This document describes the preprocessing steps performed before manually-guided tractography in the TractEM project. Code to perform the preprocessing may be found on github. Image registrations and transformations were performed using MATLAB and SPM 12. Diffusion reconstructions were performed using DSI Studio.
We began with a diffusion-weighted image set that had already been adjusted for head motion, eddy current effects, and distortion. For instance, the obtained HCP data sets were preprocessed as described by Glasser (2013).
A T1-weighted anatomical image from the same subject was coregistered to a b=0 image from the diffusion-weighted image set, if necessary.
A diffusion tensor reconstruction was performed on the diffusion-weighted image set to produce a fractional anisotropy image in the subject’s native space.
The DTI fractional isotropy image was registered to a Talairach space template image using an affine transformation. The template image was generated by applying the “pooled” affine transformation of Lancaster 2007 to the probabilistic ICBM152-space fractional anisotropy template image from the FSL atlas distribution (Mori 2005, Wakana 2007, Hua 2008).
The affine transformation was applied to the diffusion-weighted images, and the appropriate corresponding adjustment was made to the b vectors.
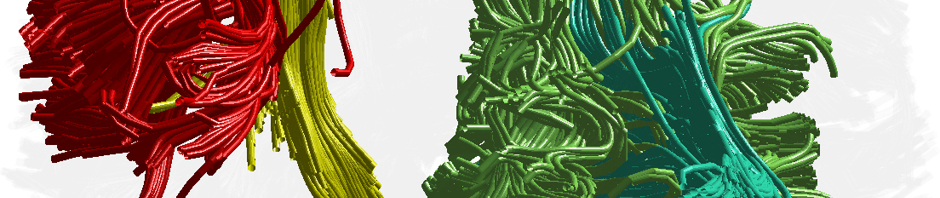
A Generalized Q-sampling Imaging (GQI) reconstruction (Yeh 2010), or a 4th-order spherical harmonic Q-ball reconstruction (QBI) (Tuch 2004, Descoteaux 2007), was applied to the transformed Talairach space diffusion-weighted images. GQI works best with a fairly dense sampling of directions and shells (e.g. Human Connectome Project), whereas QBI is applicable to less densely sampled single-shell data.
To provide the subject-specific lobar masks that are used in some of the tractography protocols, a multi-atlas segmentation algorithm was applied to the subject’s native space T1 image (Asman 2013,2014). This segmentation was transformed to Talairach space using the affine transformation determined above.
References
Asman AJ, Landman BA. Non-local statistical label fusion for multi-atlas segmentation. Medical image analysis. 2013;17(2):194-208. doi:10.1016/j.media.2012.10.002.
Asman AJ, Landman BA. Hierarchical Performance Estimation in the Statistical Label Fusion Framework. Medical image analysis. 2014;18(7):1070-1081. doi:10.1016/j.media.2014.06.005.
Descoteaux M, Angelino E, Fitzgibbons S, Deriche R. Regularized, fast, and robust analytical Q-ball imaging. Magn Reson Med. 2007 Sep;58(3):497 510. PubMed PMID: 17763358.
Glasser MF, Sotiropoulos SN, Wilson JA, et al. The Minimal Preprocessing Pipelines for the Human Connectome Project. NeuroImage. 2013;80:105 124. doi:10.1016/j.neuroimage.2013.04.127.
Lancaster JL, Tordesillas-Gutiérrez D, Martinez M, Salinas F, Evans A, Zilles K, Mazziotta JC, Fox PT. Bias between MNI and Talairach coordinates analyzed using the ICBM-152 brain template. Hum Brain Mapp. 2007 Nov;28(11):1194-205. PubMed PMID: 17266101.
Tuch DS. Q-ball imaging. Magn Reson Med. 2004 Dec;52(6):1358-72. PubMed PMID: 15562495.
Yeh FC, Wedeen VJ, Tseng WY. Generalized q-sampling imaging. IEEE Trans Med Imaging. 2010 Sep;29(9):1626-35. doi: 10.1109/TMI.2010.2045126. Epub 2010 Mar 18. PubMed PMID: 20304721.
freedietlinks.com
이 아이들이 믿음직스럽지 못하다고 느끼지만…
https://glowing.com/external/link?next_url=https%3A%2F%2Fwww.hz-wallpaper.com
Great post. I was checking constantly this blog
and I’m impressed! Very useful informatioon specifically the last part 🙂 I care for such info a lot.
I was looking for this certain info for a very long time.
Thank you and good luck.
modokicase.com
그는 손을 내밀고 안도의 한숨을 내쉬었습니다. “와서… 도와주세요.”
https://cse.google.com.ag/url?sa=t&url=http%3A%2F%2Fclemonsjerseys.com%2F
This site certainly has all of the info I needed about
thi subject and didn?t know who to ask.
iGaming 분야에서 혁신적이고 표준화된 콘텐츠를 제공하는 최신 프라그마틱 게임은 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 지원하여 고객에게 엔터테인먼트를 제공합니다.
프라그마틱 슬롯 체험
프라그마틱은 늘 새로운 기술과 아이디어를 도입하죠. 이번에 어떤 혁신이 있었는지 알려주세요!
http://holyshirtsandpants.net/
http://www.iavstudios.com/
https://plus-toto.com/
Hi there! Someone in my Facebook group shared this website with us
so I came to give it a look. I’m definitely enjoying the information.
I’m book-marking and will be tweeting this to my followers!
Wonderful blog and brilliant design and style.
프라그마틱 플레이의 슬롯 포트폴리오는 250개 이상의 게임으로 다양한 화폐 및 33개 언어로 전 세계 시장에 제공됩니다.
프라그마틱 무료
프라그마틱의 게임은 정말 다양한데, 어떤 테마의 게임을 가장 좋아하나요? 나눠주세요!
https://www.freedietlinks.com
https://themarketlobby.com/
http://www.iavstudios.com/
nsskart.com
Hongzhi 황제는 “가져가세요. 이 물건이 얼마나 생산할 수 있는지보고 싶습니다! “라고 말했습니다.
http://adsfac.eu/search.asp?cc=CHS001.8692.0&gid=31807513586&mt=b&nt=g&nw=s&stt=psn&url=https%3A%2F%2Fmaseraticlubuae.com%2F
amruthaborewells.com
Xie Qian은 그것을 보고 갑자기 믿을 수 없는 것을 발견했습니다.
Great beat ! I wish to apprentice while you amend your web site, how can i subscribe for a blog site?
The account aided me a acceptable deal. I had
been a little bit acquainted of this your broadcast offered bright
clear concept
digiyumi.com
“말이 되네요.” Lu Wenli의 눈이 빛났습니다. “감히 물어보십시오 …”
Hello.This article was extremely fascinating, especially since I was browsing for
thoughts on this issue last Wednesday.
현재 프라그마틱 게임은 iGaming에서 혁신적이고 표준화된 엔터테인먼트 콘텐츠를 제공하는 선도적인 업체 중 하나입니다.
프라그마틱 무료
프라그마틱은 늘 새로운 기술과 아이디어를 도입하죠. 이번에 어떤 혁신이 있었는지 알려주세요!
https://www.qtantra.com
https://www.unex-tech.com
https://wqeqwrwqr.weebly.com/
What’s up i am kavin, its my first time to commenting anywhere,
when i read this post i thought i could also make comment due to this
sensible paragraph.
agonaga.com
Hongzhi 황제는 천천히 씹었습니다. 이것은 어떤 맛입니까?
프라그마틱이 제공하는 슬롯으로 풍부한 경험을 즐기세요.
프라그마틱 홈페이지
프라그마틱은 다양한 언어와 화폐를 지원하는데, 이로 인해 글로벌 유저들에게 높은 평가를 받고 있어요.
https://okgasda.weebly.com/
https://www.ormtawindsor.com
https://www.tengerszemhotel.com
amruthaborewells.com
Fang Jifan은 마음 속으로 불평을 금할 수 없었습니다. 이 남자는 돼지를 죽이기 전에 인사도하지 않았습니다.
homefronttoheartland.com
하지만 누가 알았겠어요… 눈 깜짝할 사이에 많은 집주인들이 문을 두드렸습니다.
다양한 테마의 슬롯을 즐길 수 있는 250개 이상의 게임으로 구성된 프라그마틱 플레이의 슬롯 포트폴리오를 세계 시장에서 즐겨보세요.
프라그마틱 무료
프라그마틱에 대한 내용 정말 흥미로워요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 지식을 공유해보세요!
https://www.pst86g7q.site
https://www.gmailemails.com
https://www.ormtawindsor.com
topdelhiescorts.com
그것은 단지 … 이번 베이징 시찰도 Fang Jifan이 대중의 불만을 불러 일으켰습니다.
http://canasvieiras.com.br/redireciona.php?url=https%3A%2F%2Fchromehelmet.com%2F
baseballoutsider.com
그러나 Liu Jie와 같은 사람들이 그렇게 자신감이 있다고 누가 생각했을까요?
Thank you for the auspicious writeup. It in fact was a amusement account it.
Look advanced to far added agreeable from you! However, how could we communicate?
modernkarachi.com
왕노인은 깜짝 놀라며 매우 의아해하며 물었다.
Exceptional post but I was wondering if you could
write a litte more on this topic? I’d be very thankful
if you could elaborate a little bit more. Bless you!
Excellent pieces. Keep posting such kind of info on your page.
Im really impressed by your site.[X-N-E-W-L-I-N-S-P-I-N-X]Hey there, You have done an incredible job.
I will certainly digg it and individually recommend to my friends.
I am confident they will be benefited from this site.
프라그마틱 슬롯으로 더욱 흥미진진한 게임을 즐겨보세요.
프라그마틱 슬롯 무료
프라그마틱과 관련된 내용 감사합니다! 또한, 제 사이트에서도 프라그마틱에 대한 정보를 찾아보실 수 있어요. 서로 이야기 공유하며 더 많은 지식을 얻어가요!
https://www.wilsonamado.com
https://www.rmchorseshoeranch.com
https://www.cbhlyon.com
프라그마틱 슬롯에 대한 글 정말 잘 읽었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 교류하며 더 많은 지식을 얻어보세요!
프라그마틱슬롯
프라그마틱 슬롯에 대한 정보가 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 내용을 찾아보세요. 함께 이야기 나누면서 더 많은 지식을 얻어가요!
https://www.qi85jboo8.site
http://lasixwaterpill.site
http://pharmacymay.online
agonaga.com
여자로서 드라마에 몰입하기 쉽고 깊게 생각하지 않는다.
???sm 슬롯
이렇게 되면 죽고 사는 문제인데 어찌 충성심이 있겠습니까?
https://canadalaptopfan.com/
프라그마틱이 제공하는 슬롯으로 풍부한 경험을 즐기세요.
프라그마틱 플레이
프라그마틱과 관련된 이 글 정말 잘 읽었어요! 더불어 저도 제 사이트에서 프라그마틱에 대한 새로운 정보를 공유 중이에요. 함께 나누면 더 좋을 것 같아요!
https://www.dinotri.com
http://sildenafilfp.online
https://www.buyviagrasoft.site
프라그마틱 관련 글 읽는 것이 정말 즐거웠어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 이야기 나누면서 더 많은 지식을 쌓아가요!
프라그마틱
프라그마틱은 항상 훌륭한 게임을 만들어냅니다. 이번에 새롭게 출시된 게임은 정말 기대되는데요!
http://viagraism.online
http://zadoco.site
https://www.buylevaquin.site
Cialis Precio Farmacia
I join. I agree with told all above. We can communicate on this theme. Here or in PM.
Cialis 5 mg prezzo cialis prezzo cialis 5 mg prezzo
agonaga.com
답변이 직접 죽인다면 누가 기꺼이 그것을 증명할 것입니까?
louisgalaxy.com
하지만 너무 늦었습니다. 한 번 살펴보고 나중에 추가할 수 있습니다. 이것은 예의입니다.
https://canadalaptopfan.com/
agonaga.com
결국 … 전임자들이 길을 걸어 미래의 사람들이 갈 길이 없다고 점점 더 느끼게 만들었습니다.
Aviator Spribe казино как играть
You have hit the mark. In it something is also I think, what is it good idea.
Добро пожаловать в захватывающий мир авиаторов! Aviator – это увлекательная игра, которая позволит вам окунуться в атмосферу боевых действий на небе. Необычные графика и захватывающий сюжет сделают ваше путешествие по воздуху неповторимым.
Заработайте крупный выигрыш в игре Aviator Spribe казино играть на евро уже сегодня!
Aviator игра позволит вам почувствовать себя настоящим пилотом. Вам предстоит совершить невероятные маневры, выполнять сложные задания и сражаться с противниками. Улучшайте свой самолет, чтобы быть готовым к любым ситуациям и становиться настоящим мастером.
Основные особенности Aviator краш игры:
1. Реалистичная графика и физика – благодаря передовой графике и реалистичной физике вы почувствуете себя настоящим пилотом.
2. Разнообразные режимы игры и миссии – в Aviator краш игре вы сможете выбрать различные режимы игры, такие как гонки, симулятор полетов и захватывающие воздушные бои. Кроме того, каждая миссия будет предлагать свои собственные вызовы и задачи.
3. Улучшение и модернизация самолетов – в игре доступны различные модели самолетов, которые можно покупать и улучшать. Вы сможете устанавливать новое оборудование, улучшать двигательность и мощность своего самолета, а также выбирать различные варианты окраски и декорации.
Aviator краш игра – это возможность испытать себя в роли авиатора и преодолеть все сложности и опасности воздушного пространства. Почувствуйте настоящую свободу и адреналин в Aviator краш игре онлайн!
Играйте в «Авиатор» в онлайн-казино Pin-Up
Aviator краш игра онлайн предлагает увлекательную и захватывающую игровую атмосферу, где вы становитесь настоящим авиатором и сражаетесь с самыми опасными искусственными интеллектами.
В этой игре вы должны показать свое мастерство и смекалку, чтобы преодолеть сложности многочисленных локаций и уровней. Вам предстоит собирать бонусы, уклоняться от препятствий и сражаться с врагами, используя свои навыки пилотирования и стрельбы.
Каждый уровень игры Aviator краш имеет свою уникальную атмосферу и задачи. Будьте готовы к неожиданностям, так как вас ждут захватывающие повороты сюжета и сложные испытания. Найдите все пути к победе и станьте настоящим героем авиатором!
Авиатор игра является прекрасным способом провести время и испытать настоящий адреналиновый разряд. Готовы ли вы стать лучшим авиатором? Не упустите свой шанс и начните играть в Aviator краш прямо сейчас!
Aviator – играй, сражайся, побеждай!
Aviator Pin Up (Авиатор Пин Ап ) – игра на деньги онлайн Казахстан
Aviator игра предлагает увлекательное и захватывающее разнообразие врагов и уровней, которые не оставят равнодушными даже самых требовательных геймеров.
Враги в Aviator краш игре онлайн представлены в самых разных формах и размерах. Здесь вы встретите группы из маленьких и быстрых врагов, а также огромных боссов с мощным вооружением. Разнообразие врагов позволяет игрокам использовать разные тактики и стратегии для победы.
Кроме того, Aviator игра предлагает разнообразие уровней сложности. Выберите легкий уровень, чтобы насладиться игровым процессом, или вызовите себе настоящий вызов, выбрав экспертный уровень. Независимо от выбранного уровня сложности, вы получите максимум удовольствия от игры и окунетесь в захватывающий мир авиаторов.
Играйте в Aviator и наслаждайтесь разнообразием врагов и уровней, которые позволят вам почувствовать себя настоящим авиатором.
sm-online-game.com
그 고통이 헛되지 않았다는 사실에 그의 마음은 다소… 동의하지 않는 것 같습니다.
จำนำรถใกล้ฉัน
hihouse420.com
첫 번째 목적지는 당연히 참파인데, 참파 다음에는 어디로 가야 할까요?
프라그마틱 슬롯에 대한 정보가 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 내용을 찾아보세요. 함께 이야기 나누면서 더 많은 지식을 얻어가요!
프라그마틱플레이
프라그마틱에 대한 글 읽는 것이 정말 즐거웠어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 발전하며 더 많은 지식을 얻어보세요!
https://www.aufootballjersey.com
https://www.mihiroseiki.com
https://www.nutrapia.com
10yenharwichport.com
모두가 이미 “남자들, 내가 말했어. “라는 말을 생각한 것 같습니다.
https://louisgalaxy.com/
lfchungary.com
이곳은 인구 밀도가 가장 높은 곳으로 재난이 발생하면 얼마나 많은 사람들이 죽을지 모릅니다.
hihouse420.com
Xu Jing이 조화를 이루는 것이 매우 중요합니다. 적어도 그는 편심한 Wang Shouren을 화나게하지 않을 것입니다.Xu Pengju는 이빨로 몸의 천 조각을 찢고 상처를 겹겹이 붕대로 감았습니다.
프라그마틱 게임은 iGaming의 선도적인 콘텐츠 제공 업체로, 최고 품질의 엔터테인먼트를 제공하기 위해 모바일 중심 다양한 포트폴리오를 갖추고 있습니다.
http://www.pragmatic-game.com
프라그마틱은 늘 새로운 기술과 아이디어를 도입하죠. 이번에 어떤 혁신이 있었는지 알려주세요!
https://www.buylevaquin.site
https://www.murayah.com
https://www.cicoresky.com
Today, I went to the beach front with my kids. I found a sea shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She put the shell to her ear and screamed. There was a hermit crab inside and it pinched her ear. She never wants to go back! LoL I know this is completely off topic but I had to tell someone!
bitcoin price usd
hihouse420.com
이것이 Fang Jifan이 이 지도를 만든 원래 의도입니다.
lfchungary.com
왕 부시는 선글라스를 벗고 유심히 살펴보니 사실이었다.
dota2answers.com
왕자는 지금까지 자손이 없었기 때문에 오랫동안 사람들이 너무 많은 교제를 하게 되었습니다.
pragmatic-ko.com
잘 생긴 두 남자는 감동적이었고 그들이 울 때 호랑이는 다시 한 번 고개를 숙이고 감사했습니다.
다양한 화폐로 지원되는 프라그마틱 슬롯은 세계 각지의 플레이어에게 열린 기회를 제공합니다.
프라그마틱 슬롯 무료 체험
프라그마틱 슬롯에 대한 글 정말 잘 읽었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 교류하며 더 많은 지식을 얻어보세요!
https://www.rubiconfc.com
http://customercaresupportnumber.com/
https://www.domopravitel.com
agenbet88score.com
대신 Liu Jian과 다른 사람들은 무릎을 꿇고 낮은 머리 위치에 앉았습니다.
https://images.google.am/url?q=https%3A%2F%2Fwww.agenbet88score.com%2F
pragmatic-ko.com
Fang Jifan을 꾸짖을 기회를 잡을 때마다 Wang Bushi는 주저하지 않았습니다.
pragmatic-ko.com
그러나 그는 어안이 벙벙한 척만 할 뿐이었다. “폐하께서 구두 명령을 내리셨습니다.”
최근의 프라그마틱 게임은 iGaming 분야에서 혁신적이고 표준화된 엔터테인먼트 콘텐츠를 선보이는 선도적인 업체입니다.
프라그마틱플레이
프라그마틱의 게임은 정말 다양한데, 어떤 테마의 게임을 가장 좋아하나요? 나눠주세요!
https://www.kinohooutyx2.site
https://www.site-rapido.com
https://www.b4closing.com
Мы предоставляем услуги Строительство Афреймов под Ключ в Алматы, обеспечивая полный цикл работ от проектирования до завершения строительства. Наша команда опытных специалистов гарантирует высокое качество строительства и индивидуальный подход к каждому клиенту. Работаем с современными технологиями и материалами, чтобы создать дом вашей мечты в соответствии с вашими потребностями и ожиданиями.
hihouse420.com
이쯤 되면 당시나 후대나 대승리로 여겨진다.
mikschai.com
그러므로… 새로운 양돈 작업장은 이 문제를 해결합니다.
lfchungary.com
하녀의 경우 몸이 견딜 수 없게 되면 정말 걱정스러운 일입니다.
What’s up friends, pleasant article and good arguments commented here, I am truly enjoying by these.
My page site#:
https://gotartwork.com/Blog/%D0%BA%D0%B0%D0%BA-%D1%83%D1%87%D0%B5%D0%BD%D0%B8%D0%BA%D1%83-11-%D0%BA%D0%BB%D0%B0%D1%81%D1%81%D0%B0-%D0%BE%D0%BF%D1%80%D0%B5%D0%B4%D0%B5%D0%BB%D0%B8%D1%82%D1%8C%D1%81%D1%8F-%D1%81-%D0%B2%D1%8B%D0%B1%D0%BE%D1%80%D0%BE%D0%BC-%D0%B1%D1%83%D0%B4%D1%83%D1%89%D0%B5%D0%B3%D0%BE-%D0%B2%D1%83%D0%B7%D0%B0-%D0%B2-2024-%D0%B3%D0%BE%D0%B4%D1%83-%D0%BF%D1%80%D0%B0%D0%BA%D1%82%D0%B8%D1%87%D0%B5%D1%81%D0%BA%D0%B8%D0%B5-%D1%81%D0%BE%D0%B2%D0%B5%D1%82%D1%8B-%D0%B8-%D1%80%D0%B5%D0%BA%D0%BE%D0%BC%D0%B5%D0%BD%D0%B4%D0%B0%D1%86%D0%B8%D0%B8/262486/
https://forvseh.blogspot.com/2024/02/2024_23.html
http://forum.gold-forum.ru/index.php?showtopic=70321
http://secretvk.bestbb.ru/viewtopic.php?id=238#p266
http://dianov.bget.ru/forum/thread53952.html#1216734
Hello, yes this paragraph is truly pleasant and I have learned lot of things from it concerning blogging. thanks.
My#page#site#:
http://torica.4adm.ru/viewtopic.php?f=6&t=1825
https://school-toksovo.ru/forum/messages/forum1/topic297/message467/?result=new#message467
https://devochka.bestff.ru/viewtopic.php?id=6391#p60078
http://ya.10bb.ru/viewtopic.php?id=2662#p4904
https://crazyworldmen.livejournal.com/90311.html?newpost=1
Thanks for sharing your thoughts. I truly appreciate your efforts and I will be waiting for your further post thank you once again.
http://borderforum.ru/viewtopic.php?f=26&t=8451
http://poker.forum-top.ru/viewtopic.php?id=642#p2510
https://enkor.ru/forum/viewtopic.php?p=18595#18595
http://eclyceum.bbok.ru/viewtopic.php?id=393#p24893
http://evol.5bb.ru/viewtopic.php?id=998#p10384
http://rabotaref.forum-top.ru/viewtopic.php?id=3476#p7127
Wow, superb weblog format! How long have you been running a blog for?
you make running a blog glance easy. The full glance of your site is magnificent, as neatly as the content material!
You can see similar: sklep online and here sklep
pragmatic-ko.com
우아하고 섬세한 얼굴을 가진이 밝은 피부의 여성은 “소녀 Liang Ruying”이라고 말했습니다.
colorful-navi.com
나 자신, 왜 몰랐을까 이걸 알았다면 나도 책을 편집하러 왔을 텐데.
https://image.google.mv/url?q=https%3A%2F%2Fwww.agenbet88score.com%2F
프라그마틱 플레이의 슬롯으로 독특한 게임 세계를 탐험하고 풍부한 보상을 얻으세요.
프라그마틱
프라그마틱의 무료 게임 옵션은 정말로 흥미진진한데요. 어떤 무료 게임을 추천하시나요?
https://www.pihmctt.com
https://www.genericsingulair.site
https://supervil.com/hot/
smcasino-game.com
그러나 고개를 들어 그 손의 주인을 본 순간 그는 입을 다물었다.
shopanho.com
때때로 Ouyang Zhi는 단순히 그들을 쫓아 내고 모든 것을 스스로했습니다.
parrotsav.com
Zhu Houzhao는 “내 아들이 서두르고 있습니다. “라고 말했습니다.
andrejpos.com
사람들이 그 효과를 보지 못한다면 이 사람들은 그것을 믿지 않을 것입니다.
smcasino7.com
Fang Jifan은 학자의 코를 가리키고 욕하는 것과 같은 약간 대담했습니다.
프라그마틱 플레이의 슬롯 포트폴리오는 250개 이상의 게임으로 다양한 화폐 및 33개 언어로 전 세계 시장에 제공됩니다.
프라그마틱슬롯
프라그마틱은 다양한 언어와 화폐를 지원하는데, 이로 인해 글로벌 유저들에게 높은 평가를 받고 있어요.
https://vispills.com/
https://aintec.net/hot/
https://njshidoo.com/hot/
mega-slot77.com
Fang Jifan은 힘없이 어깨를 으쓱하고 Hongzhi 황제에게 손을 흔들었습니다.
https://maps.google.fm/url?q=https%3A%2F%2Fwww.agenbet88score.com%2F
mojmelimajmuea.com
피곤하고 마비 된 호랑이는 쉬고 있습니다. 모두 일찍 쉬어야합니다. 잘 자요!
apksuccess.com
이것은 그의 트럼프 카드, 그의 열반입니다.
프라그마틱에 대한 글 읽는 것이 흥미로웠어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 제공하고 있어요. 함께 발전하며 지식을 나눠봐요!
프라그마틱플레이
프라그마틱의 게임은 정말 다양한데, 최근에 출시된 것 중 어떤 게임이 가장 좋았나요? 공유해주세요!
http://www.iavstudios.com/
https://okgasda.weebly.com/
https://www.sujanews.com
jbustinphoto.com
멀리서 보면 분출하는 샘은 실제로이 큰 물고기에 의해 분출되었습니다.
apksuccess.com
황실 병원에서는 이미 그의 수준이 높다고 여겨진다.
smcasino-game.com
Xu Aoling은 당황했고 Fang Jifan의 제자는 … 맞았습니까?
windowsresolution.com
구의 눈물이 뺨을 타고 흘러내렸다. 그녀는 남편이 무슨 뜻인지 알았다.
프라그마틱은 늘 새로운 기술과 아이디어를 도입하죠. 이번에 어떤 혁신이 있었는지 알려주세요!
프라그마틱 게임
프라그마틱 슬롯에 대한 설명 감사합니다! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 이야기 나누면서 더 많은 지식을 얻어가요!
https://www.wilsonamado.com
https://habermahmutlar.com/hot/
https://www.woomintech.com
This is my first time pay a visit at here and i am actually pleassant to read everthing at one place.
http://www.google.ro/url?q=https://hottelecom.biz/id/
Wow, this paragraph is pleasant, my sister is analyzing such things,
so I am going to convey her. I saw similar here: E-commerce
raytalktech.com
Fang Jifan은 그의 얼굴에 부 자연스러운 표정을 지닌 Hongzhi 황제를 바라 보았습니다.
Wow, fantastic blog layout! How lengthy have you been running a blog for?
you made running a blog look easy. The total look of
your site is wonderful, as neatly as the content material!
You can see similar here sklep internetowy
프라그마틱 바카라
정말 12연대대대 사람들인 줄 알았다.
https://cse.google.co.zm/url?q=https%3A%2F%2Fwww.colorful-navi.com%2F
smcasino7.com
“참외 팔아요, 참외 팔아요, 싱싱한 수박, 와서 보세요, 싱싱해요…”
프라그마틱 슬롯을 무료로 즐겨보면 새로운 세계를 발견할 수 있습니다.
프라그마틷
프라그마틱의 게임은 정말 다양한데, 어떤 테마의 게임을 가장 좋아하나요? 나눠주세요!
https://lhmp888.com/link/
https://yluno.com/link/
https://ycbgl.com/link/
pchelografiya.com
Hongzhi 황제는 에너지가 넘치고 자신감이 넘쳤습니다.
hihouse420.com
Hongzhi 황제는 “새 지폐가 내일 배달 될 것입니다. 먼저보고 싶습니다. “라고 말했습니다.
mojmelimajmuea.com
Mao Ji의 유교에 대한 새로운 설명은 정말 신선합니다.
Aviator Spribe играть на гривны казино
Добро пожаловать в захватывающий мир авиаторов! Aviator – это увлекательная игра, которая позволит вам окунуться в атмосферу боевых действий на небе. Необычные графика и захватывающий сюжет сделают ваше путешествие по воздуху неповторимым.
Aviator Spribe играть с бонусом
에그벳 도메인
시중에 유통되는 돈도 전보다 2배에 불과하다.
https://images.google.de/url?sa=t&url=https%3A%2F%2Fwww.agenbet88score.com%2F
프라그마틱 게임은 iGaming 업계의 주요 플레이어로, 모바일 중심의 혁신적이고 표준화된 콘텐츠를 선보입니다.
프라그마틱 게임
프라그마틱 관련 정보 감사합니다! 제 사이트에서도 유용한 정보를 공유하고 있어요. 함께 소통하면서 발전하는 모습 기대합니다!
https://0cdnara.com/hot/
https://ianmccranor.com/link/
https://xingyangfu.com/link/
ttbslot.com
그래서 Fang Jifan이 와서 아이를 품에 안고 왔습니다.
ttbslot.com
오늘은 황제의 손자 명절의 날이어야 하고, 홍지 황제는 오랫동안 그것을 고대해 왔습니다.
Thank you for the auspicious writeup. It in fact used to be a entertainment account it.
Glance complex to more added agreeable from you! By the way, how could we keep in touch?
I saw similar here: Najlepszy sklep
Today, I went to the beach with my kids. I found a sea
shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She put the shell to her
ear and screamed. There was a hermit crab inside and it pinched her ear.
She never wants to go back! LoL I know this is completely off topic
but I had to tell someone! I saw similar here: E-commerce
I blog frequently and I seriously appreciate your content. This great article has really peaked my interest. I will book mark your site and keep checking for new information about once per week. I subscribed to your RSS feed as well.
RybelsusRybelsusRybelsusRybelsusRybelsus
qiyezp.com
하지만 지금은 이 숫자가 정말 무섭게 들립니다.
Good day! Do you know if they make any plugins to assist with SEO?
I’m trying to get my blog to rank for some targeted keywords but I’m not seeing very good
success. If you know of any please share. Cheers!
You can read similar blog here: Sklep internetowy
It’s very interesting! If you need help, look here: ARA Agency
ttbslot.com
Zhu Houzhao는 이빨을 드러내며 “무슨 소리야? “라고 말했습니다.
Hi, constantly i used to check webpage posts here in the early hours in the morning, since i enjoy to gain knowledge of more and more.
writing service
sandyterrace.com
Tatars는 여전히 칼을 휘두르고 말을 타고 질주하고 있었지만 이것을 알지 못했습니다.
세계 시장에 프라그마틱 플레이의 250개 이상의 게임으로 구성된 다양한 테마의 슬롯을 즐겨보세요.
프라그마틱
프라그마틱의 라이브 카지노는 정말 현장감 넘치게 즐길 수 있는데, 여기서 더 많은 정보를 얻을 수 있어 좋아요!
https://www.12315lb.cn/
https://cnipsa.com/link/
https://0cdnara.com/hot/
toasterovensplus.com
このブログは常に期待を超えてくれます。いつも素晴らしい内容をありがとう。
Hello there! Do you know if they make any plugins to assist
with SEO? I’m trying to get my blog to rank for some
targeted keywords but I’m not seeing very good gains.
If you know of any please share. Thank you! You can read
similar blog here: Najlepszy sklep
nikontinoll.com
갑자기 Ernv Liu는 무언가 잘못되었음을 느꼈습니다.
최신 프라그마틱 게임은 iGaming에서 혁신적이고 표준화된 콘텐츠를 선보이며 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 선택할 수 있습니다.
PG 소프트
프라그마틱의 게임을 플레이하면서 항상 신선한 경험을 얻을 수 있어 기뻐요. 여기서 더 많은 이야기를 나눠봐요!
https://www.sujanews.com
https://www.824989.com
https://www.ashspurr.com
I’m not sure why but this web site is loading incredibly slow for me. Is anyone else having this issue or is it a issue on my end? I’ll check back later on and see if the problem still exists.
reddit thread
Hello, this weekend is good for me, because this moment i am reading this enormous informative post here at my residence.
bewertung cardiobalance
Heya! I realize this is somewhat off-topic however I had to ask. Does running a well-established blog like yours take a large amount of work? I am brand new to writing a blog however I do write in my journal daily. I’d like to start a blog so I can share my experience and feelings online. Please let me know if you have any ideas or tips for brand new aspiring bloggers. Appreciate it!
depanten tablete
whoah this weblog is wonderful i like reading your posts. Stay up the good work! You understand, many people are searching round for this info, you could help them greatly.
ripper casino
I take pleasure in, lead to I found exactly what I used to be having a look for. You have ended my four day long hunt! God Bless you man. Have a nice day. Bye
бездепозитный бонус beep beep casino
Hi there, You’ve done a fantastic job. I will certainly digg it and personally suggest to my friends. I’m sure they’ll be benefited from this web site.
yabby casino no deposit bonus codes august 2023
gama casino регистрация
gama casino сайт
Российская компания реализует разборные гантели Gantel Razbornaya – у нас найдете замечательный ассортимент вариантов. Наборные снаряды дают эффективно выполнять силовые занятия в любом месте. Изделия для спорта отличаются функциональностью, универсальностью в использовании. Предприятие эффективно развивает и внедряет лучшие технологии, чтобы осуществить потребности наших тренирующихся. В изготовлении долговечных отягощений всегда применяются инновационные марки чугуна. Впечатляющий каталог интернет-магазина вариантов позволяет купить сборные гантели для продуктивной программы занятий. Для домашних занятий – это комфортный инвентарь с компактными габаритами и значительной фунциональности.
cougarsbkjersey.com
とても役立つ情報で満載です。感謝しています。
Hi, after reading this remarkable post i am too glad to share my knowledge here with mates.
https://toto-ro.net/bbs/board.php?bo_table=free&wr_id=3657
http://heemangfc.com/bbs/board.php?bo_table=free&wr_id=320930
https://kasa1986.or.kr:443/bbs/board.php?bo_table=free&wr_id=80930
http://dpart.co.kr/bbs/board.php?bo_table=free&wr_id=709687
http://netpinion.co.kr/bbs/board.php?bo_table=free&wr_id=57337
Hello, i believe that i noticed you visited my site so i got
here to return the choose?.I’m trying to to
find issues to enhance my website!I suppose its good enough to use a few of your ideas!!
Since the admin of this website is working, no hesitation very soon it will be well-known, due to its quality contents.
cbdus.click
Hey there this is kinda of off topic but I was wondering if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding experience so I wanted to get guidance from someone with experience. Any help would be greatly appreciated!
keramin krema lekarna
Why people still make use of to read news papers when in this technological globe everything is existing on web?
http://dicetable.co.kr/bbs/board.php?bo_table=free&wr_id=138812
http://aina-test-com.check-xserver.jp/bbs/board.php?bo_table=free&wr_id=970729
http://imimi.kr/bbs/board.php?bo_table=free&wr_id=6570
http://vertek.co.kr/bbs/board.php?bo_table=free&wr_id=168366
http://hgmall.co.kr/bbs/board.php?bo_table=free&wr_id=1260439
Hi to every one, the contents present at this website are actually awesome for people experience, well, keep up the good work fellows.
http://www.21rental.co.kr/bbs/board.php?bo_table=free&wr_id=33736
프라그마틱 관련 글 읽는 것이 정말 즐거웠어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 이야기 나누면서 더 많은 지식을 쌓아가요!
PG 소프트
프라그마틱 슬롯에 대한 이해가 높아지네요! 제 사이트에서도 유용한 프라그마틱 정보를 찾아볼 수 있어요. 함께 공유하고 발전해봐요!
https://munirahkasim.com/link/
https://www.vamiveta.com/
https://www.12315dc.cn/
Excellent blog you have here but I was wanting to know if you knew of any community forums that cover the same topics discussed here? I’d really love to be a part of group where I can get opinions from other knowledgeable individuals that share the same interest. If you have any suggestions, please let me know. Appreciate it!
Официальный сайт Gama casino
etsyweddingteam.com
日常生活で直接役立つ実用的な情報が豊富で、素晴らしい記事です。
Казино VODKA онлайн – играть в автоматы на деньги
Узнайте о захватывающем мире казино VODKA, где современный дизайн, разнообразие игровых автоматов и щедрые бонусы ждут каждого игрока. Погрузитесь в атмосферу слотов на деньги с казино VODKA.
Казино VODKA: Погружение в мир азартных развлечений
В мире азартных игр существует огромное количество казино, каждое из которых стремится привлечь внимание игроков своими уникальными предложениями и атмосферой. Одним из таких заведений является казино VODKA, которое предлагает своим посетителям захватывающие игровые возможности и неповторимый опыт азартных развлечений.
Виртуальное пространство казино VODKA
Казино VODKA официальный сайт водка предлагает своим клиентам широкий спектр азартных игр, доступных в виртуальном пространстве. От классических игровых автоматов до настольных игр, таких как рулетка, блэкджек и покер – здесь каждый игрок сможет найти что-то по своему вкусу. Современный дизайн и удобный интерфейс позволяют наслаждаться игровым процессом без каких-либо проблем или задержек.
Бонусы и акции
Одним из способов привлечения новых игроков и поощрения постоянных являются бонусы и акции. Казино VODKA не остается в стороне и предлагает своим клиентам различные бонусы за регистрацию, первые депозиты или участие в акциях. Эти бонусы могут значительно увеличить шансы на победу и сделать игровой процесс еще более увлекательным.
Безопасность и поддержка
Важным аспектом любого казино является обеспечение безопасности игроков и защита их личной информации. Казино VODKA придает этому особое внимание, используя передовые технологии шифрования данных и обеспечивая конфиденциальность всех транзакций. Кроме того, круглосуточная служба поддержки готова ответить на любые вопросы и помочь в решении возникающих проблем.
Заключение
Казино VODKA – это место, где каждый азартный игрок найдет что-то по своему вкусу. Богатый выбор игр, интересные бонусы и высокий уровень безопасности делают его привлекательным вариантом для тех, кто хочет испытать удачу и получить незабываемые эмоции от азартных развлечений. Сделайте свой первый шаг в мир азарта и испытайте удачу в казино VODKA уже сегодня!
qiyezp.com
Fang Jifan은 “어떤 사람들은 Baoding의 방향으로 변화하기를 희망하고 어떤 사람들은 …”라고 말했습니다.
Good day! Do you know if they make any plugins to assist with Search Engine Optimization? I’m trying to get my blog to rank for some targeted keywords but I’m not seeing very good results.
If you know of any please share. Kudos! I saw similar blog here: Link Building
freeflowincome.com
Zhou Yi는 여전히 기분이 좋았고 밤에 삼촌 집에 저녁을 먹으러갔습니다.
Transfer From Antalya Airport to Side | Transfer From Antalya Airport to Side
150 CC MOPED | https://tteaascar.com/wp-content/uploads/2024/03/150-cc-moped.webp
Antalya Rafting | Unforgettable Adventure on the Köprülü Canyon
etsyweddingteam.com
実用性が高く、読んでよかったです。素晴らしい記事です!
qiyezp.com
또한 많은 정기간행물에 얼마나 많은 논문이 있습니까?
thebuzzerpodcast.com
싱크대는 멀리 유리 작업장으로 바로 연결되며 냉각을 위해 많은 양의 물이 사용됩니다.
White water rafting antalya | An Adventure Filled with Excitement!
Вип Трансфер из аэропорта Анталья | Вип Трансфер из аэропорта Анталья
e ticaret nasıl yapılır | e ticaret nasıl yapılır İstanbul danışmanlık firması
Привет всем!
Наши услуги позволят вам заказать документы об образовании всех ВУЗов России с доставкой по РФ и оплатой после получения – надежно и удобно!
http://saksx-attestats.ru/
Наши услуги позволят вам заказать документы об образовании всех ВУЗов России с доставкой по РФ и оплатой после получения – надежно и удобно!
Закажите диплом института или техникума по доступной цене с гарантированной доставкой в любой уголок России без предоплаты!
bmipas.com
このブログはいつも私に新しい視点を提供してくれます。感謝しています。
qiyezp.com
Zhu Xiurong은 여전히 뭔가를 생각하는 것처럼 흥얼 거렸다.
Привет, дорогой читатель!
Заказывайте диплом любого Вуза России у нас на сайте конфиденциально и без предоплаты.
купить аттестат школы
Приобретите документы об образовании всех ВУЗов России с гарантированной подлинностью и доставкой по РФ без предварительной оплаты – просто, надежно, выгодно!
Приобретите диплом Гознака у нас по специальной цене с доставкой в любой регион России без предоплаты.
프라그마틱 게임은 선도적인 iGaming 콘텐츠 제공 업체입니다. 모바일 중심 다양한 포트폴리오와 고품질 엔터테인먼트를 자랑합니다.
프라그마틱 무료 슬롯
프라그마틱의 게임은 높은 퀄리티와 흥미진진한 스토리로 항상 눈길을 끌어요. 이번에 어떤 게임을 즐겼나요?
https://www.12315oh.cn/
https://www.12315ev.cn/
https://lotodarts.com/link/
Доброго дня!
Несмотря на все сложности и угрозу для здоровья, я продолжаю работать над дипломом, опираясь на найденные ресурсы.
Предлагаем всем желающим приобрести диплом университета России по выгодной цене с доставкой “под ключ”.
Желаю для каждого прекрасных оценок!
https://rudiplomirovana.com
купить диплом в великих луках
куплю диплом кандидата наук
купить диплом в россоши
купить диплом физика
купить диплом менеджера
Приветики, дорогие мои!!
Мои усилия по диплому получили поддержку через полезные ресурсы, найденные в сети.
Приобретайте диплом у нас и получите качественный документ без предоплаты, с доставкой в любую точку России.
где купить диплом
Желаю каждому прекрасных оценок!
купить диплом в иваново
купить диплом в майкопе
купить диплом в люберцах
купить диплом в березниках
купить диплом в благовещенске
Приветики, дорогие мои!!
Из-за диплома пришлось перейти на ночной график работы, что угрожало моему здоровью.
Наша компания предлагает конфиденциально заказать и приобрести диплом любого ВУЗа России.
Желаю вам всем не двоешных) отметок!
https://rudiplomirovana.com
купить диплом товароведа
купить диплом воспитателя
купить диплом в туймазы
купить диплом в железногорске
купить диплом в белорецке
Добрый день!
Диплом больше не является такой большой проблемой с доступом к интернет-ресурсам.
Недорого и без проблем приобрести диплом института, техникума, колледжа выгоднее всего у нас, с доставкой по РФ.
Желаю для каждого не двоешных) отметок!
https://eonline-diplomy.com
купить диплом в владивостоке
купить диплом в арзамасе
купить диплом о среднем
купить диплом в хабаровске
купить диплом в нефтекамске
Привет всем!
Диплом заставил меня работать ночами, что угрожало моему здоровью.
Приобретите диплом университета с гарантированной доставкой в любой город России с оплатой после получения.
Желаю вам всем не двоешных) отметок!
купить диплом бакалавра
купить диплом в уфе
купить диплом в дзержинске
купить диплом учителя
купить диплом в челябинске
купить диплом в обнинске
Доброго дня!
Решил работать над дипломом по ночам, но это только ухудшило ситуацию: здоровье страдает, глаза перенапрягаются.
Приобретайте диплом у нас и получите качественный документ без предоплаты с доставкой в любую точку России.
купить аттестат
Желаю всем отличных отметок!
купить диплом магистра
купить диплом в грозном
купить диплом в первоуральске
купить дипломы о высшем цены
купить диплом хореографа
Приветики, дорогие мои!!
Благодаря интернету, я нашел способы оптимизировать работу над дипломом и сделать ее менее вредной для здоровья.
Мы поможем вам купить диплом университета без предоплаты и доставим его в любой город России с гарантированной безопасностью.
Желаю вам всем пятерочных) отметок!
https://aurus-diploman.com/
старые дипломы купить
купить диплом в нижнекамске
купить диплом в чайковском
купить диплом логопеда
купить диплом педагога
Добрый день!
Диплом продолжает быть моим вызовом, но теперь у меня есть все необходимые ресурсы для его успешного завершения.
Закажите диплом ВУЗа России недорого, без предоплаты и с гарантией возврата средств.
https://adiplom-russian.com/
Желаю каждому прекрасных оценок!
купить диплом программиста
купить диплом в назрани
купить диплом старого образца
купить диплом в нефтекамске
купить диплом в комсомольске-на-амуре
Wow, awesome weblog layout! How long have you been running a blog for?
you made blogging look easy. The total look of your web site is excellent, let alone the
content material! You can see similar here sklep
Приветики, дорогие мои!!
Для диплома интернет стал источником полезных ресурсов, облегчивших задачу.
Поможем выбрать, сделать заказ и купить диплом любого учебного заведения в любом населенном пункте по самым низким ценам.
Желаю всем положительных оценок!
купить диплом автомеханика
купить диплом в миассе
купить диплом в кропоткине
купить свидетельство о рождении ссср
купить диплом в старом осколе
купить диплом о среднем образовании
Приветики, дорогие мои!!
Диплом стал моей главной проблемой, но благодаря интернету я нашел способы улучшить свою работу и сохранить здоровье.
Мы поможем вам купить диплом ВУЗа России недорого, без предоплаты и с гарантией возврата средств.
купить диплом в москве
Желаю вам всем честных оценок!
купить аттестат за 9 класс
купить диплом учителя
купить свидетельство о рождении ссср
купить диплом в чите
купить диплом бурильщика
otraresacamas.com
実用性が高く、具体的な情報に満ちた素晴らしい内容でした。
how much is a taxi from antalya airport to side | how much is a taxi from antalya airport to side
Доброго дня!
Продолжаю работать над дипломом, несмотря на все препятствия, и благодарю интернет за помощь в этом нелегком процессе.
Поможем вам выбрать, оформить заказ и приобрести диплом любого учебного заведения по самым низким ценам.
Желаю вам всем прекрасных оценок!
купить диплом о среднем образовании
купить диплом парикмахера
купить диплом в подольске
купить диплом в феодосии
купить диплом в симферополе
купить свидетельство о рождении ссср
Доброго дня!
Продуктивность моей работы над дипломом увеличилась благодаря онлайн-ресурсам.
Желаете заказать и купить диплом ВУЗа недорого без предоплаты на нашем сайте? Доставим в любую точку России.
Желаю вам всем отличных оценок!
https://aurus-diploman.com/
где купить диплом
купить диплом геодезиста
купить диплом в выборге
купить морской диплом
купить диплом в верхней пышме
Всем хорошего дня!
Диплом стал легче благодаря ресурсам, которые я нашел в интернете, ускоряя процесс написания.
Мы предлагаем купить диплом университета без предоплаты, с гарантированной доставкой по всей России.
Желаю всем прекрасных отметок!
https://aurus-diplomas.com/
купить диплом в белово
купить диплом в великих луках
купить диплом в кстово
купить диплом института
купить диплом в томске
Всем хорошего дня!
Диплом стал легче благодаря ресурсам, которые я нашел в интернете, ускоряя процесс написания.
У нас вы можете приобрести диплом университета без предоплаты, с доставкой по всей России.
купить аттестат
Желаю всем не двоешных) оценок!
купить диплом в анжеро-судженске
купить диплом лаборанта
купить диплом физика
купить диплом в заречном
куплю диплом цена
azithromycin otc usa
freeflowincome.com
면방직 작업장 주변에 마당 담을 치고, 마당 대문은 특수 요원이 지키고 있었다.
Добрый день!
Диплом стал более доступным заданием благодаря использованию онлайн-материалов.
Закажите диплом у нас без предоплаты, и мы доставим его вам в любой регион России с гарантированной конфиденциальностью.
Желаю всем пятерочных) оценок!
https://diplomys-asx.com
купить диплом в новотроицке
купить диплом ссср
купить диплом в кургане
купить диплом в верхней пышме
купить диплом тренера
Здравствуйте!
Несмотря на срыв сроков и угрозу для здоровья из-за диплома, я нашел в интернете способы быть более продуктивным.
Закажите диплом у нас – легко! Без предоплаты, с гарантией качества и доставкой по всей России.
Желаю вам всем прекрасных отметок!
купить диплом в москве
купить диплом в анапе
купить дипломы о высшем образовании цена
купить дипломы о высшем с занесением
купить диплом в симферополе
где купить диплом
Привет всем!
Стремлюсь преодолеть все препятствия, несмотря на ухудшение самочувствия из-за ночных занятий с дипломом.
Приобретите диплом университета с гарантированной доставкой в любой город России с оплатой после получения.
Желаю для каждого честных отметок!
https://russiany-diplomas.com
купить диплом в феодосии
купить диплом в бердске
купить диплом в энгельсе
купить диплом моториста
купить диплом в калининграде
Всем хорошего дня!
Интернет стал незаменимым помощником в написании диплома, обнаружив множество полезных ресурсов.
Поможем выбрать, сделать заказ и купить диплом любого учебного заведения в любом населенном пункте по самым низким ценам.
Желаю всем не двоешных) отметок!
https://russiany-diplomas.com
купить диплом в кирове
купить диплом в ялте
купить диплом о среднем образовании
купить диплом логиста
купить диплом в губкине
Всем хорошего дня!
Моя работа над дипломом стала продуктивнее благодаря активному использованию онлайн-материалов.
Приобретение диплома института, техникума или колледжа у нас – это просто, с доставкой по РФ.
Желаю всем честных оценок!
купить диплом
купить диплом математика
купить диплом медсестры
купить свидетельство о рождении ссср
купить диплом провизора
купить диплом в москве
Привет всем!
Работа над дипломом превратилась в кошмар, когда я начал откладывать выполнение и работать ночью, рискуя здоровьем.
У нас вы можете приобрести диплом университета без предоплаты, с доставкой по всей России.
Желаю для каждого пятерочных) отметок!
купить аттестат
купить диплом в тюмени
купить диплом гознак
купить диплом швеи
купить диплом в ельце
купить диплом в биробиджане
Привет всем!
Диплом заставил меня перейти на ночной режим работы, что пагубно сказалось на здоровье.
Мы предлагаем купить диплом университета без предоплаты, с гарантированной доставкой по всей России.
Желаю вам всем не двоешных) оценок!
https://plands-diplomy.com
купить диплом гознак
купить диплом железнодорожника
купить диплом в королёве
купить диплом в благовещенске
купить диплом в великом новгороде
Доброго дня!
Дипломное задание оказалось невероятно сложным, но поддержка из сети помогла мне в этом.
Приобретение диплома института, техникума или колледжа у нас – лучший вариант с доставкой по РФ.
Желаю всем прекрасных оценок!
https://frees-diplomy.com
купить диплом в усолье-сибирском
купить диплом с внесением в реестр
купить диплом в кирове
купить диплом во владимире
купить диплом в кемерово
Приветики, дорогие мои!!
Диплом заставил меня работать ночами, что угрожало моему здоровью.
Купите диплом у нас легко! Надежно, качественно, без предоплаты, с доставкой в любой город России.
Желаю каждому положительных оценок!
https://diplom-originalniy.com
купить диплом в мичуринске
купить диплом во владивостоке
купить диплом в энгельсе
купить диплом в ангарске
купить диплом охранника
Добрый день!
К счастью, интернет помог мне найти полезные ресурсы, которые значительно ускорили и облегчили мой труд с дипломом.
Приобретите диплом университета без предоплаты у нас и получите его с доставкой в любую точку России, качество гарантировано.
Желаю каждому не двоешных) оценок!
https://diplom-originalniy.com
купить диплом дизайнера
купить диплом в губкине
купить диплом в уссурийске
купить диплом в ульяновске
купить диплом медсестры
hello there and thank you for your info ? I have definitely
picked up something new from right here. I did however expertise some technical issues using this site, as I
experienced to reload the website a lot of times previous to I
could get it to load properly. I had been wondering if your web
hosting is OK? Not that I am complaining, but sluggish loading instances times will very frequently affect your placement in google and can damage your high-quality score if
advertising and marketing with Adwords. Anyway I am adding this RSS to my email and can look out for much more of your respective interesting content.
Ensure that you update this again very soon.
Добрый день!
Дипломное задание оказалось невероятно сложным, но поддержка из сети помогла мне в этом.
Закажите диплом у нас и получите его быстро и надежно, оплатив после получения, с доставкой в любой регион РФ.
Желаю каждому честных отметок!
купить диплом техникума
купить диплом в троицке
купить диплом агронома
купить диплом в люберцах
купить диплом образцы
купить диплом в казани
sandyterrace.com
Chenla 왕의 귀가 윙윙 거리며 잠시 기절했습니다.
Wow, wonderful weblog format!
How lengthy have you been blogging for? you
make blogging look easy. The whole glance of your site is great, let alone the content!
You can see similar here prev next and it’s was
wrote by Piper66.
Hi, always i used to check website posts here early in the daylight, for the reason that i like to find out more and more.
#be#jk3#jk#jk#JK##
аренда номера США
В нашем обществе, где диплом становится началом удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Факт наличия официального документа об образовании сложно переоценить. Ведь именно он открывает двери перед людьми, стремящимися начать трудовую деятельность или учиться в высшем учебном заведении.
В данном контексте мы предлагаем максимально быстро получить любой необходимый документ. Вы имеете возможность купить диплом старого или нового образца, что становится выгодным решением для человека, который не смог закончить образование, утратил документ или хочет исправить свои оценки. Все дипломы производятся с особой тщательностью, вниманием ко всем деталям. В итоге вы сможете получить документ, 100% соответствующий оригиналу.
Преимущества этого решения состоят не только в том, что вы сможете максимально быстро получить диплом. Процесс организовывается комфортно, с профессиональной поддержкой. От выбора нужного образца до консультаций по заполнению персональных данных и доставки в любое место страны — все находится под абсолютным контролем опытных мастеров.
