One of our primary research interest is in cell membrane structure. Over the past 15 years, a model has emerged that emphasizes the role of lateral heterogeneities in regulating membrane structure and function. This model, termed the lipid raft hypothesis, postulates that cholesterol- and sphingolipid-enriched membrane microdomains act as platforms that function to organize proteins, concentrating some proteins together within the same raft while segregating raft from non-raft proteins. Lipid rafts are now thought to participate in a wide range of cellular functions, ranging from membrane trafficking, to cell signaling, to pathogen exit and entry from cells. Despite the widespread implications of this model for human health, many of the fundamental properties of lipid rafts, such as their structure, composition, and lifetime are still poorly understood. To gain better insights into the features of these elusive domains, we are using a combination of cell biology and quantitative fluorescence microscopy to study the spatial distribution and dynamics of raft-associated proteins and lipids in cells.
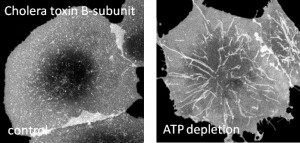
One of our research interest is on mechanisms regulating clathrin-independent endocytosis of bacterial toxins. This will enable us to understand how proteins and lipids cooperate to build functional domains and help us decipher the rules that dictate how domains are formed. Further, we are also interested in endocytic consequences of caveolin mutants on caveolae structure and function. Understanding these mutations in caveolin-1 and defects in caveolae would help us in understanding the causes of human diseases such as breast cancer and pulmonary arterial hypertension. In collaboration with Chuck Sanders at Vanderbilt, we are also interested in understanding the contribution of cholesterol to the progression of Alzheimer’s disease through regulation of the affinity of amyloid precursor protein for lipid rafts.
Another interest of our group is intracellular trafficking and protein dynamics, basic processes that underlie all cellular functions. With the advent of GFP, it is possible to directly visualize proteins within their native environment of the cell. While much can be learned from simply watching proteins move using fluorescence microscopy, advanced techniques such as fluorescence recovery after photobleaching (FRAP) and photoactivation hold the potential to provide quantitative measurements of trafficking kinetics, the on and off rates of the interactions of proteins with membranes or other cellular structures, and the diffusion rate of the bound and unbound species. However, the in vivo analysis of such kinetic constants is still in its relative infancy due to a lack of consensus as to how best extract these parameters from data obtained by FRAP and related techniques. We are therefore currently working in collaboration with mathematicians to develop new methods and models with which to calibrate, measure and quantify protein dynamics in living cells.
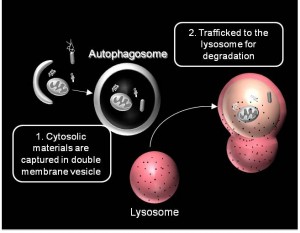
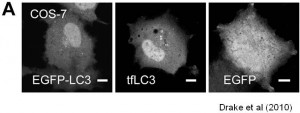
We are working on extending the analytical approaches that we have developed to generate spatially and temporally resolved maps of protein complexes involved in the autophagy pathway, which is a lysosomal degradation pathway that helps cells combat stress, starvation, aging, and disease. Specifically, we are interested in the central protein in the autophagy pathway, LC3. LC3 associates with complexes in the nucleus, suggesting it may have novel nuclear functions We recently identified a novel macromolecular complex associated with LC3 using FRAP and FCS . Along with biochemical techinques we are working on understanding the kinetics of these autophagy related complexes.