Congratulations to Kirsten Diggins and co-authors!
Diggins KE, Greenplate AR, Leelatian N, Wogsland CE, Irish JM. Characterizing cell subsets in heterogeneous tissues using marker enrichment modeling. Nature Methods PMC5330853. Pubmed. DOI.
► This peer-reviewed methods research followed Diggins et al., Methods 2015 and created a new algorithm for machine learning cell identity and coined the term “cytotype” to describe a quantified cell identity. Marker Enrichment Modeling (MEM) generates human- and machine-readable cytotype labels and works with a wide range of data, including fluorescence and mass cytometry, gene expression profiles, and single cell RNA-seq. R code for MEM is available free for academic use. We demonstrate the use of MEM in different types of cytometry data (fluorescence and mass cytometry) and in identifying cells in human brain tumors.
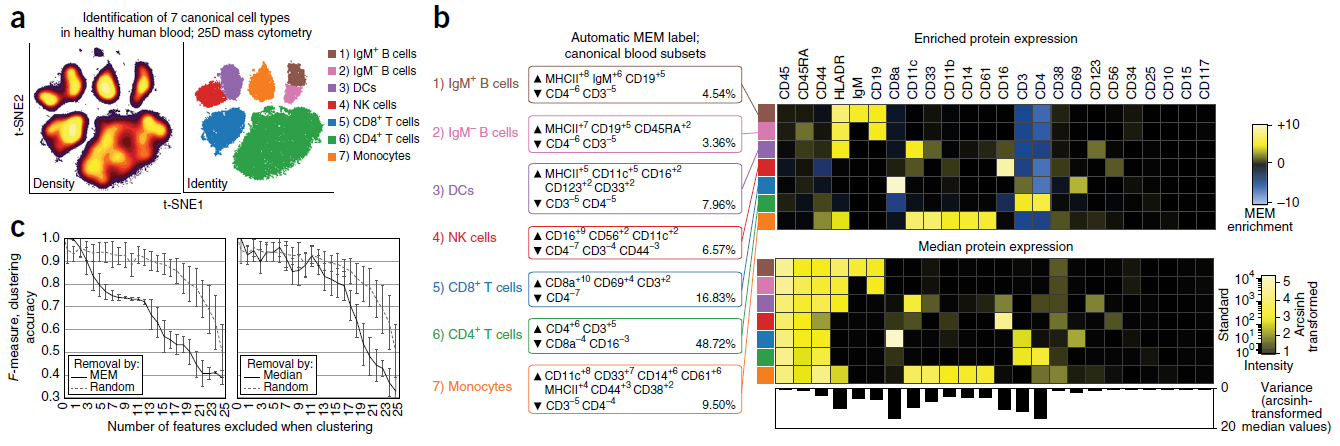
Diggins et al – Nature Methods 2017 MEM Fig 1
This work was covered by Vanderbilt and a lay summary is available here:
Marker Enrichment Modeling (MEM) code is available for academic and non-profit use and can be licensed for commercial use through VUeInnovations: https://mem.vueinnovations.com/
Update: we have published a detailed MEM protocol, scripts, and updated code (Diggins et al., Current Protocols in Cytometry 2018).
Diggins KE, Gandelman JS, Roe CE, Irish JM. Generating Quantitative Cell Identity Labels with Marker Enrichment Modeling (MEM). Current Protocols in Cytometry 2017. NIHMS912131. PMC in progress. Pubmed. DOI.