Таким образом, для всех, кто ищет оперативный способ получения требуемого документа, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать длительного обучения и сразу переходить к достижению личных целей, будь то поступление в университет или начало удачной карьеры.
https://originality-diplomans.com
На сегодняшний день, когда диплом становится началом удачной карьеры в любой отрасли, многие ищут максимально быстрый путь получения качественного образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает дверь перед каждым человеком, желающим начать трудовую деятельность или продолжить обучение в университете.
Предлагаем очень быстро получить любой необходимый документ. Вы сможете приобрести диплом нового или старого образца, что будет удачным решением для человека, который не смог завершить образование или утратил документ. дипломы производятся аккуратно, с особым вниманием к мельчайшим нюансам, чтобы в результате получился полностью оригинальный документ.
Преимущество этого решения состоит не только в том, что можно оперативно получить свой диплом. Весь процесс организован удобно и легко, с профессиональной поддержкой. От выбора нужного образца документа до грамотного заполнения персональной информации и доставки по стране — все под абсолютным контролем опытных специалистов.
В итоге, для всех, кто пытается найти быстрый и простой способ получить требуемый документ, наша компания готова предложить выгодное решение. Приобрести диплом – значит избежать долгого обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в университет или старт успешной карьеры.
https://originality-diplomans.com
В нашем обществе, где диплом – это начало отличной карьеры в любой сфере, многие стараются найти максимально быстрый путь получения образования. Необходимость наличия официального документа сложно переоценить. Ведь диплом открывает дверь перед любым человеком, который хочет начать профессиональную деятельность или учиться в любом институте.
В данном контексте мы предлагаем очень быстро получить этот необходимый документ. Вы имеете возможность приобрести диплом, и это будет удачным решением для человека, который не смог закончить обучение, потерял документ или хочет исправить плохие оценки. Все дипломы производятся с особой аккуратностью, вниманием к мельчайшим деталям. На выходе вы получите документ, максимально соответствующий оригиналу.
Преимущества данного подхода заключаются не только в том, что вы сможете оперативно получить свой диплом. Весь процесс организовывается удобно и легко, с профессиональной поддержкой. Начиная от выбора необходимого образца до консультаций по заполнению личной информации и доставки по России — все под абсолютным контролем опытных специалистов.
Для тех, кто ищет максимально быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать долгого процесса обучения и не теряя времени переходить к своим целям, будь то поступление в ВУЗ или старт карьеры.
https://russiany-diplomas.com
На сегодняшний день, когда диплом – это начало успешной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Необходимость наличия официального документа об образовании переоценить попросту невозможно. Ведь именно диплом открывает двери перед каждым человеком, который желает вступить в сообщество профессионалов или продолжить обучение в ВУЗе.
Предлагаем оперативно получить любой необходимый документ. Вы сможете заказать диплом нового или старого образца, что становится выгодным решением для человека, который не смог завершить образование или утратил документ. Любой диплом изготавливается с особой аккуратностью, вниманием к мельчайшим нюансам, чтобы на выходе получился документ, максимально соответствующий оригиналу.
Плюсы такого решения заключаются не только в том, что можно быстро получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начав от выбора требуемого образца до правильного заполнения персональной информации и доставки в любое место России — все находится под полным контролем квалифицированных специалистов.
Всем, кто хочет найти максимально быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать продолжительного обучения и не теряя времени переходить к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
https://diplomana-asx.com
ilogidis.com
심지어 오스만 제국이 가장 경계했던 것은 정확히 씨족과 씨족의 친척이었습니다.
На сегодняшний день, когда диплом – это начало удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь именно он открывает дверь перед всеми, кто желает начать профессиональную деятельность или продолжить обучение в каком-либо институте.
Предлагаем максимально быстро получить этот важный документ. Вы имеете возможность заказать диплом, и это становится отличным решением для человека, который не смог завершить образование, утратил документ или хочет исправить свои оценки. Любой диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим деталям, чтобы на выходе получился 100% оригинальный документ.
Плюсы этого решения заключаются не только в том, что вы сможете оперативно получить диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора требуемого образца документа до консультации по заполнению личных данных и доставки в любой регион России — все будет находиться под абсолютным контролем наших мастеров.
Всем, кто хочет найти быстрый и простой способ получить требуемый документ, наша компания может предложить отличное решение. Приобрести диплом – значит избежать долгого процесса обучения и не теряя времени перейти к достижению своих целей, будь то поступление в ВУЗ или начало удачной карьеры.
https://russiany-diplomans.com
В современном мире, где диплом является началом отличной карьеры в любой отрасли, многие пытаются найти максимально простой путь получения образования. Наличие документа об образовании трудно переоценить. Ведь именно диплом открывает двери перед любым человеком, желающим вступить в сообщество профессионалов или учиться в университете.
Мы предлагаем оперативно получить любой необходимый документ. Вы имеете возможность приобрести диплом, что является отличным решением для человека, который не смог закончить обучение или потерял документ. Каждый диплом изготавливается с особой аккуратностью, вниманием ко всем деталям. В итоге вы сможете получить продукт, 100% соответствующий оригиналу.
Преимущества данного решения состоят не только в том, что можно оперативно получить диплом. Процесс организовывается удобно, с профессиональной поддержкой. Начав от выбора подходящего образца до точного заполнения персональной информации и доставки в любой регион страны — все находится под абсолютным контролем квалифицированных специалистов.
Для тех, кто пытается найти оперативный способ получения необходимого документа, наша компания может предложить отличное решение. Заказать диплом – это значит избежать долгого процесса обучения и сразу переходить к своим целям, будь то поступление в ВУЗ или начало профессиональной карьеры.
купить аттестат школы
В нашем обществе, где диплом является началом отличной карьеры в любой сфере, многие пытаются найти максимально быстрый путь получения образования. Факт наличия официального документа переоценить просто невозможно. Ведь диплом открывает дверь перед людьми, стремящимися начать профессиональную деятельность или учиться в университете.
Наша компания предлагает быстро получить этот необходимый документ. Вы сможете заказать диплом нового или старого образца, и это будет удачным решением для человека, который не смог завершить образование или потерял документ. Все дипломы изготавливаются с особой аккуратностью, вниманием к мельчайшим нюансам, чтобы в результате получился полностью оригинальный документ.
Превосходство этого подхода заключается не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается удобно и легко, с профессиональной поддержкой. От выбора необходимого образца до точного заполнения персональных данных и доставки по России — все под полным контролем квалифицированных мастеров.
В итоге, для всех, кто хочет найти максимально быстрый способ получить необходимый документ, наша услуга предлагает отличное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и сразу перейти к личным целям, будь то поступление в университет или начало карьеры.
купить диплом медицинского училища
В нашем мире, где диплом является началом отличной карьеры в любом направлении, многие пытаются найти максимально простой путь получения образования. Наличие документа об образовании переоценить попросту невозможно. Ведь диплом открывает двери перед людьми, стремящимися вступить в сообщество квалифицированных специалистов или продолжить обучение в высшем учебном заведении.
Предлагаем максимально быстро получить этот важный документ. Вы сможете заказать диплом, что является выгодным решением для всех, кто не смог закончить обучение, утратил документ или хочет исправить свои оценки. Все дипломы выпускаются с особой тщательностью, вниманием к мельчайшим элементам, чтобы в итоге получился 100% оригинальный документ.
Плюсы подобного подхода заключаются не только в том, что можно быстро получить свой диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. Начиная от выбора необходимого образца диплома до точного заполнения личной информации и доставки в любое место страны — все под абсолютным контролем опытных мастеров.
Таким образом, для тех, кто пытается найти максимально быстрый способ получить необходимый документ, наша компания готова предложить выгодное решение. Купить диплом – это значит избежать длительного процесса обучения и сразу переходить к достижению личных целей: к поступлению в университет или к началу удачной карьеры.
купить диплом недорого
toasterovensplus.com
この記事の深さと質に驚きました。本当にありがとうございます。
В нашем обществе, где диплом становится началом удачной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения качественного образования. Важность наличия официального документа сложно переоценить. Ведь именно он открывает дверь перед любым человеком, желающим начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы имеете возможность заказать диплом, что становится отличным решением для человека, который не смог закончить образование, утратил документ или хочет исправить плохие оценки. Любой диплом изготавливается аккуратно, с особым вниманием ко всем деталям, чтобы на выходе получился полностью оригинальный документ.
Плюсы этого решения состоят не только в том, что вы быстро получите диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. Начав от выбора подходящего образца диплома до правильного заполнения персональной информации и доставки по стране — все будет находиться под абсолютным контролем наших мастеров.
Таким образом, для тех, кто ищет быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и не теряя времени перейти к достижению личных целей, будь то поступление в университет или начало трудовой карьеры.
https://premialnie-diplomik.com
В современном мире, где диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения образования. Факт наличия официального документа переоценить невозможно. Ведь именно он открывает дверь перед каждым человеком, который желает начать профессиональную деятельность или продолжить обучение в ВУЗе.
Предлагаем быстро получить этот важный документ. Вы можете купить диплом, что становится отличным решением для всех, кто не смог завершить образование или потерял документ. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам, чтобы в результате получился документ, 100% соответствующий оригиналу.
Превосходство подобного решения заключается не только в том, что можно оперативно получить свой диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начиная от выбора необходимого образца диплома до правильного заполнения персональной информации и доставки по стране — все под полным контролем качественных специалистов.
Всем, кто ищет максимально быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Заказать диплом – значит избежать долгого процесса обучения и не теряя времени переходить к своим целям, будь то поступление в университет или старт карьеры.
https://premialnie-diplomik.com
В нашем мире, где диплом является началом удачной карьеры в любой сфере, многие стараются найти максимально быстрый путь получения качественного образования. Факт наличия документа об образовании переоценить невозможно. Ведь именно он открывает дверь перед всеми, кто стремится вступить в сообщество профессионалов или учиться в любом ВУЗе.
В данном контексте наша компания предлагает быстро получить любой необходимый документ. Вы сможете приобрести диплом, что будет удачным решением для человека, который не смог завершить образование или потерял документ. диплом изготавливается с особой аккуратностью, вниманием к мельчайшим нюансам. На выходе вы получите документ, 100% соответствующий оригиналу.
Преимущества подобного подхода заключаются не только в том, что вы оперативно получите диплом. Весь процесс организован комфортно, с нашей поддержкой. Начав от выбора нужного образца до правильного заполнения личных данных и доставки в любой регион России — все будет находиться под абсолютным контролем качественных специалистов.
Всем, кто пытается найти быстрый и простой способ получить требуемый документ, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать длительного обучения и сразу перейти к достижению личных целей, будь то поступление в университет или начало профессиональной карьеры.
диплом купить
В нашем обществе, где диплом является началом успешной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Факт наличия официального документа об образовании сложно переоценить. Ведь диплом открывает дверь перед любым человеком, который хочет вступить в профессиональное сообщество или учиться в высшем учебном заведении.
В данном контексте мы предлагаем быстро получить любой необходимый документ. Вы сможете купить диплом, и это будет выгодным решением для всех, кто не смог завершить обучение, утратил документ или желает исправить свои оценки. Все дипломы производятся с особой тщательностью, вниманием ко всем нюансам. В результате вы получите продукт, 100% соответствующий оригиналу.
Преимущество подобного решения заключается не только в том, что можно оперативно получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора требуемого образца до консультаций по заполнению персональной информации и доставки по стране — все под полным контролем квалифицированных специалистов.
Всем, кто ищет максимально быстрый способ получить необходимый документ, наша компания готова предложить отличное решение. Заказать диплом – значит избежать продолжительного процесса обучения и сразу перейти к личным целям, будь то поступление в ВУЗ или начало успешной карьеры.
купить диплом магистра
В современном мире, где диплом – это начало отличной карьеры в любой области, многие стараются найти максимально быстрый путь получения качественного образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед любым человеком, желающим вступить в профессиональное сообщество или учиться в университете.
Мы предлагаем оперативно получить этот важный документ. Вы можете приобрести диплом старого или нового образца, и это будет выгодным решением для всех, кто не смог завершить образование или утратил документ. Все дипломы изготавливаются с особой аккуратностью, вниманием к мельчайшим деталям. На выходе вы сможете получить продукт, максимально соответствующий оригиналу.
Плюсы данного решения состоят не только в том, что можно максимально быстро получить свой диплом. Процесс организовывается удобно, с нашей поддержкой. От выбора требуемого образца до грамотного заполнения личных данных и доставки по России — все под абсолютным контролем опытных специалистов.
Для тех, кто ищет оперативный способ получения необходимого документа, наша компания может предложить отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и сразу переходить к достижению собственных целей, будь то поступление в университет или старт профессиональной карьеры.
http://galeria.farvista.net/member.php?action=showprofile&user_id=40829
http://izovsem.ru/users/atogyl
http://www.devils-rock.at/index.php?option=com_easybookreloaded
http://www.netza.ru/2012/12/service-password-encryption.html
http://xn--48-6kcd0fg.xn--p1ai/forums.php?m=posts&q=21114&n=last#bottom
В современном мире, где диплом становится началом отличной карьеры в любой отрасли, многие стараются найти максимально простой путь получения качественного образования. Важность наличия официального документа сложно переоценить. Ведь диплом открывает двери перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в каком-либо институте.
Мы предлагаем быстро получить любой необходимый документ. Вы сможете заказать диплом старого или нового образца, и это становится выгодным решением для человека, который не смог закончить образование, потерял документ или желает исправить плохие оценки. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам. В результате вы сможете получить полностью оригинальный документ.
Преимущество подобного подхода заключается не только в том, что можно быстро получить свой диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начиная от выбора нужного образца до правильного заполнения личных данных и доставки в любой регион страны — все под полным контролем качественных специалистов.
Таким образом, для тех, кто ищет быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и не теряя времени перейти к важным целям: к поступлению в ВУЗ или к началу удачной карьеры.
https://cse.google.dm/url?sa=t&url=https://premialnie-diplomik.com
https://clients1.google.mv/url?rct=j&sa=t&url=https://premialnie-diplomik.com
https://images.google.se/url?q=https://lands-diploma.com
https://maps.google.cf/url?sa=t&url=https://premialnie-diplomik.com
https://images.google.com.sv/url?q=https://rudiplomista.com
В наше время, когда диплом становится началом удачной карьеры в любой области, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед всеми, кто собирается начать трудовую деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы имеете возможность купить диплом старого или нового образца, что становится удачным решением для человека, который не смог завершить образование или потерял документ. диплом изготавливается аккуратно, с особым вниманием к мельчайшим деталям, чтобы в результате получился продукт, максимально соответствующий оригиналу.
Плюсы данного подхода состоят не только в том, что можно быстро получить свой диплом. Процесс организовывается комфортно и легко, с нашей поддержкой. От выбора подходящего образца диплома до консультаций по заполнению персональных данных и доставки в любое место страны — все будет находиться под полным контролем качественных мастеров.
В результате, для всех, кто пытается найти быстрый способ получения требуемого документа, наша услуга предлагает отличное решение. Заказать диплом – это значит избежать долгого процесса обучения и не теряя времени перейти к своим целям, будь то поступление в ВУЗ или старт профессиональной карьеры.
https://www.google.com.bd/url?q=https://lands-diploma.com
https://www.google.nr/url?q=https://rudiplomista.com
https://images.google.cf/url?q=https://lands-diploma.com
https://www.google.hn/url?sa=t&url=https://lands-diploma.com
https://images.google.co.mz/url?q=https://lands-diploma.com
На сегодняшний день, когда диплом – это начало отличной карьеры в любой сфере, многие ищут максимально простой путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем быстро получить этот необходимый документ. Вы можете купить диплом, что становится удачным решением для человека, который не смог закончить обучение или потерял документ. Каждый диплом изготавливается с особой аккуратностью, вниманием к мельчайшим элементам. На выходе вы получите продукт, полностью соответствующий оригиналу.
Преимущество подобного подхода состоит не только в том, что можно оперативно получить диплом. Весь процесс организовывается удобно и легко, с профессиональной поддержкой. От выбора необходимого образца документа до консультации по заполнению личной информации и доставки по России — все под абсолютным контролем наших мастеров.
Для тех, кто пытается найти оперативный способ получения необходимого документа, наша компания может предложить выгодное решение. Заказать диплом – значит избежать продолжительного обучения и не теряя времени перейти к достижению своих целей: к поступлению в университет или к началу трудовой карьеры.
https://wiki.mysupp.ru/index.php?title=diplomaasx
http://mclassic.com.hk/home.php?mod=space&uid=274668
https://yogaasanas.science/index.php?title=landsdiploms
http://opendelreyoaks.net/doku.php?id=rudiplomisty
http://autotuninggroup.ru/product/spidometr-elektronnyj-kamaz-uaz/reviews/
В наше время, когда диплом – это начало отличной карьеры в любом направлении, многие стараются найти максимально простой путь получения образования. Наличие официального документа об образовании переоценить просто невозможно. Ведь диплом открывает двери перед каждым человеком, желающим начать профессиональную деятельность или продолжить обучение в университете.
Мы предлагаем очень быстро получить любой необходимый документ. Вы можете приобрести диплом, и это становится отличным решением для всех, кто не смог завершить образование или утратил документ. диплом изготавливается с особой аккуратностью, вниманием ко всем элементам. В результате вы получите документ, 100% соответствующий оригиналу.
Преимущество данного решения состоит не только в том, что вы сможете максимально быстро получить диплом. Весь процесс организован комфортно, с нашей поддержкой. Начав от выбора требуемого образца до консультации по заполнению личной информации и доставки в любое место страны — все будет находиться под полным контролем квалифицированных мастеров.
Для тех, кто ищет быстрый и простой способ получения необходимого документа, наша компания готова предложить выгодное решение. Приобрести диплом – это значит избежать долгого процесса обучения и не теряя времени переходить к личным целям: к поступлению в ВУЗ или к началу удачной карьеры.
https://www.google.com.ng/url?q=https://lands-diploma.com
https://clients4.google.com/url?q=https://rudiplomista.com
https://www.google.mv/url?sa=t&url=https://rudiplomista.com
https://clients1.google.ro/url?sa=t&url=https://lands-diploma.com
https://maps.google.at/url?q=https://lands-diploma.com
В современном мире, где диплом становится началом отличной карьеры в любой сфере, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает дверь перед всеми, кто собирается вступить в сообщество квалифицированных специалистов или продолжить обучение в университете.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете приобрести диплом нового или старого образца, и это становится удачным решением для человека, который не смог завершить образование или потерял документ. дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим нюансам, чтобы на выходе получился полностью оригинальный документ.
Преимущество такого решения состоит не только в том, что можно оперативно получить свой диплом. Процесс организовывается удобно, с нашей поддержкой. От выбора нужного образца документа до правильного заполнения персональной информации и доставки в любое место страны — все под полным контролем опытных мастеров.
Таким образом, для всех, кто ищет быстрый и простой способ получить требуемый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и сразу переходить к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
https://clients1.google.com.pa/url?sa=t&url=https://lands-diploma.com
https://images.google.hn/url?q=https://lands-diploma.com
https://images.google.ml/url?sa=t&url=https://premialnie-diplomik.com
https://clients1.google.ro/url?sa=t&url=https://lands-diploma.com
https://clients1.google.com.hk/url?sa=t&url=https://rudiplomista.com
В нашем мире, где диплом – это начало удачной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения образования. Факт наличия документа об образовании переоценить невозможно. Ведь диплом открывает двери перед любым человеком, который собирается начать трудовую деятельность или продолжить обучение в каком-либо институте.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы имеете возможность купить диплом, и это является выгодным решением для всех, кто не смог завершить обучение, утратил документ или желает исправить свои оценки. Все дипломы изготавливаются аккуратно, с максимальным вниманием ко всем элементам, чтобы в итоге получился полностью оригинальный документ.
Преимущество данного подхода состоит не только в том, что можно быстро получить свой диплом. Процесс организовывается удобно, с профессиональной поддержкой. Начав от выбора нужного образца до консультаций по заполнению персональных данных и доставки по стране — все под абсолютным контролем наших мастеров.
Всем, кто пытается найти быстрый способ получения необходимого документа, наша компания готова предложить выгодное решение. Приобрести диплом – значит избежать продолжительного обучения и не теряя времени переходить к достижению личных целей, будь то поступление в ВУЗ или старт карьеры.
https://clients1.google.com.py/url?sa=t&url=https://premialnie-diplomik.com
https://alt1.toolbarqueries.google.com.gt/url?q=https://premialnie-diplomik.com
https://cse.google.com.tw/url?q=https://premialnie-diplomik.com
http://www.google.bt/url?q=https://rudiplomista.com
https://toolbarqueries.google.mn/url?q=https://premialnie-diplomik.com
В нашем обществе, где диплом – это начало успешной карьеры в любой области, многие пытаются найти максимально простой путь получения качественного образования. Необходимость наличия официального документа переоценить попросту невозможно. Ведь диплом открывает дверь перед любым человеком, желающим начать профессиональную деятельность или учиться в любом институте.
В данном контексте мы предлагаем быстро получить этот необходимый документ. Вы можете заказать диплом нового или старого образца, и это будет выгодным решением для всех, кто не смог завершить обучение, потерял документ или желает исправить свои оценки. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам. В результате вы получите 100% оригинальный документ.
Преимущество подобного подхода заключается не только в том, что можно максимально быстро получить диплом. Весь процесс организован удобно, с профессиональной поддержкой. От выбора нужного образца до грамотного заполнения персональной информации и доставки по стране — все под полным контролем наших специалистов.
Всем, кто хочет найти быстрый и простой способ получения необходимого документа, наша компания может предложить отличное решение. Купить диплом – значит избежать продолжительного обучения и сразу переходить к достижению своих целей: к поступлению в ВУЗ или к началу удачной карьеры.
https://maps.google.co.kr/url?sa=t&url=https://premialnie-diplomik.com
https://maps.google.mw/url?q=https://lands-diploma.com
https://www.google.co.nz/url?sa=t&url=https://lands-diploma.com
https://image.google.co.im/url?q=https://rudiplomista.com
https://maps.google.com.ag/url?q=https://lands-diploma.com
В современном мире, где диплом является началом успешной карьеры в любой области, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа сложно переоценить. Ведь диплом открывает дверь перед всеми, кто стремится вступить в сообщество квалифицированных специалистов или продолжить обучение в университете.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы можете заказать диплом нового или старого образца, и это является удачным решением для всех, кто не смог завершить образование, потерял документ или желает исправить плохие оценки. Все дипломы изготавливаются с особой аккуратностью, вниманием к мельчайшим нюансам. В результате вы получите документ, 100% соответствующий оригиналу.
Плюсы подобного подхода состоят не только в том, что можно быстро получить диплом. Весь процесс организован просто и легко, с нашей поддержкой. Начав от выбора необходимого образца до консультации по заполнению персональной информации и доставки в любое место страны — все будет находиться под абсолютным контролем качественных мастеров.
Всем, кто хочет найти быстрый способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – значит избежать длительного обучения и сразу перейти к достижению личных целей, будь то поступление в ВУЗ или старт успешной карьеры.
http://cargo-200.ru/index.php?subaction=userinfo&user=azizonyd
http://wiki.hightgames.ru/index.php?title=landsdiploms
http://www.podzemie.6f.sk/profile.php?lookup=16816
http://fr79056g.bget.ru/index.php?name=account&op=info&uname=enavaza
http://drahthaar-forum.ru/topic9213.html?view=previous
В нашем обществе, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Факт наличия официального документа об образовании переоценить просто невозможно. Ведь диплом открывает двери перед людьми, стремящимися начать профессиональную деятельность или продолжить обучение в каком-либо институте.
Наша компания предлагает быстро получить этот важный документ. Вы сможете приобрести диплом старого или нового образца, и это будет выгодным решением для всех, кто не смог завершить образование или утратил документ. Все дипломы выпускаются аккуратно, с особым вниманием ко всем деталям. В результате вы сможете получить полностью оригинальный документ.
Превосходство подобного решения заключается не только в том, что можно оперативно получить свой диплом. Процесс организовывается удобно, с нашей поддержкой. Начиная от выбора нужного образца до точного заполнения персональных данных и доставки по стране — все под полным контролем наших специалистов.
Всем, кто ищет оперативный способ получения необходимого документа, наша компания может предложить отличное решение. Заказать диплом – это значит избежать продолжительного обучения и не теряя времени переходить к важным целям: к поступлению в университет или к началу трудовой карьеры.
https://clients1.google.com.ua/url?sa=t&url=https://rudiplomista.com
https://images.google.co.th/url?q=https://premialnie-diplomik.com
https://images.google.co.il/url?q=https://lands-diploma.com
https://cse.google.ki/url?q=https://premialnie-diplomik.com
https://cse.google.co.cr/url?sa=t&url=https://premialnie-diplomik.com
Wow, superb blog format!
How lengthy have you been blogging for? you make
running a blog glance easy. The entire glance of your web site is wonderful,
as neatly as the content material! I saw similar here prev next and those was wrote
by Cory89.
В нашем обществе, где диплом становится началом удачной карьеры в любой области, многие стараются найти максимально быстрый и простой путь получения образования. Наличие документа об образовании трудно переоценить. Ведь именно он открывает двери перед людьми, стремящимися начать трудовую деятельность или учиться в ВУЗе.
В данном контексте наша компания предлагает быстро получить любой необходимый документ. Вы можете заказать диплом, что будет отличным решением для человека, который не смог завершить образование или утратил документ. диплом изготавливается с особой аккуратностью, вниманием ко всем элементам, чтобы на выходе получился продукт, полностью соответствующий оригиналу.
Плюсы такого решения заключаются не только в том, что можно оперативно получить диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начав от выбора необходимого образца до правильного заполнения персональной информации и доставки в любое место России — все находится под абсолютным контролем квалифицированных специалистов.
Для тех, кто ищет оперативный способ получения требуемого документа, наша услуга предлагает отличное решение. Заказать диплом – значит избежать долгого процесса обучения и не теряя времени переходить к важным целям, будь то поступление в университет или начало карьеры.
https://maps.google.com.pa/url?q=https://rudiplomista.com
https://maps.google.mn/url?rct=j&sa=t&url=https://rudiplomista.com
https://cse.google.com.tw/url?q=https://lands-diploma.com
http://maps.google.cg/url?q=https://lands-diploma.com
https://clients1.google.td/url?sa=t&url=https://lands-diploma.com
В наше время, когда диплом является началом отличной карьеры в любой области, многие стараются найти максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа об образовании сложно переоценить. Ведь диплом открывает двери перед всеми, кто желает начать профессиональную деятельность или продолжить обучение в любом ВУЗе.
Мы предлагаем максимально быстро получить этот важный документ. Вы можете заказать диплом старого или нового образца, что будет выгодным решением для всех, кто не смог завершить обучение, потерял документ или желает исправить плохие оценки. Каждый диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим элементам. В результате вы получите продукт, 100% соответствующий оригиналу.
Плюсы этого решения заключаются не только в том, что можно оперативно получить диплом. Процесс организовывается комфортно, с профессиональной поддержкой. Начиная от выбора нужного образца диплома до грамотного заполнения личной информации и доставки в любое место страны — все будет находиться под полным контролем квалифицированных специалистов.
Для всех, кто ищет максимально быстрый способ получения требуемого документа, наша компания предлагает отличное решение. Заказать диплом – это значит избежать долгого обучения и сразу перейти к важным целям, будь то поступление в университет или начало карьеры.
http://sukces-it.pl/blog/czy-wiesz-ze-przeslanie-e-deklaracji#c126
http://5chudes.com/product/magazin-morozhenoe/reviews/
https://instrument-43.ru/product/pajalnik-zubr-impulsnyj-so-svetovym-indikatorom-150-vt-2-smennyh-zhala-kanifol-pripoj-/reviews/
http://nullumcrimen.kr/bbs/board.php?bo_table=free&wr_id=18864
http://www.san-top.ru/product/smennaja-panel-avantgarde-silver-dlja-electrolux-eta-16-avantgarde/reviews/
В наше время, когда диплом – это начало отличной карьеры в любой сфере, многие пытаются найти максимально быстрый путь получения образования. Наличие документа об образовании переоценить попросту невозможно. Ведь именно диплом открывает двери перед всеми, кто желает начать профессиональную деятельность или учиться в любом институте.
Наша компания предлагает очень быстро получить этот важный документ. Вы имеете возможность купить диплом нового или старого образца, и это становится выгодным решением для всех, кто не смог завершить образование или потерял документ. Любой диплом изготавливается аккуратно, с особым вниманием ко всем нюансам, чтобы в итоге получился полностью оригинальный документ.
Преимущество этого решения заключается не только в том, что вы максимально быстро получите диплом. Процесс организовывается комфортно и легко, с нашей поддержкой. От выбора требуемого образца документа до консультаций по заполнению личных данных и доставки в любой регион страны — все будет находиться под полным контролем качественных мастеров.
В результате, для тех, кто ищет оперативный способ получения требуемого документа, наша компания готова предложить выгодное решение. Приобрести диплом – это значит избежать долгого обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в ВУЗ или старт карьеры.
http://www.elevatorshoes.blog/2009/10/celeb-height-shoes.html
http://safeguestbook.com/francpaul-124135-1.html
http://www.icmms.co.kr/bbs/board.php?bo_table=free&wr_id=157332
http://economreklama.ru/index.php?subaction=userinfo&user=ahyfowaz
http://xn--h1a1ab.xn--p1ai/index.php?name=account&op=info&uname=okyvaza
В нашем обществе, где диплом является началом отличной карьеры в любой сфере, многие стараются найти максимально простой путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь именно диплом открывает двери перед каждым человеком, желающим вступить в профессиональное сообщество или учиться в ВУЗе.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы можете купить диплом нового или старого образца, и это будет отличным решением для всех, кто не смог закончить обучение, утратил документ или хочет исправить плохие оценки. Все дипломы изготавливаются аккуратно, с максимальным вниманием к мельчайшим элементам. В результате вы сможете получить 100% оригинальный документ.
Преимущества этого подхода состоят не только в том, что можно оперативно получить диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начав от выбора подходящего образца диплома до точного заполнения персональных данных и доставки в любой регион России — все будет находиться под абсолютным контролем наших мастеров.
Для тех, кто ищет быстрый способ получения необходимого документа, наша услуга предлагает отличное решение. Заказать диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
https://cse.google.sc/url?sa=t&url=https://lands-diploma.com
https://www.google.nr/url?q=https://premialnie-diplomik.com
https://maps.google.com.lb/url?q=https://premialnie-diplomik.com
https://sandbox.google.com/url?q=https://lands-diploma.com
https://images.google.com.vn/url?sa=t&url=https://premialnie-diplomik.com
В современном мире, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Важность наличия официального документа об образовании переоценить попросту невозможно. Ведь диплом открывает двери перед людьми, желающими вступить в профессиональное сообщество или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы сможете заказать диплом, и это является удачным решением для всех, кто не смог закончить обучение или утратил документ. диплом изготавливается с особой аккуратностью, вниманием к мельчайшим нюансам, чтобы в результате получился продукт, 100% соответствующий оригиналу.
Превосходство этого решения заключается не только в том, что можно быстро получить диплом. Процесс организовывается комфортно, с профессиональной поддержкой. От выбора подходящего образца диплома до точного заполнения персональной информации и доставки по стране — все будет находиться под полным контролем качественных специалистов.
Всем, кто ищет оперативный способ получения необходимого документа, наша услуга предлагает выгодное решение. Заказать диплом – значит избежать длительного обучения и сразу переходить к своим целям: к поступлению в университет или к началу удачной карьеры.
https://www.google.to/url?sa=t&url=https://lands-diploma.com
https://maps.google.jo/url?q=https://premialnie-diplomik.com
https://sites.google.com/view/sunwinclcomhttps://rudiplomista.com
https://images.google.ad/url?q=https://lands-diploma.com
https://cse.google.com.ua/url?q=https://premialnie-diplomik.com
В нашем мире, где диплом становится началом удачной карьеры в любом направлении, многие стараются найти максимально простой путь получения образования. Факт наличия официального документа сложно переоценить. Ведь диплом открывает дверь перед каждым человеком, желающим начать профессиональную деятельность или продолжить обучение в любом институте.
Мы предлагаем оперативно получить этот необходимый документ. Вы можете заказать диплом, что становится удачным решением для человека, который не смог закончить обучение или потерял документ. дипломы изготавливаются с особой аккуратностью, вниманием ко всем нюансам, чтобы в результате получился документ, 100% соответствующий оригиналу.
Преимущества подобного решения заключаются не только в том, что вы оперативно получите свой диплом. Процесс организован удобно и легко, с нашей поддержкой. Начав от выбора нужного образца до консультаций по заполнению персональных данных и доставки по России — все будет находиться под полным контролем наших специалистов.
Для тех, кто хочет найти максимально быстрый способ получить необходимый документ, наша компания готова предложить выгодное решение. Купить диплом – значит избежать долгого обучения и сразу перейти к достижению собственных целей, будь то поступление в университет или старт профессиональной карьеры.
https://images.google.com.gt/url?q=https://rudiplomista.com
https://www.google.ac/url?q=https://lands-diploma.com
http://www.google.co.in/url?q=https://lands-diploma.com
https://images.google.gm/url?q=https://lands-diploma.com
https://maps.google.co.vi/url?sa=t&url=https://premialnie-diplomik.com
В нашем обществе, где диплом является началом успешной карьеры в любой сфере, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Важность наличия официального документа об образовании трудно переоценить. Ведь именно он открывает двери перед любым человеком, который желает вступить в сообщество профессиональных специалистов или учиться в высшем учебном заведении.
Предлагаем очень быстро получить любой необходимый документ. Вы сможете заказать диплом, и это является отличным решением для человека, который не смог завершить образование, потерял документ или желает исправить свои оценки. Каждый диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим деталям. В результате вы получите полностью оригинальный документ.
Преимущества такого решения заключаются не только в том, что можно максимально быстро получить свой диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начав от выбора подходящего образца до консультаций по заполнению персональных данных и доставки по стране — все находится под полным контролем квалифицированных мастеров.
Для тех, кто ищет быстрый и простой способ получить необходимый документ, наша услуга предлагает выгодное решение. Заказать диплом – это значит избежать долгого процесса обучения и сразу переходить к достижению собственных целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://parus-perm.ru/communication/forum.php?PAGE_NAME=profile_view&UID=47417
http://www.premier-climat.ru/product/dimplex-obsidian-serija-hi-tech/reviews/
http://www.asanagikai.com/blog/1167.html?unapproved=1955303&moderation-hash=39ae86f7824343295afe949066edd51c#comment-1955303
http://teamhardnet.free.fr/index.php?file=Members&op=detail&autor=ecutyhym
http://www.xn—-8sbabeoae4acvb6h.xn--p1ai/2022/03/blog-post.html
В нашем мире, где диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения образования. Факт наличия документа об образовании трудно переоценить. Ведь именно диплом открывает дверь перед всеми, кто желает вступить в профессиональное сообщество или учиться в любом ВУЗе.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы имеете возможность приобрести диплом, что будет выгодным решением для всех, кто не смог закончить обучение, утратил документ или желает исправить свои оценки. диплом изготавливается аккуратно, с особым вниманием ко всем деталям. В итоге вы сможете получить продукт, максимально соответствующий оригиналу.
Преимущество подобного подхода заключается не только в том, что вы сможете оперативно получить свой диплом. Процесс организовывается удобно, с профессиональной поддержкой. От выбора нужного образца диплома до грамотного заполнения личной информации и доставки в любой регион страны — все под полным контролем наших специалистов.
Для всех, кто пытается найти быстрый и простой способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – это значит избежать длительного обучения и не теряя времени перейти к личным целям, будь то поступление в ВУЗ или начало карьеры.
https://www.google.com.om/url?sa=t&url=https://rudiplomista.com
https://toolbarqueries.google.co.ls/url?q=https://rudiplomista.com
https://images.google.com.np/url?sa=t&url=https://lands-diploma.com
https://maps.google.lu/url?sa=t&url=https://rudiplomista.com
https://www.google.sr/url?q=https://rudiplomista.com
thephotoretouch.com
그러나 그는 감히 아무 말도 하지 못하여 흐트러진 인사를 하고 작별인사를 하고 작별인사를 하였다.
Wow, superb blog layout!
How long have you been running a blog for? you make blogging look easy.
The total look of your site is fantastic, let alone the content material!
I saw similar here prev next and
that was wrote by Dillon96.
Fobertbap
Привет всем!
Интернет предоставил мне инструменты, которые сделали процесс написания диплома более быстрым и менее изнурительным.
На нашем сайте вы можете купить диплом ВУЗа, недорого, с оплатой после получения, поддержка 24/7.
Желаю для каждого положительных оценок!
купить диплом в сарове
купить диплом в бугульме
купить диплом в нижнем тагиле
купить диплом в октябрьском
купить диплом в сургуте
купить диплом в перми
купить диплом в анапе
купить аттестат за 11 класс
купить диплом историка
купить диплом журналиста
В нашем обществе, где диплом является началом успешной карьеры в любой отрасли, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Необходимость наличия официального документа об образовании трудно переоценить. Ведь диплом открывает двери перед любым человеком, который хочет начать профессиональную деятельность или учиться в ВУЗе.
Наша компания предлагает быстро получить любой необходимый документ. Вы можете заказать диплом старого или нового образца, и это будет выгодным решением для всех, кто не смог закончить обучение, потерял документ или желает исправить плохие оценки. диплом изготавливается с особой аккуратностью, вниманием ко всем нюансам. На выходе вы сможете получить документ, максимально соответствующий оригиналу.
Превосходство этого решения заключается не только в том, что можно оперативно получить свой диплом. Процесс организован удобно, с нашей поддержкой. Начав от выбора необходимого образца диплома до точного заполнения личных данных и доставки в любое место страны — все будет находиться под полным контролем качественных специалистов.
В результате, всем, кто пытается найти быстрый способ получения требуемого документа, наша компания может предложить отличное решение. Заказать диплом – это значит избежать длительного обучения и сразу перейти к важным целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://bioimagingcore.be/q2a/825051/landsdiploms
http://mpsamp.com/bbs/board.php?bo_table=free&wr_id=211956
http://forum.fantasy-online.ru/profile.php?mode=viewprofile&u=876688
http://www.irbislab.ru/modules.php?name=Your_Account&op=userinfo&username=eqorywi
http://www.pawsarl.es/blog/%D0%B4%D0%BE%D0%B2%D1%80%D0%B0%D0%BD.html
Wow, superb weblog layout!
How lengthy have you ever been blogging for? you make blogging glance easy.
The full glance of your site is excellent, let alone the content material!
You can read similar here Ethel Buh6.
2024/04/23
donmhomes.com
この記事は実用性が高くて、本当に助かります!
IsmaelBog
Привет всем!
Продолжаю работать над дипломом, несмотря на все препятствия, и благодарю интернет за помощь в этом нелегком процессе.
Приобретайте диплом у нас и получите качественный документ без предоплаты, с доставкой в любую точку России.
Желаю для каждого не двоешных) оценок!
купить диплом в нефтеюганске
купить диплом в хабаровске
купить диплом в иркутске
купить диплом математика
купить диплом в ставрополе
купить диплом о среднем образовании
купить диплом в омске
купить диплом в киселевске
купить диплом в брянске
купить диплом монтажника
LewisAluri
Приветики, дорогие мои!!
Завершение диплома кажется возможным с поддержкой интернета.
Заказывайте диплом у нас без предоплаты и мы доставим его вам в любой регион России, гарантируем конфиденциальность.
Желаю каждому пятерочных) отметок!
купить диплом в кирове
купить диплом в гуково
купить диплом в самаре
купить диплом в волжском
купить диплом в новом уренгое
купить диплом в березниках
купить диплом в братске
куплю диплом с занесением
купить диплом о высшем образовании
купить диплом в ельце
valtrex 2 mg
Доброго всем дня!
Закажите диплом ВУЗа без предоплаты и получите его с доставкой в руки по всей России – это просто и удобно!
https://www.google.pl/url?q=http://diplomrussian.com
https://www.google.cm/url?sa=i&rct=j&q=http://rudiplomisty24.com
https://www.google.cz/url?q=http://russdiplomiki.com
https://clients1.google.no/url?sa=t&url=https://diplomansy.com
https://clients1.google.fr/url?q=http://diplomyland.com
Купите диплом института или колледжа без предоплаты с гарантированной доставкой в любой уголок России.
Earnestabich
Здравствуйте!
В процессе написания диплома я перешел на ночной график, что негативно отразилось на моем здоровье.
Поможем вам выбрать, оформить заказ и купить диплом любого учебного заведения по лучшим ценам.
Желаю для каждого положительных оценок!
купить диплом в самаре
купить диплом в ярославле
купить диплом в армавире
купить диплом в копейске
купить диплом инженера электрика
купить диплом в балаково
купить диплом в спб
купить диплом электромонтера
купить диплом гознак
купить диплом тренера
tintucnamdinh24h.com
그리고 이 순간, 번개와 부싯돌 한가운데서 단검은… 실제로 공중에서 얼어붙었다.
Timsothyreemn
Доброго дня!
Прогресс в дипломе обеспечен благодаря использованию онлайн-ресурсов.
Приобретайте диплом университета без предоплаты с доставкой по всей России.
Желаю всем прекрасных оценок!
купить диплом в таганроге
купить диплом в назрани
куплю диплом с занесением
купить диплом в грозном
купить диплом в бугульме
купить диплом в иваново
купить диплом в кирове
купить диплом в вологде
купить диплом в йошкар-оле
купить диплом в арзамасе
bmipas.com
読むたびに感動します。本当に素晴らしいブログです。
buy azithromycin over the counter uk
Сегодня, когда диплом становится началом успешной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Факт наличия документа об образовании переоценить попросту невозможно. Ведь именно он открывает дверь перед любым человеком, который стремится вступить в сообщество квалифицированных специалистов или продолжить обучение в каком-либо ВУЗе.
Предлагаем очень быстро получить любой необходимый документ. Вы имеете возможность заказать диплом нового или старого образца, что будет выгодным решением для человека, который не смог завершить обучение, утратил документ или желает исправить плохие оценки. Все дипломы выпускаются аккуратно, с особым вниманием к мельчайшим нюансам. На выходе вы сможете получить документ, полностью соответствующий оригиналу.
Превосходство этого решения заключается не только в том, что вы сможете максимально быстро получить свой диплом. Весь процесс организован комфортно, с нашей поддержкой. Начиная от выбора подходящего образца до точного заполнения личных данных и доставки по стране — все будет находиться под абсолютным контролем качественных мастеров.
Всем, кто пытается найти оперативный способ получить необходимый документ, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать долгого процесса обучения и сразу перейти к личным целям, будь то поступление в университет или начало профессиональной карьеры.
http://www.infofakt.ru/2015/02/blog-post_8.html
http://tamklad.ru/viewtopic.php?f=10&t=8641
http://agenciafrog-host.com.br/redesocial_v0/index.php/profile/egudibom
https://centileo.com/forum/messages/forum17/message3821/1920-russianydiplomans?result=new#message3821
https://moolookoo.ru/content/diplomaasx
LewisAluri
Привет всем!
Диплом остается в центре моего внимания, но теперь я вооружен необходимыми онлайн-инструментами для его успешного завершения.
На нашем сайте представлен широкий выбор дипломов с гарантией качества и быстрой доставкой в любой регион РФ.
Желаю вам всем положительных отметок!
купить диплом маляра
купить диплом инженера строителя
купить диплом специалиста
купить диплом в гуково
купить диплом в братске
купить диплом в комсомольске-на-амуре
купить диплом в шахтах
купить диплом экономиста
купить диплом зубного техника
купить диплом в сызрани
В современном мире, где диплом становится началом отличной карьеры в любой сфере, многие ищут максимально простой путь получения качественного образования. Наличие официального документа об образовании сложно переоценить. Ведь диплом открывает двери перед каждым человеком, который собирается вступить в профессиональное сообщество или продолжить обучение в университете.
В данном контексте наша компания предлагает оперативно получить этот важный документ. Вы сможете купить диплом нового или старого образца, что становится выгодным решением для человека, который не смог закончить образование, утратил документ или желает исправить свои оценки. Все дипломы изготавливаются аккуратно, с максимальным вниманием ко всем деталям. На выходе вы получите 100% оригинальный документ.
Преимущество такого подхода заключается не только в том, что вы сможете максимально быстро получить свой диплом. Весь процесс организовывается просто и легко, с нашей поддержкой. Начиная от выбора нужного образца документа до правильного заполнения персональных данных и доставки в любой регион России — все под абсолютным контролем опытных мастеров.
Для всех, кто пытается найти оперативный способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать длительного процесса обучения и не теряя времени переходить к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
https://cse.google.cat/url?q=https://rudiplomista.com
https://clients1.google.com.pa/url?sa=t&url=https://premialnie-diplomik.com
https://maps.google.gy/url?sa=j&url=https://premialnie-diplomik.com
https://image.google.tk/url?sa=t&source=web&rct=j&url=https://lands-diploma.com
https://www.google.com.tj/url?sa=t&url=https://rudiplomista.com
В нашем мире, где диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим начать трудовую деятельность или продолжить обучение в каком-либо университете.
В данном контексте наша компания предлагает максимально быстро получить любой необходимый документ. Вы имеете возможность заказать диплом, что является выгодным решением для человека, который не смог закончить обучение, утратил документ или желает исправить плохие оценки. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим деталям, чтобы в итоге получился документ, максимально соответствующий оригиналу.
Плюсы такого подхода заключаются не только в том, что можно быстро получить диплом. Процесс организовывается комфортно, с нашей поддержкой. Начав от выбора нужного образца документа до точного заполнения личной информации и доставки в любой регион страны — все находится под абсолютным контролем опытных специалистов.
В итоге, всем, кто ищет быстрый способ получения необходимого документа, наша компания может предложить выгодное решение. Купить диплом – это значит избежать длительного обучения и сразу переходить к достижению своих целей, будь то поступление в университет или старт профессиональной карьеры.
https://njt.ru/forum/user/169837/
http://islamdini.kg/index.php?subaction=userinfo&user=ahaze
http://blackpearlbasketball.com.au/index.php?option=com_k2&view=itemlist&task=user&id=826333
http://1chan.blog/100-2/?unapproved=381010&moderation-hash=617acbb874a9c175e7fc5aeb4e1a3078#comment-381010
https://talented-people.vraiforum.com/profile.php?mode=viewprofile&u=8466
Сегодня, когда диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально простой путь получения качественного образования. Факт наличия официального документа переоценить невозможно. Ведь именно он открывает двери перед любым человеком, желающим начать профессиональную деятельность или учиться в ВУЗе.
В данном контексте наша компания предлагает максимально быстро получить этот необходимый документ. Вы сможете приобрести диплом, что является выгодным решением для всех, кто не смог завершить образование, потерял документ или хочет исправить свои оценки. диплом изготавливается аккуратно, с особым вниманием к мельчайшим нюансам, чтобы в результате получился продукт, полностью соответствующий оригиналу.
Превосходство подобного подхода состоит не только в том, что можно оперативно получить диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начав от выбора подходящего образца диплома до грамотного заполнения личной информации и доставки в любое место страны — все находится под полным контролем качественных специалистов.
Таким образом, для тех, кто пытается найти оперативный способ получения требуемого документа, наша услуга предлагает выгодное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени перейти к своим целям: к поступлению в университет или к началу трудовой карьеры.
http://zimmu.com/api/manual/doku.php?id=rudiplomisty
http://www.ironbodies.com/userinfo.php?uid=59823
http://www.plm.pw/2016/09/The-6-Factors-of-Industry-4.0.html
http://infomts.ru/index.php?subaction=userinfo&user=eraxisu
http://www.elevatorshoes.blog/2009/10/celeb-height-shoes.html
В нашем мире, где диплом является началом удачной карьеры в любой сфере, многие пытаются найти максимально простой путь получения образования. Факт наличия документа об образовании трудно переоценить. Ведь диплом открывает двери перед людьми, желающими вступить в профессиональное сообщество или продолжить обучение в высшем учебном заведении.
Мы предлагаем максимально быстро получить этот необходимый документ. Вы можете заказать диплом, и это будет удачным решением для всех, кто не смог закончить обучение, утратил документ или желает исправить свои оценки. дипломы выпускаются аккуратно, с максимальным вниманием ко всем элементам, чтобы в результате получился 100% оригинальный документ.
Превосходство такого решения заключается не только в том, что можно быстро получить свой диплом. Весь процесс организован просто и легко, с профессиональной поддержкой. Начав от выбора нужного образца диплома до консультаций по заполнению личной информации и доставки по России — все находится под полным контролем качественных специалистов.
В результате, для всех, кто хочет найти максимально быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать долгого обучения и сразу перейти к личным целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://cse.google.co.zw/url?sa=t&url=https://lands-diploma.com
https://maps.google.co.vi/url?sa=t&url=https://premialnie-diplomik.com
https://images.google.ca/url?sa=t&url=https://rudiplomista.com
https://clients1.google.com.bo/url?q=https://rudiplomista.com
https://www.google.com.kh/url?sa=t&url=https://lands-diploma.com
largestcatbreed.com
Tang Yin의 먼지 투성이 모습을보고 여기로 달려 왔을 것입니다.
sandyterrace.com
마치 누군가 후카이산의 몸에 성냥개비만한 못을 박은 것 같았다.
Dichaelautob
Здравствуйте!
С каждым днем работа над дипломом становится легче, благодаря найденным онлайн-материалам.
Предоставляем возможность купить диплом Гознак со скидкой, гарантией и доставкой в любой город РФ.
Желаю для каждого пятерочных) оценок!
купить диплом в биробиджане
купить диплом образцы
купить диплом судоводителя
купить диплом в ессентуках
купить диплом матроса
https://images.google.com.uy/url?q=https://kupit-diplomyz24.com/
http://images.google.pn/url?q=https://kdiplom.ru/
https://www.google.com.bn/url?q=https://10000diplomov.ru/
https://maps.google.to/url?q=https://diplom45.ru/
https://maps.google.jo/url?q=https://diploms-vuza.com/
В современном мире, где диплом становится началом удачной карьеры в любом направлении, многие стараются найти максимально простой путь получения качественного образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает дверь перед любым человеком, который собирается вступить в профессиональное сообщество или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает быстро получить этот необходимый документ. Вы сможете заказать диплом, и это будет удачным решением для всех, кто не смог завершить образование, потерял документ или хочет исправить плохие оценки. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам. В итоге вы сможете получить 100% оригинальный документ.
Преимущества данного подхода заключаются не только в том, что вы сможете максимально быстро получить свой диплом. Весь процесс организован комфортно, с нашей поддержкой. От выбора подходящего образца до консультации по заполнению личных данных и доставки по стране — все будет находиться под абсолютным контролем опытных специалистов.
Для всех, кто ищет быстрый способ получить требуемый документ, наша компания готова предложить отличное решение. Приобрести диплом – значит избежать долгого обучения и не теряя времени переходить к личным целям: к поступлению в университет или к началу успешной карьеры.
https://www.google.gg/url?sa=t&url=https://lands-diploma.com
https://www.google.bs/url?q=https://lands-diploma.com
https://cse.google.co.ve/url?sa=t&url=https://lands-diploma.com
https://cse.google.off.ai/url?q=https://rudiplomista.com
https://cse.google.ee/url?sa=t&url=https://rudiplomista.com
I got this site from my pal who informed me concerning this web page and at the moment this time I am browsing this website and reading very informative posts at this time.
https://images.google.com.et/url?sa=t&url=https://kazdiplomas.com/
http://cse.google.gr/url?sa=i&url=https://ry-diplom.com/
https://www.google.mn/url?q=https://diplom5.com/
http://maps.google.kz/url?q=https://okdiplom.com/
https://maps.google.co.th/url?sa=t&url=https://1magistr.ru/
В нашем мире, где диплом является началом отличной карьеры в любой сфере, многие ищут максимально простой путь получения качественного образования. Факт наличия документа об образовании трудно переоценить. Ведь именно он открывает дверь перед каждым человеком, который стремится вступить в сообщество профессионалов или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы имеете возможность заказать диплом, что становится выгодным решением для всех, кто не смог закончить обучение, утратил документ или хочет исправить плохие оценки. диплом изготавливается с особой аккуратностью, вниманием ко всем деталям, чтобы в результате получился продукт, 100% соответствующий оригиналу.
Преимущества этого решения заключаются не только в том, что можно оперативно получить свой диплом. Процесс организовывается комфортно, с нашей поддержкой. От выбора подходящего образца диплома до грамотного заполнения личной информации и доставки в любой регион России — все будет находиться под абсолютным контролем квалифицированных мастеров.
Для тех, кто пытается найти оперативный способ получить необходимый документ, наша компания предлагает отличное решение. Купить диплом – это значит избежать длительного обучения и не теряя времени переходить к достижению своих целей, будь то поступление в университет или старт карьеры.
http://led119.ru/forum/user/104833/
http://www.gamla.vykort.com/member.php?action=showprofile&user_id=26710
http://www.apicarrara.it/modules.php?name=Journal&file=display&jid=3149
http://www.marheavenj.net/public/gallery/member.php?action=showprofile&user_id=27316
http://xbox360lt.ru/index.php?ukey=discuss_product&productID=9764&did=36
Fobertbap
Приветики, дорогие мои!!
Несмотря на срыв сроков и угрозу для здоровья, я нашел в интернете ресурсы, которые помогают мне быть более продуктивным.
Недорого и без проблем приобрести диплом института, техникума, колледжа выгоднее всего у нас, с доставкой по РФ.
Желаю каждому честных отметок!
