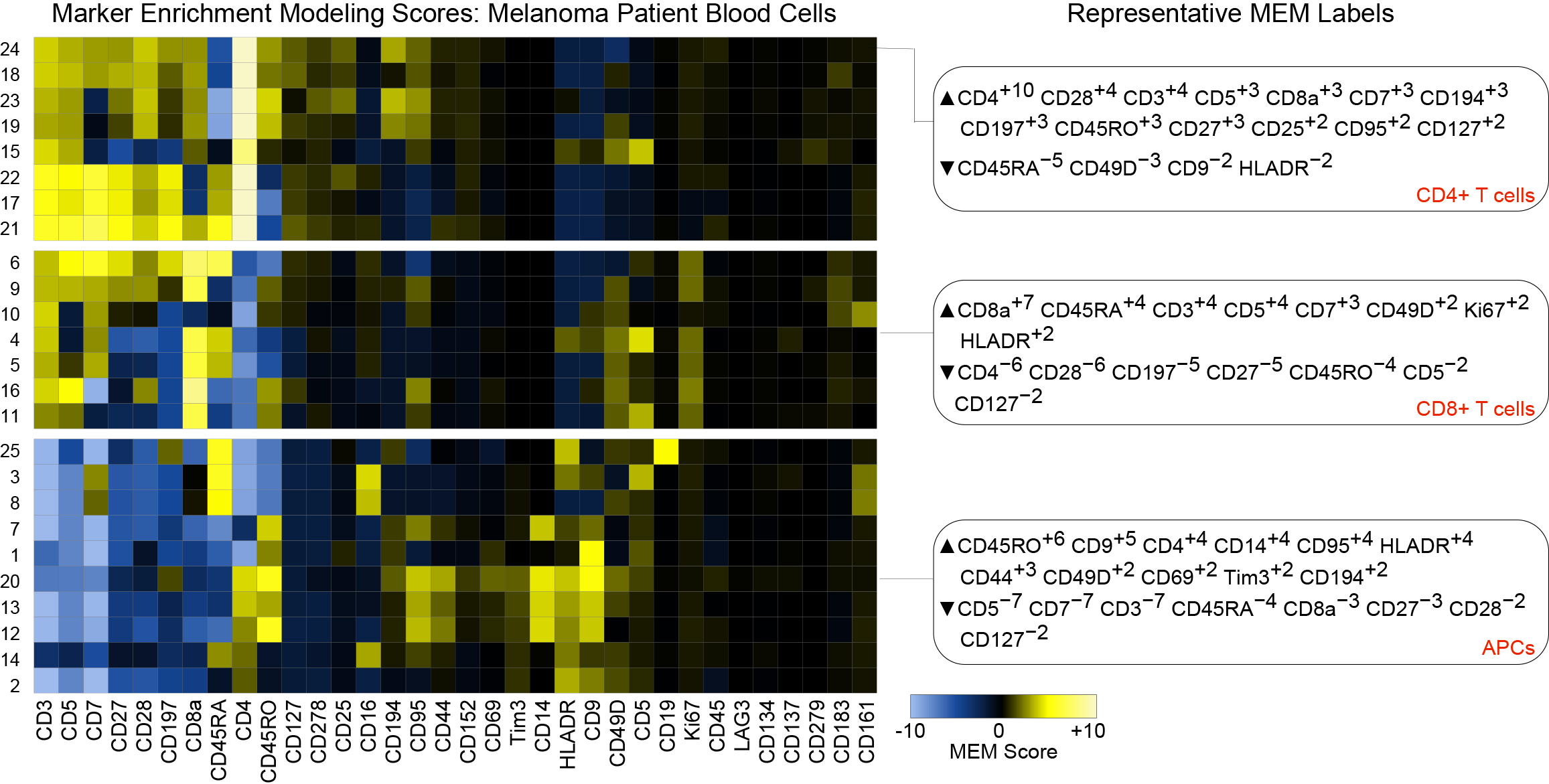
► This peer-reviewed protocol is targeted for a biology background audience and describes in detail the steps needed to obtain and run code for Marker Enrichment Modeling (MEM) in R/flowCore using text, CSV, or flow cytometry standard (FCS) files. MEM was originally described in Diggins et al., Nature Methods 2017 (DOI). Since publication, MEM has been successfully applied to identify subgroups in a wide variety of data types, including fluorescence cytometry, mass cytometry, single cell RNA-seq, quantitative imaging, gene expression profiling, and clinical phenotyping of patients.
Diggins et al., Current Protocols in Cytometry – Measure and share cell identity with a MEM label