Research

Our goals are to understand the biology of genomes and to advance genomic medicine.
We are interested in elucidating the genetic basis of human traits and diseases using novel statistical & computational methodologies, integrative analyses of heterogeneous multi-omics data, and innovative experimental approaches. A primary research focus is the development of methods in statistical and population genetics. We pursue deep learning methods in genomics. The knowledge gained can be the basis for a genetics-driven forward pharmacology that enables identification of molecules with a desired phenotypic effect or mechanism of action. We leverage functional genomics to dissect the transcriptional regulatory logic and its context specificity in order to advance our understanding of biological mechanisms.
Here is a list of select publications and the latest news. See also this talk series, which reflects some of our research interests.
We are developing approaches and methodologies that are broadly applicable, but we may evaluate these with specific disease areas, cell types, or genes (or related genomes). Our goals are to understand the biology of genomes and to advance genomic medicine. Some active areas of research are described below, but see our publications for a more complete picture of our research activities.
 FUNCTIONAL GENOMICS & PRINCIPLES OF COMPLEX TRAIT GENETICS
FUNCTIONAL GENOMICS & PRINCIPLES OF COMPLEX TRAIT GENETICS
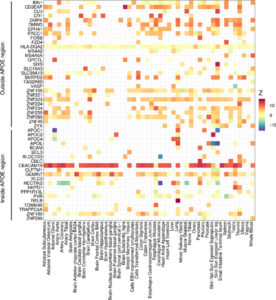
We are interested in multi-disciplinary approaches to mapping the role of genetic variation in the development and function of the nervous system and elucidating the precise molecular and cellular mechanisms underlying brain disease phenotypes. Ongoing research includes modeling the transcriptional regulatory programs in the brain, in collaboration with human geneticists, neuroscientists, and evolutionary biologists. This work has obvious relevance to understanding the molecular basis of neuropsychiatric and neurodegenerative disorders.
 BRIDGING GENOMICS and PHENOMICS
BRIDGING GENOMICS and PHENOMICS
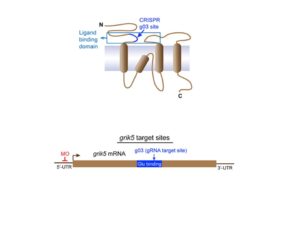
Recent advances have enabled precise genome editing with high efficiency and minimal off-target effects. Meanwhile, EHR-linked DNA biobanks have been unprecedented in their efficiency for human genetic discovery. We are conducting studies and developing methods across systems. In recent collaboration with colleagues at Vanderbilt, we integrated human phenomic data and in vivo loss-of-function models generated using CRISPR-Cas9 genome editing strategies and morpholino (MO) oligonucleotide-based protein knockdown to characterize a mechanism that implicates vascular biology in the eye disease phenome.
 iPSCs, FUNCTIONAL GENOMICS, and DISEASE
iPSCs, FUNCTIONAL GENOMICS, and DISEASE
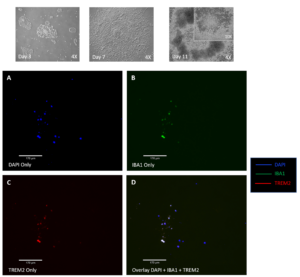
Current research interests in complex traits genetics include functionally characterizing the genetic architecture of complex diseases, using induced pluripotent stem cells as a platform.
 SINGLE-CELL GENOMICS
SINGLE-CELL GENOMICS
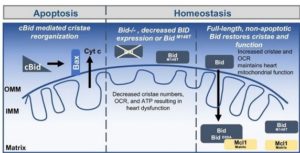
In collaboration with Dr Sandra Zinkel, we also combine human genetics, phenomics, bulk and single-cell transcriptomics, hematopoietic cell culture systems, mouse models, immunofluorescence, electron microscopy, mass cytometry, and cell death assays to understand apoptosis and necrosis and their role in human disease.
We are also developing single-cell computational approaches for causal inference.
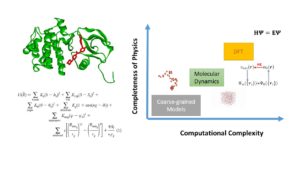 STRUCTURE, DYNAMICS, and GENOMICS
STRUCTURE, DYNAMICS, and GENOMICS
In recent highly interdisciplinary work, Dr Gamazon is developing computational approaches to studying the structure, dynamics, and stability of biological molecules within Density Functional Theory (DFT), molecular dynamics, and coarse-grained modelling and using experimental techniques (e.g., X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy).
He is co-founder of the Chemistry and Disease Genomics Group (CDGG) with David Collison (Manchester) and Lalarukh Akhter (Cambridge).
He is part of the GTEx Consortium and the T2D-GENES Consortium. He was also a member of the International Warfarin Pharmacogenetics Consortium GWAS team. As part of the Pharmacogenomics of Anticancer Agents Research (PAAR) Group at the University of Chicago, he developed and applied computational approaches to the study of the genetic basis of cancer susceptibility and of response to a wide array of molecularly diverse chemotherapeutics.
Funding
 We gratefully acknowledge research support from these institutions and agencies.
We gratefully acknowledge research support from these institutions and agencies.
Research Interests
Prediction ; Bayesian methods ; Statistical Genetics ; Regulation of gene expression / transcription ; Computational Biology ; Functional Genomics ; Single-cell Omics ; Data Science ; Cancer Biology ; Hematopoiesis ; Stem cells ; High-throughput sequencing ; Machine Learning ; Omics data management storage and annotation ; Methylation ; Bioinformatics ; Big Data in Biology ; Evolutionary Genomics ; Next Generation Sequencing ; Pharmacogenomics ; Neuropsychiatric Genetics ; Biobank and Phenomics ; Epigenome ; Molecular Dynamics ; X-ray Crystallography ; NMR Spectroscopy ; Structural Biology ; Coarse-grained Models ; Density Functional Theory ; Neural Networks ; Deep Learning