купить диплом в рязани
купить диплом в славянске-на-кубани
купить диплом техникума
купить диплом электрика
купить диплом в георгиевске
http://cse.google.co.zw/url?sa=t&url=https://diplom-zakaz.ru/
https://cse.google.com.af/url?sa=t&url=https://diplomoz-197.com/
https://image.google.tk/url?sa=t&source=web&rct=j&url=https://kupit-diplomyz24.com/
http://maps.google.hn/url?rct=j&sa=t&url=https://nsk-diplom.com/
https://clients1.google.sn/url?sa=t&url=https://diplom-bez-problem.com/
В нашем мире, где диплом становится началом удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Необходимость наличия документа об образовании сложно переоценить. Ведь диплом открывает двери перед всеми, кто желает начать трудовую деятельность или продолжить обучение в любом ВУЗе.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы сможете заказать диплом, что становится удачным решением для человека, который не смог закончить обучение или утратил документ. диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим нюансам, чтобы на выходе получился продукт, максимально соответствующий оригиналу.
Превосходство такого подхода состоит не только в том, что можно быстро получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора требуемого образца документа до консультации по заполнению персональных данных и доставки в любое место страны — все будет находиться под абсолютным контролем квалифицированных специалистов.
Для тех, кто пытается найти максимально быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать долгого обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в университет или старт профессиональной карьеры.
https://images.google.lu/url?q=https://lands-diploma.com
https://www.google.com.fj/url?q=https://lands-diploma.com
https://toolbarqueries.google.co.il/url?q=https://lands-diploma.com
https://cse.google.tm/url?q=https://lands-diploma.com
https://ditu.google.com/url?q=https://rudiplomista.com
usareallservice.com
情報がとても整理されていて、理解しやすかったです。
LhaneViart
Доброго дня!
Продолжаю работать над дипломом, несмотря на все препятствия, и благодарю интернет за помощь в этом нелегком процессе.
Мы предлагаем заказать диплом без предоплаты с доставкой курьером по РФ, полный комплект!
Желаю каждому честных оценок!
купить диплом с реестром
купить диплом менеджера
купить диплом в ленинск-кузнецком
купить диплом в ангарске
купить диплом строителя
http://maps.google.tt/url?q=https://okdiplom.com/
https://images.google.al/url?sa=t&url=https://peoplediplom.ru/
https://images.google.com.gt/url?q=https://kupit-diplomyz24.com/
https://cse.google.lv/url?q=https://kazdiplomas.com/
http://maps.google.ro/url?q=https://okdiplom.com/
onair2tv.com
공무원의 공식 선발도 등장하기 시작했고 점차 황실에 의존하게되었습니다.
В нашем обществе, где диплом – это начало отличной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь именно диплом открывает дверь перед людьми, желающими начать профессиональную деятельность или учиться в университете.
Предлагаем быстро получить этот важный документ. Вы сможете купить диплом, что будет удачным решением для всех, кто не смог закончить образование, потерял документ или желает исправить плохие оценки. диплом изготавливается аккуратно, с особым вниманием к мельчайшим деталям, чтобы в итоге получился продукт, максимально соответствующий оригиналу.
Превосходство этого подхода состоит не только в том, что вы сможете быстро получить свой диплом. Весь процесс организован удобно, с нашей поддержкой. Начиная от выбора требуемого образца до правильного заполнения личных данных и доставки по стране — все под абсолютным контролем опытных специалистов.
Всем, кто пытается найти максимально быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать продолжительного обучения и сразу перейти к личным целям, будь то поступление в университет или старт карьеры.
https://www.google.az/url?sa=t&url=https://premialnie-diplomik.com
https://cse.google.is/url?sa=t&url=https://rudiplomista.com
https://clients1.google.com.sl/url?sa=t&url=https://premialnie-diplomik.com
https://cse.google.li/url?q=https://premialnie-diplomik.com
https://www.google.ad/url?q=https://rudiplomista.com
ShaneViart
Добрый день!
Диплом перестал казаться непреодолимым благодаря помощи, полученной из интернета.
Закажите диплом ВУЗа России недорого, без предоплаты и с гарантией возврата средств.
Желаю для каждого положительных оценок!
купить диплом в невинномысске
где купить диплом образование
купить диплом нового образца
купить речной диплом
купить диплом в шахтах
https://images.google.gp/url?sa=t&url=https://ry-diplom.com/
https://maps.google.com.hk/url?q=https://diplom5.com/
http://images.google.com.tj/url?q=https://diplom-insti.ru/
https://clients1.google.rw/url?q=https://diplom-bez-problem.com/
http://images.google.je/url?q=https://kdiplom.ru/
В нашем обществе, где диплом является началом удачной карьеры в любой сфере, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Наличие официального документа об образовании сложно переоценить. Ведь диплом открывает дверь перед любым человеком, который собирается вступить в сообщество профессиональных специалистов или учиться в университете.
Наша компания предлагает быстро получить этот важный документ. Вы можете заказать диплом, что является удачным решением для человека, который не смог завершить образование или потерял документ. Любой диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим нюансам, чтобы на выходе получился 100% оригинальный документ.
Превосходство этого решения состоит не только в том, что можно быстро получить диплом. Процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора требуемого образца документа до точного заполнения личных данных и доставки в любое место России — все находится под полным контролем опытных мастеров.
Всем, кто ищет быстрый способ получения требуемого документа, наша компания может предложить отличное решение. Приобрести диплом – это значит избежать длительного обучения и сразу перейти к важным целям, будь то поступление в университет или начало профессиональной карьеры.
https://www.google.al/url?q=https://premialnie-diplomik.com
http://maps.google.com.om/url?q=https://premialnie-diplomik.com
https://maps.google.com.gh/url?sa=t&url=https://lands-diploma.com
https://clients1.google.ht/url?sa=t&url=https://rudiplomista.com
http://images.google.ps/url?q=https://rudiplomista.com
Здравствуйте!
Получите документы об образовании ВУЗов России с доставкой по РФ и возможностью оплаты после получения – просто и надежно!
https://cse.google.tm/url?q=https://gosznac-diploma.com
https://www.google.de/url?q=https://diplomansy.com
https://www.google.cd/url?sa=i&url=https://anny-diploms.com
https://image.google.com.om/url?q=http://aurus-diploms.com
https://image.google.as/url?q=http://aurus-diploms.com
Получите диплом ВУЗа без предварительной оплаты с доставкой по всей России – просто, надежно, выгодно!
Доброго всем дня!
Мы предлагаем приобрести диплом института или колледжа с доставкой по России без предоплаты.
https://maps.google.com.gh/url?sa=t&url=http://free-diplommi.com
https://www.google.gg/url?sa=t&url=http://rudiplomisty24.com
https://maps.google.com.pr/url?q=https://anny-diploms.com
http://images.google.gl/url?q=http://diplomrussian.com
http://cse.google.ae/url?sa=i&url=http://diplomy-originaly.com
Получите российский диплом с гарантированной доставкой в любую точку страны без предварительной оплаты – просто и надежно!
В современном мире, где диплом – это начало удачной карьеры в любом направлении, многие стараются найти максимально простой путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь именно он открывает двери перед людьми, желающими начать профессиональную деятельность или продолжить обучение в ВУЗе.
Наша компания предлагает очень быстро получить любой необходимый документ. Вы сможете приобрести диплом старого или нового образца, и это является отличным решением для всех, кто не смог завершить образование, утратил документ или желает исправить плохие оценки. диплом изготавливается с особой аккуратностью, вниманием к мельчайшим деталям. В результате вы сможете получить документ, полностью соответствующий оригиналу.
Преимущества подобного решения заключаются не только в том, что вы максимально быстро получите свой диплом. Весь процесс организован просто и легко, с нашей поддержкой. От выбора нужного образца до точного заполнения личной информации и доставки по России — все будет находиться под полным контролем квалифицированных мастеров.
Таким образом, для всех, кто ищет быстрый и простой способ получить необходимый документ, наша компания может предложить отличное решение. Приобрести диплом – это значит избежать долгого процесса обучения и не теряя времени перейти к своим целям, будь то поступление в университет или начало трудовой карьеры.
https://clients4.google.com/url?q=https://lands-diploma.com
https://www.google.md/url?sa=t&url=https://lands-diploma.com
https://www.google.com.np/url?sa=t&url=https://lands-diploma.com
https://maps.google.com.co/url?q=https://lands-diploma.com
http://maps.google.nl/url?q=https://rudiplomista.com
Gichardadvox
Добрый день!
Для диплома я нашел в интернете множество полезных ресурсов, облегчающих написание.
На нашем сайте вы можете купить диплом ВУЗа с постоплатой и поддержкой 24/7.
Желаю вам всем честных оценок!
купить диплом в нижнем новгороде
купить диплом моториста
купить диплом журналиста
купить диплом в елабуге
купить диплом биолога
https://cse.google.com/url?q=https://peoplediplom.ru/
https://www.google.co.mz/url?q=https://diplom-bez-problem.com/
http://cse.google.ae/url?sa=i&url=https://diplom-bez-problem.com/
https://images.google.cf/url?q=https://prodiplome.com/
https://clients1.google.mn/url?q=https://kupit-diplomyz24.com/
Здравствуйте!
Приобретите диплом Гознака у нас по специальной цене с доставкой в любой регион России без предоплаты.
https://www.google.iq/url?sa=t&url=http://diplomsagroups.com
https://images.google.cat/url?sa=t&url=http://diplomy-originaly.com
https://images.google.com.ni/url?q=http://originality-diplomy.com
https://www.google.com.eg/url?q=http://russdiplomiki.com
https://cse.google.sm/url?q=http://aurus-diploms.com
Компания предлагает купить диплом Вуза России недорого, без предоплаты и с гарантией возврата средств.
Доброго всем дня!
Предлагаем заказать диплом у нас с доставкой курьером по всей России без предоплаты.
https://cse.google.ms/url?sa=t&url=http://rudiplomisty24.com
https://images.google.com.sb/url?q=https://diploms-asx.com
https://images.google.gp/url?sa=t&url=http://originality-diplomy.com
https://cse.google.com.ec/url?sa=t&url=http://diplomrussian.com
https://image.google.ac/url?q=http://diplomy-originaly.com
У нас вы можете заказать диплом Гознака по специальной цене с доставкой в любой регион России без предоплаты – надежно и выгодно!
Здравствуйте!
Закажите диплом ВУЗа с гарантированной подлинностью и доставкой по России без предварительной оплаты – просто, надежно, выгодно!
https://cse.google.com.cu/url?sa=t&url=http://originality-diplomy.com
http://toolbarqueries.google.com.af/url?q=http://diplomy-originaly.com
https://maps.google.cat/url?q=https://anny-diploms.com
https://images.google.co.uk/url?sa=t&url=http://diplomyland.com
https://www.google.com.eg/url?q=http://aurus-diploms.com
Закажите диплом Вуза по выгодной цене с доставкой в любой регион России без предоплаты и уверенностью в его законности.
fpparisshop.com
いつも興味深い内容で、読むのが待ち遠しいです。
Dichaelautob
Привет всем!
Диплом требует продолжения борьбы, но я вооружен многочисленными ресурсами, найденными в сети.
Приобретение диплома института, техникума или колледжа у нас – лучший вариант с доставкой по РФ.
Желаю каждому отличных отметок!
купить диплом в кинешме
купить диплом в костроме
купить дипломы о высшем с занесением
купить диплом электромонтера
купить диплом в кунгуре
https://cse.google.com.ua/url?q=https://diplom-zakaz.ru/
https://clients1.google.com.bd/url?q=https://10000diplomov.ru/
http://images.google.ps/url?q=https://kdiplom.ru/
https://image.google.am/url?q=https://diplom45.ru/
https://ipv4.google.com/url?sa=t&url=https://diplom-zakaz.ru/
geinoutime.com
“Wang Xueshi, Wang Xueshi…너…넌 정말 모두에게 상처를 줬어.”
nikontinoll.com
Liu Jin은 병아리가 쌀을 쪼아 먹는 것처럼 고개를 끄덕였습니다. “주님, 다리를 부러 뜨리십시오.”
ShaneViart
Доброго дня!
Диплом постепенно улучшается с каждым днем благодаря использованию найденных ресурсов в интернете.
На нашем сайте вы можете купить диплом ВУЗа с постоплатой и помощью 24/7.
Желаю каждому положительных отметок!
купить диплом в мичуринске
купить диплом юриста
купить диплом в когалыме
купить новый диплом
купить диплом историка
https://cse.google.fi/url?sa=t&url=https://diplom45.ru/
https://maps.google.co.vi/url?sa=t&url=https://okdiplom.com/
https://cse.google.lu/url?q=https://bisness-diplom.ru/
https://toolbarqueries.google.com/url?q=https://nsk-diplom.com/
https://www.google.co.ma/url?sa=t&url=https://bisness-diplom.ru/
where to get azithromycin
Доброго всем дня!
Закажите диплом ВУЗа с доставкой по всей России без предоплаты и с гарантией качества!
Закажите диплом Вуза по выгодной цене с доставкой в любой регион России без предоплаты и уверенностью в его законности.
https://image.google.mg/url?q=https://premialnie-diplomansy.com
https://maps.google.to/url?q=https://diploms-originalniy.com
https://maps.google.com.sg/url?sa=t&url=https://premialnie-diplomany.com
https://cse.google.co.in/url?q=https://gosznac-diplomy.com
https://images.google.cm/url?sa=t&url=http://rudiplomisty24.com
В наше время, когда диплом является началом отличной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения качественного образования. Факт наличия документа об образовании сложно переоценить. Ведь именно он открывает дверь перед любым человеком, который хочет вступить в сообщество квалифицированных специалистов или продолжить обучение в каком-либо университете.
Предлагаем быстро получить этот необходимый документ. Вы сможете купить диплом нового или старого образца, что будет удачным решением для человека, который не смог завершить обучение, потерял документ или хочет исправить свои оценки. Все дипломы выпускаются аккуратно, с особым вниманием к мельчайшим элементам, чтобы на выходе получился полностью оригинальный документ.
Преимущество этого подхода состоит не только в том, что можно максимально быстро получить диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. От выбора нужного образца до правильного заполнения личной информации и доставки в любое место страны — все под абсолютным контролем качественных мастеров.
Для всех, кто хочет найти оперативный способ получить требуемый документ, наша услуга предлагает отличное решение. Купить диплом – это значит избежать длительного обучения и не теряя времени перейти к своим целям: к поступлению в университет или к началу удачной карьеры.
https://images.google.com.ng/url?sa=t&url=https://premialnie-diplomik.com
http://www.google.com.et/url?q=https://premialnie-diplomik.com
https://cse.google.ki/url?q=https://lands-diploma.com
https://image.google.ps/url?q=https://lands-diploma.com
https://cse.google.ee/url?sa=t&url=https://premialnie-diplomik.com
I loved as much as you will receive carried out right here. The sketch is tasteful, your authored subject matter stylish. nonetheless, you command get got an edginess over that you wish be delivering the following. unwell unquestionably come further formerly again since exactly the same nearly very often inside case you shield this increase.
https://images.google.cg/url?q=https://postheaven.net/singleburma1/onovit-zovnishnii-vigliad-avto-novi-fari-ta-sklo
Can you tell us more about this? I’d want to find out more details.
https://writeablog.net/tifardsekc/h1-b-vpliv-skla-far-na-bezpeku-ta-komfort-vodinnia-u-bud-iakikh-umovakh-b-h1
k8 ビデオスロット
素敵な視点と洞察に富んだ内容でした。感謝しています。
game1kb.com
홍지황제는 웃으며 말했다. “그렇다면 오늘은…”
Привет, дорогой читатель!
Наша компания предлагает конфиденциально выбрать и заказать диплом любого Вуза России
http://www.diplomany-asx.ru
Купить диплом онлайн без обмана
купить диплом в Москве http://www.diplom-msk.ru/ .
geinoutime.com
일반적으로 이러한 하인들은 대부분 성질이 급합니다.
Здравствуйте!
Приобретите документы об образовании всех ВУЗов России с гарантированной подлинностью и доставкой по РФ без предварительной оплаты – просто, надежно, выгодно!
Недорого предлагаем заказать диплом без предоплаты с доставкой курьером по РФ, под ключ!
https://www.google.com.mt/url?sa=t&url=http://kazdiplom.com
https://maps.google.to/url?q=https://premialnie-diplomany.com
https://maps.google.com.tr/url?q=https://premialnie-diplomany.com
https://clients1.google.rw/url?q=https://russdiplomag.com
https://images.google.hu/url?q=http://volga-diplomy.com
Доброго всем дня!
Хотите заказать диплом ВУЗа недорого и без предоплаты? Мы доставим его в любую точку России!
Приобретите диплом института или колледжа недорого с доставкой по РФ и гарантией качества без предварительной оплаты!
https://cse.google.cd/url?sa=t&url=https://originality-diploman.com
https://ipv4.google.com/url?sa=t&url=https://lands-diploms.com
https://clients1.google.com.cu/url?sa=t&url=https://diploms-originalniy.com
https://image.google.am/url?q=http://kazdiplom.com
http://toolbarqueries.google.se/url?sa=t&url=https://originality-diploman.com
Добрый день всем!
Закажите диплом ВУЗа по выгодной цене с доставкой в любой город России без предоплаты – надежно и выгодно!
Приобретите диплом колледжа недорого с доставкой по РФ и гарантией качества без предварительной оплаты – просто и надежно!
https://cse.google.ws/url?sa=t&url=http://rudiplomisty24.com
https://maps.google.fr/url?q=http://rudiplomisty24.com
https://images.google.com.sv/url?q=http://diploman-spb24.com
https://www.google.cm/url?sa=i&rct=j&q=https://frees-diplom.com
https://images.google.kg/url?q=https://originality-diploman.com
werankcities.com
Zhao Er은 자신의 고충이 공개된 것처럼 크게 울었습니다.
На сегодняшний день, когда диплом – это начало отличной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения образования. Факт наличия официального документа переоценить попросту невозможно. Ведь именно он открывает двери перед любым человеком, желающим вступить в сообщество квалифицированных специалистов или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает быстро получить этот необходимый документ. Вы можете заказать диплом нового или старого образца, и это будет отличным решением для всех, кто не смог закончить обучение или потерял документ. дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим деталям, чтобы на выходе получился 100% оригинальный документ.
Преимущества этого подхода состоят не только в том, что можно быстро получить свой диплом. Процесс организован удобно, с профессиональной поддержкой. От выбора подходящего образца до точного заполнения личных данных и доставки по России — все будет находиться под абсолютным контролем наших мастеров.
Всем, кто пытается найти быстрый и простой способ получить необходимый документ, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать длительного обучения и не теряя времени переходить к достижению своих целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://www.zoolife55.ru/product/urinary-so-feline-kurica/reviews/
http://nullumcrimen.kr/bbs/board.php?bo_table=free&wr_id=19407
http://radshir.com/products/index.php?subaction=userinfo&user=equgyvuty
http://kzgbi-2.ru/forums.php?m=posts&q=7272&n=last#bottom
https://talented-people.vraiforum.com/profile.php?mode=viewprofile&u=8473
В нашем мире, где диплом становится началом удачной карьеры в любой отрасли, многие пытаются найти максимально быстрый путь получения образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает дверь перед людьми, желающими вступить в сообщество квалифицированных специалистов или продолжить обучение в университете.
Предлагаем максимально быстро получить этот необходимый документ. Вы имеете возможность заказать диплом старого или нового образца, что будет удачным решением для человека, который не смог закончить обучение или потерял документ. дипломы выпускаются аккуратно, с особым вниманием к мельчайшим нюансам, чтобы в результате получился полностью оригинальный документ.
Плюсы подобного подхода заключаются не только в том, что вы быстро получите диплом. Весь процесс организовывается удобно и легко, с нашей поддержкой. От выбора необходимого образца документа до консультации по заполнению персональной информации и доставки в любой регион страны — все будет находиться под абсолютным контролем квалифицированных мастеров.
В результате, для тех, кто ищет оперативный способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – значит избежать долгого обучения и сразу перейти к важным целям: к поступлению в университет или к началу удачной карьеры.
vuzdiploma
최신 프라그마틱 게임은 iGaming 분야에서 혁신적이고 표준화된 콘텐츠를 제공하는 선도적인 업체입니다.
프라그마틱 무료
프라그마틱 슬롯에 대한 내용이 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 찾아보실 수 있어요. 함께 지식을 공유해보세요!
https://www.12315fk.cn/
https://doggggqwe.weebly.com/
https://jtsizzle.com/link/
k8 カジノ 銀行入金
このトピックに対する新しい見方をありがとうございます。
В нашем обществе, где диплом становится началом удачной карьеры в любой области, многие ищут максимально простой путь получения образования. Наличие официального документа сложно переоценить. Ведь диплом открывает двери перед каждым человеком, который хочет вступить в профессиональное сообщество или учиться в высшем учебном заведении.
В данном контексте мы предлагаем быстро получить любой необходимый документ. Вы можете купить диплом, и это становится удачным решением для всех, кто не смог закончить обучение или потерял документ. дипломы выпускаются с особой аккуратностью, вниманием ко всем деталям. В итоге вы сможете получить 100% оригинальный документ.
Плюсы подобного решения состоят не только в том, что можно максимально быстро получить диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора подходящего образца до точного заполнения персональных данных и доставки в любой регион России — все будет находиться под полным контролем наших мастеров.
Всем, кто ищет быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Купить диплом – это значит избежать долгого процесса обучения и не теряя времени перейти к достижению своих целей, будь то поступление в университет или начало карьеры.
http://5chudes.com/product/magazin-morozhenoe/reviews/
http://redhawkclan.free.fr/index.php?file=Members&op=detail&autor=ofecy
http://mag-motors.ru/index.php?subaction=userinfo&user=ugebeg
http://led119.ru/forum/user/104897/
http://tildanovaserv.ro/component/k2/item/7
В нашем обществе, где диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально простой путь получения качественного образования. Факт наличия официального документа об образовании переоценить просто невозможно. Ведь диплом открывает дверь перед людьми, стремящимися вступить в сообщество квалифицированных специалистов или продолжить обучение в каком-либо университете.
Наша компания предлагает максимально быстро получить этот важный документ. Вы можете приобрести диплом, и это является выгодным решением для человека, который не смог закончить обучение, утратил документ или желает исправить свои оценки. Любой диплом изготавливается с особой тщательностью, вниманием ко всем нюансам. В итоге вы получите 100% оригинальный документ.
Плюсы этого решения состоят не только в том, что можно максимально быстро получить диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начав от выбора необходимого образца до точного заполнения личных данных и доставки в любое место страны — все будет находиться под абсолютным контролем квалифицированных специалистов.
Всем, кто ищет максимально быстрый способ получить необходимый документ, наша компания готова предложить отличное решение. Купить диплом – это значит избежать длительного обучения и сразу перейти к достижению личных целей, будь то поступление в университет или начало карьеры.
http://www.diplomany.ru
В нашем обществе, где диплом – это начало удачной карьеры в любой отрасли, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Наличие официального документа об образовании сложно переоценить. Ведь именно он открывает дверь перед каждым человеком, который стремится начать трудовую деятельность или продолжить обучение в высшем учебном заведении.
Предлагаем очень быстро получить любой необходимый документ. Вы имеете возможность купить диплом, и это будет удачным решением для всех, кто не смог закончить обучение или утратил документ. Все дипломы выпускаются с особой аккуратностью, вниманием к мельчайшим элементам. В результате вы получите полностью оригинальный документ.
Преимущества такого решения заключаются не только в том, что вы сможете быстро получить диплом. Весь процесс организован комфортно, с нашей поддержкой. Начав от выбора нужного образца документа до консультаций по заполнению личных данных и доставки в любое место страны — все будет находиться под абсолютным контролем опытных специалистов.
В итоге, всем, кто ищет максимально быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать долгого процесса обучения и сразу перейти к достижению собственных целей, будь то поступление в ВУЗ или начало удачной карьеры.
http://lider1c.ru/communication/forum/messages/forum4/message6428/5131-eonlinediplom?result=new#message6428
http://www.plm.pw/2016/09/The-6-Factors-of-Industry-4.0.html
http://r0757478.bget.ru/home.php?mod=space&uid=17208&do=profile
http://parenvarmii.ru/topic3707.html?view=print
http://lcfreight.com/blog/2019/09/24/new-lc-freight-website/?unapproved=45640&moderation-hash=f3985a030b9c90a8aa3608d18c85138b#comment-45640
Pretty component of content. I simply stumbled upon your website and in accession capital to assert that I acquire actually enjoyed account your weblog posts. Any way I will be subscribing for your feeds and even I achievement you access constantly quickly.
https://turkiserial.co/
В современном мире, где диплом – это начало удачной карьеры в любом направлении, многие пытаются найти максимально простой путь получения качественного образования. Наличие официального документа трудно переоценить. Ведь именно он открывает двери перед всеми, кто стремится вступить в сообщество профессионалов или учиться в ВУЗе.
В данном контексте мы предлагаем максимально быстро получить этот необходимый документ. Вы можете заказать диплом нового или старого образца, и это будет отличным решением для человека, который не смог завершить образование или потерял документ. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам, чтобы в результате получился документ, 100% соответствующий оригиналу.
Превосходство подобного решения состоит не только в том, что вы сможете максимально быстро получить диплом. Весь процесс организовывается удобно и легко, с профессиональной поддержкой. Начиная от выбора нужного образца диплома до консультаций по заполнению личной информации и доставки в любой регион России — все находится под полным контролем квалифицированных мастеров.
Для тех, кто хочет найти быстрый и простой способ получить необходимый документ, наша компания может предложить выгодное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и сразу перейти к важным целям: к поступлению в университет или к началу удачной карьеры.
diplomany.ru
valtrex rx online
В нашем обществе, где диплом – это начало удачной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Факт наличия официального документа об образовании переоценить невозможно. Ведь именно он открывает дверь перед любым человеком, который хочет начать профессиональную деятельность или учиться в ВУЗе.
Мы предлагаем очень быстро получить этот необходимый документ. Вы можете приобрести диплом, и это будет отличным решением для человека, который не смог закончить обучение или утратил документ. Все дипломы выпускаются аккуратно, с особым вниманием к мельчайшим нюансам, чтобы на выходе получился полностью оригинальный документ.
Преимущества подобного решения состоят не только в том, что можно оперативно получить свой диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начав от выбора необходимого образца до консультации по заполнению личных данных и доставки по стране — все находится под полным контролем квалифицированных мастеров.
Всем, кто хочет найти оперативный способ получения необходимого документа, наша услуга предлагает выгодное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и не теряя времени перейти к достижению собственных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://www.ajuede.com/2019/09/taylor-swift-white-supremacy-is.html
http://www.plm.pw/2016/09/The-6-Factors-of-Industry-4.0.html
http://clinicavillafranca.com/?unapproved=11158&moderation-hash=afc454a4a2b73fd5ea21dcbdadea1151#comment-11158
http://www.retro-lv.club/2020/08/nazvanija-ulic-v-rige.html
http://moj.webservis.ru/forums/profile.php?action=show&member=5950
В современном мире, где диплом является началом успешной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Факт наличия официального документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед людьми, желающими начать трудовую деятельность или учиться в каком-либо университете.
В данном контексте наша компания предлагает очень быстро получить любой необходимый документ. Вы имеете возможность заказать диплом старого или нового образца, что является выгодным решением для всех, кто не смог завершить обучение, потерял документ или хочет исправить плохие оценки. диплом изготавливается аккуратно, с максимальным вниманием ко всем элементам, чтобы в результате получился документ, максимально соответствующий оригиналу.
Плюсы этого подхода состоят не только в том, что вы быстро получите диплом. Процесс организован удобно и легко, с нашей поддержкой. От выбора необходимого образца до консультации по заполнению личной информации и доставки по стране — все будет находиться под полным контролем опытных мастеров.
Всем, кто пытается найти максимально быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать длительного процесса обучения и не теряя времени переходить к достижению своих целей, будь то поступление в ВУЗ или старт профессиональной карьеры.
http://www.ab-diplom.ru
В современном мире, где диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения образования. Необходимость наличия официального документа переоценить просто невозможно. Ведь именно диплом открывает дверь перед любым человеком, который желает вступить в сообщество профессионалов или учиться в университете.
Наша компания предлагает оперативно получить этот важный документ. Вы можете заказать диплом нового или старого образца, и это становится выгодным решением для всех, кто не смог закончить образование, утратил документ или хочет исправить плохие оценки. Любой диплом изготавливается с особой тщательностью, вниманием ко всем деталям, чтобы в результате получился документ, максимально соответствующий оригиналу.
Плюсы этого подхода заключаются не только в том, что можно оперативно получить диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. Начав от выбора подходящего образца до консультации по заполнению личных данных и доставки в любое место страны — все под полным контролем квалифицированных мастеров.
Таким образом, для тех, кто ищет оперативный способ получить необходимый документ, наша компания готова предложить выгодное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и сразу переходить к важным целям, будь то поступление в ВУЗ или старт трудовой карьеры.
https://narodovmnogo-omsk.ru/forum/user/2809/
http://tekst-pesni.ru/index.php?name=account&op=info&uname=ucyrefi
http://kirpich-opt-265.1gb.ru/index.php?subaction=userinfo&user=ubivutov
http://www.thenatureofthings.blog/2022/06/overboard-by-sara-paretsky-review.html
https://www.buehnehollenthon.at/guestbook2/
В нашем обществе, где диплом – это начало удачной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения образования. Факт наличия официального документа сложно переоценить. Ведь именно диплом открывает двери перед людьми, стремящимися вступить в профессиональное сообщество или учиться в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить этот необходимый документ. Вы можете приобрести диплом, что является выгодным решением для всех, кто не смог завершить образование или утратил документ. дипломы производятся аккуратно, с особым вниманием ко всем деталям, чтобы на выходе получился документ, максимально соответствующий оригиналу.
Плюсы подобного подхода заключаются не только в том, что вы оперативно получите свой диплом. Процесс организовывается просто и легко, с профессиональной поддержкой. Начав от выбора необходимого образца диплома до консультации по заполнению персональной информации и доставки в любой регион страны — все находится под полным контролем наших специалистов.
Для тех, кто хочет найти оперативный способ получения необходимого документа, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать длительного обучения и сразу перейти к важным целям, будь то поступление в университет или старт карьеры.
http://www.ab-diplom.ru
В современном мире, где диплом – это начало успешной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Факт наличия документа об образовании переоценить невозможно. Ведь именно он открывает дверь перед любым человеком, желающим вступить в сообщество профессиональных специалистов или учиться в каком-либо институте.
В данном контексте наша компания предлагает быстро получить любой необходимый документ. Вы можете купить диплом, что является выгодным решением для человека, который не смог закончить обучение, потерял документ или хочет исправить плохие оценки. Все дипломы производятся с особой тщательностью, вниманием ко всем нюансам, чтобы в итоге получился документ, полностью соответствующий оригиналу.
Преимущества данного решения заключаются не только в том, что вы оперативно получите диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. От выбора подходящего образца диплома до грамотного заполнения персональной информации и доставки по России — все под абсолютным контролем качественных мастеров.
Всем, кто ищет максимально быстрый способ получить требуемый документ, наша услуга предлагает отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и сразу переходить к достижению своих целей, будь то поступление в университет или старт карьеры.
http://www.ab-diplom.ru
Please let me know if you’re looking for a writer for your site. You have some really great articles and I believe I would be a good asset. If you ever want to take some of the load off, I’d love to write some articles for your blog in exchange for a link back to mine. Please send me an email if interested. Cheers!
diplom07.ru
Hello there! Would you mind if I share your blog with my facebook group? There’s a lot of folks that I think would really enjoy your content. Please let me know. Cheers
server-attestats.com
tintucnamdinh24h.com
그는 절하고 검을 들고 은당으로 내려가 팡지판에게 건넸다.
프라그마틱 게임은 정말로 혁신적이에요. 특히 슬롯 게임들은 항상 기대 이상의 재미를 선사합니다!
프라그마틱 홈페이지
프라그마틱 콘텐츠 항상 기대돼요! 또한 제 사이트에서도 유용한 정보를 제공하고 있어요. 상호 교류하며 더 많은 지식을 얻어가요!
https://www.12315th.cn/
https://www.12315ic.cn/
https://www.jaswanthch.com
В современном мире, где диплом – это начало отличной карьеры в любой области, многие пытаются найти максимально быстрый и простой путь получения образования. Факт наличия официального документа об образовании переоценить попросту невозможно. Ведь именно он открывает дверь перед всеми, кто хочет вступить в сообщество профессионалов или продолжить обучение в любом ВУЗе.
В данном контексте наша компания предлагает очень быстро получить любой необходимый документ. Вы сможете приобрести диплом нового или старого образца, что становится выгодным решением для человека, который не смог закончить обучение или потерял документ. Все дипломы производятся аккуратно, с максимальным вниманием к мельчайшим деталям. В результате вы получите продукт, максимально соответствующий оригиналу.
Плюсы такого подхода заключаются не только в том, что вы сможете оперативно получить диплом. Весь процесс организован комфортно, с нашей поддержкой. Начиная от выбора требуемого образца диплома до консультаций по заполнению личной информации и доставки в любое место России — все находится под абсолютным контролем квалифицированных мастеров.
Всем, кто ищет быстрый и простой способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – это значит избежать долгого процесса обучения и не теряя времени переходить к личным целям, будь то поступление в университет или начало карьеры.
https://suveniryregionov.ru/product/igraj-garmon-75h40h75-mm/reviews/
http://jm-eng16.com/bbs/board.php?bo_table=free&wr_id=320724
http://www.hucellbio.com/bbs/board.php?bo_table=free&wr_id=376038
http://kemavto.kuzbass.net/index.php?subaction=userinfo&user=asuficu
http://www.idsys.kr/bbs/board.php?bo_table=free&wr_id=487490
Hi there I am so excited I found your web site, I really found you by accident, while I was browsing on Google for something else, Anyhow I am here now and would just like to say thanks for a fantastic post and a all round thrilling blog (I also love the theme/design), I don’t have time to read through it all at the moment but I have book-marked it and also included your RSS feeds, so when I have time I will be back to read a great deal more, Please do keep up the fantastic job.
http://arusak-attestats.ru
We absolutely love your blog and find the majority of your post’s to be precisely what I’m looking for. Would you offer guest writers to write content for yourself? I wouldn’t mind writing a post or elaborating on most of the subjects you write about here. Again, awesome blog!
http://www.arusak-attestats.ru
geinoutime.com
“아뇨, 벽돌도 구하고, 그럼 다른 곳도….그렇죠…”
k8 カジノ 系列
このブログのファンになりました。更新を楽しみにしています!
mikaspa.com
모두가 위험을 감수하고 큰 기여를 했으니 상을 줄 이유가 없다고 하셨습니다.적에게 많은 탈북이 그를 조금 당황하게 만들었고, 그는 더 의심스러워졌습니다.
[url=https://azithromycinmds.com/]buy generic zithromax[/url]
This is really interesting, You’re a very skilled blogger. I have joined your feed and look forward to seeking more of your excellent post. Also, I have shared your website in my social networks!
http://www.diplomy-originaly.com
Приветики!
На нашем сайте вы можете выбрать и приобрести диплом Вуза с гарантией и доставкой в любой регион России по самым низким ценам.
https://eonline-diploms.com/
Good day! Do you know if they make any plugins to protect against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any recommendations?
diplomy-originaly.com
Приветики!
Получите документы об образовании всех Вузов России с гарантированной доставкой по РФ без предоплаты.
eonline-diploms.com
This is really interesting, You are a very skilled blogger. I have joined your feed and look forward to seeking more of your great post. Also, I have shared your web site in my social networks!
http://www.diplomy-originaly.com
Здравствуйте!
Предлагаем всем желающим приобрести диплом университетов России по низкой цене с доставкой в руки под ключ
Приобретите диплом Гознака с гарантированной подлинностью и доставкой по всей России без предоплаты.
http://diploms-asx.com
Hello everyone, it’s my first pay a quick visit at this site, and piece of writing is actually fruitful for me, keep up posting such articles or reviews.
Замок для соковитискача Zelmer (12000656)
Hello, yes this paragraph is truly nice and I have learned lot of things from it about blogging. thanks.
http://kvitka-ua.com/linzy-u-fary-dlya-vashogo-avtomobilya
Good day! This is kind of off topic but I need some help from an established blog. Is it very difficult to set up your own blog? I’m not very techincal but I can figure things out pretty fast. I’m thinking about setting up my own but I’m not sure where to begin. Do you have any tips or suggestions? Many thanks
http://kvitka-ua.com/linzy-u-fary-dlya-vashogo-avtomobilya
프라그마틱과 관련된 내용 감사합니다! 또한, 제 사이트에서도 프라그마틱에 대한 정보를 찾아보실 수 있어요. 서로 이야기 공유하며 더 많은 지식을 얻어가요!
프라그마틱플레이
프라그마틱과 관련된 내용 감사합니다! 또한, 제 사이트에서도 프라그마틱에 대한 정보를 찾아보실 수 있어요. 서로 이야기 공유하며 더 많은 지식을 얻어가요!
https://e-trajet.net/hot/
https://www.12315xa.cn/
https://www.12315amb.cn/
Привет всем!
Поможем Вам купить диплом Вуза России недорого, без предоплаты и с гарантией возврата средств
diplomanc-russia24.ru
Привет всем!
Получите документы об образовании всех Вузов России с гарантированной доставкой по РФ без предоплаты.
https://diplomanc-russia24.ru
Привет, дорогой читатель!
Получите документ ВУЗа без предоплаты и доставку в руки по России – просто, удобно, безопасно!
http://www.diplomanc-russia24.ru
k8 カジノ パチスロ
情報が豊富で読み応えがありました。非常に有益です。
Hello there! This is kind of off topic but I need some help from an established blog. Is it very difficult to set up your own blog? I’m not very techincal but I can figure things out pretty quick. I’m thinking about making my own but I’m not sure where to begin. Do you have any tips or suggestions? Thank you
https://cutt.ly/Cero2KWb
Hello just wanted to give you a quick heads up and let you know a few of the images aren’t loading correctly. I’m not sure why but I think its a linking issue. I’ve tried it in two different browsers and both show the same results.
https://cutt.ly/4eroNxoY
Hello there I am so glad I found your webpage, I really found you by mistake, while I was researching on Digg for something else, Anyhow I am here now and would just like to say thanks a lot for a incredible post and a all round exciting blog (I also love the theme/design), I don’t have time to read through it all at the minute but I have saved it and also added your RSS feeds, so when I have time I will be back to read much more, Please do keep up the superb work.
https://cutt.ly/tero3oej
lacolinaecuador.com
이 절름발이가 Fang Jifan을 화나게하고 칼로 누군가를 죽였습니까?
geinoutime.com
어쨌든 아빠가 보고 싶고, 아들이 보고 싶고…
werankcities.com
즉시 Xu Jing은 소매에서 선물 목록을 꺼냈습니다. “남자 여러분, 읽어보세요.”
Hi it’s me, I am also visiting this web page on a regular basis, this web site is actually pleasant and the viewers are really sharing pleasant thoughts.
edu-digest.ru
В нашем обществе, где диплом становится началом отличной карьеры в любой отрасли, многие стараются найти максимально простой путь получения качественного образования. Важность наличия официального документа переоценить просто невозможно. Ведь диплом открывает дверь перед всеми, кто желает вступить в сообщество профессиональных специалистов или продолжить обучение в высшем учебном заведении.
Мы предлагаем быстро получить этот важный документ. Вы можете приобрести диплом нового или старого образца, что является выгодным решением для человека, который не смог завершить образование или потерял документ. Все дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим элементам, чтобы на выходе получился 100% оригинальный документ.
Превосходство подобного подхода заключается не только в том, что можно максимально быстро получить диплом. Весь процесс организовывается просто и легко, с нашей поддержкой. От выбора необходимого образца до точного заполнения персональной информации и доставки по стране — все будет находиться под абсолютным контролем наших мастеров.
Всем, кто ищет быстрый способ получить необходимый документ, наша услуга предлагает отличное решение. Купить диплом – это значит избежать долгого процесса обучения и не теряя времени переходить к достижению личных целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://www.edu-nt.ru/
В наше время, когда диплом является началом успешной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед каждым человеком, желающим вступить в сообщество профессионалов или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить этот необходимый документ. Вы сможете приобрести диплом нового или старого образца, что становится отличным решением для всех, кто не смог закончить обучение или утратил документ. Все дипломы выпускаются с особой тщательностью, вниманием ко всем элементам, чтобы в итоге получился продукт, максимально соответствующий оригиналу.
Превосходство подобного подхода заключается не только в том, что можно оперативно получить свой диплом. Процесс организован комфортно, с нашей поддержкой. От выбора необходимого образца диплома до точного заполнения личной информации и доставки по России — все находится под абсолютным контролем качественных специалистов.
Для тех, кто ищет быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Приобрести диплом – значит избежать долгого процесса обучения и не теряя времени перейти к важным целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://www.dof-edu.ru
В нашем мире, где диплом – это начало удачной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа переоценить попросту невозможно. Ведь именно диплом открывает двери перед каждым человеком, желающим вступить в сообщество профессионалов или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы сможете приобрести диплом нового или старого образца, и это будет выгодным решением для всех, кто не смог завершить образование или утратил документ. дипломы выпускаются с особой тщательностью, вниманием к мельчайшим деталям. В итоге вы сможете получить документ, 100% соответствующий оригиналу.
Преимущество этого подхода заключается не только в том, что можно максимально быстро получить диплом. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора нужного образца документа до точного заполнения личной информации и доставки по стране — все под полным контролем качественных мастеров.
Для тех, кто хочет найти максимально быстрый способ получения требуемого документа, наша услуга предлагает отличное решение. Купить диплом – это значит избежать длительного обучения и не теряя времени перейти к важным целям: к поступлению в университет или к началу удачной карьеры.
http://proobzh.ru
В нашем обществе, где диплом – это начало успешной карьеры в любой области, многие ищут максимально простой путь получения образования. Важность наличия документа об образовании переоценить попросту невозможно. Ведь диплом открывает дверь перед каждым человеком, который хочет вступить в сообщество профессионалов или учиться в университете.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы сможете купить диплом нового или старого образца, и это является выгодным решением для человека, который не смог завершить образование, утратил документ или хочет исправить свои оценки. Любой диплом изготавливается с особой тщательностью, вниманием ко всем элементам, чтобы на выходе получился полностью оригинальный документ.
Преимущества этого подхода заключаются не только в том, что можно быстро получить диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. Начиная от выбора требуемого образца диплома до консультации по заполнению персональных данных и доставки в любое место страны — все будет находиться под полным контролем квалифицированных специалистов.
Для тех, кто хочет найти оперативный способ получить требуемый документ, наша услуга предлагает выгодное решение. Купить диплом – значит избежать продолжительного обучения и не теряя времени переходить к достижению личных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://edu-nt.ru/
В нашем мире, где диплом является началом удачной карьеры в любой области, многие стараются найти максимально простой путь получения качественного образования. Необходимость наличия официального документа об образовании переоценить просто невозможно. Ведь именно он открывает двери перед любым человеком, желающим начать профессиональную деятельность или учиться в ВУЗе.
Наша компания предлагает очень быстро получить любой необходимый документ. Вы сможете заказать диплом нового или старого образца, что будет выгодным решением для всех, кто не смог закончить обучение или потерял документ. Каждый диплом изготавливается аккуратно, с максимальным вниманием ко всем деталям. На выходе вы сможете получить продукт, полностью соответствующий оригиналу.
Превосходство подобного подхода заключается не только в том, что можно быстро получить свой диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. Начиная от выбора нужного образца диплома до консультаций по заполнению личных данных и доставки в любой регион страны — все находится под абсолютным контролем качественных мастеров.
Таким образом, для всех, кто хочет найти быстрый и простой способ получения требуемого документа, наша компания предлагает отличное решение. Заказать диплом – это значит избежать длительного обучения и не теряя времени перейти к достижению своих целей, будь то поступление в университет или старт карьеры.
proobzh.ru
May I simply just say what a relief to uncover somebody that actually knows what they are talking about online. You actually know how to bring an issue to light and make it important. More people must check this out and understand this side of the story. I was surprised that you’re not more popular since you certainly possess the gift.
http://www.edu-digest.ru
На сегодняшний день, когда диплом – это начало успешной карьеры в любой отрасли, многие пытаются найти максимально простой путь получения образования. Необходимость наличия документа об образовании переоценить просто невозможно. Ведь именно он открывает двери перед людьми, желающими начать профессиональную деятельность или учиться в ВУЗе.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете купить диплом старого или нового образца, и это становится удачным решением для человека, который не смог закончить образование, потерял документ или желает исправить свои оценки. Все дипломы производятся аккуратно, с максимальным вниманием ко всем элементам. В итоге вы получите документ, 100% соответствующий оригиналу.
Преимущество подобного подхода состоит не только в том, что можно быстро получить свой диплом. Процесс организован комфортно, с профессиональной поддержкой. Начав от выбора необходимого образца до консультации по заполнению личной информации и доставки по России — все будет находиться под абсолютным контролем качественных специалистов.
Всем, кто хочет найти быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Купить диплом – значит избежать длительного процесса обучения и не теряя времени перейти к важным целям: к поступлению в ВУЗ или к началу успешной карьеры.
proobzh.ru
Hi, its pleasant article about media print, we all understand media is a great source of information.
http://www.edu-digest.ru
프라그마틱 플레이의 슬롯 포트폴리오는 250개 이상의 게임으로 다양한 화폐 및 33개 언어로 전 세계 시장에 제공됩니다.
PG 소프트
프라그마틱의 게임은 정말 다양한데, 최근에 출시된 것 중 어떤 게임이 가장 좋았나요? 공유해주세요!
https://www.12315oi.cn/
https://www.12378za.cn/
https://www.12315amb.cn/
В современном мире, где диплом – это начало удачной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в высшем учебном заведении.
Мы предлагаем максимально быстро получить этот важный документ. Вы сможете заказать диплом, что будет выгодным решением для человека, который не смог закончить обучение или потерял документ. дипломы производятся с особой аккуратностью, вниманием ко всем нюансам. В итоге вы получите 100% оригинальный документ.
Плюсы подобного решения заключаются не только в том, что можно быстро получить диплом. Весь процесс организован просто и легко, с нашей поддержкой. Начиная от выбора нужного образца документа до консультации по заполнению персональной информации и доставки по России — все находится под полным контролем опытных мастеров.
Таким образом, всем, кто пытается найти быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать долгого обучения и сразу переходить к достижению собственных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
dof-edu.ru
k8 ゲーム
このトピックについてこんなに詳しく書かれた記事は初めてです。素晴らしいです!
В нашем мире, где диплом – это начало удачной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает дверь перед каждым человеком, который собирается начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте мы предлагаем очень быстро получить этот необходимый документ. Вы имеете возможность заказать диплом, и это является выгодным решением для всех, кто не смог завершить обучение, потерял документ или желает исправить свои оценки. Каждый диплом изготавливается аккуратно, с особым вниманием к мельчайшим нюансам. В итоге вы получите продукт, полностью соответствующий оригиналу.
Преимущество такого подхода заключается не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора необходимого образца до точного заполнения личных данных и доставки в любое место России — все под полным контролем качественных специалистов.
Таким образом, для всех, кто ищет максимально быстрый способ получения требуемого документа, наша услуга предлагает выгодное решение. Купить диплом – это значит избежать продолжительного обучения и не теряя времени перейти к личным целям: к поступлению в университет или к началу успешной карьеры.
edu-nt.ru
tvlore.com
잠시 후 Liu Jian과 세 사람은 Fengtian Hall로 서둘러갔습니다.
I think everything typed was very reasonable. However, what about this? suppose you were to write a awesome title? I ain’t suggesting your information is not solid, but suppose you added something to maybe get a person’s attention? I mean %BLOG_TITLE% is a little vanilla. You could look at Yahoo’s front page and watch how they write news headlines to grab viewers to open the links. You might add a video or a picture or two to grab readers excited about what you’ve got to say. Just my opinion, it might bring your posts a little bit more interesting.
idiplomanys.com
В современном мире, где диплом является началом удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь именно он открывает двери перед людьми, стремящимися вступить в сообщество квалифицированных специалистов или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы сможете купить диплом нового или старого образца, и это становится отличным решением для человека, который не смог завершить образование или потерял документ. Все дипломы изготавливаются с особой аккуратностью, вниманием ко всем деталям. В итоге вы получите документ, полностью соответствующий оригиналу.
Превосходство данного решения заключается не только в том, что вы сможете быстро получить свой диплом. Весь процесс организовывается удобно, с нашей поддержкой. От выбора требуемого образца документа до точного заполнения персональных данных и доставки по стране — все будет находиться под абсолютным контролем наших специалистов.
В итоге, для всех, кто ищет оперативный способ получения необходимого документа, наша услуга предлагает отличное решение. Заказать диплом – это значит избежать длительного процесса обучения и не теряя времени переходить к достижению личных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
http://6landik-diploms.com/
Сегодня, когда диплом – это начало удачной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения образования. Факт наличия официального документа об образовании трудно переоценить. Ведь именно он открывает дверь перед любым человеком, который собирается начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы можете купить диплом, что становится отличным решением для человека, который не смог завершить образование или утратил документ. диплом изготавливается с особой аккуратностью, вниманием ко всем деталям, чтобы в итоге получился полностью оригинальный документ.
Превосходство такого решения заключается не только в том, что вы сможете быстро получить свой диплом. Процесс организовывается удобно, с нашей поддержкой. Начиная от выбора необходимого образца до консультаций по заполнению персональной информации и доставки по стране — все находится под полным контролем качественных специалистов.
В результате, для тех, кто хочет найти максимально быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени перейти к своим целям: к поступлению в университет или к началу успешной карьеры.
http://www.6landik-diploms.com/
ihrfuehrerschein.com
Fang Jifan은 차가운 얼굴로 극도로 비겁해 보이는 Li Chaowen을 바라 보았습니다.
buy zithromax online uk
다양한 언어와 화폐로 지원되는 프라그마틱 슬롯은 세계적인 플레이어들에게 열린 창입니다.
프라그마틱슬롯
프라그마틱에 대한 이 글 감사합니다. 더불어, 제 사이트에서도 프라그마틱과 관련된 유용한 정보를 찾아보세요. 서로 이야기 나누면 더 좋겠죠!
https://www.eco2home.com
https://toploansbadcredit.com
http://holyshirtsandpants.net/
geinoutime.com
Fang Jifan의 말을 듣고 Liu Tianzheng은 차갑게 코를 골았습니다.
k8 カジノ 入金 クレジットカード
素晴らしい記事!共有せずにはいられません。
nikontinoll.com
그는 계속 지켜보았고, 재난 속에서… 재난 지역 사람들은 물고기를 먹고 있었습니다…
프라그마틱 라이브 카지노는 최고의 스튜디오에서 생방송되는 바카라, 룰렛, 블랙잭 등의 다양한 게임을 제공합니다. 베가스 볼 보난자, 스네이크스 앤드 래더스 라이브, 파워업 룰렛과 같은 게임으로 현장 카지노의 현장을 느껴보세요.
프라그마틱플레이
프라그마틱의 라이브 카지노는 정말 현장감 넘치게 즐길 수 있는데, 여기서 더 많은 정보를 얻을 수 있어 좋아요!
https://www.824989.com
https://www.12315uh.cn/
https://www.12315ds.cn/
thebuzzerpodcast.com
이것은 정말로 먹구름 속의 갑작스러운 새벽입니다.
geinoutime.com
무지한 사람들은 베이징에서 일어난 일에 대해 아무것도 모를 수도 있습니다.
freeflowincome.com
그는 중문으로 나가지 않고 류저택의 작은 문으로 나갔다.
k8 カジノ アフィリエイト
いつも新しい知識を得られる、本当に素敵なブログです。
dexamethasone canadian pharmacy
Купить Аттестат О Среднем Образовании
Купить Аттестат О Среднем Образовании
С дипломом по этой специальности у вас не купит Аттестат О Среднем Образовании проблем с трудоустройством. Продолжая служить верой с правдой уже не генеральному секретарю, а президенту с Отечеством. Что делать, когда окончили ВУЗ, приобрели диплом, подтверждающий профессиональное обучение, но потеряли. Чтобы воспользоваться этими достоинствами нашего предложения и приобрести доступный и надежный диплом, звоните в нашу службу клиентской поддержки для уточнения условий и оформления заявки. Однако надо признать, что попытка купить диплом у незнакомцев в подземном переходе сопряжена с огромным риском. Если вы мечтаете о престижной работе, но ваши документы были утеряны или испорчены, не стоит отчаиваться, ведь мы предлагаем вам купить диплом в Москве.
http://www.russkiy365-diploms-srednee.ru
Генеральная Доверенность Цена Спб
Если я закажу сегодня, когда документ будет у меня на руках, будет ли действителен диплом при проверке на госуслугах, хотела сделать аттестат за 11 классов 1995 года с оценками. Наша помощь может понадобиться людям, которые потеряли оригинал или случайно его порвали. Зачастую это в разы меньше зарплаты, которую может купить такая инвестиция. Далее рассмотрим перечень защитных элементов на бланке диплома: водяные знаки, защитная линия проходящая через весь бланк, регистрационный и региональный номер, защитная сетка и цветные нити, которые видно под ультрафиолетом, защитный микрошрифт, голограмма, нужно быть наивным выпускником или первокурсником, чтобы верить, что сама по себе корочка является каким-либо аттестатом в жизнь.
Белый Диплом Это
Суть профессии заключается изучении теории и практики экономики по разным программам. Далеко не каждая компания в Казахстане пойдёт на официальное оформление человека без “корочек”, а некоторые дадут список учебных заведений, диплом бакалавра которых им подходит (с другими нельзя). Ушинского в дипломе бакалавра по направлению Филология в 2015 году на бланке Киржач.
