Challenge name
All models are wrong: are yours useful?
Purpose and relevance of the challenge
With this challenge, we aim to understand the current ability of the MRI field in modelling white matter (WM) tissue microstructure. The challenge will consist of a number of simulated WM digital environments (or “substrates”), on the scale of individual voxels, generated by changing in a controlled fashion a range of microstructural features, including:
- self-diffusivity (D, um2/ms)
- intra-fibre volume fraction (f, unitless)
- fibre radius index [1] (a, um)
- permeability (k, um/ms).
Participants will be given the simulated signal acquired from the acquisition protocols PGSE, DDE and DODE of sub-challenge #1 and #3. Participants are then asked to estimate any (or all) of the above microstructural features, according to their proposed model of WM microstructure. Participants are also encouraged to submit potential biomarkers that may be indicative of these features even if they do not directly estimate them. The outcome of this challenge will be an evaluation of the accuracy and sensitivity of microstructural parameter estimation.
4 winners will be determined by the most accurate estimates of each of the four indices.
Datasets
The dataset for this challenge is composed of 3D substrates of WM tissues designed following the methods in [2], which provide flexibility and control on microstructural features. The substrates are combined with the Monte Carlo based simulator in Camino [3] to provide synthetic dMRI signal.
The challenge data will be based upon 256 substrates representative of an array of white matter voxels. For each substrate, each of the 4 tissue parameters is swept through a range of physiologically realistic values (for example, D from 0.7-3 um2/ms, f from 0.4-0.7, etc.), and six different levels of signal-to-noise-ratio(SNR is hidden from participants).
The signal is simulated using the three acquisition protocols from sub-challenges #1 and #3: a PGSE acquisition with 1300 unique datapoints using a multi-shell strategy, a DDE with 2 different diffusion times, and a DODE with 5 different frequencies, 5 b-values, and 72 directions each. [In an extended version of this sub-challenge, planned for next year, we will work to make available any user-defined acquisition, in which each participant will be able to create and submit a “scheme” file describing the diffusion sensitizing gradients of any experimental protocol they would like to simulate.]
Link to Data: See Registration and Data Access tab
Participation (Data given to the participants)
The task will be to estimate any (or all) of the microstructural parameters of interest.
Participants are free to use all or any subset of the generated signals from any set of acquisitions.
Participants will be provided with the following files. Sample scripts to load the data will be provided for popular working environments (MATLAB, Python, C/C++).
Note that for each acquisition type, we include files for (1) a protocol description, (2) the acquisition parameters, and (3) the MR signal. For example, the dataset for each sequence is provided in a text file and consists of M columns, where M = 1536 is the total number of simulated voxels to be analysed (256 voxels * 6 noise levels) and N rows, where N is the number of measurements. [N=3011 measurements for PGSE, N=2000 measurements for DODE, N=800 for DDE).
- PGSE_ProtocolDescription.txt: Description of the acquisition parameters for the PGSE sequences
- PGSE_AcqParams.txt: Acquisition parameters for all measurements of the PGSE shell dataset. It is a NxA matrix with N rows and A columns, where N is the number of measurements and A is the number of sequence parameters.
- PGSE_Simulations.txt: Dataset for PGSE shell sequence
- DDE_ProtocolDescription.txt: Description of the acquisition parameters for the DDE sequences
- DDE_AcqParams.txt: Acquisition parameters for all measurements of the DDE dataset.
- DDE_Simulations.txt: Dataset for DDE sequences
- DODE_ProtocolDescription.txt: Description of the acquisition parameters for the DODE sequences
- DODE_AcqParams.txt: Acquisition parameters for all measurements of the DODE dataset.
- DODE_Simulations.txt: Dataset for DODE sequences
Submission
Participants are asked to submit any or all of the following for each of the 256 environments:
- self-diffusivity (D, um2/ms);
- intra-fibre volume fraction (f, unitless);
- fibre radius index [1] (a, um) computed as:
assuming a Gamma distribution P(a) of axonal radii a.
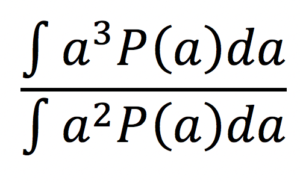
- permeability (k, um/ms).
Submission for each metric estimated must be submitted as a single compressed folder. All metrics will be submitted separately (for example separate submission for volume fraction and for permeability)
Within this folder will be two files.
- {SEQ}.txt, where SEQ = {’PGSE’, ’DDE’, ’DODE’} describes the acquisition used to estimate the index of choice. This file should contain a 1×1536 matrix. 1536 is the number of simulated substrates.
- Info.txt: this file contains relevant information for the challenge organizers. For all sub-challenges, this file must contain: the (1) submission name, (2), submission abbreviation, (3), team name, (4) team members who made meaningful contributions, (5) member affiliation, (6) brief one sentence submission description, (7) extended submission description (to enable reproducibility of methods) (8) all relevant citations, (9) observations (optional), and (9) relevant discussion points (optional). For sub-challenge #2 specifically, we also ask for: (10) number of free model parameters, (11) the type of model (signal/tissue), (12) noise assumptions, (13) model parameters estimation and algorithm optimization strategies, (14) pre-processing (outlier strategies), (15) data and subsets of data used to form predictions.
We want tp emphasize that f you wish to submit a potential biomarker that is not a direct estimator of the four parameters, but wish for it to be included in evaluation (we will assess correlation of markers both with each other and with ground truth data, and assess robustness to SNR) you may still submit this as a standard submission. This will simply be a 1×1536 matrix, Example submissions files will be made available with the data
Evaluation
Reference “gold standard” for this sub-challenge will be ground truth values for the four specific microstructural metrics, known by design of the numerical simulation study.
We will evaluate accuracy (or bias) and precision (or statistical dispersion) of dMRI measured metrics related to the microstructural features, compared to ground truth values known by design from numerical simulations. Specifically, accuracy for each estimated microstructural feature fj|j=1..4will be quantified by mean normalized error across the Ncmicrostructural scenarios taken into account:
Precision will be evaluated as interquartile range of the normalized error:

Overall score of accuracy and precision will be quantified by the mean squared error:
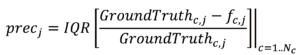
The code for the evaluation metrics will be open source. There will be a winning submission for each microstructural measure j, defined by the submission (for that feature) with the lowest scorej.

How to get the data
Please see “Registration and Data Access” Page.
Sub-challenge Chair
Marco Palombo <University College London>
Daniel Alexander <University College London>
[1] Alexander, D. C., Hubbard, P. L., Hall, M. G., Moore, E. A., Ptito, M., Parker, G. J., & Dyrby, T. B. (2010). Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage, 52(4), 1374-1389.
[2] Hall, M. G., & Alexander, D. C. (2009). Convergence and parameter choice for Monte-Carlo simulations of diffusion MRI. IEEE transactions on medical imaging, 28(9), 1354-1364.
[3] Y. B. P. A. Cook, S. Nedjati-Gilani, K. K. Seunarine, M. G. Hall, G. J. Parker, D. C. Alexander,, “Camino: Open-Source Diffusion-MRI Reconstruction and Processing,” in 14th Scientific Meeting of the International Society for Magnetic Resonance in Medicine, Seattle, WA, USA, May 2006, p. 2759.