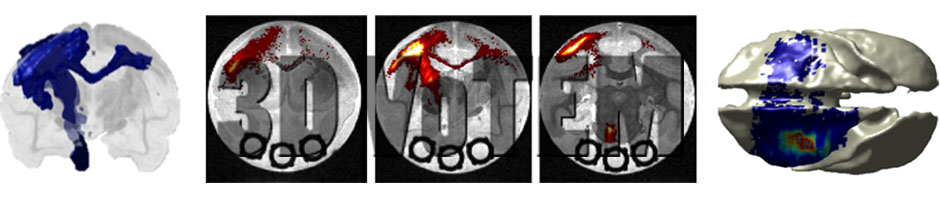
Diffusion MRI fiber tractography has become an extremely useful tool for mapping white matter pathways in-vivo. However, the anatomical accuracy of tractography results must be carefully validated to facilitate clinical usability and utility. Currently, neurosurgeons and scientists face the challenge of selecting the appropriate diffusion MRI acquisition and tractography methods without prior knowledge of the true anatomical connectivity of the brain. The main challenge facing tractography validation is the lack of anatomical ground truths. Here, we present three separate ground truth datasets, with which diffusion tractography can be compared and evaluated against. The first sub-challenge centers on an Anisotropic Diffusion Phantom, created by Synaptive Medical, which contains complex geometries and anisotropic microstructure intended to mimic linear, curving, crossing, and “kissing” fiber bundles. The second sub-challenge centers on ground truth dataset of the squirrel monkey brain, with MRI and BDA histological tracer injections on the same brain. The third sub-challenge is based on the dataset featured in [1], which features a high-angular and –spatial resolution dataset acquired on ex vivo Rhesus macaque monkey over 3 days of scanning, which can be compared to a wealth of knowledge from tracer studies on the same species.
High angular resolution diffusion MRI is acquired on all challenge datasets, which enables evaluation and comparisons of a variety of reconstruction and tracking techniques, ranging from Diffusion Tensor Imaging to more advanced, multi-shell methods. The task of the challenge, then, is to provide the most accurate reconstruction of fiber pathways in both a physical phantom and real brain tissue.
Previously, diffusion MRI challenges have focused on recovering intra-voxel fiber geometries using synthetic data (HARDI Reconstruction Challenge [2][6]) and 2D physical phantoms (Sparse Reconstruction Challenge [3]). Similarly, tractography challenges have provided insight into the effects of different acquisition settings, reconstruction techniques, and tracking parameters on tract validity by comparing results to ground truth physical phantom fiber configurations (Tractometer [4] and Fiberfox [5]). Recently, more clinically relevant challenges have been proposed, for example, challenge benchmarks DTI tractography of the pyramidal tract against tracts reconstructed by neurosurgeons and radiologists.
The proposed challenge is the first diffusion tractography challenge to include a three-dimensional physical phantom (sub-challenge #1), which models the intricate details of brain white matter microstructure, such as crossing and bending fibers, and allows testing of an array of acquisition conditions and tractography methods intended to extract these features. Additionally, this is the first tractography challenge to include histologically derived white matter tracts (sub-challenge #2 and #3). Histology adds an extra layer of realism to both data acquisition and potential tractographic challenges –including not only the true anatomical complexity of white matter pathways, but also tracer sensitivity on the scale of microns. The histological challenges, then, are to most accurately replicate the anatomical connections as identified by the histological tracers. Together, these three sub-challenges present a unique three-dimensional opportunity to validate diffusion tractography. These are offered in combination with our industry sponsor, Synaptive Medical, as well as NIH funded research (Anderson, Landman).
References
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C. Anatomical Accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proceedings of the National Academy of Sciences. 2016; 111(46): 16574-16579.
- Daducci A, Canales-Rodriguez EJ, Descoteaux M, Garyfallidis E, Gur Y, Lin YC, et al. Quantitative comparison of reconstruction methods for intra-voxel fiber recovery from diffusion MRI. IEEE transactions on medical imaging. 2014;33(2):384-99. doi: 10.1109/TMI.2013.2285500. PubMed PMID: 24132007.
- Ning L, Laun F, Gur Y, DiBella EV, Deslauriers-Gauthier S, Megherbi T, et al. Sparse Reconstruction Challenge for diffusion MRI: Validation on a physical phantom to determine which acquisition scheme and analysis method to use? Med Image Anal. 2015;26(1):316-31. doi: 10.1016/j.media.2015.10.012. PubMed PMID: 26606457; PubMed Central PMCID: PMCPMC4679726.
- Cote MA, Girard G, Bore A, Garyfallidis E, Houde JC, Descoteaux M. Tractometer: towards validation of tractography pipelines. Med Image Anal. 2013;17(7):844-57. doi: 10.1016/j.media.2013.03.009. PubMed PMID: 23706753.
- Neher PF, Laun FB, Stieltjes B, Maier-Hein KH. Fiberfox: facilitating the creation of realistic white matter software phantoms. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2014;72(5):1460-70. doi: 10.1002/mrm.25045. PubMed PMID: 24323973.
- https://www.biorxiv.org/content/early/2016/11/07/084137.article-metrics