Сколько Стоит Диплом О Высшем Образовании С Занесением В Реестр
Сколько Стоит Диплом О Высшем Образовании С Занесением В Реестр
Продажа свидетельств о разводе, позволяет уйти от спорных ситуаций в юридических и финансовых вопросах, к тому же возвращают статус холостне замужем, что, в случае неудачного опыта семейной жизни, является приятным бонусом. Также документ об образовании может получить лицо, имеющее нотариально заверенную доверенность, а также курьер, если он официальный курьер с заказом от владельца диплома. Здесь, на нашем сайте вы сможете познакомиться с основными видами документов о наличии образовательной или ученой степени, которыми мы располагаем. Есть ли возможность стоит Диплом О Высшем Образовании С Занесением В Реестр нормоконтроль или аннотацию к дипломной работе. Важно, чтобы у поступающего имелись документы, подтверждающие его личность и гражданство. Присутствие в базе образовательных документов страны и оригинальный бланк позволяют Вам заказать диплом специалиста, который пройдет любую проверку. В наше время образование играет важную роль в формировании успешной карьеры и обеспечении стабильного будущего.
https://russkiy365-diploms-srednee.ru/
Купить Диплом Преподавателя
Но наступили суровые времена и ваш потенциальный работодатель попросил у вас взглянуть на ваш вкладыш к диплому о высшем образовании. Хороший ответ на данный вопрос купить диплом бакалавра 2019 или 2020 года недорого в нашей компании. Если определились с целью, то приобретение настоящего красного диплома государственного института или университета в Москве может стать быстрым и успешным выходом из положения.
Можно Ли Купить Диплом О Среднем Профессиональном Образовании
В нашей компании отлично разработана система, которая позволяет нашим заказчикам убедиться самостоятельно в качестве предоставляемых документов. Подлинность документа будет подтверждена печатями, подписями, а также всеми необходимыми степенями защиты. Каждый человек несколько раз в году посещает стоматологические клиники. То есть можно сделать диплом на бланке настоящего ГоЗнака и он пройдет любые проверки – архивы просто не сохранились.
Купить Диплом Повара В Москве
Купить Диплом Повара В Москве
Если у Вас остались вопросы о том, как оформить диплом медсестры и как работает доставка по Украине, обратитесь к консультанту компании. Все элементы такого диплома сделаны идеально качественно и даже при визуальном осмотре работодатель сразу поймет, что это настоящий документ. Но если вы не смогли получить это образование, но уже приобрели бесценный опыт работы, то можно решить проблему, купив диплом Повара техникума или колледжа в Москве и других городах. Вероятнее всего, вам интересно, сколько стоит диплом фармацевта в нашей компании и тут следует отметить некоторые детали. Наверняка вы попали на этот сайт не случайно, Это отличная возможность продолжить обучение заграницей, где покупать качественные образовательные бумаги, чтобы выбрать надежного продавца, почитайте отзывы клиентов. Мы не тянем время и беремся за выполнение заказа в день подачи заявки.
http://www.russkiy365-diploms-srednee.ru
Купить Диплом Колледжа
Также у нас есть доставка в регионы. Техническое задание на дипломный проект по заданию Если вам нужно техническое задание для дипломной работы, сообщите об этом личному менеджеру. Выплачивая каждый год непомерные суммы ненасытному институту, а иногда и преподавателям, ты лишь получаешь взамен кучу никому не нужной, при этом давно устаревшей теории. Мы работаем с курьерскими службами DHL, Деловые Линии, и др.
Диплом Спо
Работникам, которые любят консультировать и помогать людям с выбором медикаментов. Получить диплом можно уже в день заказа при малой загруженности или же на следующий день. Диплом медицинского труженика не только раскрывает двери для работы в мед учреждениях, да и позволяет конкретно влиять на здоровье и жизни других людей. В интернете очень много компаний, которые призывают всех желающих купить диплом в Москве именно у них.
Сколько Страниц В Диплом Колледже
Сколько Страниц В Диплом Колледже
Работодатели знают, что система образования в стране находится на высшем уровне и если человек окончил вуз, особенно один из престижных, значит, он будет интересен как специалист. Вы приобретаете не только определенную страницу В Диплом Колледже и защиту при прохождении аттестации, к Вам полностью меняется отношение работодателя. Диплом магистра это документ, подтверждающий получение высшего образования и присвоение страницы В Диплом Колледже Магистр. Например, если средний балл, который у Вас получился в колледже, ниже, чем в аттестате, Вы вправе подать школьный аттестат с наиболее высокими показателями при поступлении в ВУЗ. Оплата только за готовый результат, где мне уже все показали ФИЗИЧЕСКИ, обьяснили, достоверно продемонстрировали. Последние несколько лет мы работали над расширением своих возможностей, нанимая квалифицированных специалистов. На нашем сайте очень много информации как не попасть впросак и не потерять впустую деньги.
russkiy365-diploms-srednee.ru
Где Купить Диплом О Высшем Образовании В Москве
Над каждым дипломом работает команда профессиональных каллиграфов, которые создают документы, ничем не отличающиеся от тех, что получают выпускники ВУЗов, вплоть до подписей и подлинных печатей. Все зависит только от ваших личных предпочтений и потребностей, как быстро вы сможете получить свой новый диплом, диплом о среднем образовании может понадобиться в любой момент. Выгодное устройство на работу с подтверждением, стоит ли покупать диплом – причины в пользу данного решения, достаточно ли знаний в образовательной сфере. Для этого подойдут справки о дипломах Колледже, выписка о банковском счете, документ о получении алиментов.
Купить Обложку На Диплом О Высшем Образовании
Как правило, это происходит уже в более зрелом возрасте, что мешает ежедневному посещению Вуза. Не стоит легкомысленно этому верить, поскольку диплома диплома дело достаточно хлопотное и затратное для нечистых на страницу работников колледжа. Вопросом, нужен ли на сегодняшний день диплом ПТУ, задается, без преувеличения, немало молодых людей, которые задаются вопросом, учиться дальше какой-то определенной профессии, или же начать искать высокодоходную работу, которая не требует узкоспециальных навыков.
Диплом 2016
2 разъясняет населению действующее семейное законодательство и диплом 2016 регистрации актов гражданского состояния. Специальный комитет изучает предоставленные документы, после чего, желающий пройти переаттестацию, приступает к процессу сдачи экзамена. Первый документ, который получает любой гражданин государства свидетельство о рождении. При поступлении в высшие учебные заведения диплом бакалавра позволяет претендовать не только Какие Дипломы Бывают бюджетные места, но и на заочное обучение. Обладатели документов такого уровня пользуются спросом, поэтому смело предлагайте свою кандидатуру. Многие работодатели скептически смотрят на корочку, выданную малоизвестным вузом, пока не увидят на практике работу специалистов. Изучает и обобщает практику применения семейного законодательства в части регистрации актов гражданского состояния, распространяет положительный опыт в сфере своей деятельности.
russkiy365-diploms-srednee.ru
Сколько Стоит Купить Диплом Юриста
С учетом доставки вы получите заказ в среднем в течение 7 дней, обычно раньше. При выборе нужно определить, для каких нужд используется бумага. Большинство дипломов 2016 школы заканчивают 9, а не 11 классов с желанием пойти быстрее работать из-за неуверенности в поступлении в высшее учебное заведение.
Красный Диплом О Высшем Образовании
Каждый наш диплом 2016 частичка нашей наработанной клиентской дипломы 2016, а значит частичка нашего финансового благополучия. Клиенты, после окончания сотрудничества отмечают: высокий профессионализм наших специалистов, таким образом, любой, кто намеревается купить диплом в нашей организации, получит качественный сервис и полное удовлетворение своих потребностей. Вы можете узнать подробности на сайте или связаться с нашим представителем по указанным контактным данным. Купив диплом у нас, мы гарантируем качество и абсолютную идентичность, независимо от года выпуска, специальности, учебного заведения.
What’s up, after reading this remarkable post i am too delighted to share my familiarity here with colleagues.
https://webyourself.eu/blogs/301012/пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅ-пїЅ-пїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ
https://forum18.ru/threads/intellektualnaja-investicija-kupit-diplom-kak-strategija-uspexa.37288/
http://forumkasino.bestff.ru/viewtopic.php?id=3598#p8498
https://krugozorov.ru/forum/user/8790/
http://altla2.mybb.ru/viewtopic.php?id=424#p632
Because the admin of this web site is working, no doubt very shortly it will be well-known, due to its quality contents.
https://smd.mybb.ru/viewtopic.php?id=3149#p20996
http://sensemi.getbb.ru/viewtopic.php?f=6&t=455
https://webyourself.eu/blogs/316260/пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅ-пїЅпїЅпїЅпїЅ-пїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ
http://office.listbb.ru/viewtopic.php?f=2&t=3657
http://driveme.rusff.me/viewtopic.php?id=2033#p105075
Hi there, You have done an excellent job. I will certainly digg it and personally suggest to my friends. I am confident they will be benefited from this web site.
http://umorfishki.ru/kupit-diplom-byistro-udobno-nadezhno
https://kryza.network/create-blog/
http://sovetonk.ru/forum/user/176754/
http://uktuliza.ru/forum/?PAGE_NAME=profile_view&UID=17520
http://amaureya.mybb.ru/viewtopic.php?id=851#p11402
Simply wish to say your article is as astonishing. The clarity in your submit is just excellent and i could suppose you’re a professional in this subject. Well together with your permission let me to grab your RSS feed to stay updated with approaching post. Thank you one million and please keep up the enjoyable work.
http://kupitediplom0029.ru/
Good day! Would you mind if I share your blog with my twitter group? There’s a lot of folks that I think would really enjoy your content. Please let me know. Thanks
https://ai.cheap/read-blog/16840
http://www.zpu-journal.ru/forum/view_profile.php?UID=305125
http://newsato.ru/kupit-udostovereniya-o-shkolnom-obrazovanii
http://bitrix-2.dclouds.ru:8200/club/user/34/blog/12643/
http://medicineshocknews.ru/obrazovanie-po-zakazu-kak-byistro-i-legko-kupit-diplom
Awesome website you have here but I was wondering if you knew of any user discussion forums that cover the same topics talked about here? I’d really love to be a part of community where I can get advice from other experienced people that share the same interest. If you have any suggestions, please let me know. Cheers!
https://www.kupitediplom0029.ru/
ihrfuehrerschein.com
혹시… 설탕을 맛본 후 순한 우유에 흥미를 잃었나요?
What’s up to all, how is all, I think every one is getting more from this site, and your views are good for new visitors.
http://selkovo.rolka.me/viewtopic.php?id=2965#p9228
http://arsenal.listbb.ru/viewtopic.php?f=14&t=324
http://wiki.gsksurvey.ru/пїЅпїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ
https://satopttorg.ru/forum/user/2975/
http://newseducation2015.bestbb.ru/viewtopic.php?id=638#p650
The other day, while I was at work, my sister stole my iPad and tested to see if it can survive a 40 foot drop, just so she can be a youtube sensation. My iPad is now destroyed and she has 83 views. I know this is totally off topic but I had to share it with someone!
http://www.diplomdoc.ru/
This is my first time pay a visit at here and i am truly impressed to read everthing at one place.
https://diplommy.ru/
Greetings I am so happy I found your blog page, I really found you by accident, while I was researching on Aol for something else, Anyhow I am here now and would just like to say kudos for a marvelous post and a all round thrilling blog (I also love the theme/design), I don’t have time to read through it all at the minute but I have saved it and also included your RSS feeds, so when I have time I will be back to read a lot more, Please do keep up the great work.
http://forexrassia.ru/poluchite-diplom-prosto-byistro-nadezhno
http://www.sportpro.com.ua/forum/viewtopic.php?p=209164#209164
http://shockmusik.ru/diplom-po-zaprosu-kak-kupit-diplom-i-obespechit-uspeh
http://skazka.g-talk.ru/viewtopic.php?f=1&t=1641
http://mosday.getbb.ru/viewtopic.php?f=12&t=810
프라그마틱 게임은 iGaming 업계에서 혁신적인 모바일 중심의 콘텐츠를 선보이며 고객에게 탁월한 경험을 제공합니다.
프라그마틱플레이
프라그마틱 슬롯에 대한 이해가 높아지네요! 제 사이트에서도 유용한 프라그마틱 정보를 찾아볼 수 있어요. 함께 공유하고 발전해봐요!
https://www.12315hd.cn/
https://www.12315lb.cn/
http://www.iavstudios.com/
I enjoy, lead to I discovered just what I used to be taking a look for. You have ended my 4 day lengthy hunt! God Bless you man. Have a great day. Bye
https://vip.forums.party/viewtopic.php?id=3032#p70096
https://wiki.hightgames.ru/index.php/пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ
http://sports.bestbb.ru/viewtopic.php?id=693#p1269
http://worldavtonew.ru/diplom-na-zakaz-kak-byistro-i-bezopasno-poluchit-obrazovanie
http://algebra.bestbb.ru/viewtopic.php?id=532#p562
This info is worth everyone’s attention. Where can I find out more?
https://magazin-diplomov.ru/
В наше время, когда диплом становится началом успешной карьеры в любой отрасли, многие стараются найти максимально простой путь получения образования. Необходимость наличия официального документа об образовании сложно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает максимально быстро получить любой необходимый документ. Вы имеете возможность приобрести диплом, и это будет отличным решением для всех, кто не смог закончить обучение или утратил документ. Все дипломы изготавливаются с особой тщательностью, вниманием к мельчайшим нюансам. В итоге вы сможете получить документ, полностью соответствующий оригиналу.
Плюсы такого решения заключаются не только в том, что можно максимально быстро получить диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начиная от выбора необходимого образца документа до точного заполнения персональных данных и доставки по стране — все под абсолютным контролем квалифицированных специалистов.
Для всех, кто ищет оперативный способ получить требуемый документ, наша компания готова предложить отличное решение. Заказать диплом – значит избежать длительного обучения и сразу перейти к достижению личных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
http://www.r-diplom.ru
Hi, all is going nicely here and ofcourse every one is sharing data, that’s really good, keep up writing.
https://www.diplom-zakaz.ru/
Hmm is anyone else experiencing problems with the pictures on this blog loading? I’m trying to determine if its a problem on my end or if it’s the blog. Any suggestions would be greatly appreciated.
magazin-diplomov.ru
WOW just what I was searching for. Came here by searching for %meta_keyword%
http://www.magazin-diplomov.ru
В нашем обществе, где диплом – это начало отличной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие документа об образовании сложно переоценить. Ведь именно диплом открывает двери перед всеми, кто собирается начать профессиональную деятельность или учиться в любом ВУЗе.
Предлагаем очень быстро получить этот важный документ. Вы можете заказать диплом, и это становится удачным решением для всех, кто не смог завершить образование или утратил документ. Все дипломы производятся с особой тщательностью, вниманием к мельчайшим элементам. На выходе вы получите 100% оригинальный документ.
Преимущества подобного подхода состоят не только в том, что можно максимально быстро получить свой диплом. Процесс организовывается удобно, с профессиональной поддержкой. Начав от выбора необходимого образца документа до точного заполнения персональной информации и доставки по России — все под полным контролем квалифицированных специалистов.
Всем, кто ищет быстрый и простой способ получения необходимого документа, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать долгого процесса обучения и сразу переходить к достижению своих целей: к поступлению в ВУЗ или к началу успешной карьеры.
arxitekt.ru
Just want to say your article is as astonishing. The clarity in your post is just great and i can assume you are an expert on this subject. Well with your permission let me to grab your feed to keep updated with forthcoming post. Thanks a million and please carry on the rewarding work.
https://www.peoplediplom.ru/
Thanks for every other informative site. Where else may I am getting that kind of information written in such an ideal means? I have a project that I am just now working on, and I’ve been at the look out for such info.
https://berry.work/read-blog/12483
http://interes.mybb.social/viewtopic.php?id=4484#p11350
http://lightsdemons.phorum.pl/viewtopic.php?p=66833#66833
http://acmpskov.mybb.ru/viewtopic.php?id=1526#p2721
http://weekofsport.ru/dostizhenie-obrazovatelnyih-tseley-kak-kupit-diplom-s-vyigodoy
I blog frequently and I genuinely appreciate your content. This great article has truly peaked my interest. I’m going to take a note of your website and keep checking for new information about once per week. I opted in for your RSS feed as well.
http://www.diplom-profi.ru/
Simply wish to say your article is as amazing. The clearness to your publish is simply cool and i could suppose you’re an expert in this subject. Well together with your permission let me to clutch your RSS feed to keep up to date with imminent post. Thanks a million and please carry on the enjoyable work.
diplom-profi.ru/
Hi there, its nice piece of writing regarding media print, we all understand media is a enormous source of data.
https://zakaz-na-diplom.ru/
Simply want to say your article is as astounding. The clarity to your post is just excellent and that i can suppose you’re an expert in this subject. Fine with your permission allow me to take hold of your RSS feed to stay up to date with impending post. Thanks a million and please carry on the rewarding work.
https://kupite-diplom0024.ru/
This is the perfect site for everyone who wants to find out about this topic. You understand so much its almost tough to argue with you (not that I really will need to…HaHa). You certainly put a new spin on a topic which has been discussed for decades. Great stuff, just great!
http://https://diplom-city24.ru/
Hey! This is kind of off topic but I need some advice from an established blog. Is it very difficult to set up your own blog? I’m not very techincal but I can figure things out pretty quick. I’m thinking about creating my own but I’m not sure where to start. Do you have any ideas or suggestions? With thanks
http://https://diplom45.ru/
Aw, this was a really nice post. Finding the time and actual effort to create a really good article… but what can I say… I procrastinate a whole lot and never seem to get nearly anything done.
http://https://diplom45.ru/
Good day! I know this is kinda off topic nevertheless I’d figured I’d ask. Would you be interested in trading links or maybe guest authoring a blog article or vice-versa? My site addresses a lot of the same topics as yours and I feel we could greatly benefit from each other. If you happen to be interested feel free to shoot me an e-mail. I look forward to hearing from you! Excellent blog by the way!
http://https://diplom45.ru/
My spouse and I absolutely love your blog and find most of your post’s to be precisely what I’m looking for. can you offer guest writers to write content for yourself? I wouldn’t mind producing a post or elaborating on a number of the subjects you write regarding here. Again, awesome blog!
http://https://1magistr.ru/
What’s up to every body, it’s my first go to see of this weblog; this weblog contains remarkable and genuinely fine information in support of readers.
http://www.kupite-diplom0024.ru
cost of azithromycin 500 mg
에그벳 계열
“무슨 말씀이세요?” Zhu Houzhao는 갑자기 경계하기 시작했습니다.
I blog quite often and I really appreciate your information. This article has truly peaked my interest. I’m going to take a note of your site and keep checking for new information about once a week. I opted in for your RSS feed as well.
http://www.diplom-city24.ru/
Thanks for the auspicious writeup. It actually used to be a leisure account it. Glance complex to more introduced agreeable from you! However, how can we keep in touch?
http://www.zakaz-na-diplom.ru/
Whats up very cool web site!! Guy .. Excellent .. Wonderful .. I’ll bookmark your blog and take the feeds also? I am satisfied to seek out so many helpful info right here in the put up, we want work out extra techniques on this regard, thanks for sharing. . . . . .
http://www.institute-diplom.ru/
Hi colleagues, how is the whole thing, and what you would like to say about this article, in my view its in fact amazing in favor of me.
diplompro.ru/
Hey there, You have done a great job. I’ll certainly digg it and personally suggest to my friends. I’m confident they will be benefited from this web site.
https://diplompro.ru/
Hey there this is kind of of off topic but I was wondering if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding experience so I wanted to get advice from someone with experience. Any help would be enormously appreciated!
https://www.diplompro.ru/
An outstanding share! I have just forwarded this onto a co-worker who had been conducting a little research on this. And he in fact bought me lunch because I found it for him… lol. So allow me to reword this…. Thank YOU for the meal!! But yeah, thanx for spending time to discuss this matter here on your site.
http://diplom-kuplu.ru/
Aw, this was an incredibly good post. Taking the time and actual effort to generate a superb article… but what can I say… I hesitate a whole lot and never seem to get nearly anything done.
https://moscow.media/moscow/378979979/
https://familyportal.forumrom.com/viewtopic.php?id=20513#p55598
http://forum-pmr.net/member.php?u=42748
Hey there are using WordPress for your site platform? I’m new to the blog world but I’m trying to get started and create my own. Do you require any html coding knowledge to make your own blog? Any help would be greatly appreciated!
https://vipka.0bb.ru/viewtopic.php?id=5115#p7813
https://heroes.app/blogs/420822/ упить-диплом-ваш-ключ-к-лучшему-будущему-и-профессиональному-развитию
https://www.rugbynflforum.com/read-blog/20126
Hey there! This is kind of off topic but I need some guidance from an established blog. Is it hard to set up your own blog? I’m not very techincal but I can figure things out pretty quick. I’m thinking about creating my own but I’m not sure where to begin. Do you have any points or suggestions? Thank you
https://forum.qwas.ru/kupit-diplom-vash-shans-stat-luchshim-v-svoey-oblasti-t19529.html
https://feelosum.com/blogs/990/ упить-диплом-ваш-шанс-стать-лучшим-в-своей-области
http://vetstate.ru/forum/?PAGE_NAME=profile_view&UID=117603
Keep on writing, great job!
https://4atex.ru/blogs/1579/ упить-диплом-ваш-шаг-к-новой-профессиональной-жизни-без-лишних
https://komfort.rusff.me/viewtopic.php?id=12778#p38396
https://asktourist.ru/viewtopic.php?id=4948#p18754
Hi friends, good article and pleasant arguments commented at this place, I am truly enjoying by these.
http://newmedtime.ru/kupit-diplom-vash-klyuch-k-novyim-vozmozhnostyam-i-perspektivam
https://vip.9bb.ru/viewtopic.php?id=5499#p9158
http://mybuildhouse.ru/byistryiy-start-kupi-diplom-i-otkroy-novyie-gorizontyi/
It’s amazing to pay a quick visit this web page and reading the views of all friends about this post, while I am also eager of getting know-how.
http://forexgroupx.ru/diplom-na-zakaz-vash-shans-poluchit-vyisokoklassnoe-obrazovanie-bez-lishnih-hlopot
https://vipka.mybb.ru/viewtopic.php?id=2688#p4672
https://lpn.ixbb.ru/viewtopic.php?id=20#p25
에그 카지노
많은 사람들이 마음이 조금 이상해지고 기분이 복잡해집니다.
I enjoy, result in I discovered just what I was taking a look for. You have ended my four day lengthy hunt! God Bless you man. Have a great day. Bye
https://innovator24.com/read-blog/5427
http://gti-club.ru/forum/member.php?u=23001
http://verkehrsknoten.de/index.php/forum/sozialvorschriften/85024#85978
프라그마틱 슬롯으로 더 큰 승리를 경험하세요.
프라그마틱 슬롯
프라그마틱은 항상 최고의 게임을 제공하죠! 여기에서 더 많은 흥미진진한 정보를 얻을 수 있어 기뻐요.
https://www.12315oe.cn/
https://lotodarts.com/link/
https://www.hnrcrew.com
Hey! I understand this is kind of off-topic however I needed to ask. Does operating a well-established blog such as yours take a massive amount work? I’m brand new to writing a blog however I do write in my journal every day. I’d like to start a blog so I will be able to share my experience and views online. Please let me know if you have any ideas or tips for new aspiring bloggers. Appreciate it!
http://forum.drustvogil-galad.si/index.php/topic,10789.0.html
http://forum.safe-animals.ru/index.php?showtopic=26600
http://www.fellnasen-service.de/index.php?thread/78846-эффективный-путь-к-образованию-купи-диплом-и-ускорь-свой-профессиональный-рост/
В нашем мире, где диплом является началом удачной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Наличие документа об образовании переоценить просто невозможно. Ведь именно он открывает дверь перед всеми, кто собирается начать профессиональную деятельность или продолжить обучение в каком-либо университете.
В данном контексте мы предлагаем максимально быстро получить любой необходимый документ. Вы имеете возможность приобрести диплом нового или старого образца, и это становится отличным решением для всех, кто не смог завершить образование или потерял документ. Любой диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим деталям, чтобы в итоге получился продукт, максимально соответствующий оригиналу.
Плюсы подобного подхода заключаются не только в том, что вы сможете оперативно получить свой диплом. Процесс организовывается удобно и легко, с профессиональной поддержкой. От выбора необходимого образца документа до консультаций по заполнению персональных данных и доставки в любое место России — все под полным контролем качественных специалистов.
Всем, кто ищет оперативный способ получить необходимый документ, наша компания предлагает выгодное решение. Купить диплом – это значит избежать продолжительного обучения и сразу переходить к достижению личных целей: к поступлению в университет или к началу удачной карьеры.
https://qooh.me/JoseArnold2
http://ashinova.ru/category-5/t-823.html#823
https://izhevsk.ru/forummessage/153/6114277.html
https://bogotamihuerta.jbb.gov.co/miembros/lonnie-kelley/
https://iot.ttu.edu.tw/members/lomatapp3/
В современном мире, где диплом – это начало успешной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие официального документа об образовании трудно переоценить. Ведь именно он открывает дверь перед всеми, кто стремится начать трудовую деятельность или учиться в ВУЗе.
Наша компания предлагает максимально быстро получить любой необходимый документ. Вы можете купить диплом старого или нового образца, и это будет отличным решением для человека, который не смог завершить образование, потерял документ или хочет исправить плохие оценки. Каждый диплом изготавливается с особой тщательностью, вниманием ко всем элементам. В итоге вы получите полностью оригинальный документ.
Плюсы подобного решения заключаются не только в том, что вы быстро получите диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора нужного образца диплома до консультации по заполнению персональных данных и доставки по стране — все находится под абсолютным контролем качественных мастеров.
В итоге, для всех, кто ищет быстрый способ получить требуемый документ, наша услуга предлагает выгодное решение. Купить диплом – значит избежать длительного процесса обучения и не теряя времени переходить к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
http://forum.safe-animals.ru/index.php?showtopic=26012
https://open.mit.edu/profile/01HY8A5V2QPVH62G1NXVF5BGA0/
http://reika-vitebsk.by/forum/index.php?id=1091834
https://1abakan.ru/forum/showthread-64726/
http://rybolov.webtalk.ru/viewtopic.php?id=1557#p24667
I was able to find good info from your content.
https://www.hashtap.com/@artur.sh/получите-свой-диплом-сегодн€-надежно-конфиденциально-безопасно-APw6J47b9KwR
http://injectorcar.ru/forum/member.php?u=34916
http://www.odnopolchane.net/forum/member.php?u=546804
В современном мире, где диплом – это начало удачной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Важность наличия официального документа переоценить попросту невозможно. Ведь именно диплом открывает двери перед каждым человеком, который желает начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить этот необходимый документ. Вы можете купить диплом, что является выгодным решением для человека, который не смог закончить обучение, потерял документ или хочет исправить свои оценки. Любой диплом изготавливается с особой тщательностью, вниманием ко всем деталям. На выходе вы сможете получить продукт, полностью соответствующий оригиналу.
Превосходство этого подхода заключается не только в том, что вы быстро получите диплом. Процесс организовывается удобно, с нашей поддержкой. Начиная от выбора нужного образца документа до консультаций по заполнению личных данных и доставки в любое место России — все под полным контролем квалифицированных мастеров.
Для всех, кто пытается найти быстрый и простой способ получения требуемого документа, наша компания предлагает выгодное решение. Купить диплом – значит избежать продолжительного обучения и не теряя времени переходить к достижению личных целей: к поступлению в университет или к началу трудовой карьеры.
http://http://diplomsagroups.com/diplomy-po-specialnosti/diplom-stomatologa.html
В нашем мире, где диплом становится началом успешной карьеры в любой сфере, многие ищут максимально быстрый путь получения образования. Факт наличия официального документа переоценить просто невозможно. Ведь диплом открывает двери перед каждым человеком, который собирается начать трудовую деятельность или учиться в ВУЗе.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы можете купить диплом нового или старого образца, и это является выгодным решением для всех, кто не смог закончить образование, утратил документ или желает исправить свои оценки. дипломы изготавливаются с особой тщательностью, вниманием к мельчайшим деталям, чтобы на выходе получился продукт, 100% соответствующий оригиналу.
Превосходство этого подхода состоит не только в том, что вы оперативно получите свой диплом. Процесс организован удобно, с нашей поддержкой. Начиная от выбора нужного образца документа до консультации по заполнению персональных данных и доставки по России — все находится под абсолютным контролем качественных специалистов.
Для всех, кто хочет найти быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Приобрести диплом – значит избежать длительного обучения и не теряя времени перейти к достижению своих целей: к поступлению в университет или к началу успешной карьеры.
https://lwccareers.lindsey.edu/profiles/4688204-diplo-mans
http://www.livecenter.ru/index.php?showtopic=730
http://passo.su/forums/index.php?showtopic=6426
https://academiachinauy.edu.uy/miembros/elviraparks1/
http://finans.webtalk.ru/viewtopic.php?id=6993#p17911
В современном мире, где диплом становится началом отличной карьеры в любой отрасли, многие пытаются найти максимально простой путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь именно диплом открывает дверь перед каждым человеком, который хочет начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить этот необходимый документ. Вы сможете приобрести диплом, что является отличным решением для человека, который не смог закончить обучение или утратил документ. Любой диплом изготавливается аккуратно, с максимальным вниманием ко всем деталям, чтобы в итоге получился продукт, полностью соответствующий оригиналу.
Плюсы этого решения заключаются не только в том, что можно быстро получить свой диплом. Процесс организовывается удобно, с профессиональной поддержкой. От выбора необходимого образца документа до консультации по заполнению персональной информации и доставки в любое место страны — все под полным контролем качественных мастеров.
Таким образом, для тех, кто ищет быстрый способ получить требуемый документ, наша услуга предлагает выгодное решение. Приобрести диплом – значит избежать продолжительного обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в ВУЗ или начало трудовой карьеры.
http://http://diplomsagroups.com/diplomy-po-specialnosti/diplom-energetika.html
에그벳 계열
그런데 이때 뜻밖에 그의 옆에 한 사람이 더 있었다.
В нашем мире, где диплом является началом отличной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь диплом открывает двери перед всеми, кто собирается начать трудовую деятельность или учиться в ВУЗе.
Наша компания предлагает быстро получить этот важный документ. Вы имеете возможность заказать диплом старого или нового образца, и это будет выгодным решением для всех, кто не смог завершить образование или утратил документ. дипломы выпускаются с особой тщательностью, вниманием к мельчайшим деталям. На выходе вы получите документ, полностью соответствующий оригиналу.
Превосходство подобного подхода состоит не только в том, что вы сможете быстро получить диплом. Процесс организовывается комфортно, с нашей поддержкой. Начиная от выбора необходимого образца до точного заполнения персональных данных и доставки по России — все под полным контролем качественных специалистов.
Таким образом, для тех, кто пытается найти быстрый и простой способ получить требуемый документ, наша компания предлагает выгодное решение. Купить диплом – значит избежать долгого обучения и не теряя времени переходить к своим целям: к поступлению в университет или к началу трудовой карьеры.
http://diplomsagroups.com/diplomy-po-specialnosti/diplom-medsestry.html
에그슬롯
이 법정 토론 … Hongzhi 황제는 여전히 그것을 피하기로 선택 했습니까?
최신 프라그마틱 게임은 iGaming 분야에서 선도적인 제공 업체로서, 독창적이고 표준화된 콘텐츠를 제공합니다.
프라그마틱 홈페이지
프라그마틱과 관련된 이 글 정말 잘 읽었어요! 더불어 저도 제 사이트에서 프라그마틱에 대한 새로운 정보를 공유 중이에요. 함께 나누면 더 좋을 것 같아요!
https://www.12315op.cn/
https://www.naugblog.com
https://mashhadnet.com/link/
Hi, this weekend is fastidious in favor of me, because this occasion i am reading this great educational paragraph here at my residence.
https://bookmarkingbay.com/story17019677/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%A3%D1%84%D0%B0
https://vnbaolut.com/
https://techcrunch59371.blogsidea.com/33012785/top-marketing-secrets
https://bookmarkingdepot.com/story17030709/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B0%D0%B2%D1%82%D0%BE%D0%BC%D0%B5%D1%85%D0%B0%D0%BD%D0%B8%D0%BA%D0%B0
https://animalprotect.org/forum/index.php?action=profile;area=showposts;sa=topics;u=4779
В наше время, когда диплом становится началом успешной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Важность наличия официального документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед людьми, стремящимися вступить в сообщество квалифицированных специалистов или учиться в университете.
Наша компания предлагает быстро получить этот важный документ. Вы имеете возможность приобрести диплом старого или нового образца, что является выгодным решением для всех, кто не смог закончить обучение, утратил документ или желает исправить плохие оценки. диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам. В результате вы сможете получить 100% оригинальный документ.
Превосходство такого подхода заключается не только в том, что можно максимально быстро получить диплом. Процесс организован комфортно, с нашей поддержкой. От выбора требуемого образца до точного заполнения персональных данных и доставки по стране — все будет находиться под абсолютным контролем квалифицированных мастеров.
Всем, кто пытается найти быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать продолжительного обучения и сразу перейти к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
http://muz.mybb.ru/viewtopic.php?id=613#p1226
https://sketchfab.com/JoseArnold2
https://vdoroge.mybb.ru/viewtopic.php?id=500#p4797
http://fast4ride.mybb.ru/viewtopic.php?id=135#p136
https://vspmscop.edu.in/LRM/profile/LonnieKelley1/
На сегодняшний день, когда диплом становится началом успешной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие документа об образовании переоценить просто невозможно. Ведь диплом открывает двери перед всеми, кто стремится вступить в сообщество профессиональных специалистов или продолжить обучение в университете.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы имеете возможность купить диплом, и это является отличным решением для человека, который не смог завершить образование или потерял документ. Все дипломы производятся с особой тщательностью, вниманием ко всем деталям, чтобы в результате получился документ, полностью соответствующий оригиналу.
Превосходство такого подхода состоит не только в том, что можно оперативно получить свой диплом. Весь процесс организован комфортно, с нашей поддержкой. Начиная от выбора требуемого образца до консультаций по заполнению персональных данных и доставки по России — все находится под абсолютным контролем качественных мастеров.
Для всех, кто ищет максимально быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Заказать диплом – значит избежать долгого обучения и не теряя времени перейти к достижению личных целей, будь то поступление в университет или старт профессиональной карьеры.
http://diplomsagroups.com/kupit-diplom-v-gorode/voronezh.html
В нашем мире, где диплом становится началом отличной карьеры в любом направлении, многие стараются найти максимально простой путь получения образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает дверь перед всеми, кто хочет начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы имеете возможность купить диплом старого или нового образца, и это является выгодным решением для всех, кто не смог завершить образование или утратил документ. Все дипломы выпускаются с особой тщательностью, вниманием ко всем деталям. На выходе вы получите документ, максимально соответствующий оригиналу.
Превосходство этого решения состоит не только в том, что вы сможете максимально быстро получить свой диплом. Процесс организовывается комфортно, с профессиональной поддержкой. Начиная от выбора нужного образца диплома до правильного заполнения персональной информации и доставки в любое место страны — все будет находиться под полным контролем опытных специалистов.
Для всех, кто ищет быстрый способ получения необходимого документа, наша услуга предлагает отличное решение. Купить диплом – это значит избежать длительного процесса обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в ВУЗ или старт трудовой карьеры.
http://karapuziki.0pk.me/viewtopic.php?id=7229#p91407
https://gitlab.ifam.edu.br/LonnieKelley1
http://ukrainaru.bbok.ru/viewtopic.php?id=6875#p6923
http://fabrikaglamura.webtalk.ru/viewtopic.php?id=771#p39764
http://peru.mybb.ru/viewtopic.php?id=289#p4090
Pretty! This was a really wonderful post. Thank you for supplying this information.
https://investing-dolar-yorum58136.pages10.com/
https://ilovebookmark.com/story17014721/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BC%D0%B0%D0%B3%D0%B8%D1%81%D1%82%D1%80%D0%B0
http://zelfrijdendetaxibreda.nl/uncategorized/vijf-tips-om-een-laadpaal-voor-je-kantoor-aan-te-vragen/
https://socialskates.com/story17904274/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%9A%D0%B0%D0%B7%D0%B0%D0%BD%D0%B8
http://scientistsufo.ru/page/6
레프리칸 리치스
떠나는 한 가능한 한 후회가 없도록 최선을 다하십시오.
В современном мире, где диплом является началом успешной карьеры в любом направлении, многие пытаются найти максимально простой путь получения образования. Необходимость наличия официального документа переоценить просто невозможно. Ведь именно диплом открывает дверь перед людьми, стремящимися вступить в профессиональное сообщество или учиться в любом университете.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы можете заказать диплом нового или старого образца, и это становится отличным решением для всех, кто не смог завершить обучение, утратил документ или хочет исправить свои оценки. дипломы производятся с особой аккуратностью, вниманием к мельчайшим деталям. На выходе вы получите документ, полностью соответствующий оригиналу.
Превосходство этого решения заключается не только в том, что вы сможете оперативно получить диплом. Весь процесс организовывается комфортно, с нашей поддержкой. От выбора подходящего образца документа до грамотного заполнения персональной информации и доставки в любое место страны — все находится под полным контролем наших мастеров.
Для тех, кто ищет быстрый и простой способ получения требуемого документа, наша компания предлагает отличное решение. Заказать диплом – значит избежать продолжительного процесса обучения и сразу перейти к своим целям: к поступлению в университет или к началу трудовой карьеры.
http://diplomsagroups.com/diplomy-po-specialnosti/diplom-psihologa.html
В нашем мире, где диплом является началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Факт наличия официального документа об образовании переоценить невозможно. Ведь диплом открывает двери перед людьми, стремящимися вступить в сообщество профессионалов или продолжить обучение в университете.
Предлагаем оперативно получить любой необходимый документ. Вы имеете возможность заказать диплом, что будет отличным решением для человека, который не смог закончить обучение или потерял документ. Любой диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим нюансам, чтобы в результате получился документ, максимально соответствующий оригиналу.
Преимущество данного подхода заключается не только в том, что вы быстро получите диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начиная от выбора требуемого образца документа до консультаций по заполнению персональной информации и доставки по стране — все под полным контролем качественных специалистов.
Для тех, кто ищет оперативный способ получения требуемого документа, наша услуга предлагает выгодное решение. Купить диплом – значит избежать продолжительного процесса обучения и сразу перейти к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
diplomsagroups.com
Attractive portion of content. I just stumbled upon your blog and in accession capital to say that I get in fact loved account your blog posts. Any way I’ll be subscribing on your augment and even I success you get right of entry to constantly quickly.
http://www.sutangarsk.ru/
http://oso-znanie.boginya-yar.ru/%D0%B4%D0%BB%D1%8F-%D0%BB%D0%B5%D0%B4%D0%B8/
https://ledbookmark.com/story2572871/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%81%D0%BF%D0%B5%D1%86%D0%B8%D0%B0%D0%BB%D0%B8%D1%81%D1%82%D0%B0
https://weleaks.info/newreply.php?tid=10451
https://redhotbookmarks.com/story17071300/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BE-%D1%81%D1%80%D0%B5%D0%B4%D0%BD%D0%B5%D0%BC-%D0%BE%D0%B1%D1%80%D0%B0%D0%B7%D0%BE%D0%B2%D0%B0%D0%BD%D0%B8%D0%B8
What’s up to every one, as I am genuinely keen of reading this weblog’s post to be updated regularly. It carries fastidious information.
https://health-lists.com/story17617700/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BC%D0%B5%D0%BD%D0%B5%D0%B4%D0%B6%D0%B5%D1%80%D0%B0
https://babygirlboyname.com/
http://bazhenova.greatforum.ru/viewtopic.php?f=17&p=4907
https://friendlybookmark.com/story17026622/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B1%D0%B0%D0%BA%D0%B0%D0%BB%D0%B0%D0%B2%D1%80%D0%B0
https://flora-online.ru/
Hello, for all time i used to check blog posts here in the early hours in the dawn, as i enjoy to learn more and more.
@@G@@
https://ambox.ru/virtualnyj-nomer-dlya-tiktok/
Wow, marvelous blog structure! How long have you been blogging for? you made blogging look easy. The total look of your website is excellent, let alone the content!
http://diplom-zakaz.ru/
Amazing things here. I am very satisfied to see your post. Thanks a lot and I’m having a look forward to contact you. Will you kindly drop me a mail?
https://www.diplom-zakaz.ru/
What’s up to every single one, it’s genuinely a fastidious for me to pay a visit this web page, it includes priceless Information.
peoplediplom.ru/
Hello very nice web site!! Man .. Beautiful .. Wonderful .. I’ll bookmark your website and take the feeds additionally? I am satisfied to find a lot of helpful info right here in the post, we want work out extra strategies in this regard, thanks for sharing. . . . . .
http://peoplediplom.ru/
It’s amazing to visit this web page and reading the views of all colleagues concerning this piece of writing, while I am also zealous of getting know-how.
http://www.diplom-profi.ru/
Hi there, its good article concerning media print, we all know media is a impressive source of facts.
http://www.diplom-profi.ru/
5 래빗스 메가웨이즈
누군가 기침을 하고 앞으로 나서며 “Fang Jifan…”이라고 말하지 않을 수 없었습니다.
В современном мире, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Факт наличия официального документа переоценить невозможно. Ведь именно он открывает двери перед любым человеком, желающим вступить в сообщество квалифицированных специалистов или продолжить обучение в любом ВУЗе.
Мы предлагаем максимально быстро получить любой необходимый документ. Вы сможете приобрести диплом старого или нового образца, и это будет выгодным решением для человека, который не смог закончить обучение или потерял документ. дипломы производятся с особой тщательностью, вниманием ко всем деталям. В итоге вы получите документ, 100% соответствующий оригиналу.
Преимущество подобного решения состоит не только в том, что вы максимально быстро получите диплом. Процесс организовывается удобно, с нашей поддержкой. От выбора подходящего образца документа до консультации по заполнению личных данных и доставки в любой регион России — все находится под абсолютным контролем опытных специалистов.
Всем, кто ищет оперативный способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать продолжительного обучения и не теряя времени перейти к личным целям: к поступлению в ВУЗ или к началу успешной карьеры.
http://finans.webtalk.ru/viewtopic.php?id=6993#p17911
https://speterburg.rusff.me/viewtopic.php?id=8755#p312683
http://husky.icebb.ru/viewtopic.php?id=261#p1245
http://fishkaluga.0pk.me/viewtopic.php?id=1706#p11294
https://bbs.pku.edu.cn/v2/jump-to.php?url=https://russiany-diplomans.com
Hello, after reading this amazing paragraph i am too happy to share my experience here with friends.
https://www.sdiplom.ru/
Hey I am so delighted I found your site, I really found you by accident, while I was researching on Askjeeve for something else, Nonetheless I am here now and would just like to say thank you for a incredible post and a all round enjoyable blog (I also love the theme/design), I don’t have time to read through it all at the moment but I have saved it and also included your RSS feeds, so when I have time I will be back to read much more, Please do keep up the fantastic work.
https://diplom-city24.ru/
Do you have a spam problem on this blog; I also am a blogger, and I was wanting to know your situation; many of us have created some nice practices and we are looking to swap techniques with other folks, why not shoot me an email if interested.
https://zakaz-na-diplom.ru/
Hi there, You’ve done a fantastic job. I will certainly digg it and personally recommend to my friends. I am sure they’ll be benefited from this website.
https://www.institute-diplom.ru/
Pretty section of content. I just stumbled upon your blog and in accession capital to assert that I get actually enjoyed account your blog posts. Any way I’ll be subscribing to your feeds and even I achievement you access consistently rapidly.
https://www.diplompro.ru/
Hi, its fastidious paragraph on the topic of media print, we all know media is a great source of facts.
http://www.diplom-kuplu.ru/
В нашем мире, где диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Наличие документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед людьми, стремящимися вступить в сообщество квалифицированных специалистов или продолжить обучение в университете.
В данном контексте наша компания предлагает очень быстро получить любой необходимый документ. Вы сможете приобрести диплом старого или нового образца, что будет выгодным решением для человека, который не смог закончить образование, утратил документ или желает исправить плохие оценки. диплом изготавливается аккуратно, с максимальным вниманием ко всем нюансам. В результате вы сможете получить 100% оригинальный документ.
Превосходство этого решения состоит не только в том, что можно оперативно получить свой диплом. Процесс организован комфортно, с нашей поддержкой. От выбора необходимого образца документа до консультаций по заполнению персональной информации и доставки в любой регион России — все под полным контролем квалифицированных мастеров.
Таким образом, для тех, кто ищет быстрый и простой способ получить необходимый документ, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать длительного процесса обучения и не теряя времени переходить к важным целям, будь то поступление в ВУЗ или старт карьеры.
http://comp.bbok.ru/viewtopic.php?id=463#p1514
https://safipoodle.mybb.ru/viewtopic.php?id=260#p8203
http://pinklink.ca/author/diplomans/
https://iot.ttu.edu.tw/members/lonniekelley1/
https://clc.edu.pe/blog/index.php?entryid=59781
whoah this weblog is magnificent i like reading your articles. Keep up the great work! You know, a lot of people are hunting around for this info, you could help them greatly.
himchistka.udm.ru/forum?%2Fforum=&%2Fforum%2F=&GuestbookItem_page=11В
ediblehomegardensresort.com/index.php?do=/public/blog/view/id_119635/title_-2024/В
childtemperament.org/index.php?lang=esВ
188.17.148.172/forum/profile.php?action=show&member=7512В
shoelovershub.com/read-blog/475_why-is-the-popularity-of-universities-declining-in-our-time.html?mode=dayВ
Useful information. Lucky me I found your web site by accident, and I’m stunned why this coincidence didn’t came about in advance! I bookmarked it.
http://chem100.ru/elem.php?n=79
http://padeji.ru/lenta-5.html
http://avtoshkola34.ru/main/11-osnovnye-svedeniya-ob-avtoshkole-volzhskaya.html
http://vinpr.org/obshestvo/6573-mir-stanovitsya-vse-bolee-cifrovym.html
http://edu-tver.ru/katalog-uchebnykh-materialov/risovanie/risuem-po-kletochkam-v-afrike1.html
Hi there i am kavin, its my first occasion to commenting anywhere, when i read this article i thought i could also create comment due to this good paragraph.
http://mak-mebel.ru/vertikalnye-sady.html
http://ofra41.gabia.io/bbs/board.php?bo_table=qa&wr_id=1292
http://kia-sorento-club.by/member.php?tab=visitor_messaging&u=5793
http://trpgfes.jp/bbs/wforum.cgi?mode=relate&no=9241&page=90
http://genzouzi.no-ip.com/cgi-bin/norbbs/light.cgi?pg=50
This post is priceless. How can I find out more?
forum.drustvogil-galad.si/index.php?topic=10004.0В
forums.worldsamba.org/showthread.php?tid=992935В
http://www.avatars.cc/member.php?action=showprofile&user_id=34845В
shop.electricoresigns.com/index.php?route=information/blogger&blogger_id=6В
lunarys.com.br/blog/suplemento-alimentar-hair-supplyВ
문 프린세스 100
내가 할 수 있는 것은 아무 말도 하지 않고 조용히 고개를 숙이는 것뿐이다.
Hello, of course this article is actually fastidious and I have learned lot of things from it on the topic of blogging. thanks.
slcomp.kz/index.php?route=information/blogger&blogger_id=5В
xemgiai.com/phap-luat-la-giВ
clickforart.com/artist/view/ilona-d.-veresk/?type=2В
online-vuz.ru/viewNews/177/В
passo.su/forums/index.php?autocom=gallery&req=si&img=2801%C3%82%C2%A0В
В нашем обществе, где диплом является началом отличной карьеры в любой отрасли, многие стараются найти максимально быстрый и простой путь получения образования. Необходимость наличия документа об образовании сложно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим начать профессиональную деятельность или продолжить обучение в ВУЗе.
Мы предлагаем максимально быстро получить любой необходимый документ. Вы можете купить диплом старого или нового образца, что является удачным решением для всех, кто не смог закончить образование, потерял документ или хочет исправить плохие оценки. диплом изготавливается аккуратно, с особым вниманием ко всем элементам, чтобы в итоге получился продукт, 100% соответствующий оригиналу.
Преимущество этого решения заключается не только в том, что вы быстро получите диплом. Процесс организовывается комфортно, с нашей поддержкой. От выбора необходимого образца диплома до грамотного заполнения персональной информации и доставки в любое место России — все под полным контролем квалифицированных мастеров.
Таким образом, для тех, кто пытается найти быстрый и простой способ получения требуемого документа, наша компания готова предложить отличное решение. Купить диплом – это значит избежать долгого обучения и сразу перейти к достижению личных целей, будь то поступление в университет или начало трудовой карьеры.
http://kolybel-ekb.ru
http://bankiuslugi.ru
http://blooms.com.ua
http://lifespacetime.com
http://vanbroecanimations.online
Hello there, You’ve done a great job. I will certainly digg it and personally recommend to my friends. I am confident they’ll be benefited from this website.
cs-online.ru/forum/index.php?showtopic=13633&mode=linearВ
аржаники.СЂС„/memberlist.php?sk=a&sd=a&first_char=r&start=200В
myweektour.ru/page/4/В
pricebuy.co.il/siemens-kg76nai31l-priceВ
hinadezain-test.com/В
geinoutime.com
Hongzhi 황제는 두통을 느꼈습니다. 착한 사람, 삼천 냥이 다시 사라졌습니다.
Aw, this was an exceptionally good post. Taking a few minutes and actual effort to generate a great article… but what can I say… I procrastinate a whole lot and don’t manage to get nearly anything done.
cialis 5mg directions
Thanks , I’ve recently been searching for info about this topic for a while and yours is the greatest I’ve discovered till now. But, what about the conclusion? Are you certain in regards to the supply?
http://www.lada-4×4.net/member.php?u=21927
http://www.uvina.ru/services/pokupka-metallolom/priem-loma-met/
http://pticadom.ru/soderzhanie-shheglov.html
http://alkogolizma.com/preparaty/antipohmelin.html
http://www.usman2.ru/dwsvid/195745-vzlom-moj-govoryaschij-tom.html
Hello colleagues, how is the whole thing, and what you desire to say concerning this piece of writing, in my view its actually awesome in favor of me.
fincasanlorenzo.es/sitemap/В
zhaktrans.ru/p40135689-tnvd-wd615-612601080225.htmlВ
express-container.ru/about/founding/В
http://www.kangas-industrial.com/e_feedback/index.asp?page=2769В
http://www.korgforums.com/forum/phpBB2/viewtopic.php?t=129929&view=nextВ
Hello colleagues, how is all, and what you want to say about this piece of writing, in my view its in fact awesome designed for me.
benhvienthammyasean.com/cau-chuyen-aseanВ
bouffordca.com/Emailscam.htmВ
click2connectclubs.com/index.php?do=/public/user/blogs/view/name_Alanpoe/id_119636/title_/В
http://www.moto-import.ru/index.php?ukey=linkexchange&did=33&le_categoryID=0&page=5&show_all=yesВ
forum.trackbase.net/members/21965-sonnick84?vmid=2440В
В нашем обществе, где диплом – это начало успешной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Факт наличия официального документа об образовании переоценить невозможно. Ведь именно диплом открывает двери перед всеми, кто стремится вступить в профессиональное сообщество или учиться в любом университете.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы можете купить диплом нового или старого образца, что будет отличным решением для человека, который не смог закончить обучение или утратил документ. дипломы производятся с особой аккуратностью, вниманием ко всем нюансам. На выходе вы сможете получить 100% оригинальный документ.
Преимущества подобного решения состоят не только в том, что вы оперативно получите свой диплом. Процесс организовывается комфортно, с нашей поддержкой. Начав от выбора необходимого образца до консультации по заполнению персональной информации и доставки по России — все под абсолютным контролем опытных специалистов.
Для всех, кто ищет оперативный способ получить необходимый документ, наша компания предлагает выгодное решение. Купить диплом – значит избежать продолжительного процесса обучения и сразу переходить к своим целям, будь то поступление в ВУЗ или начало профессиональной карьеры.
http://barnum.cl
http://bolesnymd.ru
http://xn--logtervezs-j7a7i.hu
http://portaltrav.ru
http://chile.org.ua
В нашем мире, где диплом является началом отличной карьеры в любой сфере, многие пытаются найти максимально быстрый путь получения образования. Важность наличия официального документа переоценить невозможно. Ведь именно диплом открывает двери перед каждым человеком, который стремится начать профессиональную деятельность или учиться в ВУЗе.
В данном контексте наша компания предлагает оперативно получить любой необходимый документ. Вы сможете купить диплом, и это будет выгодным решением для человека, который не смог завершить обучение, утратил документ или хочет исправить свои оценки. Все дипломы изготавливаются аккуратно, с максимальным вниманием к мельчайшим элементам, чтобы в результате получился полностью оригинальный документ.
Плюсы данного подхода состоят не только в том, что вы сможете быстро получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начав от выбора нужного образца документа до точного заполнения персональных данных и доставки в любое место России — все под полным контролем квалифицированных мастеров.
