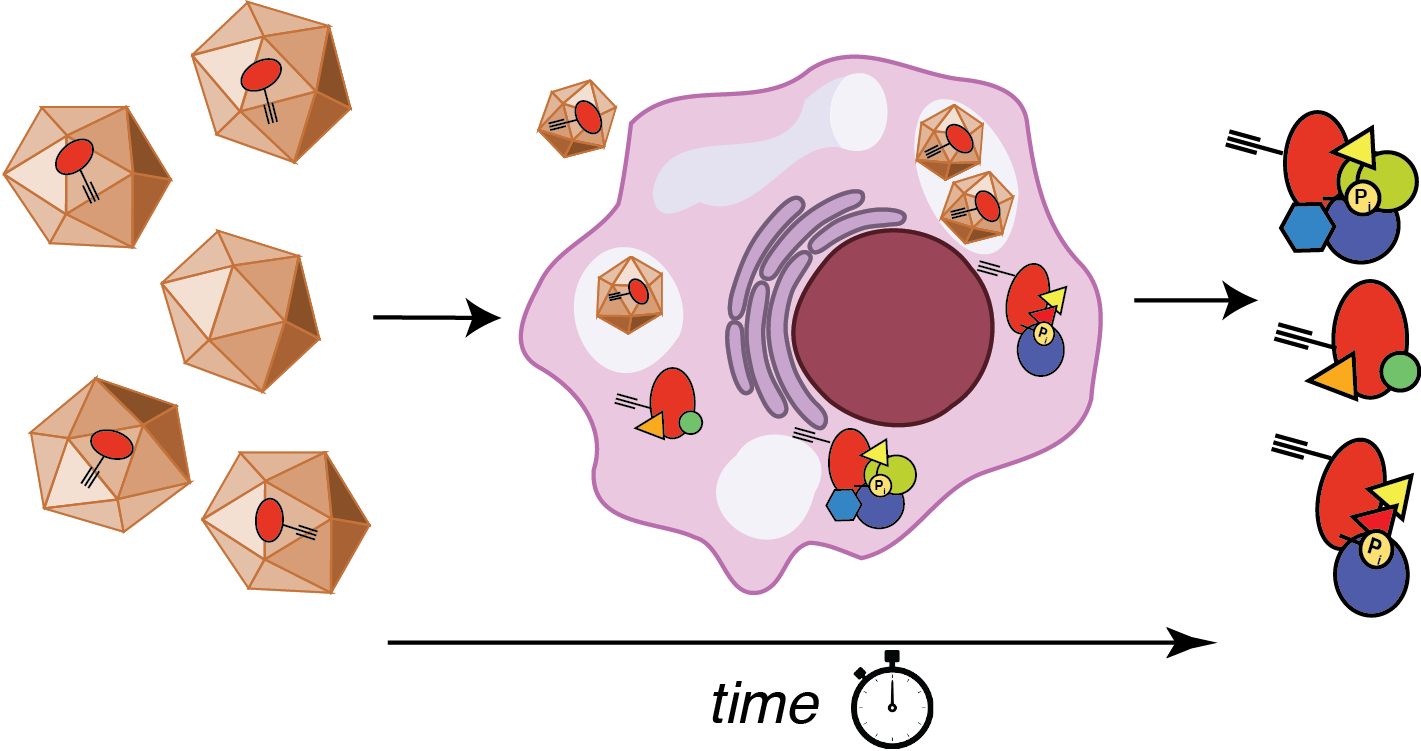
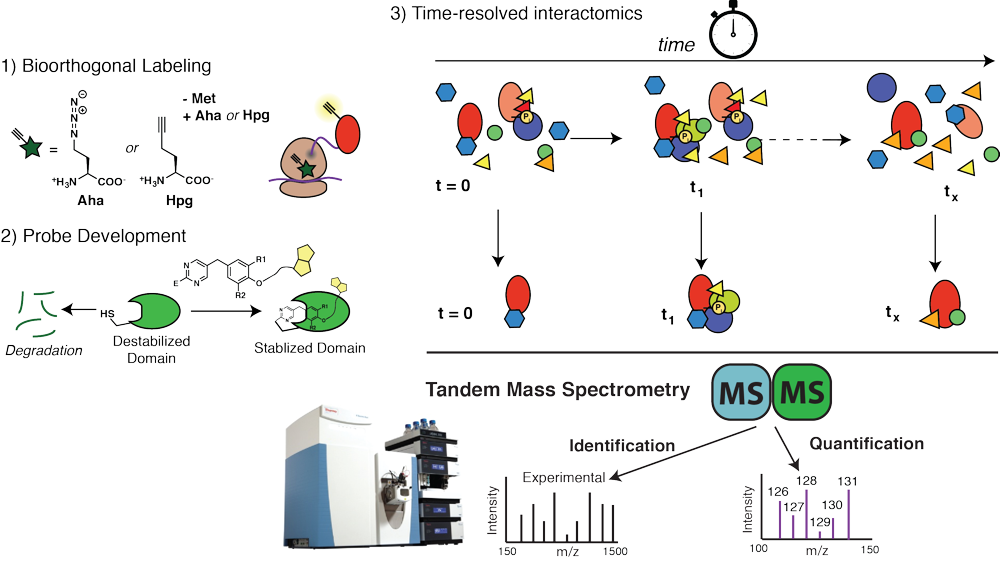
Dynamics of chaperone interactions with client proteins during folding and secretion
Protein folding and assembly inside cells rely on the coordinated action of chaperones, co- chaperones and other quality control factors that comprise the proteostasis network. Our goal is to monitor the sequential engagement and interaction dynamics between client proteins and proteostasis network components during cellular protein folding. We are developing a mass spectrometry-based platform for time-resolved interactomics as well as new chemical probes and bioorthogonal protein labeling approaches. Using these tools, we explore how altered progression of client proteins through the proteostasis pathways can affect the quality of the folded proteins and lead to protein misfolding diseases. This will reveal previously invisible mechanisms responsible for the quality control defects in protein misfolding diseases and identify new strategies for disease intervention.
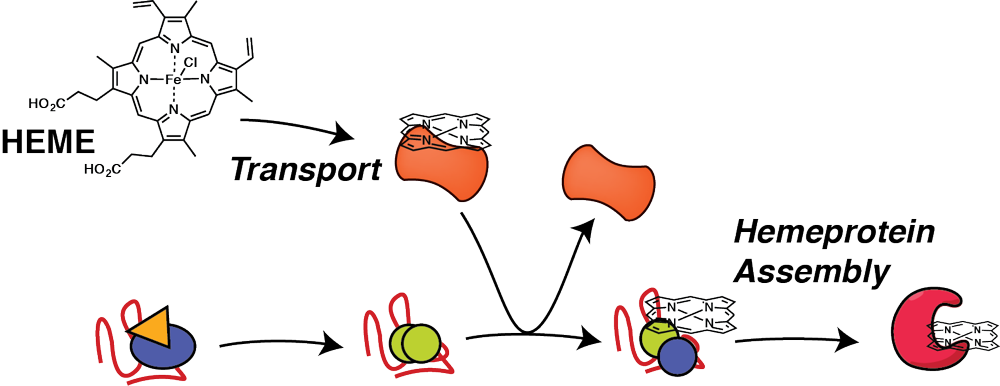
Hemeprotein proteostasis and dysregulation by stress and disease
Biogenesis of cofactor containing proteins requires careful timing of cofactor insertion and coordination with proteostasis network components that aid in the protein folding process. Heme is one of the most common and most versatile cofactors, but free heme is highly toxic due to generation of reactive oxygen species. Therefore, heme has to be securely chaperoned and trafficked to the sites of heme protein synthesis in different organelles, yet specific heme transport factors are largely unknown. Our research identifies intracellular heme transport factors and heme chaperones, and defines how these interact and coordinate with the proteostasis network to facilitate timely insertion of heme during protein folding. Investigating how heme transport and and hemeprotein assembly is regulated during stress and disease will identify new targets to mitigate oxidative damage during cardiovascular disease or to intervene with malaria infections.
Coordination of host pathogen protein-protein interactions during flavivirus infections
Viruses have evolved sophisticated strategies to co-opt host cellular processes to facilitate viral replication. Host-pathogen protein-protein interactions govern all processes of the viral life cycle including viral entry, disassembly, replication, assembly and suppression of the cellular immune response. In particular, hijacking of cytosolic and ER proteostasis pathways has emerged as a common strategy for viral replication that might extend beyond simply facilitating the folding of viral proteins. Our research elucidates the timing and coordination of host pathogen protein interactions throughout the viral life cycle with a particular focus on uncovering novel dependencies on the host proteostasis pathways. These discoveries will point to new host processes as therapeutic targets to combat infection of several widespread viruses that pose global health threats, such as Dengue, Hepatitis C, or Zika Virus.