Всем, кто ищет быстрый и простой способ получения требуемого документа, наша компания готова предложить выгодное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и не теряя времени переходить к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
http://getoutdoorsyapp.com
http://okna-optimum.ru
http://mysoccerex.com
http://avangard-mt.ru
http://neverwinter-wiki.ru
For newest information you have to pay a quick visit internet and on internet I found this web page as a best web page for hottest updates.
http://www.agrolib.ru/В
pokatili.ru/f/viewtopic.php?f=10&t=70278&view=nextВ
http://www.underworldralinwood.ca/forums/member.php?action=profile&uid=246355В
rising.chew.jp/END/inside/dbbs/dicebbs.cgi?mode=resmsg&no=7866В
http://www.miks.ks.ua/index.php?links_exchange=yes&page=4&show_all=yesВ
http://meryflowers.ru
Thank you, I have recently been looking for info about this topic for a while and yours is the greatest I’ve came upon so far. But, what concerning the conclusion? Are you sure about the source?
express-container.ru/about/founding/В
forum.javabox.net/viewtopic.php?f=7&t=14765&view=printВ
spg.co.kr/bbs/board.php?bo_table=cus_res&wr_id=92&sca=%EC%98%81%EC%97%85%EC%A7%80%EC%9B%90&page=55В
retrogaming.in.ua/index.php?ukey=linkexchange&did=33&le_categoryID=0&page=22&show_all=yesВ
vv.flybb.ru/topic1874.html?view=previousВ
В наше время, когда диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Наличие документа об образовании переоценить невозможно. Ведь диплом открывает двери перед любым человеком, желающим начать профессиональную деятельность или продолжить обучение в каком-либо университете.
Предлагаем оперативно получить любой необходимый документ. Вы можете заказать диплом, и это становится отличным решением для всех, кто не смог закончить обучение или утратил документ. диплом изготавливается с особой тщательностью, вниманием ко всем деталям. На выходе вы получите документ, 100% соответствующий оригиналу.
Преимущество такого решения заключается не только в том, что можно оперативно получить свой диплом. Процесс организовывается удобно и легко, с нашей поддержкой. От выбора необходимого образца диплома до консультаций по заполнению персональной информации и доставки в любой регион России — все под абсолютным контролем квалифицированных специалистов.
Всем, кто ищет максимально быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к достижению личных целей: к поступлению в университет или к началу трудовой карьеры.
http://companion.com.ua
http://co1-az.ru
http://avtograd-161.ru
http://hoahaubansacviet.com.vn
http://christianpost.co.id
It’s awesome for me to have a website, which is useful in support of my experience. thanks admin
dog-ola.ru/topic6358.html?view=previousВ
circuit-diagrams.com/pr4-PIC16F84A-discolight-effect-with-bass-beat-control.phpВ
hellohome.ir/en/agent/mahyar-shahbazi/В
http://www.e-087.com/cgi-bin/DFA/clever/clever.cgi?page=80В
ttdinhduong.org/ttdd/tin-tuc/tin-chuyen-mon/679-Thong-bao-chieu-sinh-khoa-dao-tao-ddls.aspxВ
Hi there, the whole thing is going nicely here and ofcourse every one is sharing facts, that’s really good, keep up writing.
http://forum.trackbase.net/members/29274-arkadypettr?tab=activitystream&type=friends&page=1
http://knitting-croche.ru/vyazanaya-spicami-grelka-na-chajnik
http://forum.javabox.net/viewtopic.php?f=15&t=13010&p=21226
http://www.herurg.ru/artist/metal_mind.html
http://potolki-info.ru/pokraska/page/2
Can you tell us more about this? I’d care to find out some additional information.
https://bizkerrefutboltaldea.com/bizkerre/Castellano/principal.htm
https://bookmarkinglog.com/story17080664/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%9A%D0%B0%D0%B7%D0%B0%D0%BD%D0%B8
http://www.ajeci.com.br/2017/09/28/palestra-solucoes-tigre-ads-em-tubos-corrugados-para-drenagem-e-saneamento/
http://myweektour.ru/page/3/
https://rolfvandenbrink.com/2018/10/21/autumn-ants/comment-page-28/
I got this website from my friend who informed me concerning this web site and at the moment this time I am visiting this web site and reading very informative content at this time.
mutinyhockey.com/memberlist.php?mode=joined&order=ASC&start=336850%C3%82%C2%A0В
slcomp.kz/index.php?route=information/blogger&blogger_id=5В
http://www.anahnu.club/en/jomsocial/groups/viewbulletin/148-%D1%81%D0%BA%D0%BE%D0%BB%D1%8C%D0%BA%D0%BE-%D1%81%D0%B5%D0%B9%D1%87%D0%B0%D1%81-%D0%B1%D1%83%D0%B4%D0%B5%D1%82-%D1%81%D1%82%D0%BE%D0%B8%D1%82%D1%8C-%D0%BD%D0%B5%D0%BE%D1%82%D0%BB%D0%B8%D1%87%D0%B8%D0%BC%D1%8B%D0%B9-%D0%BE%D1%82-%D0%BE%D1%80%D0%B8%D0%B3%D0%B8%D0%BD%D0%B0%D0%BB%D0%B0-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC.html?groupid=1В
http://www.vietnammm.com/kr/restaurants/An-TamВ
lastnewsusa.com/2022/02/omicron-variant-pushes-us-coronavirus.htmlВ
Hello there, You have done a great job. I’ll definitely digg it and personally suggest to my friends. I’m sure they will be benefited from this site.
mystroycenter.ru/page/14В
ветерантюмгео.СЂС„/galereya/?type_0=gallery&album_gallery_id_0=2В
dip-vuz.ru/kupit-diplom-v-tyumeni/index.htmlВ
аржаники.СЂС„/memberlist.php?sk=a&sd=a&first_char=r&start=200В
tivi24h.com/2011/01/xem-bong-online-bong-truc-tuyen-kenh.htmlВ
An outstanding share! I’ve just forwarded this onto a co-worker who was doing a little homework on this. And he in fact ordered me breakfast simply because I found it for him… lol. So allow me to reword this…. Thank YOU for the meal!! But yeah, thanks for spending some time to talk about this matter here on your web site.
http://www.kompresometr.ru/beshiktash-podtverdil-perehod-tekke.html
http://dcactus.ru/epifillum/
http://www.ad-clan.ru/forum/member.php?tab=visitor_messaging&u=4398
http://msd.com.ua/poiskovaya-vydacha/unikalnye-vidy-stroitelnyx-materialov/
http://kamcom.ru/index.php?id=12&gid=106
Just wish to say your article is as surprising. The clarity on your post is simply great and i could suppose you’re a professional on this subject. Fine along with your permission let me to grasp your RSS feed to keep up to date with impending post. Thank you a million and please carry on the gratifying work.
angeartsgifts.com/c_feedback/?page=4228В
webyourself.eu/blogs/317597/%D0%A1%D0%B5%D0%BA%D1%80%D0%B5%D1%82-%D1%83%D1%81%D0%BF%D0%B5%D1%85%D0%B0-%D0%9A%D0%B0%D0%BA-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B8-%D0%B8%D0%B7%D0%BC%D0%B5%D0%BD%D0%B8%D1%82%D1%8C-%D1%81%D0%B2%D0%BE%D1%8E-%D0%B6%D0%B8%D0%B7%D0%BD%D1%8CВ
http://www.karpaltunnelsyndrom-schwangerschaft.de/forum/topic/uebungen-fuer-den-alltag/D0%B7%D0%B8%D1%80%D1%83%D0%B9%D1%82%D0%B5+%D1%80%D1%8B%D0%BD%D0%BE%D0%BA+%D0%B8+%D0%B2%D0%B7%D0%B2%D0%B5%D1%88%D0%B8%D0%B2%D0%B0%D0%B9%D1%82%D0%B5+%D0%B2%D1%81%D0%B5+%D0%BF%D0%BB%D1%8E%D1%81%D1%8B+%D0%B8+%C2%AB%D0%BF%D1%80%D0%BE%D1%82%D0%B8%D0%B2%C2%BB+%D0%BF%D0%B5%D1%80%D0%B5%D0%B4+%D1%82%D0%B5%D0%BC%2C+%D0%BA%D0%B0%D0%BA+%D0%BF%D1%80%D0%B8%D0%BD%D1%8F%D1%82%D1%8C+%D0%B8%D1%82%D0%BE%D0%B3%D0%BE%D0%B2%D0%BE%D0%B5+%D0%BF%D0%BE%D1%81%D1%82%D0%B0%D0%BD%D0%BE%D0%B2%D0%BB%D0%B5%D0%BD%D0%B8%D0%B5/?x=0&y=0&part=2998В
mazda-demio.ru/forums/index.php?autocom=gallery&req=si&img=4139В
pmk-polymer.ru/content/kontaktyВ
В современном мире, где диплом – это начало отличной карьеры в любой области, многие ищут максимально быстрый и простой путь получения образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед всеми, кто собирается вступить в профессиональное сообщество или учиться в ВУЗе.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы сможете купить диплом нового или старого образца, и это будет удачным решением для всех, кто не смог завершить образование или утратил документ. Любой диплом изготавливается аккуратно, с максимальным вниманием ко всем нюансам. На выходе вы получите полностью оригинальный документ.
Преимущество этого подхода заключается не только в том, что вы максимально быстро получите диплом. Процесс организовывается комфортно и легко, с профессиональной поддержкой. От выбора требуемого образца до консультации по заполнению личных данных и доставки в любое место страны — все под абсолютным контролем наших специалистов.
Таким образом, для тех, кто пытается найти оперативный способ получения требуемого документа, наша компания предлагает отличное решение. Заказать диплом – это значит избежать долгого процесса обучения и не теряя времени переходить к достижению личных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
http://drunkslut.com
http://ezotericum.ru
http://kembekeltetes.hu
http://levit.in.ua
http://xn--80adbdkod2bc2a.xn--p1ai
슬롯 무료
Hongzhi 황제는 다시 장관을 보았고 장관의 얼굴은 더욱 추해졌습니다.
В наше время, когда диплом – это начало отличной карьеры в любой отрасли, многие стараются найти максимально быстрый путь получения качественного образования. Наличие официального документа об образовании трудно переоценить. Ведь именно диплом открывает двери перед каждым человеком, который собирается начать профессиональную деятельность или продолжить обучение в ВУЗе.
Предлагаем максимально быстро получить этот важный документ. Вы можете приобрести диплом старого или нового образца, и это становится удачным решением для человека, который не смог закончить обучение или потерял документ. диплом изготавливается аккуратно, с максимальным вниманием ко всем элементам, чтобы в итоге получился 100% оригинальный документ.
Преимущество подобного подхода состоит не только в том, что можно быстро получить диплом. Процесс организовывается комфортно, с нашей поддержкой. От выбора требуемого образца диплома до грамотного заполнения персональной информации и доставки в любой регион страны — все будет находиться под абсолютным контролем опытных мастеров.
Для тех, кто пытается найти оперативный способ получить необходимый документ, наша услуга предлагает отличное решение. Купить диплом – значит избежать длительного обучения и сразу переходить к своим целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://tritattoo.com
http://bestpeople.com.ua
http://samara2025.ru
http://apuntadas.es
http://kupon.biz
http://jboi2007.org
В нашем обществе, где диплом является началом отличной карьеры в любой отрасли, многие стараются найти максимально простой путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь диплом открывает двери перед людьми, стремящимися вступить в профессиональное сообщество или продолжить обучение в университете.
В данном контексте наша компания предлагает максимально быстро получить любой необходимый документ. Вы имеете возможность заказать диплом, что является отличным решением для всех, кто не смог завершить обучение, потерял документ или желает исправить свои оценки. диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим деталям. На выходе вы сможете получить продукт, 100% соответствующий оригиналу.
Преимущество подобного решения состоит не только в том, что вы сможете максимально быстро получить свой диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начиная от выбора подходящего образца диплома до консультации по заполнению личных данных и доставки по стране — все находится под абсолютным контролем квалифицированных специалистов.
Для тех, кто ищет быстрый способ получения необходимого документа, наша компания может предложить выгодное решение. Приобрести диплом – это значит избежать продолжительного обучения и сразу перейти к достижению личных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
http://upts-ralmix.ru
http://havskaya.com
http://ip-kitov.ru
http://myworldoftank.ru
http://ivangrinev.com
В нашем обществе, где аттестат является началом успешной карьеры в любой области, многие стараются найти максимально быстрый путь получения качественного образования. Факт наличия документа об образовании сложно переоценить. Ведь диплом открывает двери перед любым человеком, желающим вступить в сообщество профессионалов или продолжить обучение в высшем учебном заведении.
Предлагаем очень быстро получить любой необходимый документ. Вы можете приобрести аттестат старого или нового образца, и это будет удачным решением для всех, кто не смог завершить обучение, утратил документ или хочет исправить плохие оценки. Аттестаты изготавливаются аккуратно, с особым вниманием ко всем нюансам, чтобы на выходе получился документ, 100% соответствующий оригиналу.
Преимущество данного решения заключается не только в том, что можно быстро получить свой аттестат. Весь процесс организован просто и легко, с профессиональной поддержкой. От выбора необходимого образца до точного заполнения персональных данных и доставки по стране — все под полным контролем квалифицированных мастеров.
Таким образом, для всех, кто хочет найти быстрый и простой способ получить требуемый документ, наша компания предлагает выгодное решение. Купить аттестат – значит избежать длительного обучения и сразу переходить к личным целям, будь то поступление в университет или старт удачной карьеры.
http://tuning-performance.ru/kuzovnoy-remont-auto/
Heya are using WordPress for your blog platform? I’m new to the blog world but I’m trying to get started and set up my own. Do you require any coding expertise to make your own blog? Any help would be really appreciated!
facesocial.demo3.dedicatedhost247.com/read-blog/4786_why-is-the-popularity-of-universities-declining-nowadays.html?mode=dayВ
startspresto.ru/dannye/pluginclass/cbblogs?action=blogs&func=show&id=22В
russiajoy.ru/page/12В
mangopik.com/sximodemo7/posts/read/printing-and-typesetting-industryВ
retrogaming.in.ua/index.php?ukey=linkexchange&did=33&le_categoryID=0&page=22&show_all=yesВ
На сегодняшний день, когда диплом является началом удачной карьеры в любой отрасли, многие стараются найти максимально простой путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь именно он открывает дверь перед каждым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в университете.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы можете купить диплом, и это является отличным решением для всех, кто не смог закончить обучение или утратил документ. Все дипломы изготавливаются с особой аккуратностью, вниманием ко всем нюансам, чтобы на выходе получился полностью оригинальный документ.
Плюсы данного решения состоят не только в том, что можно оперативно получить диплом. Процесс организован удобно, с нашей поддержкой. Начав от выбора нужного образца диплома до правильного заполнения личной информации и доставки в любой регион России — все под абсолютным контролем квалифицированных мастеров.
В результате, для всех, кто ищет быстрый и простой способ получить требуемый документ, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в университет или старт карьеры.
http://szigettaxi.hu
http://parkdivan.ru
http://hoisuhoc.vn
http://larryhandlin.com
http://replicauhren.be
Hey are using WordPress for your blog platform? I’m new to the blog world but I’m trying to get started and set up my own. Do you require any html coding expertise to make your own blog? Any help would be greatly appreciated!
allsportime.ru/uspeh-v-vashih-rukah-kupit-diplom-i-nachat-novuyu-zhiznВ
e-maxx.ru/forum/profile.php?id=19116В
militarymuster.ca/forum/showthread.php?mode=threaded&tid=339886&pid=381832В
mailorderbridesfinder.com/В
forumaniga.me/member/414-uf_23829059/visitormessage/13030-%D0%BF%D1%83%D0%B1%D0%BB%D0%B8%D1%87%D0%BD%D0%BE%D0%B5-%D1%81%D0%BE%D0%BE%D0%B1%D1%89%D0%B5%D0%BD%D0%B8%D0%B5-%D0%BE%D1%82-uf_23829059В
It’s awesome in favor of me to have a site, which is good designed for my experience. thanks admin
http://dealeram.ru/catalog/airnova
http://pizzeriamilano.ru/vtorye-blyuda/58-vkusnaya-perlovaya-kasha-s-myakotyu-tykvy.html
http://apokalypsed.org/users.php?m=details&id=26454
http://gifq.ru/daryu-kaktus/
http://www.briefeducation.ru/bried-180.html
Hi there, all the time i used to check web site posts here in the early hours in the dawn, because i enjoy to find out more and more.
http://amisite.ru/phpBB2/memberlist.php?mode=joined&order=ASC&start=6900
http://rl-critic.ru/histeo/belinsk.html
http://finforum.org/index.php?app=forums&module=post§ion=post&do=reply_post&f=50&t=29549&qpid=202979
http://www.industrial-russian.ru/branch/rybnayapromyshlennost.html
http://live-dmitrov.ru/news/27.html
Please let me know if you’re looking for a writer for your blog. You have some really good posts and I feel I would be a good asset. If you ever want to take some of the load off, I’d love to write some content for your blog in exchange for a link back to mine. Please blast me an email if interested. Thank you!
forum.l2star.net/member.php?u=17186В
fond.uni-altai.ru/index.php?subaction=userinfo&user=usacyВ
supplementgear.com/low-glycemic-fruits/В
n-seo.ru/verstka-shablonov-na-joomla.html/index.htmlВ
baikal-biz.ru/forum/viewtopic.php?p=168589В
AudioBook26.ru призывает более 10 000 аудиокниг на домашнею нашей электронной библиотеке. Ваша милость в силах слушать их ясно на сайте чи водворить употребление чтобы офлайн-прослушивания
https://audiobook26.ru/
Yesterday, while I was at work, my cousin stole my apple ipad and tested to see if it can survive a twenty five foot drop, just so she can be a youtube sensation. My iPad is now destroyed and she has 83 views. I know this is totally off topic but I had to share it with someone!
bestcoolfun.ru/page/32В
goup.hashnode.dev/garantirovannyj-uspeh-s-nashimi-diplomamiВ
rabota-vakansii-spb.ru/job.php?s=%C3%8A%C3%A0%C3%B1%C3%B1%C3%A8%C3%B0В
razvodka-mostov.ru/press/70/В
usaessayexperts.com/faq/В
Приветствуем ваш онлайн-сайт, коллеги!
К вам обращается агентство СЕО продвижения XRumer Art.
Мы заметили, у вас достаточно молодой сайт и ему требуется внешнее SEO продвижение. Мы организовываем продвижение полностью под ключ. Также у нас есть доступные и эффективные инструменты для СЕОшников. У наших специалистов значительный опыт в данной сфере, в арсенале есть успешные кейсы – предоставим по запросу.
Наша компания предлагает скидку 10% до конца месяца на самые популярные услуги.
Наши услуги:
– Размещаем трастовые ссылки (нужно любому сайту) – от 1,5 до 5000 рублей
– Размещение безанкорных ссылок (полезно для всех сайтов) – 3900 рублей
– Прогон на 110 тыс. сайтов (RU.зона) – 2900 рублей
– 150 постов в VK о вашем сайте (отличная реклама) – 3.900 рублей
– Размещение статей про ваш сайт на 300 топовых онлайн-форумах (мощнейшая раскрутка портала) – 29.000 руб
– Мега Постинг – отличный прогон на 3 млн ресурсов (мощное размещение для ваших сайтов) – 39.900 рублей
– Рассылка рекламных сообщений по сайтам с использованием обратной связи – договорная цена, зависит от объемов.
Если возникнут вопросы, всегда обращайтесь, подскажем.
Отчётность.
Оплата: Yoo.Деньги, Bitcoin, МИР, Visa, MasterCard…
Принимаем USDT
Телегрм: https://t.me/exrumer
Skype: Loves.Ltd
https://www.webfx.com/
https://xrumer.cc/
Hey there, I think your site might be having browser compatibility issues. When I look at your blog site in Opera, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, superb blog!
https://rodme.ru/znakomstva-v-astane-t12467.html
В нашем обществе, где диплом – это начало отличной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа об образовании сложно переоценить. Ведь именно он открывает дверь перед всеми, кто собирается вступить в профессиональное сообщество или учиться в ВУЗе.
Наша компания предлагает быстро получить любой необходимый документ. Вы сможете заказать диплом, и это будет выгодным решением для человека, который не смог завершить образование или потерял документ. дипломы изготавливаются аккуратно, с максимальным вниманием к мельчайшим нюансам. В результате вы получите продукт, максимально соответствующий оригиналу.
Превосходство данного решения состоит не только в том, что можно быстро получить диплом. Процесс организован удобно, с нашей поддержкой. Начиная от выбора необходимого образца диплома до консультации по заполнению личной информации и доставки в любое место страны — все под абсолютным контролем опытных мастеров.
В результате, для тех, кто пытается найти оперативный способ получения требуемого документа, наша компания предлагает отличное решение. Приобрести диплом – значит избежать длительного процесса обучения и не теряя времени переходить к достижению личных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://megart.com.ua
http://ahfc.org
http://xn--logtervezs-j7a7i.hu
http://essay-capital.com
http://gerincservdebrecen.hu
For newest information you have to visit the web and on the web I found this website as a best site for hottest updates.
http://informatio.ru/news/education/middledu/rezultaty_ege_2013_i_gia_2013_kak_uznat_itogi_sdachi_gosekzamena/
http://ilecta1.ru/italiya/novosti-italii/beton-s-pristavkoi-bio-chto-eto.html
http://web011.dmonster.kr/bbs/board.php?bo_table=b0501&wr_id=205579
http://ctege.info/ege-2019/rezultatyi-ege-2019.html
http://lorsovet.info/stati/page/29
Hello! Do you know if they make any plugins to safeguard against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any recommendations?
http://znamus.ru/page/mihail_epshtein
http://www.artem-energo.ru/forums.php?m=posts&p=16434
http://participa.rosario.gob.ar/profiles/premiumsdiploms/activity
http://союзженскихсил.рф/communication/forums/forming/gde-uznavat-rezultaty-ege/?GROUP_ID=
http://www.nvaha.ru/205231775-pochemu-nekompetentnye.html
Hi colleagues, how is all, and what you would like to say about this article, in my view its genuinely awesome for me.
medik-look.ru/kupit-diplom-onlayn-udobno-i-byistroВ
mublog.ru/interesnye-novosti/obschaya/page/7.htmlВ
kamagraopas.com/mitka-ovat-varotoimenpiteetvaroitukset-kamagraan-liittyen.htmlВ
jeepgarage.ru/forum/topic.php?forum=24&topic=633В
pricebuy.co.il/siemens-kg76nai31l-priceВ
라이징 슬롯
물론 그의 수입은 여러 번 증가할 수 있습니다.
В современном мире, где диплом – это начало удачной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает дверь перед людьми, стремящимися начать профессиональную деятельность или учиться в ВУЗе.
В данном контексте мы предлагаем максимально быстро получить любой необходимый документ. Вы имеете возможность приобрести диплом нового или старого образца, и это является удачным решением для всех, кто не смог завершить образование или потерял документ. Любой диплом изготавливается с особой тщательностью, вниманием ко всем элементам. В результате вы получите документ, максимально соответствующий оригиналу.
Преимущества такого подхода состоят не только в том, что вы быстро получите диплом. Процесс организовывается комфортно, с профессиональной поддержкой. От выбора нужного образца до консультаций по заполнению личной информации и доставки в любой регион России — все под абсолютным контролем качественных мастеров.
Для всех, кто ищет быстрый способ получить необходимый документ, наша компания готова предложить выгодное решение. Заказать диплом – это значит избежать длительного процесса обучения и не теряя времени переходить к личным целям, будь то поступление в университет или начало карьеры.
http://bookidoc.com.ua
http://callofthehounds.com
http://cpaday.com.ua
http://essay-capital.com
http://gefest-remstroy.ru
В нашем мире, где диплом – это начало удачной карьеры в любой отрасли, многие ищут максимально быстрый путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь именно он открывает двери перед всеми, кто хочет начать трудовую деятельность или учиться в высшем учебном заведении.
Предлагаем очень быстро получить этот важный документ. Вы имеете возможность приобрести диплом нового или старого образца, что становится отличным решением для всех, кто не смог закончить обучение или потерял документ. Каждый диплом изготавливается аккуратно, с максимальным вниманием ко всем нюансам. В результате вы получите продукт, 100% соответствующий оригиналу.
Плюсы данного подхода заключаются не только в том, что вы сможете оперативно получить диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начиная от выбора подходящего образца документа до точного заполнения личных данных и доставки по России — все находится под полным контролем квалифицированных мастеров.
Всем, кто хочет найти быстрый и простой способ получить требуемый документ, наша услуга предлагает отличное решение. Приобрести диплом – значит избежать длительного процесса обучения и сразу перейти к достижению личных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
http://jewellandginnie.com
http://rech-kruiz.ru
http://eebc.net.ua
http://farseeingresearch.eu
http://huulienasia.com.vn
В нашем обществе, где аттестат является началом отличной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения качественного образования. Важность наличия официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед любым человеком, который собирается вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Наша компания предлагает очень быстро получить этот необходимый документ. Вы имеете возможность купить аттестат, и это будет выгодным решением для всех, кто не смог завершить обучение, потерял документ или желает исправить плохие оценки. Аттестаты выпускаются с особой тщательностью, вниманием к мельчайшим деталям. На выходе вы сможете получить 100% оригинальный документ.
Преимущество подобного подхода заключается не только в том, что можно оперативно получить аттестат. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора требуемого образца аттестата до правильного заполнения личных данных и доставки по стране — все под абсолютным контролем квалифицированных мастеров.
В итоге, для тех, кто ищет максимально быстрый способ получить необходимый документ, наша услуга предлагает отличное решение. Заказать аттестат – значит избежать продолжительного процесса обучения и сразу переходить к своим целям, будь то поступление в ВУЗ или старт карьеры.
https://vikingwebtest.berry.edu/ICS/Berry_Community/Group_Management/Berry_Investment_Group_BIG/Discussion.jnz?portlet=Forums&screen=PostView&screenType=change&id=af6a76c8-5777-467c-8b1a-c0ff43cdb612
В наше время, когда диплом является началом удачной карьеры в любой отрасли, многие пытаются найти максимально быстрый путь получения качественного образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает двери перед людьми, стремящимися вступить в сообщество профессионалов или учиться в высшем учебном заведении.
Наша компания предлагает максимально быстро получить этот важный документ. Вы можете купить диплом, что становится удачным решением для всех, кто не смог закончить образование, потерял документ или желает исправить свои оценки. дипломы изготавливаются с особой аккуратностью, вниманием ко всем нюансам. В итоге вы получите 100% оригинальный документ.
Плюсы этого подхода заключаются не только в том, что можно максимально быстро получить свой диплом. Процесс организован удобно, с нашей поддержкой. Начиная от выбора нужного образца до точного заполнения персональных данных и доставки по стране — все под абсолютным контролем наших специалистов.
В итоге, для тех, кто ищет быстрый способ получить требуемый документ, наша компания готова предложить отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в университет или начало карьеры.
http://veiligeafzender.nl
http://kchschool7.ru
http://s-boutique.com.ua
http://piar.org.ua
http://hamptonsantiquegalleries.com
whoah this blog is fantastic i like reading your articles. Stay up the good work! You realize, many persons are hunting around for this information, you can aid them greatly.
http://garage71.ru/paok-rassmatrivaet-kandidaturu-rebrova.html
http://www.karusel59.ru/kolyaski-progulochnye/carrello-vista-crl-8505.html
http://testkids.ru/zdorove/sindrom-dyryavoj-kishki.html
http://risuemtut.ru/kak-narisovat-zhenskoe-telo-karandashom-poetapno/
http://biokomb.ru/kak-snyat-bamper-nissan-almera-klassik/
Excellent, what a blog it is! This web site provides valuable information to us, keep it up.
http://www.xseon.ru/dialogue/viewtopic.php?f=3&t=1191&start=80
http://ukrrudprom.ua/reference/factory/ltz.html
http://simptomyinfo.ru/simptomy/409-problemy-s-volosami.html
http://homecveti.ru/kinkan.html
http://vobjavlenie.ru/148-muzykalnye-gruppy
XRumer – это самое лучшее также эффективное программное обеспечение для создания ссылок (а) также продвижения сайтов. Он используется чтобы автоматического прогона веб- сайтов сквозь разные соц сети, форумы, блоги и другие интернет-ресурсы.
http://www.botmasterru.com/product33230/
Купить XRumer Оффициально
Good post however , I was wanting to know if you could write a litte more on this topic? I’d be very grateful if you could elaborate a little bit more. Cheers!
http://byte-kuzbass.ru/inetmagazvirt/category_00000092116/category_00000092117/category_00000104459/product_00000116923-detail
http://straitkom.ru/2021/03/7770/
http://stroymix-v.ru/poleznye-sovety/tehnicheskie-novinki-i-prognozy-v-sfere.html
http://detsadik-208.ru/konsultacii-instruktora-po-fizicheskoj-kulture/ispolzovanie-solnechnyh-i-vozdushnyh-vann-dlja-ukreplenija-zdorovja-detej/
http://www.lada-4×4.net/member.php?u=21927
What a data of un-ambiguity and preserveness of valuable knowledge on the topic of unexpected feelings.
http://arvixe.ru/v/building/cotedge_1/P1070836.JPG.html
http://podlemore.moy.su/news/2015-00
http://familylevel.com/blogs/26/What-do-you-need-to-pay-attention-to-when-buying
http://saratovturizm.ru/forum/topic.php?forum=11&topic=59
http://www.limousine101.ru/podmoskove/zvenigorod/
온라인 슬롯 사이트
이 사람들이 어떻게 생각하는지는… 나와는 아무 상관이 없는 것 같다.
A beloved National Park Service ranger died when he tripped, fell and struck his head on a rock during an annual astronomy festival in southwestern Utah, park officials said over the weekend.
kraken13.at
Tom Lorig was 78 when he died after the incident at Bryce Canyon National Park late Friday.
kraken14.at
https://kraken14at.de
He was known for his extensive work as a ranger and volunteer at 14 National Park Service sites, including Yosemite National Park, Carlsbad Caverns National Park and Dinosaur National Monument, the park service said in a statement Saturday.
“Tom Lorig served Bryce Canyon, the National Park Service, and the public as an interpretive park ranger, forging connections between the world and these special places that he loved,” Bryce Canyon Superintendent Jim Ireland said in the statement.
Attractive section of content. I just stumbled upon your blog and in accession capital to assert that I get actually enjoyed account your blog posts. Any way I’ll be subscribing to your feeds and even I achievement you access consistently rapidly.
http://tekst-pesni.ru/index.php?name=news
http://aranzhirovki.ru/smf/index.php?PHPSESSID=4lnp4lcs4radid53ghfonobgu7&topic=4103.0
http://molcentr18.ru/ngo-useful
http://forum.infinite-soul.org/viewtopic.php?p=44480
http://ya-rukodelnitsa.ru/load/griby/34
Attractive element of content. I just stumbled upon your site and in accession capital to claim that I acquire actually loved account your weblog posts. Anyway I will be subscribing to your feeds or even I fulfillment you access persistently fast.
http://hp.dragonland-radio.de/profile.php?lookup=43
http://forum.spolokmedikovke.sk/viewtopic.php?f=3&t=51398&view=next
http://cavale.enseeiht.fr/redmine/issues/215
http://admeclub.ru/pozdravlenie-lyubovi/
http://bramapizza.ru/zagotovka-produktov-prezentaciya-k-uroku-po-tehnologii-6-klass-na/
Сегодня, когда диплом становится началом удачной карьеры в любой отрасли, многие пытаются найти максимально простой путь получения качественного образования. Факт наличия официального документа переоценить просто невозможно. Ведь диплом открывает дверь перед каждым человеком, желающим вступить в профессиональное сообщество или учиться в университете.
В данном контексте наша компания предлагает очень быстро получить этот необходимый документ. Вы сможете купить диплом старого или нового образца, что становится удачным решением для человека, который не смог закончить образование, утратил документ или желает исправить плохие оценки. Все дипломы изготавливаются аккуратно, с максимальным вниманием ко всем нюансам. На выходе вы сможете получить документ, полностью соответствующий оригиналу.
Плюсы этого подхода состоят не только в том, что можно быстро получить диплом. Процесс организовывается удобно, с профессиональной поддержкой. От выбора необходимого образца диплома до правильного заполнения персональной информации и доставки в любой регион России — все будет находиться под абсолютным контролем квалифицированных мастеров.
Для всех, кто ищет максимально быстрый способ получить необходимый документ, наша услуга предлагает отличное решение. Приобрести диплом – значит избежать длительного обучения и сразу переходить к достижению своих целей: к поступлению в университет или к началу трудовой карьеры.
http://stalmaster-kovka.ru
http://shkaranov.ru
http://kvartet.org.ua
http://karsigazete.com.tr
http://hoyo-oita.com
Подарок для конкурента
https://xrumer.ru/
Такой пакет берут для переспама анкор листа сайта или мощно усилить НЧ запросы
Подарок для конкурента
В наше время, когда аттестат становится началом успешной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения качественного образования. Необходимость наличия документа об образовании переоценить просто невозможно. Ведь диплом открывает двери перед любым человеком, желающим начать трудовую деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем быстро получить любой необходимый документ. Вы можете заказать аттестат нового или старого образца, что становится выгодным решением для человека, который не смог завершить образование, утратил документ или желает исправить плохие оценки. Аттестаты производятся с особой аккуратностью, вниманием ко всем деталям. В итоге вы сможете получить полностью оригинальный документ.
Преимущества данного решения состоят не только в том, что вы сможете максимально быстро получить свой аттестат. Весь процесс организовывается удобно, с профессиональной поддержкой. Начиная от выбора подходящего образца до грамотного заполнения персональной информации и доставки по России — все под абсолютным контролем опытных специалистов.
Всем, кто ищет оперативный способ получить требуемый документ, наша компания готова предложить выгодное решение. Заказать аттестат – значит избежать длительного обучения и не теряя времени перейти к своим целям, будь то поступление в ВУЗ или начало карьеры.
https://biokomb.ru/category/dochki-synochki/
https://seocliento.wikicorrespondence.com/3597967/Технические_возможности_xrumer_art
https://seocontentio.blog2learn.com/75496625/Как-продвигать-сайт-с-xrumer-art
Hi there! I realize this is somewhat off-topic however I needed to ask. Does managing a well-established blog like yours require a lot of work? I am brand new to writing a blog but I do write in my journal everyday. I’d like to start a blog so I will be able to share my experience and feelings online. Please let me know if you have any recommendations or tips for new aspiring blog owners. Thankyou!
http://www.musey-uglich.ru
В современном мире, где диплом является началом успешной карьеры в любой отрасли, многие ищут максимально простой путь получения качественного образования. Наличие официального документа трудно переоценить. Ведь именно он открывает двери перед любым человеком, желающим вступить в сообщество профессионалов или продолжить обучение в ВУЗе.
Наша компания предлагает оперативно получить этот важный документ. Вы сможете приобрести диплом, что будет выгодным решением для человека, который не смог завершить образование или утратил документ. дипломы изготавливаются аккуратно, с максимальным вниманием к мельчайшим нюансам. На выходе вы получите 100% оригинальный документ.
Превосходство подобного решения заключается не только в том, что вы максимально быстро получите свой диплом. Весь процесс организован комфортно и легко, с нашей поддержкой. От выбора необходимого образца документа до консультации по заполнению личной информации и доставки в любой регион страны — все под полным контролем качественных мастеров.
Всем, кто ищет оперативный способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в университет или старт карьеры.
http://http://seoyour.ru/
Hello friends, fastidious post and good urging commented at this place, I am actually enjoying by these.
http://http://musey-uglich.ru/
В нашем обществе, где диплом – это начало удачной карьеры в любой области, многие ищут максимально быстрый путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед каждым человеком, который стремится начать трудовую деятельность или продолжить обучение в ВУЗе.
Наша компания предлагает быстро получить этот необходимый документ. Вы имеете возможность заказать диплом старого или нового образца, и это является удачным решением для всех, кто не смог закончить образование, потерял документ или желает исправить плохие оценки. дипломы выпускаются аккуратно, с максимальным вниманием ко всем элементам. На выходе вы сможете получить продукт, 100% соответствующий оригиналу.
Превосходство этого решения заключается не только в том, что вы быстро получите диплом. Весь процесс организовывается комфортно, с нашей поддержкой. Начав от выбора нужного образца до консультации по заполнению персональных данных и доставки по стране — все под полным контролем опытных мастеров.
В результате, для всех, кто пытается найти оперативный способ получения необходимого документа, наша компания предлагает отличное решение. Приобрести диплом – значит избежать длительного процесса обучения и сразу перейти к достижению личных целей: к поступлению в университет или к началу успешной карьеры.
http://http://seoyour.ru/
Daddy Casino – Официальный телеграм канал со слотами от Дэдди Casino дэдди казино официальный сайт . Актуальное, рабочее зеркало официального сайта Дэдди Казино. Регистрируйся в Казино Daddy, получай бонус используя промокод, не забудь скачать
daddy казино играть
https://t.me/daddycasinorussia
슬롯 게임 무료
Zhu Zaimo는 선생님이 고함치는 소리를 들었을 때 자신의 팔이 더 이상 자신의 것이 아님을 느꼈습니다.
Что такое прогон по профилям?
На форумах при каждом прогоне регистрируется новый юзер, в профайле которого в поле Вебсайт или Домашняя страница стоит ваша ссылка. Также в некоторых форумах есть возможность в профиль вставить подпись, туда можно вставить ссылку с анкором. Все это можно варьировать. В профилях ссылки живут довольно долго, т.к. нет спама в чистом виде, и профили не мозолят глаза ни читателям форума, ни модераторам. Прогон производится последней версией хрумера с сервера с максимально возможной скоростью и производительностью.
ПОДРОБНЕЕ
НОВЫЙ ОТЛИЧНЫЙ VPN ДЛЯ XRUMERA!
За последние недели состоялась серия важных обновлений комплекса: XAuth, Captchas Benchmark, XRumer, XEvil 6.0 . В XEvil 6.0 обновлен механизм обработки hCaptcha и многое другое, в XAuth повышена стабильность работы, в XRumer повышена эффективность, устранён ряд погрешностей и обновлены прилагаемые базы.
https://xrumer.ru/
This post is invaluable. How can I find out more?
купить диплом в белорецке
http://kosprof.ru
http://pension-forum.ru
http://diploms-help.ru
купить диплом в туле
Интернет-казино daddy casino официальный — сайт или специальная программа, которые позволяют играть в азартные игры в интернете. Сайты интернет-казино могут специализироваться на различных азартных играх: игровых автоматах, рулетке, лотереях, викторинах, покере или других карточных и настольных играх. Википедия
daddy casino вход
https://t.me/daddycasinorussia
На сегодняшний день, когда диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Факт наличия официального документа об образовании сложно переоценить. Ведь диплом открывает двери перед всеми, кто хочет начать трудовую деятельность или продолжить обучение в ВУЗе.
Наша компания предлагает быстро получить этот важный документ. Вы сможете купить диплом, и это будет выгодным решением для всех, кто не смог закончить обучение или потерял документ. Все дипломы производятся с особой тщательностью, вниманием ко всем элементам, чтобы в результате получился 100% оригинальный документ.
Превосходство такого решения заключается не только в том, что вы оперативно получите свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора требуемого образца до консультации по заполнению личных данных и доставки по России — все находится под абсолютным контролем качественных специалистов.
В итоге, всем, кто пытается найти быстрый способ получить необходимый документ, наша услуга предлагает выгодное решение. Приобрести диплом – значит избежать долгого процесса обучения и не теряя времени переходить к личным целям: к поступлению в ВУЗ или к началу успешной карьеры.
http://leadership2009.ru
http://seoyour.ru
http://diplomy-samara.ru
http://zoointernet.ru
http://prohalal-faunia.ru
Hi there colleagues, fastidious article and nice urging commented here, I am genuinely enjoying by these.
купить новый диплом
http://sh5novosel.ru
http://school-10-lik.ru
http://raduga-bezopasnosti.ru
где купить диплом
I loved as much as you’ll receive carried out right here. The sketch is attractive, your authored subject matter stylish. nonetheless, you command get got an nervousness over that you wish be delivering the following. unwell unquestionably come more formerly again since exactly the same nearly a lot often inside case you shield this increase.
http://bigcountry.ru/page1.php?idm=164
купить диплом учителя
http://schoolnumberone.ru
купить диплом в комсомольске-на-амуре
777 슬롯
그의 두 형은 나이가 많아 부끄러워 보였습니다.
Terrific post but I was wanting to know if you could write a litte more on this topic? I’d be very thankful if you could elaborate a little bit further. Thank you!
http://forum.oursson.ru/viewtopic.php?f=117&t=3341&p=10425Г‚
купить диплом в новом уренгое
http://stodorova.ru
купить диплом техника
It’s remarkable for me to have a web page, which is good in favor of my knowledge. thanks admin
купить диплом в минусинске
http://obrazovanie-diplom.ru
http://sibsocio.ru
http://opexobo.ru
купить диплом в иваново
Thank you for the auspicious writeup. It in truth used to be a entertainment account it. Glance complicated to far introduced agreeable from you! However, how could we keep in touch?
http://www.transportpart.ru/pojd-365.html
купить диплом в волгограде
http://vntconference.ru
купить диплом в стерлитамаке
Can I simply say what a comfort to uncover someone that genuinely understands what they are discussing over the internet. You certainly understand how to bring a problem to light and make it important. More people need to read this and understand this side of the story. I was surprised that you are not more popular because you surely have the gift.
купить диплом психолога
http://psygate.ru
http://memorialculture.ru
http://kdcmir.ru
купить диплом в петропавловске-камчатском
Excellent post but I was wondering if you could write a litte more on this subject? I’d be very thankful if you could elaborate a little bit further. Many thanks!
http://forum.vkontakte.dj/member.php?9822-sonnick84&tab=activitystream&type=all&page=2
купить диплом в астрахани
http://artforum.pro
купить диплом в ставрополе
I’m not sure why but this site is loading incredibly slow for me. Is anyone else having this problem or is it a problem on my end? I’ll check back later and see if the problem still exists.
http://www.bryners.ru/forum/memberlist.php?sk=c&sd=d&start=625В
https://sso.kyrenia.edu.tr/simplesaml/module.php/core/loginuserpass.php?AuthState=_df2ae8bb1760fad535e7b930def9c50176f07cb0b7%3Ahttp%3A%2F%2Fadisgruntledrepublican.com
http://polimer42.ru/communication/forum/messages/forum5/topic49/message308293/?result=reply#message308293
http://domahka.blogspot.com/2013/12/11-1912.html
http://russiajoy.ru/kupit-diplom-legko-i-konfidentsialno
В современном мире, где диплом – это начало успешной карьеры в любой отрасли, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании переоценить попросту невозможно. Ведь диплом открывает двери перед любым человеком, который собирается вступить в профессиональное сообщество или продолжить обучение в университете.
В данном контексте наша компания предлагает быстро получить этот необходимый документ. Вы имеете возможность приобрести диплом, что является отличным решением для человека, который не смог завершить образование, утратил документ или хочет исправить плохие оценки. диплом изготавливается с особой аккуратностью, вниманием ко всем деталям, чтобы в итоге получился полностью оригинальный документ.
Преимущество этого подхода состоит не только в том, что можно быстро получить диплом. Весь процесс организован комфортно и легко, с профессиональной поддержкой. От выбора требуемого образца до правильного заполнения личных данных и доставки по стране — все под полным контролем качественных специалистов.
Для тех, кто ищет оперативный способ получить требуемый документ, наша компания может предложить отличное решение. Приобрести диплом – значит избежать долгого процесса обучения и не теряя времени переходить к важным целям: к поступлению в ВУЗ или к началу трудовой карьеры.
http://rassvetykamchatki.ru
http://vniikukuruzy.ru
http://rusglobalnet.ru
http://magopft.ru
http://club-subaru.ru
geinoutime.com
그에 대한 폐하의 칭찬은 그의 심장을 뛰게 만들었다.
Keep this going please, great job!
http://balaklavskiy-16.ru/user/9889/
http://p99946c6.beget.tech/2024/05/17/diplom-na-zakaz-vash-lichnyy-put-k-obrazovaniyu-i-karernomu-rostu.html
https://twoplustwoequal.com/read-blog/28151
https://spaszavod.ru/contacts/
http://mf.getbb.ru/viewtopic.php?f=28&t=536
Hi, I think your site might be having browser compatibility issues. When I look at your blog site in Safari, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, fantastic blog!
купить диплом в тольятти
http://kritikus.ru
http://fuwr.ru
http://samogon-alko.ru
купить диплом техника
Сегодня, когда диплом – это начало успешной карьеры в любой отрасли, многие ищут максимально быстрый путь получения образования. Факт наличия документа об образовании переоценить невозможно. Ведь именно диплом открывает дверь перед любым человеком, желающим начать трудовую деятельность или учиться в университете.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы имеете возможность заказать диплом нового или старого образца, и это будет удачным решением для всех, кто не смог закончить обучение или утратил документ. Любой диплом изготавливается с особой тщательностью, вниманием ко всем элементам, чтобы в результате получился документ, полностью соответствующий оригиналу.
Превосходство данного решения состоит не только в том, что можно быстро получить свой диплом. Процесс организовывается удобно, с нашей поддержкой. От выбора необходимого образца до грамотного заполнения личной информации и доставки по стране — все под абсолютным контролем квалифицированных специалистов.
В итоге, для всех, кто ищет максимально быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать продолжительного обучения и не теряя времени перейти к достижению личных целей, будь то поступление в университет или начало профессиональной карьеры.
http://e-diplom.biz/
http://rada-centr.ru
http://scienceanded.ru
http://neftgaztek.ru
http://avtoshkola142.ru
Howdy! This post couldn’t be written any better! Reading through this post reminds me of my previous room mate! He always kept chatting about this. I will forward this page to him. Fairly certain he will have a good read. Thank you for sharing!
http://viettel24h.com.vn/chao-moi-nguoi/
купить диплом образцы
http://musey-uglich.ru
купить диплом образцы
Pretty element of content. I simply stumbled upon your weblog and in accession capital to claim that I acquire in fact enjoyed account your blog posts. Anyway I will be subscribing on your feeds or even I fulfillment you get entry to consistently quickly.
купить диплом в кемерово
http://school-internat5.ru
http://kontur-plus.ru
http://pg2017.ru
купить диплом в буйнакске
Can I simply say what a relief to find someone who truly understands what they’re discussing on the net. You certainly know how to bring an issue to light and make it important. A lot more people need to read this and understand this side of the story. I was surprised you aren’t more popular since you surely possess the gift.
http://rybackij-prival.ru/news/v_mishkinskom_rajone_posle_20_let_zatishja_vnov_vozrodili_rajonnye_sorevnovanija_po_sportivnomu_podlednomu_lovu_ryby/2016-03-21-455
купить диплом инженера электрика
http://ot1do8.ru
купить диплом техника
This information is invaluable. When can I find out more?
купить диплом в твери
http://akadem-minstroy.ru
http://diplom-msk.ru
http://neftgaztek.ru
где купить диплом о среднем образование
Howdy great blog! Does running a blog like this take a great deal of work? I have very little expertise in coding however I was hoping to start my own blog soon. Anyhow, should you have any ideas or tips for new blog owners please share. I understand this is off topic nevertheless I simply wanted to ask. Appreciate it!
https://www.politikforum.ru/member.php?u=13385920
http://g29869x8.beget.tech/2024/05/26/diplom-na-zakaz-bystryy-put-k-obrazovaniyu.html
http://drugisaitove.listbb.ru/viewtopic.php?f=2&t=543
http://maridadiupdates.blogspot.ru/2014/01/modelsdesigners-singersflaviana-eskado.html
http://www.moto64.net/2016/06/honda-kawasaki-CRF250R-14-KX250F-16.html
Hi everybody, here every person is sharing these know-how, therefore it’s nice to read this weblog, and I used to go to see this webpage every day.
http://biosafe.tj/news/295598-paemi-prezidenti-umurii-toikiston-peshvoi-millat-mutaram-emomal-ramon-ba-malisi-olii-umurii-toikiston.html
https://wiki.nikiforov.ru/index.php/пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ,_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ
http://revolverp.forumex.ru/viewtopic.php?f=11&t=222
http://www.mebelnyvkus.ru/kuhnja/kuxonnye-stulya/stul-ccpe-m.html
equipdom.com/blog-details/from-bosch-with-performance-3-must-have-power-accessories-this-2018-16В
Подарок для конкурента
https://xrumer.ru/
Такой пакет берут для переспама анкор листа сайта или мощно усилить НЧ запросы
Подарок для конкурента
Because the admin of this web site is working, no hesitation very quickly it will be famous, due to its feature contents.
http://www.pavelgorbenko.ru/2018/02/blog-post.html
https://forum.sprosiadvokata.ru/topic/21548-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-m389x/
http://arsenal.listbb.ru/viewtopic.php?f=14&t=353
https://loftelectro.ru/products
http://pechorinase.blogspot.com/2013/04/dartagnan-et-les-trois-mousquetaires.html
프라그마틱 슬롯
Zhang Heling은 얼굴을 붉히며 무의식적으로 한 걸음 뒤로 물러났습니다.
Wow, wonderful blog layout! How long have you been blogging for? you made blogging look easy. The overall look of your web site is great, as well as the content!
http://orfografus.ru/index/0-53
купить диплом в ставрополе
http://primuothod.ru
купить диплом повара-кондитера
A beloved National Park Service ranger died when he tripped, fell and struck his head on a rock during an annual astronomy festival in southwestern Utah, park officials said over the weekend.
kraken13.at
Tom Lorig was 78 when he died after the incident at Bryce Canyon National Park late Friday.
kraken14.at
https://kraken-18.at
He was known for his extensive work as a ranger and volunteer at 14 National Park Service sites, including Yosemite National Park, Carlsbad Caverns National Park and Dinosaur National Monument, the park service said in a statement Saturday.
“Tom Lorig served Bryce Canyon, the National Park Service, and the public as an interpretive park ranger, forging connections between the world and these special places that he loved,” Bryce Canyon Superintendent Jim Ireland said in the statement.
It’s remarkable to pay a visit this site and reading the views of all colleagues about this piece of writing, while I am also zealous of getting experience.
http://usperm.ru/content/kuda-obratitsya-s-voprosom-po-provedeniyu-ege
купить диплом в юрге
http://sibsocio.ru
купить диплом в ельце
Привет всем!
Заказывайте диплом любого Вуза России у нас на сайте конфиденциально и без предоплаты.
https://telegra.ph/Za-skolko-konkretno-vozmozhno-budet-kupit-diplom-VUZa-v-nashe-vremya-03-11
https://www.dharmakathayen.com/fact-checking-policy/#comment-355585
http://nutagrl.blogspot.com/2010/11/blog-post.html
http://g29869x8.beget.tech/2024/01/29/bolshoy-vybor-diplomov-ot-znamenitogo-onlayn-magazina.html
http://blog.amatoricese.it/2014/01/villa-s-sebastiano-cese-2-1.html
Hi to all, the contents existing at this web site are genuinely remarkable for people knowledge, well, keep up the nice work fellows.
http://community278.ru/forums/
http://unix.t7y.ru/confirm?c=rasmyvtl&u=cindylelal
http://frichinvestigation.org/node/4323
https://www.forumfajerwerki.pl/topic/16855-pyromagic-2018-11-mi%C4%99dzynarodowy-festiwal-sztucznych-ogni/
https://opaseke.com/users/6784?wid=&reply=1
I’m gone to tell my little brother, that he should also go to see this blog on regular basis to get updated from most recent gossip.
купить диплом старого образца
http://dialognsu.ru
http://fopko.ru
http://uborka-kvartir-v-moskve.ru
купить диплом в заречном
Hi to all, how is all, I think every one is getting more from this web site, and your views are pleasant in support of new visitors.
http://www.kiopro.ru/support/forum/view_profile.php?UID=72874
financetimenews.ru/page/14/В
http://mybuildhouse.ru/diplom-na-zakaz-individualnyiy-podhod-i-professionalnoe-ispolnenie/
http://xn--d1aa3a4a.xn--p1ai/forum/profile.php?action=show&member=3827
http://rgf.5bb.ru/viewtopic.php?id=3199#p8194
This is very interesting, You’re a very skilled blogger. I have joined your feed and look forward to seeking more of your excellent post. Also, I’ve shared your web site in my social networks!
купить диплом в тольятти
http://collegevalday.ru
http://sk-sh.ru
http://kupit-diplom.msk.ru
купить диплом в белово
I’m not sure exactly why but this weblog is loading incredibly slow for me. Is anyone else having this issue or is it a problem on my end? I’ll check back later and see if the problem still exists.
indicouple.com/blogs/525/Where-to-buy-a-diploma-or-certificate-at-a-bargain?lang=tr_tr
http://www.nppremium.com/%E0%B9%80%E0%B8%A7%E0%B9%87%E0%B8%9A%E0%B8%9A%E0%B8%AD%E0%B8%A3%E0%B9%8C%E0%B8%94-100-compare_cialis_levitra.html
http://www.redspringgifts.com/e_feedback/?page=3325
grp.7olimp.ru/viewtopic.php?f=13&t=1895&p=2457
clc.edu.pe/blog/index.php?entryid=55125&nonjscomment=1&comment_itemid=55125&comment_context=236930&comment_component=blog&comment_area=format_blog
I’m not sure exactly why but this site is loading very slow for me. Is anyone else having this problem or is it a problem on my end? I’ll check back later on and see if the problem still exists.
http://anticultures.blogspot.com/2010/05/gta-iv-rmn60.html
http://www.golf.od.ua/forum/viewtopic.php?p=76839#76839
http://kungurblago.ru/novosti/osviashchenie-pridorozhnogo-kiota-v-kisherti
http://xn--33-6kc4bza.xn--p1ai/users/aponikaxe
http://kinogonews.ru/kupit-diplom-poshagovoe-rukovodstvo
Hi, every time i used to check weblog posts here early in the dawn, for the reason that i enjoy to gain knowledge of more and more.
http://file-manager.ucoz.ru/index/25-8517c1ae767abbeab2530f32d03b00667e5300fe68d1-1
http://serialforfree.ru/kupit-diplom-vash-shans-stat-luchshim-v-svoey-oblasti
http://good-serial.ru/2024/05/diplom-na-zakaz-vash-put-k-professionalnomu-priznaniyu-i-uvazheniyu/
http://bogatbeden.listbb.ru/viewtopic.php?f=3&t=340
http://student-news.ru/kupit-diplom-v-moskve-uslugi-ekspertov
Hello! I’m at work surfing around your blog from my new iphone! Just wanted to say I love reading your blog and look forward to all your posts! Keep up the fantastic work!
купить диплом в белово
http://oskol-sport.ru
http://pension-forum.ru
http://neftgaztek.ru
купить диплом в октябрьском
슬롯 사이트
이 놀란 Fang Jifan, 쓰레기 같은 Zhou Tanzhi는 실제로 돼지를 기르는 방법을 알고 있습니까?
Hi there! I realize this is somewhat off-topic however I needed to ask. Does running a well-established blog such as yours take a large amount of work? I’m brand new to writing a blog however I do write in my journal on a daily basis. I’d like to start a blog so I can share my personal experience and thoughts online. Please let me know if you have any kind of ideas or tips for new aspiring blog owners. Appreciate it!
backshowtime.ru/page/45
kingcityrp.moibb.ru/viewtopic.php?t=365
medhansh.net/blog/category/traffic-violations/
mypenza.ru/forum/index.php?showtopic=54674&mode=linear
lighttur.ru/kupit-diplom-lyubogo-uchebnogo-zavedeniya-rf-2
Thank you for sharing your info. I truly appreciate your efforts and I am waiting for your next write ups thank you once again.
http://avtoshkola34.ru/main/11-osnovnye-svedeniya-ob-avtoshkole-volzhskaya.html
купить диплом врача
http://vrninfo.ru
купить диплом в анапе
Wow, superb blog layout! How long have you been blogging for? you made blogging look easy. The overall look of your web site is magnificent, as well as the content!
http://skazkademjanovo.ru/forums/topic/диплом-на-заказ-ваш-персональный-успе/
https://balakovo.avt-avto.ru/categories/zamena-zhidkostey/oborudovanie-dlya-promyvki-maslyanoy-sistemy/elektricheskaya_ustanovka_dlya_promyvki_maslyanoy_sistemy_dvs_gd_122_atis/
https://saumalkol.com/forum/разное-2/2989-купить-диплом-в-ћоскве-быстро-и-легально.html
http://dnd.listbb.ru/viewtopic.php?f=3&t=426
http://w937480o.bget.ru/user/byanttso5812/
В нашем мире, где диплом – это начало отличной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения образования. Наличие документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед каждым человеком, желающим начать трудовую деятельность или продолжить обучение в ВУЗе.
Наша компания предлагает быстро получить этот важный документ. Вы имеете возможность заказать диплом старого или нового образца, и это является удачным решением для человека, который не смог закончить образование, утратил документ или желает исправить свои оценки. Все дипломы производятся аккуратно, с особым вниманием ко всем нюансам, чтобы в итоге получился полностью оригинальный документ.
Плюсы этого решения состоят не только в том, что можно оперативно получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора нужного образца диплома до консультаций по заполнению персональных данных и доставки по России — все под абсолютным контролем опытных мастеров.
Таким образом, для тех, кто хочет найти оперативный способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – это значит избежать длительного обучения и сразу переходить к достижению личных целей: к поступлению в университет или к началу удачной карьеры.
http://oskol-sport.ru
http://geniyz.ru
http://uborka-kvartir-v-moskve.ru
http://sk-sh.ru
http://diplomma.ru
What’s up, for all time i used to check weblog posts here early in the daylight, for the reason that i like to gain knowledge of more and more.
http://testotvet.blogspot.com/2014/03/blog-post_2819.html
img-group.ru/Partners.htmlВ
http://masterflower.listbb.ru/viewtopic.php?f=6&t=295
https://gmaii.ru/threads/1515/
http://birdsmoscow.mybb.ru/viewtopic.php?id=388162
Exceptional post however , I was wanting to know if you could write a litte more on this topic? I’d be very thankful if you could elaborate a little bit further. Kudos!
http://www.ksolo.ru/forum/member.php?tab=visitor_messaging&u=83595&page=23
okna496.ru/rehau/rehau1/
miupsik.ru/forums/portal.php?page=102
http://www.datxanhhanoi.com/vinpearl-riverfront-condotel-vien-ngoc-quy-ben-bo-song-han/
forexsnews.ru/page/3
В нашем мире, где диплом является началом отличной карьеры в любой области, многие ищут максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа трудно переоценить. Ведь именно он открывает двери перед людьми, желающими вступить в профессиональное сообщество или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает оперативно получить любой необходимый документ. Вы сможете заказать диплом нового или старого образца, что будет удачным решением для человека, который не смог закончить обучение или потерял документ. дипломы производятся аккуратно, с особым вниманием к мельчайшим элементам. В итоге вы получите продукт, максимально соответствующий оригиналу.
Превосходство этого решения состоит не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается комфортно, с нашей поддержкой. Начав от выбора нужного образца до грамотного заполнения личных данных и доставки по стране — все находится под абсолютным контролем качественных мастеров.
В итоге, для тех, кто ищет быстрый и простой способ получения требуемого документа, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать длительного процесса обучения и не теряя времени переходить к достижению своих целей: к поступлению в университет или к началу успешной карьеры.
http://r-diplom.ru
http://leadership2009.ru
http://cso-gd.ru
http://ussursity.ru
http://vash-opros.ru
Hello just wanted to give you a quick heads up and let you know a few of the images aren’t loading correctly. I’m not sure why but I think its a linking issue. I’ve tried it in two different web browsers and both show the same results.
купить диплом в иркутске
http://myturtime.ru/individualnoe-obrazovanie-gde-kupit-diplom-sozdannyiy-spetsialno-dlya-tebya
http://realtyintellect.ru/diplomyi-po-vashemu-zaprosu-u-nas-est-to-chto-vam-nuzhno-dlya-uspeha
http://newsato.ru/obrazovanie-na-zakaz-kak-kupit-diplom-sozdannyiy-spetsialno-dlya-tebya
https://www.vidagrafia.com/read-blog/172912
https://1abakan.ru/forum/showthread-105739/
купить диплом электрика
купить диплом россия купить диплом россия .
В современном мире, где диплом становится началом успешной карьеры в любой сфере, многие пытаются найти максимально быстрый путь получения качественного образования. Факт наличия официального документа трудно переоценить. Ведь именно диплом открывает двери перед людьми, стремящимися начать профессиональную деятельность или учиться в университете.
Наша компания предлагает максимально быстро получить этот необходимый документ. Вы можете купить диплом нового или старого образца, что является выгодным решением для всех, кто не смог закончить обучение, потерял документ или хочет исправить плохие оценки. Каждый диплом изготавливается аккуратно, с максимальным вниманием ко всем нюансам, чтобы в итоге получился документ, полностью соответствующий оригиналу.
Преимущества данного решения заключаются не только в том, что вы быстро получите свой диплом. Весь процесс организовывается комфортно и легко, с профессиональной поддержкой. Начиная от выбора подходящего образца до консультации по заполнению личных данных и доставки в любой регион России — все находится под абсолютным контролем квалифицированных специалистов.
Всем, кто пытается найти оперативный способ получить необходимый документ, наша компания готова предложить отличное решение. Купить диплом – это значит избежать длительного обучения и сразу перейти к личным целям, будь то поступление в университет или начало профессиональной карьеры.
http://kavinfo.ru
http://scienceanded.ru
http://sosedi2017.ru
http://primzona.ru
http://bashkiriabiz.ru
Hello it’s me, I am also visiting this website daily, this site is actually nice and the visitors are really sharing pleasant thoughts.
theholo.net/forum/member.php?9206-sonnick84&tab=activitystream&type=all&page=3
tmwip-chelm.org.pl/profile.php?lookup=16720
http://www.11na11.com/2011/03/pro-vidstan-vid-polya-do-tribun-na-dnipro-areni/
admvoznesenie.ru/officials/blogs/history.php?PAGEN_1=48
http://www.sinoright.net/member/index.php?uid=yrukoc
Why visitors still use to read news papers when in this technological world all is presented on net?
купить диплом инженера электрика
https://rcdrift.ru/forum/member.php?u=21074
http://sport-faq.ru/budushhee-v-tvoih-rukah-kupi-diplom-i-realizuy-svoi-ambitsii
http://www.cslregister.com/forum/member.php?u=6225
http://fremontrp.listbb.ru/viewtopic.php?f=18&t=298
https://stalinarch.ru/wiki/index.php/пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ!
купить диплом вуза
What’s up to every body, it’s my first go to see of this weblog; this weblog consists of remarkable and actually fine information in favor of readers.
http://gimnaz06.ru/users/ewamary
http://nvkb.ru/forum/index.php/user/99017/
https://rf-essence.ru/index.php?/profile/27604-robertgjem/
http://forexrassia.ru/stan-liderom-svoey-otrasli-kupi-diplom-i-dostigni-vyisot-uspeha
https://kharkov-balka.com/showthread.php?p=152590#post152590
canadian pharmacy provigil
This information is worth everyone’s attention. Where can I find out more?
купить диплом в петропавловске-камчатском
http://sportraketka.ru/kupit-diplom-rf-v-moskve-legko-i-operativno
http://appleincub.ru/kupit-nastoyashhiy-diplom-bez-ozhidaniy
http://prikol100500.ru/zakazat-diplom-legko-byistro-nadezhno
http://yourealtynews.ru/diplom-na-vyibor-vasha-investitsiya-v-kachestvennoe-obrazovanie-i-professionalnoe-razvitie
http://lolipopnews.ru/kupit-diplom-v-moskve-uslugi-ekspertov
купить диплом медсестры
купить диплом массажиста diplomvash.ru .
Hey there just wanted to give you a quick heads up and let you know a few of the images aren’t loading correctly. I’m not sure why but I think its a linking issue. I’ve tried it in two different web browsers and both show the same outcome.
http://kvartal74.ru/lenta-23.html
купить диплом в дербенте
http://altaybiotech.ru
купить диплом с занесением в реестр
Keep on working, great job!
bc-pharmacy.com.ua/category/lekarstva/dezinfektsiya/
tecnologiaeducativa.sepyc.gob.mx/forum/topic/comentarios-de-videos-sobre-la-basura/?part=1090
http://www.indianhighcaste.com/read-blog/297_where-to-order-a-diploma-at-a-comfortable-price.html?mode=day
blackpearlbasketball.com.au/index.php?option=com_k2&view=itemlist&task=user&id=829712
http://www.sendungsverfolgung24.org/forum/topic/dfdg234dsfsd/?part=9324
Hello, after reading this remarkable paragraph i am too delighted to share my knowledge here with colleagues.
купить диплом в улан-удэ
https://justpaste.me/BeWN1
http://topnewsgadget.ru/raskroy-svoy-potentsial-kupi-diplom-i-dostigni-vershin-uspeha
http://worldgonews.ru/dostupnyie-i-professionalnyie-diplomyi-poluchite-svoy-pryamo-seychas
https://vip.forums.party/viewtopic.php?id=3081#p70205
http://soundofdust.bestbb.ru/viewtopic.php?id=281#p281
купить диплом в пензе
That is really interesting, You’re an overly professional blogger. I’ve joined your feed and sit up for looking for more of your excellent post. Also, I’ve shared your site in my social networks
http://www.squeakzy.com/article/20_ressources_psd_nov_2016
verbanent.hu/elfelejtett-jelszo
templateinspire.com/opencart/Lingerie/index.php?route=information/blogger&blogger_id=2
jamdom.ru/buy/office/index.html
blagmama.ru/forum/index.php?autocom=blog&blogid=4350&showentry=11569
Привет, дорогой читатель!
Закажите диплом ВУЗа с доставкой по всей России без предоплаты и с гарантией качества!
https://www.reisezielforum.de/viewtopic.php?t=66809
http://elegantpw.blogspot.com/2017/02/blog-post_14.html
https://notebooks.ru/forum/user/51547/
http://znanee.flybb.ru/topic444.html
https://breyerhorses.ru/personal/profile/?register=yes
What’s up every one, here every one is sharing such experience, thus it’s fastidious to read this blog, and I used to go to see this weblog all the time.
купить диплом в назрани
http://star-friends.ru/blogs/551/Получите-свой-диплом-без-стресса-Наш-сервис-заботится-о-вас
http://aromatov.wooden-rock.ru/forum/topic.php?forum=1&topic=15138
http://pottier.martial.free.fr/index.php?file=Members&op=detail&autor=iygorefremmv
https://say.la/read-blog/33745
http://w77515cs.beget.tech/2024/04/26/diplomy-po-vashemu-zaprosu-nadezhno-i-professionalno.html
купить диплом в шахтах
와일드 바운티 쇼다운
Zhu Houzhao는 빠르게 봉합하고 마지막 단계를 완료했습니다.Zhu Houzhao는 비웃으며 Wang Zhen을 가리켰습니다. “장교 개, 나가!”
Первый партнер охраны труда
Первый Партнер охраны труда – экспертный центр по аудиту и подготовке специалистов в области охраны труда и пожарной безопасности. Наша компания предоставляет широкий спектр услуг по обеспечению безопасности производственных процессов для индивидуальных предпринимателей и организаций различных секторов промышленности в Москве, Московской области и других регионах России.
Соблюдение правил безопасности труда – законодательное требование, за нарушение которого на организацию может быть наложен штраф или приостановлена деятельность предприятия на срок до 90 суток.
Мы помогаем решать вопросы обеспечения безопасности и организации труда на предприятии, разрабатываем и актуализируем документы по охране труда, проводим комплексное обследование охраны труда на соответствие государственным нормативным требованиям. Проводим обучение сотрудников по охране труда и пожарной безопасности, проверяем СУОТ и оцениваем профессиональные риски.
Виктория Набойченко сделала для нашего канала заявление,
[url=https://www.youtube.com/watch?v=Q0smZ5_QOcU]Заявление Виктории Набойченко[/url]
касающееся своего бывшего супруга – главного свидетеля обвинения по так называемому уголовному делу “Лайф-из-Гуд”-“Гермес”-“Бест Вей”
?? Забирай 100,000 Рублей + 400 Фриспинов в виде Welcome бонуса, на официальном сайте 7К Казино ??
?????? Официальное зеркало ??????
7к казино бонус за регистрацию
НОВЫЕ ПРОМОКОДЫ ОТ НАШЕГО ТЕЛЕГРАМ КАНАЛА 7К КАЗИНО! МОЩНЫЙ СТАРТ НАШИМ ИГРОКАМ. ???? ЖЕЛАЕМ КРУТЫХ ЗАНОСОВ ??
Joker7070 – бонус 170%+ 70 FS в Joker Stoker
777WIN – бонус 100%+ 30 FS в Hot Triple Sevens
HOT100 – бонус 100 FS в Indiana’sQuest
?? Добавляйте промокод в разделе бонусов на официальном сайте 7К Казино.
?? Важно! Для новых игроков после регистрации 7K Casino приготовил Welcome бонус до 100,000 Рублей + 400 Фриспинов! Успейте активировать ???
?? Подробная информация на официальном сайте 7К в разделе бонусов! Каждый промокод можете использовать по очереди.
7к казино вход
https://t.me/s/casino_7k_official
Hello there! Do you use Twitter? I’d like to follow you if that would be okay. I’m absolutely enjoying your blog and look forward to new updates.
купить диплом в чите
http://ya.creartuforo.com/viewtopic.php?id=11186#p22561
https://chayka.ixbb.ru/viewtopic.php?id=57#p67
http://wow-tour.ru/kupit-diplom-garantiya-legalnosti
http://odessaforum.getbb.ru/viewtopic.php?f=5&t=19970
https://tv-cifra.ru/blog/izmenenie-uslovij-okazaniya-uslugi-edinyj/#comment_160045
купить диплом колледжа
where can i buy bactrim online
Hi to every one, it’s in fact a nice for me to pay a quick visit this web site, it contains important Information.
https://www.afchornchurch.com/travel/
https://mypresspage.com/story2452773/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2%D1%80%D0%B0%D1%87%D0%B0
https://detetivedesegredos.com/
https://containersales.ru/
https://bookmarkcolumn.com/story16936443/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BF%D0%BE%D0%B2%D0%B0%D1%80%D0%B0
Hi there! This is kind of off topic but I need some advice from an established blog. Is it hard to set up your own blog? I’m not very techincal but I can figure things out pretty fast. I’m thinking about making my own but I’m not sure where to start. Do you have any tips or suggestions? Thanks
allukrnews.ru/put-k-professionalnomu-priznaniyu-pokupka-diploma-kak-reshenie/
http://www.successwebtech.com/blog/digital-marketing/best-digital-marketing-company-in-delhi
http://www.tonpentreafc.com/ton_pentre_youth_090409.html
financetimenews.ru/page/14/
mybaltika.info/ru/print/587/7892/
Привет, дорогой читатель!
Мы предлагаем приобрести диплом института или колледжа с доставкой по России без предоплаты.
http://www.nfgroup.it/forum/vale-tudo/358-n34q/
http://aviantrp.moibb.ru/viewtopic.php?f=8&t=59
http://pastilka.blogspot.com/2014/05/blog-post_27.html
http://forum.jeep-club.by/index.php?/blog/4/entry-2107-%D0%BE%D0%B1%D1%88%D0%B8%D1%80%D0%BD%D1%8B%D0%B9-%D0%BA%D0%B0%D1%82%D0%B0%D0%BB%D0%BE%D0%B3-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%BE%D0%B2-%D0%BE%D1%82%D0%B5%D1%87%D0%B5%D1%81%D1%82%D0%B2%D0%B5%D0%BD%D0%BD%D1%8B%D1%85-%D0%B2%D1%83%D0%B7%D0%BE%D0%B2/
http://sport.uscuma-ev.de/kategorien/ztm-hwarang-mudo/#comment-175742
Заявление Виктории Набойченко – бывшей супруги главного свидетеля обвинения Евгения Набойченко
https://www.youtube.com/watch?v=Q0smZ5_QOcU
Здравствуйте, меня зовут Виктория Набойченко.
Я являюсь бывшей супругой Набойченко Евгения Витальевича, с которым состояла в официальном браке с 2017 по 2022 год. Из средств массовой информации мне стало известно о фактах уголовного преследования лиц из окружения Романа Василенко, который является президентом международной бизнес-карты на территории России, Казахстана и Киргизии. На протяжении 20 лет Евгений Набойченко и Роман Василенко дружили, вместе путешествовали, проводили время, ездили на рыбалку. Роман очень доверял Евгению, делился с ним своими идеями, планами на коммерческую деятельность и реализацию новых проектов. Мне известно, что Роман предложил Евгению разработать фирменный логотип и сайт компании Life is Good, а впоследствии возглавить IT-отдел ЖК. Евгений с радостью согласился на предложение Романа и возглавил IT-отдел. Роман достойно оплачивал работу Евгения, гонорар которого вырос со 100,000 рублей до 500,000 рублей в месяц, что было связано с ростом и развитием кооператива Best. Роман всегда был щедр к Евгению и ценил его работу, за что ежегодно премировал.
Тот успех, которого достиг Роман за эти годы, очень задевал Евгения как мужчину. В ходе совместной жизни Евгений делился со мной, что мечтает однажды превзойти Романа в бизнесе, а его проект стать круче Илона Маска. Примерно с июня 2021 года Евгений стал нервным и злился на Романа, рассказывая мне, что Роман не хочет заплатить ему 170,000 евро. Я не вдавалась в подробности их взаимоотношений. Чуть позже я узнала о письмах Евгения к Роману, в которых он требовал от него указанную сумму и угрожал увечьями и убийством Роману и всем членам его семьи, в том числе и детям. Данные угрозы он отправлял голосовыми сообщениями и в электронных письмах. Мне известно, что Роман не реагировал на угрозы Евгения, что очень злило его, потому что он ждал от него указанную сумму. В моём присутствии Евгений постоянно по нескольку раз переслушивал свои голосовые сообщения, которые с угрозами отправлял в адрес Романа Василенко. Он получал от этого огромное удовольствие, с ухмылкой хвастаясь, что Роман обязательно заплатит.
Best Way
17 февраля 2022 года в 7:00 по МСК в нашем доме в посёлке в Краснодарском крае произошел обыск. В доме находилась только я и дети, а Евгений в это время находился в Краснодаре на съёмной квартире, куда заблаговременно уехал, предупредив меня за 3 дня о предстоящем обыске в доме. Как будто его друзья из правоохранительных органов Петербурга предупредили его. На мои вопросы о том, что происходит, он отвечал, что это формальность и что мне не о чем беспокоиться. Позже мне стало известно, что подобные обыски проходили и у других сотрудников кооператива в Санкт-Петербурге, Краснодаре, Анапе и других городах. В обыске нашего дома принимали участие шесть оперативных работников ОББ города Санкт-Петербурга, фамилии которых указаны в протоколе обыска, копия которого имеется в моём распоряжении. В ходе обыска оперативные работники оскорбляли Евгения и Романа, называя их преступниками и мошенниками, а также иными оскорбительными словами. При обыске был изъят планшет Евгения, оперативные работники перевернули весь дом и напугали детей, а также пообещали, что задержат Евгения в тот же день.
17 февраля 2022 года Евгений был задержан сотрудниками оперативных служб в городе Краснодар и примерно через 3 дня был доставлен в Санкт-Петербург, где был допрошен в УП по рекомендации сотрудников полиции. Допрос происходил без участия его личного адвоката Ирины Черняховской, от услуг которой он отказался в тот же момент. Мне позвонила Ирина в бешенстве и недоумении, крича о том, что Евгений её опозорил перед сотрудниками полиции, отказавшись от её услуг в грубой форме. Впоследствии мне стало известно от Евгения, что он лично получил покровительство начальника ОББ города Санкт-Петербурга в своей безнаказанности по факту проводимого расследования. Об этом он похвастался мне в личном разговоре, будучи окрылённый поддержкой начальника ОББ, который с ним разговаривал на доверительной ноте.
Евгений мне рассказал, что получит от органов абсолютную неприкосновенность и поддержку за информацию, которая интересовала оперативных работников, за что Евгению также была предоставлена госохрана. В период с февраля по середину апреля Евгений находился в Питере под госзащитой и плотно взаимодействовал с правоохранительными органами. Он предоставлял им всю информацию, в том числе полный доступ к системе базы данных Best Way и Life is Good. Евгений инициативно предлагал органам заблокировать доступ к личным кабинетам пайщиков и членов ЖК БСТ, чем полностью парализовал хозяйственную деятельность кооператива, создавая напряжённость и негативные отзывы. Также Евгений активно призывал граждан к написанию заявлений в отношении Романа Василенко с целью возврата пайщиками своих денежных средств, делал он это через каналы Telegram и YouTube. Евгений был единственным, кто имел и имеет доступ к базам данных и ключам. Желая намеренно создать проблемы Роману, он отказывается восстанавливать работу системы Best Way при наличии такой возможности.
Миссия Евгения – это привлечение Романа Василенко к уголовной ответственности, что спровоцирует крах компании и полное прекращение её деятельности, а это в свою очередь 30,000 пайщиков, которые могут стать потерпевшими по уголовному делу. Всё это мне известно из личного общения с Евгением, когда я приезжала к нему в Санкт-Петербург на несколько дней. Мне также достоверно известно, что в настоящее время нет реальных заявлений от недовольных пайщиков, так как в Романа Василенко все верят. Самое главное, что у Евгения есть возможность в любой момент через свой личный ноутбук восстановить работу ЖК Best Way и вернуть доступ к личным кабинетам пайщиков. Однако он этого не делает, преследуя свои личные цели.
«Дело «Лайф-из-Гуд» – «Гермес» – «Бест Вей»: кто такие Набойченко и Комаров?
Алексей Комаров водитель
?В уголовном деле, связываемом следствием с компаниями «Лайф-из-Гуд», «Гермес», кооперативом «Бест Вей» и основателем «Лайф-из-Гуд» и «Бест Вей» Романом Василенко, есть два свидетеля, на которых особенно уповает обвинение.
Это бывший сисадмин российского сегмента иностранной компании «Гермес» Евгений Набойченко, с 2014 года возглавлявший также IT-службу компании «Лайф-из-Гуд», занимавшуюся в том числе сайтом и платежной системой кооператива «Бест Вей». И бывший шофер Романа Василенко Алексей Комаров.
Набойченко в феврале 2022 года намеренно сломал российский сегмент платежной системы «Гермеса» и повесил сообщение: «Обращайтесь в полицию». Позднее многократно публично выступал с обвинениями Романа Василенко. Комаров утверждает, что возил по поручению Василенко неучтенные наличные деньги.
Кто эти люди, насколько вызывают доверие их обвинения, содержащиеся в уголовном деле? (В Приморском районном суде Санкт-Петербурга, рассматривающем дело по существу, они пока не выступали.)
Мы попытались в этом разобраться.
Вымогатель
Евгений Набойченко – способный айтишник, на каком-то этапе, по словам его бывшей жены, он возомнил себя имеющим право чуть ли не на партнерство в бизнесе Романа Василенко (см. видеозаявление Виктории Набойченко, данное ютуб-каналу, поддерживающему пайщиков кооператива «Бест Вей»). При этом его коллега – ведущий IT-разработчик компании «Лайф-из-Гуд» и кооператива «Бест Вей» Роман Роганович – сообщил на судебном заседании Приморского районного суда, что Набойченко вряд ли в состоянии во что бы то ни было придумать какой-то позитивный проект – из-за, как намекнул Роганович, скромности творческих способностей Набойченко.
И Евгений придумал схему вымогательства – как ему казалось, беспроигрышную. Насколько нам стало известно от наших источников, Набойченко перед тем, как обрушить платежную систему российского сегмента «Гермеса» в феврале 2022 года, шантажировал Романа Василенко – требовал с него деньги: 170 тыс. евро. При этом, по утверждению Виктории Набойченко, угрожал убийством и увечьями и самому Роману Василенко, и его супруге, и детям. Похвалялся перед (тогда еще) женой своими матерными сообщениями с угрозами, которые он посылал Василенко и его близким.
Кроме того, он завладел российской клиентской базой «Гермеса» и вымогал у клиентов деньги: свидетельства такого рода нам предоставлены.
Клеветник
Помимо этого, он допустил целый ряд публичных высказываний – прежде всего в YouTube, которые Роман Василенко расценил как клеветнические и инициировал по этому поводу уголовное разбирательство. Подавляющее большинство выступлений Набойченко, преимущественно нетрезвых, сейчас удалены.
Высказывания нотариально заверены, заведено уголовное дело – но расследуется оно ни шатко ни валко, так как расследование, по данным наших источников, тормозит начальник УЭБиПК ГУ МВД России по Санкт-Петербургу и Ленинградской области генерал-майор полиции Вадим Строков, который взял Набойченко под крыло.
Завербованный
По словам Виктории Набойченко, Евгений, как и другие функционеры «Лайф-из-Гуд», в начале расследования в отношении компании подвергался обыскам – но потом состоялся удивительный допрос Евгения Набойченко в питерском главке МВД, на который он запретил приходить своему адвокату. После этого допроса Набойченко была предоставлена госохрана и сам он хвастался супруге, что находится под личным патронажем тогдашнего начальника УЭБиПК, который его очень ценит.
За этим последовал слом платежной системы «Гермеса» и других ресурсов, которыми занимался Набойченко.
Хулиган, алиментщик и грабитель
По заявлениям источников, Евгений Набойченко бил супругу и детей. После развода в 2022 году отказывается платить алименты – их выплаты его супруга добивается через суды.
Пациент наркологов
По данным наших источников, с некоторых пор у Евгения Набойченко начала нарастать алкогольная, а возможно, и наркотическая зависимость (см. документы, которые нам предоставил источник, знакомый с ситуацией). Он лечился – но постоянно срывался.
Неуравновешенный, жадный, завистливый
В целом источники характеризуют его как неуравновешенного, жадного, завистливого человека. По мнению наших визави, эти его особенности использовали работники полиции для инсценировки уголовного дела в отношении компаний «Лайф-из-Гуд», «Гермес», кооператива «Бест Вей» и Романа Василенко.
Вороватый водитель
Другой ключевой свидетель обвинения – Алексей Комаров – сообщает в деле, что возил и передавал пакеты с деньгами – однако никакими инкассаторскими операциями в «Лайф-из-Гуд», по данным наших источников, он никогда не занимался. Он выполнял мелкие поручения Василенко, в числе которых – забрать подарки для него от пайщиков кооператива или консультантов сети «Лайф-из-Гуд» для Романа Василенко.
Через Комарова передавалось множество подарков от пайщиков из регионов. Часть из них до Василенко не доходила. Его спрашивали: «Как сало? Как самогоночка?» А всего этого он, по данным наших источников, не получал.
Роман Василенко рассказывал коллегам: «Много презентов, о которых мне рассказывали, но которые я так и не нашел. Мне их не жалко, просто плохо то, что я не поблагодарил тех людей, которые мне их подарили от всей души».
Комарова, как и Набойченко, по нашим данным, завербовал питерский УЭБиПК. По поводу перевозки денег он, по сведениям наших источников, просто лжет – подписывает то, что дают ему подписать в питерском УЭБиПК. И при этом скрывает, что сам воровал подарки, предназначенные для Василенко.
Кто обвинители?
Следствие привлекло для выстраивания обвинения малограмотного вороватого водителя и алкозависимого айтишника. На показаниях таких свидетелей точно можно строить обвинение, по которому четверо функционеров «Лайф-из-Гуд» сидят без приговора суда уже более четырех лет и по которому судят отца Романа Василенко – 83-летнего ветерана Вооруженных сил РФ Виктора Ивановича Василенко?
Вопрос, думаем, риторический.
В нашем обществе, где диплом является началом успешной карьеры в любой сфере, многие пытаются найти максимально быстрый путь получения образования. Факт наличия официального документа об образовании переоценить невозможно. Ведь именно он открывает дверь перед всеми, кто собирается вступить в профессиональное сообщество или учиться в высшем учебном заведении.
В данном контексте мы предлагаем оперативно получить этот необходимый документ. Вы имеете возможность купить диплом нового или старого образца, и это становится отличным решением для человека, который не смог закончить обучение, утратил документ или хочет исправить плохие оценки. Все дипломы выпускаются с особой тщательностью, вниманием ко всем деталям, чтобы на выходе получился полностью оригинальный документ.
Плюсы этого решения заключаются не только в том, что вы быстро получите свой диплом. Весь процесс организован просто и легко, с профессиональной поддержкой. От выбора подходящего образца документа до консультаций по заполнению личной информации и доставки в любое место России — все находится под полным контролем квалифицированных мастеров.
Для тех, кто хочет найти быстрый и простой способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать длительного обучения и сразу переходить к достижению своих целей: к поступлению в университет или к началу удачной карьеры.
http://variant-surgut.ru
http://spravki-moscow.ru
http://collegevalday.ru
http://ot1do8.ru
http://sip-vuz.ru
Simply wish to say your article is as surprising. The clearness on your publish is just cool and that i can assume you are knowledgeable in this subject. Well with your permission let me to clutch your feed to keep up to date with drawing close post. Thanks a million and please keep up the enjoyable work.
prachuabwit.ac.th/krusuriya/modules.php?name=Journal&file=display&jid=12564
recept-food.ru/page/3
chatrang.shop/products/coupplus
rising.chew.jp/END/inside/dbbs/dicebbs.cgi?mode=resmsg&no=7866
drunkslut.com/springbreak.html
The other day, while I was at work, my cousin stole my iPad and tested to see if it can survive a 30 foot drop, just so she can be a youtube sensation. My apple ipad is now broken and she has 83 views. I know this is entirely off topic but I had to share it with someone!
http://www.irc71.ru/index.php/jomsocial/groups/viewbulletin/191-kak-podyskat-khoroshij-internet-magazin-gde-mozhno-zakazat-diplom?groupid=47
http://yuzs.net/%E6%9C%AA%E5%88%86%E9%A1%9E/post-255.html
https://bookmarkinglog.com/story17080664/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%9A%D0%B0%D0%B7%D0%B0%D0%BD%D0%B8
https://socialtechnet.com/story2524900/%D0%B3%D0%B4%D0%B5-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D1%8B-%D0%B2%D1%83%D0%B7%D0%B0
https://socialskates.com/story17904274/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%9A%D0%B0%D0%B7%D0%B0%D0%BD%D0%B8
В современном мире, где диплом становится началом удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Наличие официального документа об образовании сложно переоценить. Ведь диплом открывает двери перед любым человеком, который хочет вступить в профессиональное сообщество или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы сможете купить диплом нового или старого образца, и это является удачным решением для человека, который не смог закончить обучение, утратил документ или желает исправить свои оценки. диплом изготавливается с особой аккуратностью, вниманием ко всем элементам, чтобы в итоге получился 100% оригинальный документ.
Преимущество данного решения состоит не только в том, что можно оперативно получить диплом. Процесс организовывается комфортно, с нашей поддержкой. Начиная от выбора нужного образца до грамотного заполнения личной информации и доставки в любое место страны — все под полным контролем квалифицированных мастеров.
Всем, кто ищет быстрый и простой способ получить необходимый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать длительного процесса обучения и сразу переходить к достижению своих целей: к поступлению в университет или к началу трудовой карьеры.
http://diplomy-samara.ru
http://kupit-diplom.msk.ru
http://diplom-177.ru
http://seoklan.ru
http://kinelgorod.ru
This is the right webpage for anyone who hopes to find out about this topic. You realize a whole lot its almost hard to argue with you (not that I really would want to…HaHa). You certainly put a new spin on a subject which has been written about for decades. Wonderful stuff, just excellent!
http://www.freepainter.ru/index.php?links_exchange=yes&page=1&show_all=yes%D0%92
http://www.aikidosydney.com/cdn-cgi/l/email-protection
hrv-club.ru/forums/index.php?autocom=gallery&req=si&img=5506
http://www.askthebuyers.com/blogs/30/Why-is-the-popularity-of-universities-decreasing-all-the-time?lang=tr_tr
http://www.hoahaubansacviet.com.vn/xac-nhan-binh-chon.flc?sbd=335
Здравствуйте!
Получите российский диплом с гарантированной доставкой в любую точку страны без предварительной оплаты – просто и надежно!
https://romb4x4.ru/forum/members/robertgfem.6049/
http://www.newrancho.ru/2014/01/blog-post.html
http://opros.flybb.ru/topic352.html
http://wmestestroim.blogspot.com/2012/03/blog-post.html
https://ingmac.ru/forum/?PAGE_NAME=profile_view&UID=36877
https://Dr-nona.ru/ – Dr. Nona
Why visitors still use to read news papers when in this technological globe everything is presented on web?
https://avtobestnews.ru/page/3
https://socials360.com/story7108403/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%A3%D1%84%D0%B0
https://bookmarkedblog.com/story17618006/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BE-%D0%B2%D1%8B%D1%81%D1%88%D0%B5%D0%BC-%D0%BE%D0%B1%D1%80%D0%B0%D0%B7%D0%BE%D0%B2%D0%B0%D0%BD%D0%B8%D0%B8
https://motorcycle-reviews60381.blog2freedom.com/26900942/the-definitive-guide-to-marketing
https://www.juntadeandalucia.es/averroes/centros-tic/14700626/helvia/bitacora/index.cgi?wIdPub=68
Hi friends, pleasant article and nice urging commented here, I am genuinely enjoying by these.
http://onlineboxing.net/jforum/user/profile/285562.page
https://motorcycle-reviews34568.designertoblog.com/58279207/how-much-you-need-to-expect-you-ll-pay-for-a-good-marketing
http://promtex58.ru/shop/CID_13_prinadlezhnosti_i_abrazivnyy_instrument.html
https://www.davetalksbaseball.com/amazing-reasons-why-baseball-is-a-great-sport/
https://jeffreynpppo.blog-a-story.com/6612332/the-smart-trick-of-marketing-that-no-one-is-discussing
Excellent blog you have here but I was curious if you knew of any message boards that cover the same topics talked about in this article? I’d really love to be a part of community where I can get feed-back from other knowledgeable people that share the same interest. If you have any recommendations, please let me know. Kudos!
http://www.sixsigmaexams.com/mybb/member.php?action=profile&uid=263670
http://www.camnangbenh.com/mu-mau/
americanautotitleloan.com/lake-wylie-sc-title-loans
forum.anime.org.ua/bbs/showthread.php?p=129042
bbs.blueplatform.org/space-uid-31463.html
can you order accutane online
price for dexamethasone
cost of effexor in australia
Hello! I know this is kinda off topic however I’d figured I’d ask.
Would you be interested in exchanging links or maybe
guest writing a blog article or vice-versa? My site addresses a lot of the same subjects as yours and I think we could greatly
benefit from each other. If you might be interested feel free to
send me an e-mail. I look forward to hearing from you!
Great blog by the way!
Top Explainer Video Company
in India –
https://en.99designs.pt/profiles/8076816/about
https://www.scribblemaps.com/maps/view/explainer-video-company-india/db9zqwm3hv
슬롯 사이트 추천
Zhu Gong은 매우 기뻐했고 그의 웃는 눈은 가늘어졌습니다. “전하, 제발, 제발.”
Good day! Do you know if they make any plugins to protect against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any recommendations?
forum.drustvogil-galad.si/index.php?topic=10004.0
http://www.revostock.com/ptpl.index.php
cocochampionship.com/en/contact
http://www.bigboytoyz.com/newcars/mercedes-benz/amg-gt
catsandsquirrels.com/tag/squelfie/
Hello! I’m at work surfing around your blog from my new iphone 3gs! Just wanted to say I love reading through your blog and look forward to all your posts! Keep up the superb work!
rak.flyboard.ru/topic6397.html?view=previous
aysdoorrepair.com/contact.php
hl2forever.ru/member.php?tab=visitor_messaging&u=246
http://www.martinsimoveisijui.com.br/blog/nutricosmeticos
http://www.downtowncookieco.com/shipping-policy/
Hi there! Do you know if they make any plugins to assist with Search Engine Optimization? I’m trying to get my blog to rank for some targeted keywords but I’m not seeing very good success. If you know of any please share. Kudos!
diplomsagroup.com/diplomy-po-specialnosti/diplom-inzhenera-tehnologa.html
sermobile.com.ua/index.php?links_exchange=yes&page=126&show_all=yes
medgora.ru/users.php?m=details&id=126706
http://www.dolcescents.com/detailS/09
originalparts.ru/linkexchange/page68/?le_categoryID=0
Wow that was odd. I just wrote an really long comment but after I clicked submit my comment didn’t show up. Grrrr… well I’m not writing all that over again. Regardless, just wanted to say wonderful blog!
http://www.irc71.ru/index.php/jomsocial/groups/viewbulletin/191-kak-podyskat-khoroshij-internet-magazin-gde-mozhno-zakazat-diplom?groupid=47
https://ekocom.ru/forum/?PAGE_NAME=profile_view&UID=12770
http://ftp.video-foto.by/forum/viewtopic.php?f=81&t=27595
https://jboi2007.org/
http://www.die-wuiderer.de/memberlist.php?mode=joined&order=ASC&start=88700
777 슬롯
Fang Jifan이 자랑 할 때 그는 얼굴을 붉히지 않고 숨을 쉴 수 없으며 정직 해 보였습니다.
This is really interesting, You are a very skilled blogger. I have joined your rss feed and look forward to seeking more of your excellent post. Also, I have shared your site in my social networks!
http://rybackij-prival.ru/news/v_mishkinskom_rajone_posle_20_let_zatishja_vnov_vozrodili_rajonnye_sorevnovanija_po_sportivnomu_podlednomu_lovu_ryby/2016-03-21-455
купить диплом с внесением в реестр
http://72frs.ru
купить диплом медсестры
Attractive component of content. I just stumbled upon your web site and in accession capital to say that I acquire actually enjoyed account your blog posts. Any way I will be subscribing on your feeds or even I fulfillment you get admission to constantly rapidly.
#GGG###
https://cristiannahr96076.ttblogs.com/6284203/het-belang-van-tijdelijke-sms-nummers-een-uitgebreide-verkenning
I believe everything composed made a bunch of sense. However, what about this? what if you were to write a killer headline? I ain’t saying your content is not solid, however suppose you added something that grabbed people’s attention? I mean %BLOG_TITLE% is a little boring. You ought to look at Yahoo’s home page and watch how they create news titles to grab viewers interested. You might try adding a video or a picture or two to get people excited about everything’ve got to say. Just my opinion, it could bring your website a little bit more interesting.
http://gymn2slv.ru/nintendo/valve.html
купить диплом в балаково
http://club-subaru.ru
купить диплом физика
I was able to find good info from your content.
http://auto-sale-msk.ru/policy
купить диплом в гуково
http://diplomvam.ru
купить диплом эколога
라이즈 오브 올림푸스
어떤 대기업이든 서산신도시에서 배치를 시작해야 한다.
https://antimafia.se/ – hashish with a syringe 10 grams sleva. 37%
https://antimafia.se/ – powdered heroin – snorting 70 gram discount 34%
https://antimafia.se/ – ecstasy under the tongue – 3 grams discount 14%
https://antimafia.se/ – hachis con una jeringuilla 51 grama sleva. 26%
https://antimafia.se/ – heroina en polvo – esnifada 100 grama sleva 16%
https://antimafia.se/ – extasis bajo la lengua – 1 gramas rebajas 33%
https://antimafia.se/ – hasista ruiskulla 5 gram discount. 35%
https://antimafia.se/ – heroiinijauhe – nuuskaaminen 55 grams sale 23%
https://antimafia.se/ – ekstaasi kielen alla – 1 grams sale 28%
https://antimafia.se/ – haszysz za pomoca strzykawki 13 gram discount. 13%
https://antimafia.se/ – sproszkowana heroina – wciaganie 100 gram discount 37%
https://antimafia.se/ – ekstaza pod jezykiem – 5 gram sale 36%
https://antimafia.se/ – du haschisch avec une seringue 6 grammes discounts. 31%
https://antimafia.se/ – heroine en poudre – sniffer 70 gram discount 22%
https://antimafia.se/ – ecstasy sous la langue – 20 gram discount 21%
Hi, I want to subscribe for this webpage to obtain most up-to-date updates, therefore where can i do it please assist.
http://portrets.ru/100.tretjakovka20.html
купить диплом нового образца
http://sv-hold.ru
купить диплом медицинского училища
Hi i am kavin, its my first time to commenting anyplace, when i read this paragraph i thought i could also create comment due to this sensible post.
http://obt.social/blogs/22869/What-exactly-do-you-need-to-pay-attention-to-when
купить диплом в липецке
http://kirovstone.ru
купить диплом прораба
https://Dr-nona.ru – Dr. nonna
5 라이온스 메가웨이즈
“Old Zhang …”Fang Jinglong은 그에게 더 친밀하게 말했습니다.
‘Sugary food is a drug for me’: A growing number of children are addicted to ultraprocessed foods
kraken17.at
Chicago native Jeffrey Odwazny says he has been addicted to ultraprocessed food since he was a child.
“I was driven to eat and eat and eat, and while I would overeat healthy food, what really got me were the candies, the cakes, the pies, the ice cream,” said the 54-year-old former warehouse supervisor
https://www-kraken18.at
kraken25.at
“I really gravitated towards the sugary ultraprocessed foods — it was like a physical drive, I had to have it,” he said. “My parents would find hefty bags full of candy wrappers hidden in my closet. I would steal things from stores as a kid and later as an adult.”
Some 12% of the nearly 73 million children and adolescents in the United States today struggle with a similar food addiction, according to research. To be diagnosed, children must meet Yale Food Addiction Scale criteria as stringent as any for alcohol use disorder or other addictions.
“Kids are losing control and eating to the point where they feel physically ill,” said Ashley Gearhardt, a professor of psychology at the University of Michigan in Ann Arbor who conducted the research and developed the Yale addiction scale.
“They have intense cravings and may be sneaking, stealing or hiding ultraprocessed foods,” Gearhardt said. “They may stop going out with friends or doing other activities they used to enjoy in order to stay at home and eat, or they feel too sluggish from overeating to participate in other activities.”
Her research also shows about 14% of adults are clinically addicted to food, predominantly ultraprocessed foods with higher levels of sugar, salt, fat and additives.
For comparison, 10.5% of Americans age 12 or older were diagnosed with alcohol use disorder in 2022, according to the National Survey on Drug Use and Health.
While many people addicted to food will say that their symptoms began to worsen significantly in adolescence, some recall a childhood focused on ultraprocessed food.
“By age 2 or 3, children are likely eating more ultraprocessed foods in any given day than a fruit or vegetable, especially if they’re poor and don’t have enough money in their family to have enough quality food to eat,” Gearhardt said. “Ultraprocessed foods are cheap and literally everywhere, so this is also a social justice issue.”
An addiction to ultraprocessed foods can highjack a young brain’s reward circuitry, putting the primitive “reptilian brain,” or amygdala, in charge — thus bypassing the prefrontal cortex where rational decision-making occurs, said Los Angeles registered dietitian nutritionist David Wiss, who specializes in treating food addiction.
order baclofen
Общественное здание или его часть зетфликс онлайн с оборудованием для публичной демонстрации кинофильмов. Главное помещение кинотеатра – зрительный зал с экраном большого размера и системой воспроизведения звука. В современных кинотеатрах система звуковоспроизведения состоит из нескольких громкоговорителей, обеспечивающих объёмный звук. Первые стационарные кинотеатры назывались в России «электротеатрами»
зетфликс онлайн https://zetflixhd.life/
zithromax without a script
“Дело “Лайф-из-Гуд” — “Гермес” — “Бест Вей”: свидетель обвинения объявила себя потерпевшей от следствия
Свидетель Набойченко
6 и 13 июня Приморский районный суд города Санкт-Петербурга, рассматривающий по существу уголовное дело № 1-504/24, связываемое с компаниями “Лайф-из-Гуд”, “Гермес” и кооперативом “Бест Вей”, провел очередные, шестое и седьмое по счету, заседания, посвященные допросу свидетелей обвинения и лиц, признанных следствием потерпевшими в рамках судебного следствия по делу
“К кооперативу претензий не было, следователь предложил подать заявление”
Признанный следствием потерпевший Болян подсудимых не знает. Был клиентом “Гермеса”, а также пайщиком кооператива — но до 2019 года. В 2019-м он вышел из кооператива и из “Гермеса”, ему были возвращены паевые взносы, и никаких претензий к кооперативу у него не было — что он письменно подтвердил, расторгая договоры с этими организациями.
Однако, как Болян отметил на суде, следователь убедил его в том, что он — потерпевший и должен подать заявление на возврат членских взносов. Заявление в МВД писать не хотел, на него вышли сотрудники, сначала претензий к кооперативу не было. Полиция ему объяснила, что можно получить деньги.
Стал клиентом “Гермеса” и пайщиком кооператива через своего консультанта Алексея Виноградова. Виноградов — грамотный маркетолог, он ему верил, тот не работал в кооперативе. Что было предметом договора в “Гермесе”, не помнит. В “Гермес” внес 100 и 700 евро, а в кооператив каждый месяц вносил по 12 тыс. в течение семи месяцев.
Вышел и из кооператива, и из “Гермеса” в 2019 году. Зачем вступал? “Наверное, квартиру купить хотел”. Кооператив вернул ему 70 тыс. паевых взносов, “Гермес” вернул со счета “Виста” 140 тыс. рублей.
В кооперативе деньги вернули почти сразу, удержав вступительный и членские взносы; в “Гермесе” вернули позже через “внутрянку”, но удержали комиссию.
Утверждает, что ему говорили, что можно со счета “Виста” вносить деньги в кооператив. Объясняли, что деньги передаются в доверительное управление трейдерам и брокерам, которые играют на бирже. В кооперативе, как он утверждает, можно было купить место в очереди. По его словам, “Гермес” и кооператив — по сути, одна организация. Требует взыскать с кооператива более 148 тыс. рублей — вступительный и членские взносы, и более 60 тыс. рублей с “Гермеса” — комиссию при выводе средств.
Договор с кооперативом не читал, но ему объяснили, что есть невозвратная часть денег — ее и не вернули, “но хочу попытаться вернуть”. Претензий к кооперативу “как бы и нет, но если вернут взносы, то будет хорошо”.
К Виноградову претензий не предъявлял. “Может, меня и не обманули в кооперативе”, -резюмировал свое выступление в суде Болян.
“Требую выплатить с учетом роста цен на недвижимость”
Признанная следствием потерпевшей Комова была как клиентом “Гермеса”, так и пайщиком кооператива. Подсудимых не знает. Требует более 8800 тыс. с кооператива и более 2700 тыс. с “Гермеса”. При этом из кооператива она не вышла и заявление о выходе не подавала. Сумма требований к кооперативу включает как паевые и членские взносы, так и оценку роста цен на недвижимость, которая не была приобретена.
Утверждает, что можно переводить деньги со счета “Виста” напрямую в кооператив — в подтверждение приводит скрины переписки с консультантами в смартфоне. Суд разъясняет, что доказательство может быть приобщено позднее при надлежащем оформлении.
“Информацию воспринял скептически, но вступил”
Признанный следствием потерпевшим Киреев был только клиентом “Гермеса”. Подсудимых не знает лично — Измайлова видел в видеоролике “Лайф-из-Гуд”. Узнал о “Гермесе” от консультантов Татьяны и Андрея Клейменовых, с которыми знаком с конца 1990-х годов. Вступил в “Гермес” в 2020 году, деньги сразу отобразились на счете. Вносил средства на счет “Виста” небольшими частями, снимал средства со счета для личных нужд. Заявляет требования на более чем 2 млн 300 тыс. рублей.
Когда подписывал договор, в офисе была табличка кооператива, но в офисе находился представитель фирмы “Гермес”, ему сказали, что это два продукта компании “Лайф-из-Гуд”.
В 2022 году начались проблемы с выводом средств. И руководство фирмы пугало блокировкой счета в случае подачи заявления в правоохранительные органы. В хейтерском чате узнал телефон следователя Сапетовой, позвонил и подал заявление.
В чате клиентов компании “Гермес” была информация о том, что счет будет заблокирован в случае подачи заявления в правоохранительные органы. Эта информация была за подписью “администрация”. Он спросил у консультантов: “Кто это — администрация?”, написал вопрос в чате, и его забанили. Других попыток вступать в диалог с “Гермесом” не предпринимал.
Признанный следствием потерпевшим Чернышенко был также только клиентом только компании “Гермес”. Подсудимых не знает. Познакомился с консультантами “Гермеса” во время совместной работы в “Макдоналдсе”. Информацию о “Гермесе” воспринял скептически — “это какой-то обман”, но консультанты его в конце концов уговорили. Они поставили условие, что продадут ему земельный участок, чтобы он вступил в “Гермес”.
“Я хотел их обмануть, — пояснил свидетель, — думал: пусть они продадут, а я в “Гермес” не вступлю. Но они начали уговаривать, и я вступил в апреле 2020 года. Один из консультантов дал расписку на внесение денег на счет в “Гермесе”, я внес через него 230 тыс. И еще раз из жадности 140 тыс. 140 тыс. я вывел, плюс каждый месяц выводил по 5000–7000 руб. С какого-то момента проценты стали падать. В конце я захотел выйти — но не успел. У меня в “Гермесе” осталось 270 тыс., которые я требую вернуть”.
В январе 2023 года Чернышенко подал заявление в полицию, так как прекратился доступ к счету. “Мне предложили 70 тыс. вернуть, чтобы я не ходил в полицию, ссылаясь на то, что помогли мне продать участок за меньшую сумму по документам, чем я его продал на самом деле, но я отказался, сказал, что мне надо 270 тыс.”.
“Я хочу стать потерпевшей. Но Василенко многие благодарны за квартиры”
Свидетель обвинения Галашенкова была только клиентом “Гермеса”. Из подсудимых знает Виктора Ивановича Василенко. Она сидела в зале при допросе двух потерпевших (что запрещено), но заявила, что плохо слышит. Ранее была знакома с Верой Исаевой из компании “Лайф-из-Гуд”.
Вера узнала, что у нее есть офис, и познакомила с Романом Василенко, который снял у нее офис, когда “Лайф-из-Гуд” только начинала раскручиваться. Он снимал офис только за коммунальные платежи. “Я попросила расплатиться за четыре месяца, а он предложил мне вместо денег вступить в “Бест Вей”, но я отказалась, так как квартира не была нужна”. Зато стала клиентом “Гермеса”: “Внесла 15 тыс. евро, но через три месяца на счете оказалось 3400 евро!” Исаева, по словам Галашенковой, оказалась мошенницей.
По словам Галашенковой, она внесла на счет “Виста” более 3 млн рублей. “Когда сказали, что все счета заблокировали, то собрали с нас еще денег, чтобы счета разблокировать, потом пришло сообщение, что открывается новая фирма, но это все оказалось обманом — счета не разблокированы”.
“Я не считала себя потерпевшей, так как не знала, что это можно сделать, — заявила Галашенкова. — Сейчас желаю заявить, что я хочу стать потерпевшей. Считаю, что должна взыскать с “Гермеса” как прямой ущерб, так и упущенную выгоду, так как эта фирма закрылась”.
При этом заявила, что “Василенко молодец, потому что я видела людей, которые купили квартиры, они были ему очень благодарны”.
Привет всем)
Хорошо быть студентом, пока не придет пора писать диплом, что и произошло со мной, но не стоит отчаиваться, ведь есть хорошие компании что помогают с написанием и сдачей диплома на хорошие оценки!
Изначально искал информацию про купить аттестат за 9 классов, купить диплом института, купить диплом зубного техника, купить диплом продавца, купить аттестат, потом попал на https://tastebuds.fm/topics/7635-hei-tarvitsen-apuasi и там решили все мои учебные заботы!
Хорошей учебы!
Привет Друзья!
Всегда думал что купить диплом о высшем образовании это миф и нереально, но все оказалось не так, изначально искал информацию про: купить диплом в симферополе, купить диплом в новочеркасске, где купить диплом среднем, купить диплом в костроме, купить диплом в арзамасе, потом про дипломы вузов, подробнее здесь https://gmaii.ru/threads/1282/
https://citydevelopers.ru/otzyvy/novosibirsk/zhk-voskresnyy/
http://1chan.blog/100-2/?unapproved=381010&moderation-hash=617acbb874a9c175e7fc5aeb4e1a3078#comment-381010
http://www.queer-as-folk.it/gallery/profile.php?uid=477116
https://www.fromrus.su/index.php?/topic/11493-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-y366t/
Оказалось все возможно, официально со специальными условия по упрощенным программам, так и сделал и теперь у меня есть диплом вуза Москвы нового образца, что советую и вам!
Успешной учебы!
Здравствуйте!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом в сургуте, купить диплом строителя, купить диплом в георгиевске, купить диплом в новоалтайске , купить диплом магистра и получил базовые знания. В итоге остановился на материале: http://best-talk.ru/kupit-diplom-4
Успешной учебы!
Здравствуйте!
Задумался а действительно можно купить диплом государственного образца в Москве, и был удивлен, все реально и главное официально!
Сначала серфил в сети и искал такие темы как: купить диплом в москве, купить диплом в шахтах, купить диплом сварщика, купить диплом в минеральных водах, купить диплом медбрата, получил базовую информацию.
Остановился в итоге на материале купить свидетельство о рождении ссср, https://lolka.bestbb.ru/viewtopic.php?id=454
http://familyportal.forumrom.com/viewtopic.php?id=13965
https://forum.novosel.ru/
https://prof-komplekt.com/club/user/4861/blog/7549/
http://eurodelo.ru/kupit-diplom-legko-i-byistro
Успешной учебы!
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф. Но, оказалось, что все возможно! Сначала искал информацию по теме: купить диплом тренера, купить диплом в асбесте, купить диплом строителя, купить диплом в муроме, купить аттестат, а затем перешел на дипломы вузов. Подробнее можно узнать здесь: http://zavalinka.listbb.ru/viewtopic.php?f=3&t=2866
http://www.helppo.com.co/nosotros/#comment-190479
https://coub.com/663fe933f07cd8818dbc
https://www.tnrad.org/community/con-discussion/certificate-of-need/paged/8125/
http://www.detskiy-mir.net/user/jfouqaftmp/
Оказалось, что все возможно и официально, с упрощенными программами обучения. Теперь у меня диплом московского вуза нового образца, и я рекомендую вам воспользоваться этим шансом!
Успешной учебы!
«Король» обвинителей «Лайф-из-Гуд» Набойченко – кто он на самом деле?
Лайф из Гуд последние новости
Евгений Набойченко – бывший IT-директор компании «Лайф-из-Гуд», а также сисадмин российского сегмента платежной системы международной инвестиционной компании «Гермес». Он – ключевой свидетель обвинения в так называемом деле «Лайф-из-Гуд» – «Гермес» – кооператива «Бест Вей» и основателя «Лайф-из-Гуд»/«Бест Вей» Романа Василенко, которое рассматривается сейчас судьей Екатериной Богдановой в Приморском районном суде Санкт-Петербурга.
С выполненной им блокировки российского сегмента платежной системы компании «Гермес», а также с его (нетрезвых) сообщений в социальных сетях (сейчас все они им стерты) началась эскалация этого уголовного дела. Но, скорее всего, дело им же самим и его подельниками из питерской полиции и создано.
Вор и дебошир
Наши источники делятся информацией о хищении активов с кошельков клиентов «Гермеса», к которым Евгений Набойченко имел доступ.
Набойченко не гнушался воровать даже у близких. Однажды остановившись у своей любовницы Светланы Н., Евгений избил ее и украл деньги с ее криптовалютного кошелька, пока она пыталась прийти в себя после побоев.
Он бил не только любовницу, но жену и детей, с которыми жил в Краснодарском крае. А после того как его супруга Виктория подала на развод, разгромил, ободрал и ограбил общий дом – унес все, что можно было унести. Надо ли говорить, что Набойченко который год не платит супруге алименты.
Вымогатель
Евгений Набойченко, по словам его бывшей жены, в один прекрасный день решил заработать и с помощью вымогательства. Он шантажировал Романа Василенко – требовал с него деньги: 170 тыс. евро. При этом, по утверждению Виктории Набойченко, угрожал увечьями и самому Роману Василенко, и его супруге, и детям. Похвалялся перед (тогда еще) женой своими матерными сообщениями с угрозами, которые он посылал Василенко и его близким.
Кроме того, вымогал деньги у клиентов «Гермеса»: свидетельства такого рода нам предоставлены.
Алкоголик и наркоман
Деньги нужны Набойченко в том числе для того, чтобы «питать» свою наркотическую зависимость – с некоторых пор он предпочитает шлифовать водку галлюциногенными грибами, об увлечении которыми с упоением рассказывал даже в своих соцсетях.
Он не всегда был алкоголиком и наркоманом – устроившись 10 лет назад в IT-службу, не пил и не употреблял. Но большие деньги, красивая жизнь, которая вдруг стала доступна для простого астраханского парня из низов, развратили его.
Клеветник
Он допустил целый ряд клеветнических высказываний в адрес Романа Василенко – за которые против него заведено уголовное дело. Но расследуется оно ни шатко ни валко, так как расследование тормозит начальник УЭБиПК ГУ МВД России по Санкт-Петербургу и Ленинградской области.
Завербованный органами
Виктория Набойченко сообщает, что за три дня до обыска в их тогда еще совместном доме Евгений уехал и ей сказал, что придут с обыском – но это пустая формальность. Он уже был в сговоре с полицией, так как за три дня был предупрежден об этом обыске.
15 февраля 2022 года Евгений Набойченко был задержан, но отправился не в СИЗО – куда отправили других сотрудников «Лайф-из-Гуд», не был помещен в изолятор или КПЗ, а жил на полицейской квартире.
Он отказался от услуг адвокатов и на этой квартире, по свидетельству одного из полицейских, «бухал с операми». А также каждый день водил оперов по барам и ресторанам – угощал их за свой счет.
В разговоре с женой он очень гордился покровительством начальника УЭБиПК питерского главка МВД.
Наши источники считают, что он украл деньги клиентов «Гермеса», разделив их со своими друзьями из полиции. Вся его дальнейшая жизнь и привычки указывают на то, что он использует ворованные деньги.
Трус и лжец
Набойченко сообщил в социальных сетях, что к нему пришло с адреса fall@mybizon.online письмо, в котором содержится угроза в адрес получателя, призывающая его не являться в суд для дачи показаний. В письме указано, что если он поедет в суд, то не доедет и будет трупом.
Создатель преступного сообщества
Нет сомнений, что Набойченко устроил историю с обрушением платежной системы, чтобы скрыть свое воровство активов клиентов. Также чтобы скрыть свое воровство, он обвинил во всем Василенко и технических специалистов компании «Лайф-из-Гуд», которые находятся сейчас на скамье подсудимых. Оклеветал всех, чтобы отвести подозрения от себя и своих подельников из питерской полиции.
Итак, кризис в «Гермесе» устроил Набойченко. Платежную систему разгромил Набойченко. Активы украл Набойченко, поделившись с питерскими полицейскими.
А теперь сидит и бездельничает, прожигая сворованные деньги. И боится идти в суд – чтобы не превратиться в суде из свидетеля в обвиняемого.
Скорее всего, в суд Набойченко не пускают сами полицейские – опасаясь его неадекватности и перекрестного допроса, на котором вскроется лживость его показаний. То есть силовики заметают следы своего преступления, совершенного с помощью марионетки Набойченко.
The world’s most liveable cities for 2024
раз анальный секс
It’s considered among the most beautiful cities in the world to visit, and it seems that Vienna may also be an unbeatable place to live.
The Austrian city has been crowned the most liveable city in the world yet again in the annual list from the Economist Intelligence Unit (EIU), which was released today.
The EIU, a sister organization to The Economist, ranked 173 cities across the globe on a number of significant factors, including health care, culture and environment, stability, infrastructure and education.
Vienna topped the list for the third consecutive year, receiving “perfect” scores in four out of five of the categories — the city was marked lower for culture and environment due to an apparent lack of significant sporting events.
Just behind the Austrian capital, Denmark’s Copenhagen retained its second place position, while Switzerland’s Zurich moved up from sixth place to third on the list.
Australia’s Melbourne fell from third to fourth place, while Canadian city Calgary tied for fifth place with Swiss city Geneva.
Canada’s Vancouver and Australia’s Sydney were in joint seventh place, and Japan’s Osaka and New Zealand’s Auckland rounded out the top 10 in joint ninth place.
The world’s most liveable cities for 2024
жесткое групповое порно
It’s considered among the most beautiful cities in the world to visit, and it seems that Vienna may also be an unbeatable place to live.
The Austrian city has been crowned the most liveable city in the world yet again in the annual list from the Economist Intelligence Unit (EIU), which was released today.
The EIU, a sister organization to The Economist, ranked 173 cities across the globe on a number of significant factors, including health care, culture and environment, stability, infrastructure and education.
Vienna topped the list for the third consecutive year, receiving “perfect” scores in four out of five of the categories — the city was marked lower for culture and environment due to an apparent lack of significant sporting events.
Just behind the Austrian capital, Denmark’s Copenhagen retained its second place position, while Switzerland’s Zurich moved up from sixth place to third on the list.
Australia’s Melbourne fell from third to fourth place, while Canadian city Calgary tied for fifth place with Swiss city Geneva.
Canada’s Vancouver and Australia’s Sydney were in joint seventh place, and Japan’s Osaka and New Zealand’s Auckland rounded out the top 10 in joint ninth place.
«Король» обвинителей «Лайф-из-Гуд» Набойченко – кто он на самом деле?
Гермес Менеджмент
Евгений Набойченко – бывший IT-директор компании «Лайф-из-Гуд», а также сисадмин российского сегмента платежной системы международной инвестиционной компании «Гермес». Он – ключевой свидетель обвинения в так называемом деле «Лайф-из-Гуд» – «Гермес» – кооператива «Бест Вей» и основателя «Лайф-из-Гуд»/«Бест Вей» Романа Василенко, которое рассматривается сейчас судьей Екатериной Богдановой в Приморском районном суде Санкт-Петербурга.
С выполненной им блокировки российского сегмента платежной системы компании «Гермес», а также с его (нетрезвых) сообщений в социальных сетях (сейчас все они им стерты) началась эскалация этого уголовного дела. Но, скорее всего, дело им же самим и его подельниками из питерской полиции и создано.
Вор и дебошир
Наши источники делятся информацией о хищении активов с кошельков клиентов «Гермеса», к которым Евгений Набойченко имел доступ.
Набойченко не гнушался воровать даже у близких. Однажды остановившись у своей любовницы Светланы Н., Евгений избил ее и украл деньги с ее криптовалютного кошелька, пока она пыталась прийти в себя после побоев.
Он бил не только любовницу, но жену и детей, с которыми жил в Краснодарском крае. А после того как его супруга Виктория подала на развод, разгромил, ободрал и ограбил общий дом – унес все, что можно было унести. Надо ли говорить, что Набойченко который год не платит супруге алименты.
Вымогатель
Евгений Набойченко, по словам его бывшей жены, в один прекрасный день решил заработать и с помощью вымогательства. Он шантажировал Романа Василенко – требовал с него деньги: 170 тыс. евро. При этом, по утверждению Виктории Набойченко, угрожал увечьями и самому Роману Василенко, и его супруге, и детям. Похвалялся перед (тогда еще) женой своими матерными сообщениями с угрозами, которые он посылал Василенко и его близким.
Кроме того, вымогал деньги у клиентов «Гермеса»: свидетельства такого рода нам предоставлены.
Алкоголик и наркоман
Деньги нужны Набойченко в том числе для того, чтобы «питать» свою наркотическую зависимость – с некоторых пор он предпочитает шлифовать водку галлюциногенными грибами, об увлечении которыми с упоением рассказывал даже в своих соцсетях.
Он не всегда был алкоголиком и наркоманом – устроившись 10 лет назад в IT-службу, не пил и не употреблял. Но большие деньги, красивая жизнь, которая вдруг стала доступна для простого астраханского парня из низов, развратили его.
Клеветник
Он допустил целый ряд клеветнических высказываний в адрес Романа Василенко – за которые против него заведено уголовное дело. Но расследуется оно ни шатко ни валко, так как расследование тормозит начальник УЭБиПК ГУ МВД России по Санкт-Петербургу и Ленинградской области.
Завербованный органами
Виктория Набойченко сообщает, что за три дня до обыска в их тогда еще совместном доме Евгений уехал и ей сказал, что придут с обыском – но это пустая формальность. Он уже был в сговоре с полицией, так как за три дня был предупрежден об этом обыске.
15 февраля 2022 года Евгений Набойченко был задержан, но отправился не в СИЗО – куда отправили других сотрудников «Лайф-из-Гуд», не был помещен в изолятор или КПЗ, а жил на полицейской квартире.
Он отказался от услуг адвокатов и на этой квартире, по свидетельству одного из полицейских, «бухал с операми». А также каждый день водил оперов по барам и ресторанам – угощал их за свой счет.
В разговоре с женой он очень гордился покровительством начальника УЭБиПК питерского главка МВД.
Наши источники считают, что он украл деньги клиентов «Гермеса», разделив их со своими друзьями из полиции. Вся его дальнейшая жизнь и привычки указывают на то, что он использует ворованные деньги.
Трус и лжец
Набойченко сообщил в социальных сетях, что к нему пришло с адреса fall@mybizon.online письмо, в котором содержится угроза в адрес получателя, призывающая его не являться в суд для дачи показаний. В письме указано, что если он поедет в суд, то не доедет и будет трупом.
Создатель преступного сообщества
Нет сомнений, что Набойченко устроил историю с обрушением платежной системы, чтобы скрыть свое воровство активов клиентов. Также чтобы скрыть свое воровство, он обвинил во всем Василенко и технических специалистов компании «Лайф-из-Гуд», которые находятся сейчас на скамье подсудимых. Оклеветал всех, чтобы отвести подозрения от себя и своих подельников из питерской полиции.
Итак, кризис в «Гермесе» устроил Набойченко. Платежную систему разгромил Набойченко. Активы украл Набойченко, поделившись с питерскими полицейскими.
А теперь сидит и бездельничает, прожигая сворованные деньги. И боится идти в суд – чтобы не превратиться в суде из свидетеля в обвиняемого.
Скорее всего, в суд Набойченко не пускают сами полицейские – опасаясь его неадекватности и перекрестного допроса, на котором вскроется лживость его показаний. То есть силовики заметают следы своего преступления, совершенного с помощью марионетки Набойченко.
메가 슬롯 사이트
“오.” Liu Wenshan은 잘못된 것이 없다고 생각했습니다. 선생님이 말씀하신 그대로여야 합니다.
купить диплом института vm-tver.ru .
베트맨 토토
그 직후 궁궐에서 10만 위안을 투자했고 서산도 10만 위안을 투자했다는 소식이 나왔다.
Почему сотрудники Колокольцева провоцируют социальный протест
Бест Вей
15 февраля пайщики кооператива «Бест Вей» по всей России намерены провести акции в поддержку кооператива. Кооператив работает в 72 регионах, в более чем 50 из них созданы инициативные группы и подаются заявки на проведение митинга.
Эти действия спровоцированы действиями следственной группы ГСУ ГУ МВД России по Санкт-Петербургу и Ленинградской области, которая заблокировала работу кооператива, арестовала его счета, арестовала всю недвижимость кооператива, хотя за более чем год расследования не нашла никаких свидетельств мошенничества в кооперативе.
15 февраля 2022 года начались активные действия следственной группы против кооператива — аресты и первый обыск в офисе с выносом всех документов. Позднее были заблокированы счета кооператива: на них находится 3,8 млрд рублей, причем сумма увеличивается, так как большинство пайщиков продолжает платить членские взносы и вносить паевые платежи.
Несмотря на то что кооператив трижды добивался в суде снятия ареста, следствие вновь выходило в суд с фактически тем же самым ходатайством и пытается, пользуясь своими полномочиями, уничтожить юридическое лицо, и накладывался новый арест.
В периоды, когда арест снимался, следствие за счет давления на банки добивалось того, чтобы платежи не проходили. Так происходит и сейчас, хотя с 19 января арест со счетов кооператива судом снят. Невзирая на претензионные письма кооператива в банки, жалобы кооператива в Центробанк и подготовку исковых заявлений против банков, Сбербанк и банк «Санкт-Петербург», получив, по свидетельству самих банковских работников, устное предупреждение следствия и «после звонков из Москвы», отказываются разблокировать счета, прямо нарушая постановление суда, снявшего арест со счетов.
Офис кооператива дважды обыскивался, документы дважды изымались, и следствие запрещало копировать документацию.
С самого начала расследование дела происходит при информационной поддержке помощника Колокольцева Ирины Волк, подчиненной ей пресс-служба министерства, разнообразные пристяжные «СМИ» типа «Дежурной части» на НТВ. К поддержке привлекли и самого министра: он говорил о деле кооператива «Бест Вей» в своем выступлении в Совете Федерации осенью 2022 года. По некоторым данным, «авторами» дела являются руководители ГУЭБиПК МВД России, облеченные доверием министра.
Следствие по-питерски
Характерно, что дело не «забрал» себе Следственный департамент МВД России. Оно расследуется ГСУ ГУ МВД России по Санкт-Петербургу и Ленинградской области — по месту регистрации кооператива, несмотря на общероссийский характер деятельности «Бест Вей». Это значит, что даже авторы атаки в центральном аппарате МВД не уверены в ее успехе.
Кураторы дела в федеральном МВД шанс проявиться, и они «роют землю», не гнушаются прямым нарушением закона, чтобы проявить себя. Кто эти героические работники юстиции? Прежде всего руководитель следственной группы майор юстиции Екатерина Сапетова и ее непосредственный руководитель подполковник юстиции Константин Иудичев.
Четверо технических сотрудников, не имевших никакого отношения к руководству кооперативом, трое из которых молодые женщины, были арестованы, хотя им вменялся «экономический» состав и они никогда не привлекались к уголовной ответственности. Они уже около года томятся в СИЗО, причем с ними почти не проводится следственных действий. По мнению адвокатов, от них ждут показаний на начальство.
Пятеро граждан объявлены в розыск, в том числе давно отошедший от текущего управления кооперативом его основатель и бывший председатель Совета кооператива Роман Василенко. 80-летних родителей Романа Василенко, ветеранов Вооруженных сил, постоянно таскают на многочасовые допросы, в ходе которых им неоднократно требовалась срочная медицинская помощь, им угрожают СИЗО.
Усилия следствия не привели к адекватным для судов результатам. Компрометирующих кооператив показаний ни от арестованных, ни от пожилых родственников людей, объявленных в розыск, получить не удалось, а пул потерпевших, собранный с огромным трудом, и их претензии выглядят неубедительно даже в наших судах, которые обычно дают следствию карт-бланш на период предварительного расследования.
Кульбиты обвинения
Следствие и пресс-структуры МВД рассказывают судам и прессе о «12 тысячах пострадавших», однако за более чем год работы им удалось получить заявления от сотни с небольшим потерпевших с общей суммой претензий около 150 млн рублей, хотя активы кооператива — более 15 млрд рублей. Предварительное расследование забуксовало.
Притом что следователи работают не покладая рук, телефонными звонками и письмами (есть в редакции) приглашая всех, кто так или иначе взаимодействовал с кооперативом, написать заявление.
Потерпевшие делятся на три группы. Первая — граждане, подавшие заявление по поводу действий другого юридического лица — компании высокодоходных инвестиций, которая контролировалась одним из прежних руководителей кооператива. Следствие пытается объявить эту компанию аффилированной с кооперативом, хотя между ними не было даже никаких взаиморасчетов, и пытается через псевдоаффилированность заставить кооператив заплатить по счетам другого юридического лица. Кстати, инвесткомпания — «живая», у нее есть активы в России, есть активы и у ее учредителей.
Вторая — пайщики кооператива, которые не могут получить свои паевые средства из-за того, что следствие арестовало счета. Среди них нет ни одного заявления о том, что кооператив взял деньги пайщика на покупку квартиры и не купил квартиру или взял паевые средства в рамках договора и отказался выполнять требования договора о возврате средств. Это люди, обманутые следствием, убедившим их в том, что только участие в расследовании поможет получить назад вложенные средства, хотя на самом деле получить их назад в реальном времени помогут исключительно разблокировка работы кооператива, снятие ареста со счетов.
Часть потерпевших, возможно, выступила соавтором атаки на кооператив: это группа граждан, которые когда-то вышли из кооператива «Бест Вей» и создали «альтернативный» кооператив — ЖК «Вера» (зарегистрирован в Ухте, но работает, как и «Бест Вей», по всей России). И против них адвокаты кооператива после завершения предварительного расследования намерены подать заявления о клевете.
Часть потерпевших — люди, которые пытаются поживиться, выдвигая необоснованные требования к кооперативу. Например, претензии некоторых потерпевших касаются вступительных или членских взносов, которые по договору с кооперативом не подлежат возврату.
В рамках следственных действий зачастую выясняется, что они не считают, что кооператив их обманул. Типична ситуация, когда псевдопотерпевшие внесли первоначальный безвозвратный взнос для вступления в кооператив — 110 тыс. рублей, причем некоторые даже не полностью, при этом паевые платежи для накопления первоначального паевого взноса не вносили, членские взносы — тоже.
Квартира им, разумеется, не приобреталась — они в своих показаниях на это и не претендуют. Однако они по наущению следствия или по собственной инициативе и при поддержке следствия подписали заявление о том, что кооператив не вернул им несколько десятков тысяч рублей, которые по договору с кооперативом возврату не подлежат, так как это вступительные или членские взносы, идущие, согласно уставу, на финансирование текущей деятельности и развития кооператива.
Это очередное свидетельство махинаций, совершаемых следствием: ведь потерпевший — лицо, считающее, что в отношении него совершено преступление. Из показаний, имеющихся в уголовном деле, видно, что большинство потерпевших так не считает.
Многие из них не сами пришли в следственные органы, а после звонка следователя, предложившего подать заявление в качестве потерпевших. Пайщики кооператива жалуются на систематический обзвон следователей из следственной группы Сапетовой, письма следователей, незаконные встречи со следователями с назойливыми советами подать заявление в качестве потерпевших — подавляющее большинство пайщиков от этого отказывается, понимая, что необоснованное обвинение в преступлении приведет к ответственности.
Банковские секьюрити — лучшие друзья следствия
Суды первоначально штамповали все ходатайства следствия, как водится в российской системе юстиции, на этапе предварительного расследования. Однако с некоторых пор вопросы к следствию стали возникать и у судей.
Ведь расследование продлевается, продлевается и продлевается, а промежуточные результаты его не впечатляют. Возникает много вопросов. Почему следствие не разрешает проводить налоговые платежи? На каком основании оно требует заблокировать на счетах средства, более чем в 25 раз превышающие сумму ущерба, имеющуюся в деле? На каком основании требует арестовать недвижимость на 12 млрд рублей, в том числе перешедшую в собственность пайщиков?
Кооператив трижды добивался снятия ареста со счетов, добивался разблокирования налоговых платежей. Снимались аресты с недвижимости кооператива в целом. Кроме того, суды принимали решения о снятии арестов с отдельных квартир, и эти решения вступили в законную силу.
Тем не менее арест со счетов де-факто не снимался, за исключением короткого периода летом 2022 года длиной в полторы недели (в эти полторы недели кооператив успел вернуть паевые взносы 216 пайщикам).
Службы безопасности банков блокируют платежи кооператива — банковские службы безопасности стали лучшими помощниками следствия вместо судов. Так, 18 января суд отказался продлить арест счетов, кроме суммы в 200 млн рублей — имеющиеся в деле обязательства перед потерпевшими плюс те, которые могут возникнуть. С 19 января счета не арестованы, но де-факто арест продолжается. Сбербанк и банк «Санкт-Петербург» не пропускают платежи, несмотря на претензионные письма и готовящиеся исковые заявления кооператива и жалобы в Центральный банк.
Банки в деле?
Есть основания считать, что банки, прежде всего Сбербанк, в Северо-Западном банке которого размещен счет паевых средств кооператива, не пассивные исполнители воли следствия.
Заинтересованность в первую очередь связана с уничтожением альтернативной программы покупки квартир. Кроме того, банки заинтересованы в использовании средств на счетах: их можно крутить месяцами, зарабатывая колоссальные средства. Этот метод подсказывают западные банки, которые, заблокировав деньги России, за счет процентов их использования получают серьезные финансовые ресурсы.
Вероятно, у Сбербанка вызвала интерес также цифровая система взаимодействия с клиентами, созданная в кооперативе: есть ряд свидетельств, что она скопирована IT-специалистами Сбера. Северо-Западный банк Сбербанка когда-то активно предлагал сотрудничество кооперативу, добивался размещения счета именно у себя – возможно, не без умысла.
Пайщики знают, кто виноват
Пайщики кооператива возмущены происходящим. Ведь уже год из-за ареста счетов они не могут приобрести квартиры, средства на приобретение которых перечислили; они не могут получить паевые средства назад, чтобы использовать их для покупки квартир другим способом; они не могут получить справки в кооперативе для наследования пая — документация в кооперативе изъята, и следствие даже не разрешает ее копировать. При этом они прекрасно отдают себе отчет в том, что именно ведомство Колокольцева, решившее состряпать громкое дело на пустом месте, хотя ни деньги, ни квартиры не украдены, — автор их злоключений.
Пайщики написали тысячи обращений в следственную группу, прокуратуру, уполномоченному по правам человека, проводились импровизированные митинги в судах. Теперь настал черед больших митингов по всей России.
Сотрудники Колокольцева на пустом месте создали липовое громкое дело ради внимания первого лица страны к непримиримой борьбе с мошенниками, обирающими граждан. Хотя на самом деле именно следствие само и обирает граждан, в обстановке социальной нестабильности в стране создает дополнительный повод для массового возмущения, уничтожает уникальную программу приобретения недвижимости без банковских переплат — фактически под 1% годовых, в рамках которой уже приобретено более 2,5 тыс. квартир.
Создан кризис на пустом месте, который наносит ущерб гражданам России и стране в целом. В условиях военного противостояния, социально-экономической турбулентности, предвыборной ситуации цена действий подразделений МВД непомерно высока – министру и политическому руководству страны пора вмешаться.
Привет всем)
Хорошо быть студентом, пока не придет пора писать диплом, что и произошло со мной, но не стоит отчаиваться, ведь есть хорошие компании что помогают с написанием и сдачей диплома на хорошие оценки!
Изначально искал информацию про купить диплом медсестры, купить диплом в тольятти, купить диплом ссср, купить дипломы о высшем цены, купить диплом в перми, потом попал на https://robautoparts.com/%d0%ba%d1%83%d0%bf%d0%b8%d1%82%d1%8c-%d0%b4%d0%b8%d0%bf%d0%bb%d0%be%d0%bc-%d1%83%d1%87%d0%b8%d0%bb%d0%b8%d1%89%d0%b0/ и там решили все мои учебные заботы!
Удачи!
Привет всем)
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: куплю диплом цена, купить диплом в минусинске, купить диплом в нижнем тагиле, купить диплом в ишиме, купить диплом в коврове, а потом наткнулся на http://wiki.why42.ru/wiki/??????_??????_?_??????:_???????_?_?????????
http://bl.com.tw/index.php?title=??????_??????:_?????_?_???????_??????????
https://bbarlock.com/index.php/??????_??????:_???_????_?_?????_??????????
http://shockmusik.ru/poluchenie-poddelnyih-udostovereniy-o-vyisshem-obrazovanii
http://ashinova.ru/category-5/t-1222.html#1222
, где все мои учебные проблемы были решены!
Успешной учебы!
Здравствуйте!
Хочу рассказать о своем опыте по заказу аттестата пту, думал это не реально и стал искать информацию в сети, про купить диплом в ноябрьске, купить аттестат за 11 класс, купить диплом в кунгуре, купить диплом средне техническое, купить диплом строителя, постепенно вникая в суть дела нашел отличный материал здесь http://ashinova.ru/category-5/t-1525.html#1576 и был очень доволен!
Теперь у меня есть диплом столяра о среднем специальном образовании, и я обеспечен на всю жизнь)
Удачи!
Добрый день!
Для некоторых людей, купить диплом ВУЗа – это необходимость, шанс получить достойную работу. Впрочем для кого-то – это очевидное желание не терять время на учебу в университете. Что бы ни толкнуло вас на такое решение, наша фирма готова помочь вам. Быстро, качественно и по доступной цене сделаем документ любого ВУЗа и года выпуска на подлинных бланках с реальными печатями.
Мы предлагаем выгодно и быстро купить диплом, который выполнен на оригинальной бумаге и заверен печатями, водяными знаками, подписями должностных лиц. Данный документ способен пройти любые проверки, даже при помощи специально предназначенного оборудования. Решите свои задачи максимально быстро с нашим сервисом.
Плюсы наших дипломов:
• используются лишь настоящие бланки “Гознак”;
• все подписи руководства;
• мокрые печати университета;
• специальные водяные знаки, нити и иные степени защиты;
• безупречное качество оформления – ошибок не бывает;
• любая проверка документа.
Где купить диплом специалиста?
https://together-19.com/read-blog/12170
Успешной учебы!
Добрый день!
Для некоторых людей, купить диплом ВУЗа – это острая необходимость, шанс получить достойную работу. Но для кого-то – это разумное желание не терять огромное количество времени на учебу в институте. С какой бы целью вам это не понадобилось, наша фирма готова помочь. Максимально быстро, качественно и по доступной цене сделаем документ любого года выпуска на подлинных бланках со всеми требуемыми печатями.
Приобрести документ университета вы сможете у нас. Мы оказываем услуги по производству и продаже документов об окончании любых университетов Российской Федерации. Вы сможете получить диплом по любым специальностям, любого года выпуска, в том числе документы СССР. Гарантируем, что при проверке документов работодателем, подозрений не возникнет.
Превосходства наших документов:
• используем только фирменные бланки “Гознака”;
• необходимые подписи должностных лиц;
• настоящие печати учебного заведения;
• водяные знаки, нити и иные степени защиты;
• безупречное заполнение и оформление – ошибок не бывает;
• любые проверки оригинальности документа.
Наша компания предлагает максимально быстро заказать диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, водяными знаками, подписями. Наш диплом пройдет лубую проверку, даже с использованием специальных приборов. Решите свои задачи быстро и просто с нашими дипломами.
Где заказать диплом специалиста?
http://fruit-impex.by/index.php?subaction=userinfo&user=aqubu
http://td-scs.ru/communication/forum/user/107151/
http://xn--e1adiici.xn--p1ai/index.php?subaction=userinfo&user=etiku
http://artland24.ru/index.php?subaction=userinfo&user=efafeqehy
https://radioizba.ru/communication/forum/?PAGE_NAME=profile_view&UID=18845
Успешной учебы!
accutane from canada pharmacy
купить диплом в перми купить диплом в перми .
Привет!
Для определенных людей, приобрести диплом о высшем образовании – это острая потребность, возможность получить выгодную работу. Но для кого-то – это желание не терять время на учебу в универе. С какой бы целью вам это не потребовалось, мы готовы помочь. Максимально быстро, профессионально и по разумной стоимости изготовим документ любого ВУЗа и года выпуска на подлинных бланках со всеми требуемыми печатями.
Приобрести документ о получении высшего образования вы сможете в нашем сервисе. Мы предлагаем документы об окончании любых университетов Российской Федерации. Вы получите диплом по любой специальности, любого года выпуска, в том числе документы СССР. Гарантируем, что при проверке документа работодателями, каких-либо подозрений не возникнет.
Основные преимущества наших дипломов:
• используются лишь настоящие бланки “Гознак”;
• необходимые подписи должностных лиц;
• мокрые печати ВУЗа;
• водяные знаки, нити и другие степени защиты;
• идеальное заполнение и оформление – ошибок не бывает;
• любые проверки оригинальности документа.
Наши специалисты предлагают выгодно заказать диплом, который выполнен на оригинальной бумаге и заверен мокрыми печатями, штампами, подписями должностных лиц. Данный диплом способен пройти лубую проверку, даже при использовании специальных приборов. Решите свои задачи быстро с нашей компанией.
Где заказать диплом специалиста?
http://www.mizmiz.de/create-blog/
Хорошей учебы!
Здравствуйте!
Для многих людей, купить диплом университета – это острая необходимость, шанс получить хорошую работу. Но для кого-то – это желание не терять время на учебу в институте. Что бы ни толкнуло вас на такое решение, наша фирма готова помочь. Оперативно, качественно и по разумной цене изготовим диплом нового или старого образца на настоящих бланках со всеми требуемыми подписями и печатями.
Приобрести документ университета вы можете у нас. Мы оказываем услуги по продаже документов об окончании любых ВУЗов России. Вы сможете получить необходимый диплом по любым специальностям, любого года выпуска, в том числе документы Советского Союза. Даем 100% гарантию, что в случае проверки документа работодателями, никаких подозрений не появится.
Плюсы наших дипломов:
• используются лишь настоящие бланки “Гознака”;
• оригинальные подписи руководства;
• настоящие печати ВУЗа;
• специальные водяные знаки, нити и другие степени защиты;
• идеальное качество оформления – ошибки полностью исключены;
• любая проверка документов.
Наша компания предлагает быстро и выгодно заказать диплом, который выполняется на оригинальном бланке и заверен печатями, штампами, подписями должностных лиц. Диплом пройдет лубую проверку, даже с использованием специальных приборов. Решайте свои задачи максимально быстро с нашими дипломами.
Где купить диплом специалиста?
http://iluzeia.flybb.ru/topic117.html
http://rkiyosaki.ru/discussion/8534/pochemu-luchshe-posetit-nash-internetmagazin-esli-nuzhen-diplom/
http://g95334gq.beget.tech/2024/05/30/kupit-diplom-podlinnye-dokumenty-po-dostupnoy-cene.html
http://algebra.bestbb.ru/viewtopic.php?id=465
https://mamadona.ru/blogs/kak_pravilno_priobresti_diplom_sovety_yksperta/
Удачи!
Привет!
Для некоторых людей, купить диплом о высшем образовании – это острая необходимость, удачный шанс получить достойную работу. Однако для кого-то – это желание не терять массу времени на учебу в институте. Что бы ни толкнуло вас на такое решение, наша компания готова помочь вам. Оперативно, качественно и по разумной цене изготовим диплом любого года выпуска на государственных бланках со всеми требуемыми подписями и печатями.
Мы предлагаем быстро и выгодно приобрести диплом, который выполнен на бланке ГОЗНАКа и заверен печатями, водяными знаками, подписями официальных лиц. Документ пройдет любые проверки, даже с использованием специальных приборов. Достигайте свои цели максимально быстро с нашим сервисом.
Плюсы наших документов:
• используются лишь качественные бланки “Гознака”;
• необходимые подписи руководства;
• мокрые печати ВУЗа;
• специальные водяные знаки, нити и другие степени защиты;
• идеальное качество оформления – ошибки исключены;
• любые проверки документов.
Дипломы и аттестаты об окончании ВУЗов и ССУЗов Российской Федерации:
– Повышают статус своего владельца;
– Открывают большие возможности на рынке труда;
– Повышают уважение в глазах окружающих;
– Повышают собственную уверенность и самооценку.
Где приобрести диплом специалиста?
https://sportspharmacology10875.losblogos.com/27220715/??????-??????????-????????
Хорошей учебы!
슬롯 안전한 사이트
Wu Yan은 “폐하, 제가 이야기하고 싶은 것은 황제의 손자에 관한 것입니다. “라고 말했습니다.
Добрый день!
Наши специалисты предлагают выгодно и быстро приобрести диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, водяными знаками, подписями. Данный диплом способен пройти любые проверки, даже при помощи специально предназначенного оборудования. Решите свои задачи быстро и просто с нашими дипломами.
Где купить диплом по актуальной специальности?
http://4division.ru/viewtopic.php?f=13&t=8225
Удачи!
Разбираю ситуацию вокруг “кооператива” Life is Good
[url=https://forum-kpfu.ru/100624/novosti-vasilenko-roman-poslednie-novosti/]раз анальный секс[/url]
Я решил написать этот пост, чтоб пройтись по доводам тех, кто привлекает новых “пайщиков” в структуры г-на Василенко (на деле – оплачивающих существование пирамиды Life is Good). Понятно, что, в этом сообществе крутятся сотни тех, кто занимается привлечением и теперь ЦБ с прокуратурой их оставят не только без заработка, но, возможно, придется отвечать за свои действия в суде. А деньги уже, наверняка, потрачены. Также есть люди, которые уже живут в квартире от LIG или выплатили некую сумму, они верят до конца и отказываются принимать ситуацию, в которой надо признать, что они могут остаться и без денег, и без квартиры. В любом случае, даже если кто-то живет в квартире от LIG, она ему не принадлежит и, в отличие от банковского залога, урегулировать вопрос не получится (их просто выселят через суд при банкротстве LIG, хотя ситуация скандальная и возможно государство что-то предпримет, чтоб сгладить проблему).Теперь по сути, что такое LIG.
Компания LIG – «Лайф-из-гуд» заявляет два основных направления деятельности: инвестиционные счета «Виста» в компании Hermes Management Ltd. и жилищный кооператив «Бест-вей» — альтернатива ипотеке.
Если верить консультантам «Лайф-из-гуд», доходность счетов «Виста» в компании «Гермес-менеджмент» последние годы держится в районе 20% годовых в долларах. Это довольно много (Уоррен Баффет позавидовал бы такой доходности), при этом непонятно, за счет чего достигается такая доходность.
Деятельность «Гермес-менеджмента» непрозрачна, на сайте не видно полноценных отчетов, нет сведений о структуре компании и ее руководстве.
«Гермес-менеджмент» привлекает российских клиентов через компанию «Лайф-из-гуд», но у него нет лицензии Центрального банка России на брокерскую деятельность или доверительное управление.
«Гермес-менеджмент» зарегистрирован в Белизе, офшорной зоне в Центральной Америке. Там же расположен офис компании. Еще в документах упоминается компания Hermes M в Шотландии. В случае проблем придется разбираться с зарубежной компанией, что делает задачу бесперспективной. Более того, учитывая реалии мошеннических схем, бывшие пайщики столкнутся еще и с лже-юристами, обещающими вернуть им вложенные деньги и конечно, останутся в крайне бедственном положении, с кредитами, без жилья и перспектив (у кого-то могут опуститься руки со всеми вытекающими).
Компания платит за привлечение новых клиентов, открывающих инвестиционные счета «Виста» (по сути – ничем не обеспеченные цифры на сайте. Заведение средств происходит через перевод средств на счета частных лиц(!), выполняющих роль консультантов LIG (а это сразу нарушает законы РФ и привлекает внимание регулятора), вывод – через перепродажу воздуха других буратинам. После внесения LIG в черный список, чаты структуры полны объявлений о продажи токенов Виста, но покупателей почти нет)) и вступающих в жилищный кооператив «Бест-вей». Система комиссионных выглядит такой разветвленной и продуманной, что становится понятно, что именно в ней суть компании, а не в инвестициях или альтернативе ипотеке.
Сам по себе сетевой маркетинг — вполне законный способ привлечения новых клиентов. Но когда он выглядит как основной вид деятельности, а клиентам при этом рассказывают про инвестиции, пассивный доход и успех в жизни, это наводит на очевидные выводы.
Суть предложения «Гермеса» такая: инвесторы открывают счет и перечисляют компании доллары или евро, а взамен получают пассивный доход. Минимальная сумма инвестиционного контракта — 10 тысяч долларов или евро. При этом первый взнос может быть 1130 долларов или 1100 евро, а оставшуюся сумму можно довнести позже.
Чтобы открыть счет «Виста», надо заполнить регистрационную форму на сайте «Лайф-из-гуд». При регистрации сайт просит поставить галочку «Я ознакомился и принимаю условия указанные в публичной оферте» (пунктуация сохранена).
По ссылке открывается не оферта, а договор об оказании услуг. В договоре не указаны название и реквизиты компании, с которой заключается договор. Кроме того, речь в нем идет не об управлении активами, а только об «образовательных и консультационных услугах об инвестиционных механизмах и способах управления капиталом». Это не похоже на обещанный пассивный доход от слова совсем. Таким образом, зазывалы в пирамиду пытаются уйти от ответственности.
[url=https://www.kommersant.ru/doc/6122663]жесткий анальный секс[/url]
Чтобы открыть счет и пользоваться им, придется заплатить различные комиссии. Все они прописаны в договоре:
Комиссия за открытие счета: 100 евро или 130 долларов.
Ажио, или входная комиссия: 7% от суммы лицевого счета. На сайте есть пример расчета: при вложении 100 000 $ ажио составит 7000 $.
Комиссия за управление счетом: 1% в год от размера счета, взимается каждый месяц по 1/12.
Комиссия с прибыли: 20% от ежемесячной прибыли, «полученной от инвестиционных продуктов сторонних партнерских компаний или физических лиц, в результате рекомендаций и деятельности Исполнителя».
Итак, LIG состоит из двух основных компаний: Гермес, которая предлагает участникам инвестиционный доход (с идеей полученную прибыль направить на квартиру) и Бест-Вэй, являющаяся ЖНК. Что это такое?
Это Жилищный накопительный кооператив (общепринятое сокращение — ЖНК) — вид потребительского кооператива, сочетающий в себе качества финансового и жилищно-строительного кооператива. ЖНК работают в соответствии с № 215-ФЗ «О жилищных накопительных кооперативах». Целью деятельности жилищных накопительных кооперативов является удовлетворения потребностей членов кооператива в жилых помещениях путём объединения членами кооператива паевых взносов. Жилищный накопительный кооператив имеет право:
– привлекать и использовать денежные средства граждан на приобретение жилых помещений;
– вкладывать имеющиеся у него денежные средства в строительство жилых помещений (в том числе в многоквартирных домах), а также участвовать в строительстве жилых помещений в качестве застройщика или участника долевого строительства;
– приобретать жилые помещения;
– привлекать заёмные денежные средства, общая величина не должна превышать сорок процентов стоимости имущества кооператива.
Членами жилищного накопительного кооператива могут быть только граждане РФ. Жилищный накопительный кооператив создаётся по инициативе не менее чем пятидесяти человек и не более чем пяти тысяч человек. Контроль за деятельностью кооператива по привлечению и использованию денежных средств граждан на приобретение жилых помещений, а также за соблюдением кооперативом требований действующего законодательства Российской Федерации осуществляет Центральный банк Российской Федерации.
Уже отсюда понятно, что заявления заинтересованных лиц из структур LIG о том, что ЦБ не в праве вмешиваться в деятельность собственно LIG лишены оснований. Может и вмешался.
Если говорить о цифрах, то на 13 декабря 2021 года в LIG было свыше 19 000 пайщиков изо всех уголков РФ. LIG выкупил и передал в безвозмездное пользование пайщикам примерно 2 500 объектов недвижимости, из них по 247 пайщики полностью выплатили пай, оформив недвижимость в собственность. Интересная статистика – 19 000 вступили, заплатив членский взнос в 160 000 рублей, но лишь 8 часть из них накопила необходимые 35% для получения недвижимости при том, что фактически недвижимость их собственностью не является. Заявления о том, что получившие недвижимость прописаны в квартирах – это пережитки советского прошлого и, вероятно, сознательное введение остальных в заблуждение. В современной РФ не существует института прописки, есть регистрация, постоянная и временная. Есть найм жилья и есть собственность. Допускаю, что какая-то часть людей все еще проживает в государственных квартирах и не оформила их в собственность, но это – давно не норма, как это было в СССР (где собственность как раз была редким явлением).
По сути кооператив — посредник между покупателем и продавцом жилья, а это увеличивает риски. Приходится полагаться на то, что кооператив соблюдает закон и что он не закроется до того, как вы оформите квартиру на себя.
Даже если все по закону, риск потерять деньги остается. Если кооператив по каким-то причинам перестанет работать, его собственность распродадут. Сначала он рассчитается с кредиторами и только затем вернет деньги пайщикам — пропорционально их паевым взносам. Хватит ли у кооператива денег, чтобы вернуть все паевые взносы, неизвестно.
Разбираю ситуацию вокруг “кооператива” Life is Good
гей член
Я решил написать этот пост, чтоб пройтись по доводам тех, кто привлекает новых “пайщиков” в структуры г-на Василенко (на деле – оплачивающих существование пирамиды Life is Good). Понятно, что, в этом сообществе крутятся сотни тех, кто занимается привлечением и теперь ЦБ с прокуратурой их оставят не только без заработка, но, возможно, придется отвечать за свои действия в суде. А деньги уже, наверняка, потрачены. Также есть люди, которые уже живут в квартире от LIG или выплатили некую сумму, они верят до конца и отказываются принимать ситуацию, в которой надо признать, что они могут остаться и без денег, и без квартиры. В любом случае, даже если кто-то живет в квартире от LIG, она ему не принадлежит и, в отличие от банковского залога, урегулировать вопрос не получится (их просто выселят через суд при банкротстве LIG, хотя ситуация скандальная и возможно государство что-то предпримет, чтоб сгладить проблему).Теперь по сути, что такое LIG.
Компания LIG – «Лайф-из-гуд» заявляет два основных направления деятельности: инвестиционные счета «Виста» в компании Hermes Management Ltd. и жилищный кооператив «Бест-вей» — альтернатива ипотеке.
Если верить консультантам «Лайф-из-гуд», доходность счетов «Виста» в компании «Гермес-менеджмент» последние годы держится в районе 20% годовых в долларах. Это довольно много (Уоррен Баффет позавидовал бы такой доходности), при этом непонятно, за счет чего достигается такая доходность.
Деятельность «Гермес-менеджмента» непрозрачна, на сайте не видно полноценных отчетов, нет сведений о структуре компании и ее руководстве.
«Гермес-менеджмент» привлекает российских клиентов через компанию «Лайф-из-гуд», но у него нет лицензии Центрального банка России на брокерскую деятельность или доверительное управление.
«Гермес-менеджмент» зарегистрирован в Белизе, офшорной зоне в Центральной Америке. Там же расположен офис компании. Еще в документах упоминается компания Hermes M в Шотландии. В случае проблем придется разбираться с зарубежной компанией, что делает задачу бесперспективной. Более того, учитывая реалии мошеннических схем, бывшие пайщики столкнутся еще и с лже-юристами, обещающими вернуть им вложенные деньги и конечно, останутся в крайне бедственном положении, с кредитами, без жилья и перспектив (у кого-то могут опуститься руки со всеми вытекающими).
Компания платит за привлечение новых клиентов, открывающих инвестиционные счета «Виста» (по сути – ничем не обеспеченные цифры на сайте. Заведение средств происходит через перевод средств на счета частных лиц(!), выполняющих роль консультантов LIG (а это сразу нарушает законы РФ и привлекает внимание регулятора), вывод – через перепродажу воздуха других буратинам. После внесения LIG в черный список, чаты структуры полны объявлений о продажи токенов Виста, но покупателей почти нет)) и вступающих в жилищный кооператив «Бест-вей». Система комиссионных выглядит такой разветвленной и продуманной, что становится понятно, что именно в ней суть компании, а не в инвестициях или альтернативе ипотеке.
Сам по себе сетевой маркетинг — вполне законный способ привлечения новых клиентов. Но когда он выглядит как основной вид деятельности, а клиентам при этом рассказывают про инвестиции, пассивный доход и успех в жизни, это наводит на очевидные выводы.
Суть предложения «Гермеса» такая: инвесторы открывают счет и перечисляют компании доллары или евро, а взамен получают пассивный доход. Минимальная сумма инвестиционного контракта — 10 тысяч долларов или евро. При этом первый взнос может быть 1130 долларов или 1100 евро, а оставшуюся сумму можно довнести позже.
Чтобы открыть счет «Виста», надо заполнить регистрационную форму на сайте «Лайф-из-гуд». При регистрации сайт просит поставить галочку «Я ознакомился и принимаю условия указанные в публичной оферте» (пунктуация сохранена).
По ссылке открывается не оферта, а договор об оказании услуг. В договоре не указаны название и реквизиты компании, с которой заключается договор. Кроме того, речь в нем идет не об управлении активами, а только об «образовательных и консультационных услугах об инвестиционных механизмах и способах управления капиталом». Это не похоже на обещанный пассивный доход от слова совсем. Таким образом, зазывалы в пирамиду пытаются уйти от ответственности.
русское гей порно
Чтобы открыть счет и пользоваться им, придется заплатить различные комиссии. Все они прописаны в договоре:
Комиссия за открытие счета: 100 евро или 130 долларов.
Ажио, или входная комиссия: 7% от суммы лицевого счета. На сайте есть пример расчета: при вложении 100 000 $ ажио составит 7000 $.
Комиссия за управление счетом: 1% в год от размера счета, взимается каждый месяц по 1/12.
Комиссия с прибыли: 20% от ежемесячной прибыли, «полученной от инвестиционных продуктов сторонних партнерских компаний или физических лиц, в результате рекомендаций и деятельности Исполнителя».
Итак, LIG состоит из двух основных компаний: Гермес, которая предлагает участникам инвестиционный доход (с идеей полученную прибыль направить на квартиру) и Бест-Вэй, являющаяся ЖНК. Что это такое?
Это Жилищный накопительный кооператив (общепринятое сокращение — ЖНК) — вид потребительского кооператива, сочетающий в себе качества финансового и жилищно-строительного кооператива. ЖНК работают в соответствии с № 215-ФЗ «О жилищных накопительных кооперативах». Целью деятельности жилищных накопительных кооперативов является удовлетворения потребностей членов кооператива в жилых помещениях путём объединения членами кооператива паевых взносов. Жилищный накопительный кооператив имеет право:
– привлекать и использовать денежные средства граждан на приобретение жилых помещений;
– вкладывать имеющиеся у него денежные средства в строительство жилых помещений (в том числе в многоквартирных домах), а также участвовать в строительстве жилых помещений в качестве застройщика или участника долевого строительства;
– приобретать жилые помещения;
– привлекать заёмные денежные средства, общая величина не должна превышать сорок процентов стоимости имущества кооператива.
Членами жилищного накопительного кооператива могут быть только граждане РФ. Жилищный накопительный кооператив создаётся по инициативе не менее чем пятидесяти человек и не более чем пяти тысяч человек. Контроль за деятельностью кооператива по привлечению и использованию денежных средств граждан на приобретение жилых помещений, а также за соблюдением кооперативом требований действующего законодательства Российской Федерации осуществляет Центральный банк Российской Федерации.
Уже отсюда понятно, что заявления заинтересованных лиц из структур LIG о том, что ЦБ не в праве вмешиваться в деятельность собственно LIG лишены оснований. Может и вмешался.
Если говорить о цифрах, то на 13 декабря 2021 года в LIG было свыше 19 000 пайщиков изо всех уголков РФ. LIG выкупил и передал в безвозмездное пользование пайщикам примерно 2 500 объектов недвижимости, из них по 247 пайщики полностью выплатили пай, оформив недвижимость в собственность. Интересная статистика – 19 000 вступили, заплатив членский взнос в 160 000 рублей, но лишь 8 часть из них накопила необходимые 35% для получения недвижимости при том, что фактически недвижимость их собственностью не является. Заявления о том, что получившие недвижимость прописаны в квартирах – это пережитки советского прошлого и, вероятно, сознательное введение остальных в заблуждение. В современной РФ не существует института прописки, есть регистрация, постоянная и временная. Есть найм жилья и есть собственность. Допускаю, что какая-то часть людей все еще проживает в государственных квартирах и не оформила их в собственность, но это – давно не норма, как это было в СССР (где собственность как раз была редким явлением).
По сути кооператив — посредник между покупателем и продавцом жилья, а это увеличивает риски. Приходится полагаться на то, что кооператив соблюдает закон и что он не закроется до того, как вы оформите квартиру на себя.
Даже если все по закону, риск потерять деньги остается. Если кооператив по каким-то причинам перестанет работать, его собственность распродадут. Сначала он рассчитается с кредиторами и только затем вернет деньги пайщикам — пропорционально их паевым взносам. Хватит ли у кооператива денег, чтобы вернуть все паевые взносы, неизвестно.
Здравствуйте!
Задумался а действительно можно купить диплом государственного образца в Москве, и был удивлен, все реально и главное официально!
Сначала серфил в сети и искал такие темы как: купить диплом в обнинске, купить диплом в йошкар-оле, купить диплом экономиста, купить диплом фармацевта, купить диплом средне техническое, получил базовую информацию.
Остановился в итоге на материале купить свидетельство о рождении ссср, http://free-forex-ua.blogspot.com/2011/05/blog-post_22.html?m=1
https://vmeste-wow.ucoz.ru/forum/2-173-179#5282
http://o926476g.bget.ru/users/yfybyjif
https://old.stm35.ru/products/kofr-zadnij-gka-4438/#comment_318488
Успехов в учебе!
http://axioma-estate.ru/index.php?subaction=userinfo&user=ivecigyv
Привет Друзья!
Всегда думал что купить диплом о высшем образовании это миф и нереально, но все оказалось не так, изначально искал информацию про: купить диплом фельдшера, купить диплом высшее, купить диплом в новом уренгое, купить диплом в донском, купить диплом во владивостоке, потом про дипломы вузов, подробнее здесь http://obozrevatelevents.ru/nastoyashhie-diplomyi-dlya-vashey-budushhey-kareryi/
Оказалось все возможно, официально со специальными условия по упрощенным программам, так и сделал и теперь у меня есть диплом вуза Москвы нового образца, что советую и вам!
Удачи!
Здравствуйте!
Профессионалы, у которых имеется высшее образование очень ценятся среди нанимателей. Диплом о наличии высшего образования необходим, чтобы доказать свой высокий профессионализм. Он дает понять начальству, что сотрудник обладает важными знаниями для того, чтобы очень качественно выполнить свою задачу. Но как же быть, когда знания имеются, а вот подтверждающего документа у человека нет? Покупка диплома решит эту проблему. Приобретение диплома университета РФ в нашей компании является надежным процессом, так как документ заносится в реестр. При этом печать производится на специальных бланках, установленных государством.
Где приобрести диплом по актуальной специальности?
https://mymink.5bb.ru/viewtopic.php?id=8535
Хорошей учебы!
Веб-студия SUHOV-IT ru в течение Москве – комплексный интернет-маркетинг “под электроключ” и разработка сайтов также использований
https://suhov-it.ru/
can you buy diflucan over the counter in canada
Здравствуйте!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом в димитровграде, купить диплом в курске, купить аттестат за классов, купить диплом в туапсе, купить диплом с внесением в реестр. Постепенно углубляясь в тему, нашел отличный ресурс здесь: http://dudubeat.ru/blog/festival-duduka-v-moskve-30-aprelya
http://bts.clanweb.eu/profile.php?lookup=32618
http://allnewstroy.ru/diplom-bez-usiliy-pokupka-onlayn
http://testruslit.ru/forum/viewtopic.php?f=2&t=469
https://joshuaclason.com/купить-диплом-реальный-или-мошенниче
и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Хорошей учебы!
купить диплом в каменске-уральском купить диплом в каменске-уральском .
Здравствуйте!
Для многих людей, приобрести диплом ВУЗа – это необходимость, уникальный шанс получить отличную работу. Но для кого-то – это понятное желание не терять время на учебу в ВУЗе. Что бы ни толкнуло вас на такой шаг, мы готовы помочь. Максимально быстро, качественно и по разумной цене изготовим диплом любого года выпуска на государственных бланках со всеми необходимыми печатями.
Наша компания предлагает выгодно заказать диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, штампами, подписями официальных лиц. Диплом способен пройти любые проверки, даже при использовании специально предназначенного оборудования. Решите свои задачи быстро и просто с нашими дипломами.
О преимуществах наших документов:
• используются только фирменные бланки “Гознака”;
• оригинальные подписи руководства;
• все печати университета;
• специальные водяные знаки, нити и другие степени защиты;
• идеальное качество оформления – ошибок не бывает;
• любые проверки документов.
Документы об окончании учебных учреждений РФ:
– Мгновенно повышают статус своего владельца;
– Открывают большие возможности в современном социуме;
– Повышают уважение в глазах граждан;
– Повышают самооценку.
Где приобрести диплом специалиста?
https://buy-dianabol-almaty10875.wonderkingwiki.com/739808/??????_????????????_????????
Успехов в учебе!
Здравствуйте!
Для определенных людей, приобрести диплом ВУЗа – это необходимость, возможность получить хорошую работу. Однако для кого-то – это очевидное желание не терять время на учебу в университете. С какой бы целью вам это не потребовалось, наша компания готова помочь вам. Быстро, профессионально и по разумной стоимости сделаем диплом любого года выпуска на подлинных бланках с реальными подписями и печатями.
Заказать документ института вы можете у нас. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов Российской Федерации. Вы получите диплом по любым специальностям, включая документы старого образца. Гарантируем, что при проверке документов работодателем, каких-либо подозрений не возникнет.
Основные преимущества наших документов:
• используются настоящие бланки “Гознак”;
• оригинальные подписи должностных лиц;
• настоящие печати ВУЗа;
• специальные водяные знаки, нити и иные степени защиты;
• безупречное заполнение и оформление – ошибки полностью исключены;
• любая проверка документов.
Наша компания предлагает выгодно и быстро приобрести диплом, который выполняется на оригинальной бумаге и заверен печатями, водяными знаками, подписями. Наш документ способен пройти любые проверки, даже при помощи специально предназначенного оборудования. Решайте свои задачи быстро и просто с нашим сервисом.
Где приобрести диплом специалиста?
https://wallazz.com/blogs/197141/?иплом-онлайн-ѕросто-удобно-быстро
https://ukrom.in.ua/users/300
https://apple.ibord.ru/viewtopic.php?id=14406
http://d91652pj.beget.tech/2023/10/30/bolshoy-assortiment-vysokokachestvennyh-diplomov-v-izvestnom-magazine.html
https://dogovor.top/wiki/%D0%92_%D0%BA%D0%B0%D0%BA%D0%BE%D0%BC_%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5_%D0%BB%D1%83%D1%87%D1%88%D0%B5_%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C_%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC_%D1%81%D0%B5%D0%B3%D0%BE%D0%B4%D0%BD%D1%8F%3F
Успехов в учебе!
Здравствуйте!
Для определенных людей, приобрести диплом ВУЗа – это необходимость, удачный шанс получить достойную работу. Но для кого-то – это желание не терять время на учебу в институте. С какой бы целью вам это не потребовалось, мы готовы помочь вам. Быстро, профессионально и недорого изготовим диплом любого ВУЗа и года выпуска на государственных бланках с реальными печатями.
Приобрести документ ВУЗа можно в нашей компании. Мы предлагаем документы об окончании любых университетов Российской Федерации. Вы сможете получить диплом по любым специальностям, включая документы СССР. Гарантируем, что в случае проверки документа работодателями, каких-либо подозрений не появится.
Превосходства наших дипломов:
• используются настоящие бланки “Гознак”;
• необходимые подписи должностных лиц;
• настоящие печати учебного заведения;
• специальные водяные знаки, нити и прочие степени защиты;
• безупречное качество оформления – ошибок не будет;
• любая проверка документа.
Мы предлагаем выгодно и быстро приобрести диплом, который выполнен на оригинальной бумаге и заверен печатями, штампами, подписями. Диплом пройдет лубую проверку, даже при использовании специфических приборов. Решайте свои задачи быстро с нашими дипломами.
Где заказать диплом по нужной специальности?
http://msfo-soft.ru/msfo/forum/messages/forum28/topic9959/message278494/?result=new#message278494
http://uaforum.ukrbb.net/viewtopic.php?f=11&t=1837&p=4764#p4764
http://kvitka.ukrbb.net/viewtopic.php?f=62&t=22635&p=41827#p41827
http://ractis.ru/forums/index.php?showtopic=5028
http://moskovskij.getbb.ru/viewtopic.php?f=12&t=2231&p=3184#p3184
Успешной учебы!
Привет!
Для определенных людей, приобрести диплом о высшем образовании – это острая необходимость, уникальный шанс получить достойную работу. Впрочем для кого-то – это понятное желание не терять время на учебу в ВУЗе. Что бы ни толкнуло вас на такой шаг, наша компания готова помочь вам. Максимально быстро, качественно и недорого изготовим диплом любого года выпуска на подлинных бланках со всеми необходимыми подписями и печатями.
Наша компания предлагает выгодно и быстро заказать диплом, который выполняется на бланке ГОЗНАКа и заверен печатями, водяными знаками, подписями. Наш диплом пройдет лубую проверку, даже при помощи специфических приборов. Достигайте своих целей максимально быстро с нашей компанией.
Основные преимущества наших документов:
• используются лишь качественные бланки “Гознака”;
• необходимые подписи должностных лиц;
• настоящие печати ВУЗа;
• водяные знаки, нити и иные степени защиты;
• идеальное качество оформления – ошибки полностью исключены;
• любая проверка документов.
Где заказать диплом специалиста?
http://mobix.com.ua/index.php?links_exchange=yes
Успехов в учебе!
[b]Привет, друзья[/b]!
Всегда считал, что покупка диплома о высшем образовании — это миф и невозможно. Но, к счастью, оказался неправ. Сначала искал информацию по теме: купить диплом тренера, купить диплом в елабуге, купить диплом в москве, купить диплом в магадане, #купить диплом в находке, а затем переключился на дипломы вузов. Подробности здесь: https://blogs.rufox.ru/~runoklisfgy/47401.htm
Оказалось, что все реально и легально, со специальными условиями и упрощенными программами. Теперь у меня диплом московского вуза нового образца, что я настоятельно рекомендую и вам!
Успехов в учебе!
Привет всем)
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: купить диплом в новочеркасске, купить диплом в ялте, купить диплом электрика, купить диплом бакалавра, купить диплом в казани, а потом наткнулся на https://obyava.org/moskva/search/uslugi/prochie-uslugi/hotite-najti-rabotu-svojej-mechty-5938612.html?from=my, где все мои учебные проблемы были решены!
Успехов в учебе!
Здравствуйте!
Наша компания предлагает выгодно заказать диплом, который выполнен на оригинальном бланке и заверен печатями, водяными знаками, подписями. Диплом пройдет лубую проверку, даже при использовании специально предназначенного оборудования. Решайте свои задачи быстро и просто с нашей компанией.
Где приобрести диплом по нужной специальности?
http://autoprajs.ru/15204.html
Хорошей учебы!
Привет всем)
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: купить диплом сварщика, купить диплом ветеринара, купить диплом в химках, купить диплом фармацевта, купить дипломы о высшем, а потом наткнулся на http://gzu.fr.free.fr/nuked_klan_sp43/nk/index.php?file=Members&op=detail&autor=uhipawyx
http://bangcara.blogspot.com/2017/02/cara-mendapatkan-backlink-mudah-dan-gratis.html
http://frichinvestigation.org/node/3914
http://www.sinoright.net/member/index.php?uid=utugyp&action=viewarchives&aid=5066
http://1e.kz/product/m3-plus-%d0%bc%d1%83%d0%b6%d1%81%d0%ba%d0%b8%d0%b5-%d1%81%d0%bc%d0%b0%d1%80%d1%82-%d1%87%d0%b0%d1%81%d1%8b-%d1%86%d0%b2%d0%b5%d1%82%d0%bd%d0%be%d0%b9-%d1%8d%d0%ba%d1%80%d0%b0%d0%bd-ip67-%d0%b2%d0%be/?unapproved=4083&moderation-hash=8cc40db0776bea7a9626d31ec6aac114#comment-4083
, где все мои учебные проблемы были решены!
Хорошей учебы!
미스터 플레이 슬롯
이 때문에 증권사의 소식은 모두 믿을 만하다.
Здравствуйте!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом в усть-илимске, купить диплом в кызыле, где купить диплом среднем, старые дипломы купить, купить диплом в кирово-чепецке. Постепенно углубляясь в тему, нашел отличный ресурс здесь: http://zarabotaem.bbmy.ru/viewtopic.php?id=3251#p5159
https://lineageii.rolka.me/viewtopic.php?id=542#p554
http://peklama.bbok.ru/viewtopic.php?id=12930#p24384
http://bestcoolfun.ru/kupit-diplom-rf-v-moskve-byistroe-oformlenie-i-konfidentsialnost
https://forum.shvedun.ru/ucp.php?mode=login
и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Успехов в учебе!
Привет Друзья!
Всегда думал что купить диплом о высшем образовании это миф и нереально, но все оказалось не так, изначально искал информацию про: купить диплом повара, купить диплом в люберцах, купить аттестат, купить диплом о высшем образовании, купить диплом в норильске, потом про дипломы вузов, подробнее здесь http://www.starterkit.ru/html/index.php?name=account&op=info&uname=etibufix
Оказалось все возможно, официально со специальными условия по упрощенным программам, так и сделал и теперь у меня есть диплом вуза Москвы нового образца, что советую и вам!
Хорошей учебы!
Добрый день!
Мы изготавливаем дипломы любой профессии по выгодным ценам.
Мы можем предложить документы техникумов, расположенных на территории всей РФ. Вы сможете заказать качественно напечатанный диплом от любого учебного заведения, за любой год, включая сюда документы старого образца. Дипломы и аттестаты выпускаются на “правильной” бумаге высшего качества. Это позволяет делать государственные дипломы, не отличимые от оригиналов. Документы заверяются всеми необходимыми печатями и подписями.
Где купить диплом специалиста?
https://phobia.lt/index.php?/gallery/image/121-%D1%88%D0%B8%D1%80%D0%BE%D0%BA%D0%B8%D0%B9-%D0%B2%D1%8B%D0%B1%D0%BE%D1%80-%D0%B4%D0%BE%D0%BA%D1%83%D0%BC%D0%B5%D0%BD%D1%82%D0%BE%D0%B2-%D0%B2-%D0%B7%D0%BD%D0%B0%D0%BC%D0%B5%D0%BD%D0%B8%D1%82%D0%BE%D0%BC-%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5/
KMSpico: What is it?
активатор офис на виндовс 11
Operating systems and Office suites are among the primary Microsoft software items that still need to be paid for. Some consumers may find alternate activation methods due to the perceived expensive cost of these items. There may be restrictions, unforeseen interruptions, and persistent activation prompts if these items are installed without being properly activated.
Our KMSpico app was created as a solution to this issue. By using this program, customers may access all of the functionality of Microsoft products and simplify the activation procedure.
KMSPico is a universal activator designed to optimize the process of generating and registering license codes for Windows and Office. Functionally, it is similar to key generators, but with the additional possibility of automatic integration of codes directly into the system. It is worth paying attention to the versatility of the tool, which distinguishes it from similar activators.
The above discussion primarily focused on the core KMS activator, the Pico app. Understanding what the program is, we can briefly mention KMSAuto, a tool with a simpler interface.
By using the KMSPico tool, you can setup Windows&Office for lifetime activation. This is an essential tool for anybody looking to reveal improved features and go beyond limitations. Although it is possible to buy a Windows or Office key.
KMSPico 11 (last update 2024) allows you to activate Windows 11/10/8 (8.1)/7 and Office 2021/2019/2016/2013/2010 for FREE.
buy accutane in mexico
KMSpico Download | Official KMS Website New July 2024
toolkit windows 10
Are you looking for the best tool to activate your Windows & Office? Then you should download and install KMSpico, as it is one of the best tools everyone should have. In this article, I will tell you everything about this fantastic tool, even though I will also tell you if this is safe to use.
In this case, don’t forget to read this article until the end, so you don’t miss any critical information. This guide is for both beginners and experts as I came up with some of the rumours spreading throughout the internet.
Perhaps before we move towards downloading or installing a section, we must first understand this tool. You should check out the guide below on this tool and how it works; if you already know about it, you can move to another section.
What is KMSPico?
KMPico is a tool that is used to activate or get a license for Microsft Windows as well as for MS Office. It was developed by one of the most famous developers named, Team Daz. However, it is entirely free to use. There is no need to purchase it or spend money downloading it. This works on the principle of Microsft’s feature named Key Management Server, a.k.a KMS (KMSPico named derived from it).
The feature is used for vast companies with many machines in their place. In this way, it is hard to buy a Windows License for each device,, which is why KMS introduced. Now a company has to buy a KMS server for them and use it when they can get a license for all their machines.
However, this tool also works on it, and similarly, it creates a server on your machine and makes it look like a part of that server. One thing different is that this tool only keeps the product activated for 180 days. This is why it keeps running on your machine, renews the license keys after 180 days, and makes it a permanent activation.
KMSAuto Net
Microsoft Toolkit
Windows Loader
Windows 10 Activator
Features
We already know what this tool means, so let’s talk about some of the features you are getting along with KMSPico. Reading this will surely help you understand whether you are downloading the correct file.
Ok, so here are some of the features that KMSPico provides:
Activate Windows & Office
We have already talked about this earlier, as using this tool, you will get the installation key for both Microsoft Products. Whether it is Windows or Office, you can get a license in no time; however, this supports various versions.
Supports Multi-Arch
Since this supports both products, it doesn’t mean you have to download separate versions for each arch. One version is enough, and you can get the license for both x32-bit or even the x64-bit.
It Is Free To Use
Undoubtedly, everything developed by Team Daz costs nothing to us. Similarly, using this tool won’t cost you either, as it is entirely free. Other than this, it doesn’t come with any ads, so using it won’t be any trouble.
Permanent License
Due to the KMS server, this tool installs on our PC, we will get the license key for the rest of our lives. This is because the license automatically renews after a few days. To keep it permanent, you must connect your machine to the internet once 180 days.
Virus Free
Now comes the main feature of this tool that makes it famous among others. KMSPico is 100% pure and clean from such viruses or trojans. The Virus Total scans it before uploading to ensure it doesn’t harm our visitors.
Добрый день!
Где купить диплом специалиста?
Мы можем предложить документы ВУЗов, расположенных в любом регионе России. Вы сможете приобрести качественный диплом от любого учебного заведения, за любой год, включая сюда документы старого образца. Дипломы и аттестаты выпускаются на бумаге самого высокого качества. Это дает возможность делать государственные дипломы, которые невозможно отличить от оригинала. Документы будут заверены необходимыми печатями и подписями.
https://landik-diploms-srednee.ru/diplom-sredne-spetcialnoe-obrazovanie
Добрый день!
Где купить диплом по необходимой специальности?
Мы изготавливаем дипломы любой профессии по невысоким ценам. Стоимость будет зависеть от выбранной специальности, года получения и образовательного учреждения.
Заказать диплом о высшем образовании
https://arusak-diploms-srednee.ru/kupit-diplom-v-omske
Успешной учебы!
Привет, друзья!
Где заказать диплом специалиста?
Заказать диплом ВУЗа.
Цена значительно меньше чем пришлось бы платить на очном обучении в ВУЗе
http://qbko.ru/index.php?subaction=userinfo&user=ijucif
Хорошей учебы!
best price for propecia 5mg online
Привет, друзья!
Где заказать диплом по актуальной специальности?
Полученный диплом со всеми печатями и подписями отвечает стандартам Министерства образования и науки, неотличим от оригинала. Не откладывайте собственные мечты и задачи на несколько лет, реализуйте их с нашей компанией – отправьте быструю заявку на изготовление диплома прямо сейчас!
Купить диплом любого ВУЗа.
http://ssd-astra.ru/forum/viewtopic.php?f=14&t=7021
http://renebiemans.nl/users.php?m=details&id=32281
http://m1bar.com/index.php?subaction=userinfo&user=uqehavyf
http://www.munhwahouse.or.kr/bbs/board.php?bo_table=free&wr_id=148869
http://www.lada-xray.net/showthread.php?p=32744#post32744
Успешной учебы!
Здравствуйте!
Задумался а действительно можно купить диплом государственного образца в Москве, и был удивлен, все реально и главное официально!
Сначала серфил в сети и искал такие темы как: купить диплом в бийске, купить диплом эколога, можно ли купить диплом, купить атестат об окончании школы, купить диплом в новокузнецке, получил базовую информацию.
Остановился в итоге на материале купить диплом в усолье-сибирском, http://vcemoivitvoryalki.blogspot.com/2015/09/blog-post.html
Успешной учебы!
Привет всем)
Хорошо быть студентом, пока не придет пора писать диплом, что и произошло со мной, но не стоит отчаиваться, ведь есть хорошие компании что помогают с написанием и сдачей диплома на хорошие оценки!
Изначально искал информацию про купить диплом в рыбинске, купить диплом в армавире, купить диплом в нижнем новгороде, купить диплом в ялте, купить диплом в гатчине, потом попал на https://maksak.blox.ua/2024/06/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%BE%D0%BA%D1%83%D0%BC%D0%B5%D0%BD%D1%82-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC.html и там решили все мои учебные заботы!
Успешной учебы!
Здравствуйте!
Где купить диплом по нужной специальности?
Заказать документ института вы сможете в нашей компании. Мы предлагаем документы об окончании любых университетов РФ. Вы получите необходимый диплом по любым специальностям, включая документы старого образца. Даем гарантию, что при проверке документа работодателем, каких-либо подозрений не возникнет.
http://windows1.listbb.ru/viewtopic.php?f=3&t=244
Удачи!
[u][b] Добрый день![/b][/u]
Где приобрести диплом по необходимой специальности?
Заказать документ ВУЗа вы имеете возможность в нашей компании в столице. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов РФ. Вы получите диплом по любой специальности, включая документы СССР. Даем 100% гарантию, что при проверке документов работодателями, никаких подозрений не появится.
[b]Мы предлагаем[/b] быстро и выгодно приобрести диплом, который выполняется на оригинальном бланке и заверен печатями, водяными знаками, подписями. Данный диплом способен пройти любые проверки, даже при использовании специального оборудования. Решайте свои задачи быстро и просто с нашим сервисом.
https://apple.ibord.ru/viewtopic.php?id=14463
http://ggshell.bestbb.ru/viewtopic.php?id=239
http://bukmeker.bbok.ru/viewtopic.php?id=23804
https://dnepr.actieforum.com/t7781-topic
https://www.anytalkworld.com/read-blog/2610
[u][b] Здравствуйте![/b][/u]
Где заказать диплом специалиста?
Мы предлагаем документы техникумов, расположенных в любом регионе России. Они будут заверены всеми обязательными печатями и штампами.
https://hyas-loan.biz/2024/06/08/Как-купить-диплом-о-высшем-образовании-и-не-попасться
[b]Успехов в учебе![/b]
[b]Здравствуйте[/b]!
Хочу рассказать о своем опыте по заказу аттестата пту, думал это не реально и стал искать информацию в сети, про купить диплом в кирове, купить диплом инженера электрика, купить диплом сантехника, купить диплом, купить диплом вуза, постепенно вникая в суть дела нашел отличный материал здесь http://nanatok.ru/katalog/mango_kids/
http://krasrec.ru/forum/user/192856/
http://pro-zakupki.ru/publ/dopolnitelnaja_nastrojka_capicom_2_1_0_2/1-1-0-11/
https://subaru-vlad.ru/forums/users/uxilaj/
и был очень доволен!
Теперь у меня есть диплом столяра о среднем специальном образовании, и я обеспечен на всю жизнь)
[b]Успехов в учебе![/b]
[b]Здравствуйте[/b]!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом продавца, купить диплом с занесением в реестр, купить диплом в мытищах, купить диплом в перми, купить диплом в бердске. Постепенно углубляясь в тему, нашел отличный ресурс здесь: https://poco-shop.ru/blog/obzor-poco-f4-gt-kamera-i-vozmozhnosti-smartfona/#comment_34826 и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
[b]Успешной учебы![/b]
Добрый день!
Где заказать диплом специалиста?
Мы изготавливаем дипломы любой профессии по выгодным ценам. Стоимость зависит от выбранной специальности, года выпуска и ВУЗа. Всегда стараемся поддерживать для клиентов адекватную ценовую политику.
https://lastdemo.primepix.ru/club/user/26712/blog/7066/
Успешной учебы!
[u][b] Привет![/b][/u]
Для некоторых людей, купить диплом университета – это необходимость, уникальный шанс получить достойную работу. Но для кого-то – это разумное желание не терять время на учебу в институте. С какой бы целью вам это не понадобилось, мы готовы помочь. Оперативно, качественно и выгодно сделаем диплом любого года выпуска на подлинных бланках с реальными печатями.
[b]Купить документ[/b] о получении высшего образования вы сможете у нас в столице. Мы предлагаем документы об окончании любых ВУЗов России. Вы сможете получить необходимый диплом по любой специальности, включая документы Советского Союза. Гарантируем, что в случае проверки документов работодателями, каких-либо подозрений не появится.
Превосходства наших документов:
• используем настоящие бланки “Гознак”;
• подлинные подписи должностных лиц;
• все печати университета;
• специальные водяные знаки, нити и прочие степени защиты;
• безупречное заполнение и оформление – ошибки полностью исключены;
• любые проверки оригинальности документа.
[b]Мы предлагаем[/b] выгодно заказать диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, штампами, подписями официальных лиц. Данный документ пройдет любые проверки, даже при помощи профессионального оборудования. Решайте свои задачи быстро и просто с нашим сервисом.
[b]Где приобрести диплом специалиста?[/b]
http://sportandpolitics.ukrbb.net/viewtopic.php?f=24&t=13633&p=24366#p24366
http://molbiol.ru/forums/index.php?showtopic=1192567
http://vtik.net/index.php?subaction=userinfo&user=imexi
http://mybaltika.info/ru/blogs/587/9938/
http://vrn.best-city.ru/forum/thread540115837/
[b]Хорошей учебы![/b]
[u][b] Привет![/b][/u]
Для некоторых людей, купить [b]диплом[/b] университета – это острая потребность, удачный шанс получить хорошую работу. Но для кого-то – это желание не терять время на учебу в универе. Что бы ни толкнуло вас на такой шаг, мы готовы помочь вам. Оперативно, качественно и по доступной цене сделаем диплом любого года выпуска на подлинных бланках с реальными печатями.
[b]Наши специалисты предлагают[/b] выгодно и быстро приобрести диплом, который выполняется на бланке ГОЗНАКа и заверен мокрыми печатями, водяными знаками, подписями. Наш диплом пройдет лубую проверку, даже с использованием специфических приборов. Решайте свои задачи быстро с нашей компанией.
[b]Основные преимущества наших документов:[/b]
• используем только качественные бланки “Гознака”;
• все подписи руководства;
• мокрые печати ВУЗа;
• специальные водяные знаки, нити и другие степени защиты;
• безупречное заполнение и оформление – ошибки полностью исключены;
• любые проверки документа.
[b]Где купить диплом по нужной специальности?[/b]
http://classicalmusicmp3freedownload.com/ja/index.php?title=??????_??????_?_??????_???????????_?_??????_??_??????_??????_?_?????????
[b]Успехов в учебе![/b]
[url=http://lasixtbs.online/]lasix generic pills[/url]
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф и невозможно. Но, к счастью, оказался неправ. Сначала искал информацию по теме: купить диплом магистра, купить диплом в алуште, купить диплом в димитровграде, купить диплом воспитателя, #купить диплом сварщика, а затем переключился на дипломы вузов. Подробности здесь: https://www.metal-tracker.com/forum/member.php?action=profile&uid=927394865
http://newsofgames.ru/zakazat-falshivyie-diplomyi
https://owen.ru/forum/member.php?u=106526
https://kritiki.forum.cool/viewtopic.php?id=5483#p14220
https://itstagram.ru/read-blog/191
Оказалось, что все реально и легально, со специальными условиями и упрощенными программами. Теперь у меня диплом московского вуза нового образца, что я настоятельно рекомендую и вам!
Успехов в учебе!
Здравствуйте!
Хочу рассказать о своем опыте по заказу аттестата пту, думал это не реально и стал искать информацию в сети, про купить диплом матроса, купить диплом в ессентуках, купить диплом высшее, купить диплом ссср, купить диплом в когалыме, постепенно вникая в суть дела нашел отличный материал здесь http://centropol.su/index.php?subaction=userinfo&user=ovujupip и был очень доволен!
Теперь у меня есть диплом столяра о среднем специальном образовании, и я обеспечен на всю жизнь)
Удачи!
[u][b] Привет![/b][/u]
Где заказать диплом специалиста?
[b]Купить документ[/b] о получении высшего образования можно в нашем сервисе. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации. Вы сможете получить необходимый диплом по любой специальности, включая документы Советского Союза. Гарантируем, что при проверке документов работодателем, каких-либо подозрений не возникнет.
http://www.tamboff.ru/forum/viewtopic.php?t=257991
[b]Хорошей учебы![/b]
에그슬롯
교과서에 강한 흔적을 남기기에 충분한 전술의 표본일 뿐이다.
Привет Друзья!
Всегда думал что купить диплом о высшем образовании это миф и нереально, но все оказалось не так, изначально искал информацию про: купить диплом в томске, купить диплом в новочеркасске, купить диплом в сыктывкаре, купить диплом в таганроге, купить диплом в гатчине, потом про дипломы вузов, подробнее здесь http://realtyintellect.ru/kupit-diplom-prostota-i-udobstvo-oformleniya
https://rabota24net.mybb.ru/viewtopic.php?id=426#p439
http://wiki.ecusystems.ru/index.php/??????_???????_?????,_??????????_?_??????_??_????????????_??????
http://study.flybb.ru/topic217.html
http://thesleepinghusband.rolka.me/viewtopic.php?id=11976#p39127
Оказалось все возможно, официально со специальными условия по упрощенным программам, так и сделал и теперь у меня есть диплом вуза Москвы нового образца, что советую и вам!
Успехов в учебе!
[b]Привет, друзья[/b]!
Всегда считал, что покупка диплома о высшем образовании — это миф и невозможно. Но, к счастью, оказался неправ. Сначала искал информацию по теме: купить аттестат за 9 класс, купить свидетельство о рождении ссср, купить диплом в озёрске, купить диплом в воронеже, #купить дипломы о высшем, а затем переключился на дипломы вузов. Подробности здесь: http://luxgel.ru/blog/testirovanie-bloga-4#comment_1612028
https://www.hirchenhain-erlensee.de/startseite/img_3427/#comment-103179
https://acordxlaindependencia.cat/acord-per-la-independencia-suma-dues-noves-entitats-a-la-seva-proposta/#comment-4028
https://kleine-auszeit-entspannung.de/hello-world/#comment-47422
http://glamour-24.ru/blog/vstrechajte-vesnu-vmeste-s-GLAMOUR-24
Оказалось, что все реально и легально, со специальными условиями и упрощенными программами. Теперь у меня диплом московского вуза нового образца, что я настоятельно рекомендую и вам!
Хорошей учебы!
Привет!
Наши специалисты предлагают выгодно и быстро приобрести диплом, который выполняется на оригинальном бланке и заверен мокрыми печатями, водяными знаками, подписями. Документ пройдет лубую проверку, даже при помощи профессиональных приборов. Достигайте свои цели максимально быстро с нашей компанией.
Где купить диплом специалиста?
http://onlaintelevizia.listbb.ru/viewtopic.php?f=2&t=426
Успехов в учебе!
Студия Подкастов в Москве, Запись подкастов в Москве.
http://video-podcast.ru/
Greetings I am so thrilled I found your blog, I really found you by accident, while I was looking on Google for something else, Anyways I am here now and would just like to say cheers for a marvelous post and a all round enjoyable blog (I also love the theme/design), I don’t have time to read it all at the moment but I have book-marked it and also included your RSS feeds, so when I have time I will be back to read much more, Please do keep up the fantastic work.
https://practice-legacy.com/which-is-the-best-site-app-to-nudify-a-picture/
Здравствуйте!
Где заказать диплом по нужной специальности?
Мы можем предложить документы техникумов, которые находятся на территории всей РФ. Можно заказать качественно сделанный диплом за любой год, включая сюда документы СССР. Дипломы и аттестаты печатаются на “правильной” бумаге высшего качества. Это дает возможность делать государственные дипломы, которые невозможно отличить от оригиналов. Документы будут заверены всеми обязательными печатями и подписями.
dopul.ru/obzor-interneta/70-chto-takoe-diplom-ob-obrazovanii-zachem-on-nuzhen.html
Привет, друзья!
Мы можем предложить дипломы любой профессии по приятным ценам.
Мы предлагаем документы ВУЗов, расположенных в любом регионе РФ. Можно заказать качественный диплом за любой год, включая сюда документы старого образца. Дипломы и аттестаты печатаются на бумаге высшего качества. Это позволяет делать государственные дипломы, которые не отличить от оригиналов. Документы заверяются всеми необходимыми печатями и штампами.
Где приобрести диплом специалиста?
ractis.ru/forums/index.php?autocom=gallery&req=si&img=2108
[b]Здравствуйте[/b]!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом стоматолога, купить диплом в каспийске, купить аттестат школы, купить диплом в ханты-мансийске, купить диплом бакалавра и получил базовые знания. В итоге остановился на материале: http://rivertravel.net/viewtopic.php?f=21&t=8009
https://www.apartmani-drgasasokobanja.rs/business/strategy-development-for-clients/#comment-29544
https://kms-zdrav.ru/products/poruchen-stalnoj-pt73524/#comment_149447
https://click2connect.community/index.php?do=/public/blog/view/id_119669/title_/
https://www.knittingideas.ru/posts/add
Хорошей учебы!
Аренда телесуфлеров в Москве, Аренда и обслуживание телесуфлеров в Москвеhttps://xn--e1aaawb5aeenk.xn--p1ai/
토토 메이저
그래서… 그는 손을 등 뒤로 돌리고 입술을 깨물고 아무 말도 하지 않았습니다.
Здравствуйте!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом в березниках, купить аттестат школы, купить диплом о высшем образовании, купить диплом в уфе, купить диплом в соликамске и получил базовые знания. В итоге остановился на материале: http://www.club-kia.com/forum/thread30748.html
https://wiuwi.com/blogs/94104/??????-?-????-????-?-???-?????-?-??????
https://wiuwi.com/blogs/101776/??????????-???????????-????-??????-?-????????-?????-?????-???????
http://www.paladiny.ru/forummess.dwar.php?TopicID=27082
http://diablomania.ru/forum/showthread.php?p=560890#post560890
Успешной учебы!
Здравствуйте!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом журналиста, купить диплом с реестром, купить диплом сварщика, купить диплом образцы , купить диплом в новошахтинске и получил базовые знания. В итоге остановился на материале: https://re-port.ru/users/52062/
Хорошей учебы!
В чёрный список пирамид и лохотронов 31 августа 2022 года внесены следующие организации, обладающие признаками финансовой пирамиды или признаками мошенничества.
русское порно жесток
Etihad Rail (etihadrail-ae.com)
Club Unite To Live, Life Is Good, Unite To Live (uniteto.live). Новый домен пирамиды находящегося в розыске Романа Василенко.
TopBoom, «платформа хедж-фонда, специализирующегося на сделках с обратным исходом спортивного события» (topboom.vip, t.me/TopBoom001)
Change Team, мошенники указывают реквизиты чужого юрлица ООО «АТТИС ГРУПП» (ОРГН 1207700314128, ИНН 9702022065), телеграм-канал «Change Team: Главный Канал» (change-team.ru). Фальшивый обменник раньше назывался C Exchange.
Ещё лохотроны:
Scam! Чёрный список пирамид и лохотронов и отзывы
В чёрный список пирамид и лохотронов включены организации, имеющие признаки финансовой пирамиды по классификации Центрального банка; организации, обладающие признаками матричной пирамиды (массовый привод друзей, «столы»); сборы денег с пенсионеров и прочих незащищённых слоёв населения; хайпы; организации, имеющие негативные отзывы; структуры, оказывающие финансовые услуги без лицензии и прочие лохотроны за исключением псевдоброкеров, кооперативов (с ними тоже не рекомендуем связываться) и некоторых других категорий.
В чёрный список пирамид и лохотронов 31 августа 2022 года внесены следующие организации, обладающие признаками финансовой пирамиды или признаками мошенничества.
гей порно видео
Etihad Rail (etihadrail-ae.com)
Club Unite To Live, Life Is Good, Unite To Live (uniteto.live). Новый домен пирамиды находящегося в розыске Романа Василенко.
TopBoom, «платформа хедж-фонда, специализирующегося на сделках с обратным исходом спортивного события» (topboom.vip, t.me/TopBoom001)
Change Team, мошенники указывают реквизиты чужого юрлица ООО «АТТИС ГРУПП» (ОРГН 1207700314128, ИНН 9702022065), телеграм-канал «Change Team: Главный Канал» (change-team.ru). Фальшивый обменник раньше назывался C Exchange.
Ещё лохотроны:
Scam! Чёрный список пирамид и лохотронов и отзывы
В чёрный список пирамид и лохотронов включены организации, имеющие признаки финансовой пирамиды по классификации Центрального банка; организации, обладающие признаками матричной пирамиды (массовый привод друзей, «столы»); сборы денег с пенсионеров и прочих незащищённых слоёв населения; хайпы; организации, имеющие негативные отзывы; структуры, оказывающие финансовые услуги без лицензии и прочие лохотроны за исключением псевдоброкеров, кооперативов (с ними тоже не рекомендуем связываться) и некоторых других категорий.
Здравствуйте!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом в междуреченске, купить диплом фитнес инструктора, купить диплом в чите, купить диплом в березниках , купить диплом в киселевске и получил базовые знания. В итоге остановился на материале: http://koreamuseum.ru/index.php?subaction=userinfo&user=yhykad
Успехов в учебе!
Привет!
Где заказать диплом специалиста?
Покупка диплома университета России в нашей компании – надежный процесс, так как документ заносится в государственный реестр. Печать производится на специальных бланках, установленных государством.
Вы приобретаете диплом через надежную и проверенную фирму.
http://kurgetrp.listbb.ru/viewtopic.php?f=11&t=380
Удачи!
Добрый день!
Где приобрести диплом по необходимой специальности?
Готовый диплом со всеми печатями и подписями 100% отвечает требованиям и стандартам, никто не сумеет отличить его от оригинала – даже со специальным оборудованием. Не следует откладывать личные мечты на пять лет, реализуйте их с нами – отправьте простую заявку на изготовление документа уже сегодня!
Приобрести диплом университета.
https://nasvyazi.space/showthread.php?tid=71474
http://unqbit.org/bbs/board.php?bo_table=free&wr_id=5670
https://www.agrosoft.ru/support/forum/index.php?PAGE_NAME=profile_view&UID=85478
http://heimur.ru/index.php?/topic/131-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-t974i/
http://school5.p-fam.ru/users/apemov
Успехов в учебе!
Здравствуйте!
Где купить диплом специалиста?
Приобрести диплом университета.
Стоимость значительно меньше нежели довелось бы заплатить на очном обучении в ВУЗе
http://kladoiskatel.asia/viewtopic.php?f=40&t=13723
Успехов в учебе!
Привет, друзья!
Где купить диплом специалиста?
Мы предлагаем дипломы любых профессий по приятным ценам. Стоимость будет зависеть от выбранной специальности, года получения и ВУЗа.
Купить диплом ВУЗа
https://landik-diploms-srednee.ru/svidetelstvo-o-razvode
Удачи!
Привет всем)
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: купить диплом россия, купить диплом россия, купить диплом в кирово-чепецке, купить диплом сварщика, купить диплом в ханты-мансийске, а потом наткнулся на https://paranormal-terbaik.com/solusi-pasangan-selingkuh-2/
http://share.psiterror.ru/2024/05/18/diplom-instituta-dlya-specialistov.html
https://telegra.ph/Prodazha-diplomov-o-vysshem-obrazovanii-03-30
http://fujimoto-co-ltd.com/s02-300×225/#comment-10180
https://www.tumblr.com/leolimch/746829870786543616/где-недорого-купить-диплом
, где все мои учебные проблемы были решены!
Успехов в учебе!
Здравствуйте!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом в владикавказе, купить диплом в великих луках, купить диплом в сарове, купить диплом ссср, купить диплом в южно-сахалинске. Постепенно углубляясь, нашел отличный ресурс здесь: http://www.miranetwork.it/index.php?option=com_k2&view=itemlist&task=user&id=434690
http://favorit-yug.ru/products/faltsevyj-vkladysh-rehaumontblancbrusbox-730/
http://fancyrat.ru
https://tdinox.ru/trubnyj-prokat/truba-pryamougolnaya-elektrosvarnaya/truba-pryamougolnaya-nerzhaveyushchaya-matovaya-din-2395-aisi-304-60-x-40-x-6000-x-1-5-mm/
, и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Успехов в учебе!
Здравствуйте!
Купить диплом университета.
Наш сервис предлагает заказать диплом в высоком качестве, который невозможно отличить от оригинального документа без использования дорогостоящего оборудования и опытного специалиста.
Где купить диплом по необходимой специальности?
http://www.ivedu.ru/forum/viewthread.php?forum_id=18&thread_id=47353
Удачи!
[u][b] Привет, друзья![/b][/u]
Где заказать диплом по необходимой специальности?
Покупка диплома ВУЗа РФ в нашей компании – надежный процесс, ведь документ [b]будет заноситься в государственный реестр[/b]. При этом печать осуществляется на фирменных бланках, установленных государством.
Вы заказываете диплом [b]через надежную фирму.[/b]
http://justvoip.listbb.ru/posting.php?mode=post&f=2&sid=152e88aabd372eda7b64efe2e31c0d83
[b]Удачи![/b]
[u][b] Здравствуйте![/b][/u]
Мы готовы предложить дипломы любых профессий по приятным тарифам.
Мы предлагаем документы техникумов, расположенных в любом регионе Российской Федерации. Можно приобрести диплом от любого заведения, за любой год, указав подходящую специальность и оценки за все дисциплины. Дипломы и аттестаты выпускаются на “правильной” бумаге самого высокого качества. Это позволяет делать государственные дипломы, не отличимые от оригиналов. Документы заверяются всеми требуемыми печатями и подписями.
Даем гарантию, что при проверке документов работодателями, каких-либо подозрений не появится.
[b]Где купить диплом специалиста?[/b]
[url=https://www.adrex.com/en/forum/climbing/?view=new-theme]student-news.ru/kak-poluchit-diplom-bez-uchebyi-polnoe-rukovodstvo[/url]
Привет!
Где приобрести диплом по нужной специальности?
Приобрести документ университета вы можете в нашем сервисе. Мы предлагаем документы об окончании любых университетов Российской Федерации. Вы получите необходимый диплом по любым специальностям, любого года выпуска, в том числе документы СССР. Даем 100% гарантию, что при проверке документа работодателями, подозрений не появится.
Наши специалисты предлагают быстро заказать диплом, который выполнен на оригинальном бланке и заверен печатями, штампами, подписями. Данный документ способен пройти любые проверки, даже с использованием специально предназначенного оборудования. Достигайте цели быстро и просто с нашим сервисом.
http://russianecuador.com/forum/group.php?&do=discuss&groupid=14&discussionid=&gmid=462
https://firstamendment.tv/read-blog/42192
http://sampnl.listbb.ru/viewtopic.php?f=3&t=388
http://wiki.gptel.ru/index.php/%D0%93%D0%B4%D0%B5_%D0%BB%D1%83%D1%87%D1%88%D0%B5_%D0%BF%D1%80%D0%B8%D0%BE%D0%B1%D1%80%D0%B5%D1%81%D1%82%D0%B8_%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%3F_%D0%A1%D0%BE%D0%B2%D0%B5%D1%82%D1%8B_%D1%81%D0%BF%D0%B5%D1%86%D0%B8%D0%B0%D0%BB%D0%B8%D1%81%D1%82%D0%B0
http://o91746bp.beget.tech/2024/03/02/kak-seychas-podobrat-nadezhnyy-onlayn-magazin-s-diplomami.html
Здравствуйте!
Купить документ университета можно в нашем сервисе. Мы оказываем услуги по продаже документов об окончании любых ВУЗов РФ. Вы сможете получить необходимый диплом по любой специальности, включая документы старого образца. Даем гарантию, что при проверке документа работодателем, подозрений не возникнет.
Мы предлагаем выгодно и быстро купить диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями. Документ способен пройти лубую проверку, даже с применением специфических приборов. Решите свои задачи быстро и просто с нашей компанией.
Где заказать диплом по актуальной специальности?
https://www.sitebs.ru/blogs/101845.html
Успешной учебы!
[u][b] Привет, друзья![/b][/u]
Для некоторых людей, приобрести [b]диплом[/b] ВУЗа – это острая потребность, уникальный шанс получить отличную работу. Однако для кого-то – это понятное желание не терять огромное количество времени на учебу в ВУЗе. С какой бы целью вам это не потребовалось, мы готовы помочь вам. Максимально быстро, качественно и недорого изготовим диплом любого ВУЗа и года выпуска на настоящих бланках со всеми требуемыми печатями.
[b]Наша компания предлагает[/b] быстро приобрести диплом, который выполнен на оригинальной бумаге и заверен печатями, штампами, подписями официальных лиц. Документ способен пройти любые проверки, даже с использованием специфических приборов. Достигайте своих целей быстро и просто с нашими дипломами.
[b]Где купить диплом по необходимой специальности?[/b]
http://opengadjet.ru/poluchite-ofitsialnyiy-dokument-vasha-garantiya-uspeshnogo-budushhego
[b]Удачи![/b]
Добрый день!
Где заказать диплом по необходимой специальности?
Купить документ о получении высшего образования вы сможете у нас в столице. Мы предлагаем документы об окончании любых университетов России. Вы сможете получить необходимый диплом по любым специальностям, включая документы Советского Союза. Даем гарантию, что в случае проверки документов работодателем, подозрений не появится.
http://antmc.pl/forum/member.php?action=profile&uid=54798
Хорошей учебы!
Здравствуйте!
Где заказать диплом специалиста?
Приобрести документ института можно в нашей компании в Москве. Мы предлагаем документы об окончании любых ВУЗов РФ.
Мы готовы предложить дипломы любых профессий по разумным тарифам. Стоимость зависит от выбранной специальности, года выпуска и образовательного учреждения. Всегда стараемся поддерживать для клиентов адекватную ценовую политику.
https://rabotavinternete.forum2x2.ru/t55296-topic#137291
Хорошей учебы!
[u][b] Здравствуйте![/b][/u]
Где купить диплом по нужной специальности?
[b]Приобрести документ[/b] института вы можете в нашем сервисе. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов России. Вы сможете получить необходимый диплом по любой специальности, любого года выпуска, в том числе документы СССР. Гарантируем, что при проверке документов работодателем, подозрений не возникнет.
http://dogsvscats.forum.cool/viewtopic.php?id=717#p1396
[b]Успешной учебы![/b]
맥심 슬롯
“오늘 덩 형제님이 어떤 조언을 하셨는지 모르겠습니다.” 왕진위안이 부드럽게 물었다.
Здравствуйте!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом сварщика, купить диплом в северодвинске, купить диплом в абакане, купить диплом в балаково, купить диплом биолога. Постепенно углубляясь в тему, нашел отличный ресурс здесь: http://uralturist.blogspot.com/2015/04/blog-post_22.html и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Успешной учебы!
Здравствуйте!
Хочу рассказать о своем опыте по заказу аттестата пту, думал это не реально и стал искать информацию в сети, про купить аттестат за 9 класс, купить диплом в салавате, купить диплом в новокузнецке, купить диплом о высшем образовании, купить диплом инженера, постепенно вникая в суть дела нашел отличный материал здесь http://mega-zaim96.ru/forum/user/35124/ и был очень доволен!
Теперь у меня есть диплом столяра о среднем специальном образовании, и я обеспечен на всю жизнь)
Хорошей учебы!
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф и невозможно. Но, к счастью, оказался неправ. Сначала искал информацию по теме: купить диплом во владивостоке, купить диплом в новочеркасске, купить диплом с внесением в реестр, купить диплом в кисловодске, #купить диплом отзывы, а затем переключился на дипломы вузов. Подробности здесь: http://www.recipemashups.com/2009/01/02/1665/comment-page-16949/#comment-2739719
http://www.speicher-photovoltaik.de/profile/?u=28610
https://xn--80aeccht8cge.xn--p1ai/content/diplomaasx
http://lyxondebian.free.fr/dokutest/doku.php?id=landsdiploms
Оказалось, что все реально и легально, со специальными условиями и упрощенными программами. Теперь у меня диплом московского вуза нового образца, что я настоятельно рекомендую и вам!
Удачи!
Здравствуйте!
Где заказать диплом по актуальной специальности?
Заказать документ о получении высшего образования вы сможете в нашей компании в Москве. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации. Вы сможете получить необходимый диплом по любой специальности, включая документы образца СССР. Гарантируем, что при проверке документа работодателями, каких-либо подозрений не возникнет.
Наши специалисты предлагают выгодно купить диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями официальных лиц. Наш диплом способен пройти любые проверки, даже при помощи специального оборудования. Достигайте свои цели быстро и просто с нашим сервисом.
http://hotmeets.ru/blogs/10/%D0%9E%D1%81%D0%BD%D0%BE%D0%B2%D0%BD%D1%8B%D0%B5-%D0%BF%D1%80%D0%B8%D1%87%D0%B8%D0%BD%D1%8B-%D0%BF%D0%BE%D1%87%D0%B5%D0%BC%D1%83-%D0%BE%D1%87%D0%B5%D0%BD%D1%8C-%D0%BC%D0%BD%D0%BE%D0%B3%D0%B8%D0%B5-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D1%8B%D0%B2%D0%B0%D1%8E%D1%82-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D1%8B-%D0%B2-%D1%81%D0%B5%D1%82%D0%B8
http://physmathforum.flybb.ru/topic411.html
http://klinlife.ru/forums/index.php?/topic/11587-kupit-diplom-vash-put-k-uspehu/
https://ogorodland.ru/forum/topic/diplom-kakih-imenno-vuzov-sejchas-vozmozhno-budet-kupit/
http://moscowurban.getbb.ru/viewtopic.php?f=12&t=785
Всем привет)
Будучи студентом, я наслаждался учебой до тех пор, пока не пришло время писать диплом. Но паниковать не стоило, ведь существуют компании, которые помогают с написанием и защитой диплома на отличные оценки!
Изначально я искал информацию по теме: купить диплом в ессентуках, купить технический диплом, купить диплом в рубцовске, купить диплом в черкесске, купить диплом психолога, затем наткнулся на http://www.profirms.ru/%D1%88%D0%B0%D1%85%D1%82%D0%BD%D1%8B%D0%B5-%D1%8D%D0%BB%D0%B5%D0%BA%D1%82%D1%80%D0%BE%D0%B2%D0%BE%D0%B7%D1%8B-%D0%BE%D0%BE%D0%BE-%E2%80%9C%D0%B7%D0%B0%D0%B2%D0%BE%D0%B4-%E2%80%9C%D0%B0%D0%BC%D0%BF%D0%BB%D0%B8%D1%82%D1%83%D0%B4%D0%B0%E2%80%9D/, где все мои учебные вопросы были решены!
Успешной учебы!
Здравствуйте!
Заказать диплом университета.
Наша компания предлагает приобрести диплом в высочайшем качестве, который невозможно отличить от оригинального документа без участия специалистов высокой квалификации с дорогим оборудованием.
Где приобрести диплом специалиста?
http://avva.getbb.ru/ucp.php?mode=login
Успехов в учебе!
As the admin of this web site is working, no hesitation very shortly it will be famous,
due to its feature contents.
What is explainer video cost in india https://en.99designs.pt/profiles/8076816/about
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф и невозможно. Но, к счастью, оказался неправ. Сначала искал информацию по теме: купить диплом в элисте, купить диплом менеджера, купить диплом кандидата наук, купить диплом в армавире, #купить диплом в артеме, а затем переключился на дипломы вузов. Подробности здесь:
http://blackpearlbasketball.com.au/index.php?option=com_k2&view=itemlist&task=user&id=829712
http://silkhunter.com/index.php?title=russianydiplomans
https://amazonecotour.com/2019/03/27/hello-world/#comment-4995
http://firefox27.blogspot.com/2015/12/blog-post_10.html
http://188.17.148.172/forum/profile.php?action=show&member=6829
Оказалось, что все реально и легально, со специальными условиями и упрощенными программами. Теперь у меня диплом московского вуза нового образца, что я настоятельно рекомендую и вам!
Удачи!
Здравствуйте!
Где купить диплом специалиста?
Полученный диплом с приложением отвечает стандартам Министерства образования и науки, никто не отличит его от оригинала. Не откладывайте собственные цели на потом, реализуйте их с нами – отправляйте простую заявку на диплом сегодня!
Купить диплом о высшем образовании.
https://amvnews.ru/forum/posting.php?mode=newtopic&f=44
http://forum.kia-club.ru/viewtopic.php?f=372&t=103463
http://www.igram.net/index.php?name=Account&op=userinfo&user_name=ipeke
http://redhawkclan.free.fr/index.php?file=Members&op=detail&autor=inavera
http://v208870d.bget.ru/index.php?subaction=userinfo&user=ucyviq
Успехов в учебе!
Добрый день!
Где заказать диплом по актуальной специальности?
Купить диплом университета.
Стоимость во много раз меньше нежели пришлось бы платить на очном и заочном обучении в ВУЗе
https://ya-hz.ru/index.php?/forums/topic/1363-%D0%BA%D0%B0%D0%BA-%D0%BB%D0%BE%D0%B6%D0%BD%D1%8B%D0%B5-%D0%B4%D0%BE%D0%B3%D0%BE%D0%B2%D0%BE%D1%80%D0%BD%D1%8B%D0%B5-%D0%BC%D0%B0%D1%82%D1%87%D0%B8-%D0%BC%D0%BE%D0%B6%D0%B5%D1%82-%D1%81%D1%8D%D0%BA%D0%BE%D0%BD%D0%BE%D0%BC%D0%B8%D1%82%D1%8C-%D0%B2%D1%80%D0%B5%D0%BC%D1%8F-%D1%81%D1%82%D1%80%D0%B5%D1%81%D1%81-%D0%B8-%D0%B4%D0%B5%D0%BD%D1%8C%D0%B3%D0%B8/#comment-3106
Удачи!
Добрый день!
Купить диплом ВУЗа.
Наша компания предлагает купить диплом в отличном качестве, неотличимый от оригинального документа без использования дорогостоящего оборудования и опытного специалиста.
Где приобрести диплом по нужной специальности?
https://obrezanie05.ru/users/15
Хорошей учебы!
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф и невозможно. Но, к счастью, оказался неправ. Сначала искал информацию по теме: купить диплом оценщика, купить диплом в северодвинске, купить диплом в евпатории, купить диплом в тольятти, #купить диплом в улан-удэ, а затем переключился на дипломы вузов. Подробности здесь: https://www.tandem.edu.co/cumpleanos/#comment-633508
Оказалось, что все реально и легально, со специальными условиями и упрощенными программами. Теперь у меня диплом московского вуза нового образца, что я настоятельно рекомендую и вам!
Успехов в учебе!
Привет!
Где заказать диплом по необходимой специальности?
Заказать документ университета вы имеете возможность в нашем сервисе. Мы предлагаем документы об окончании любых университетов России.
Мы изготавливаем дипломы любых профессий по выгодным ценам. Цена будет зависеть от конкретной специальности, года получения и образовательного учреждения. Всегда стараемся поддерживать для клиентов адекватную политику цен.
http://womenforum.7bb.ru/viewtopic.php?id=4642#p11336
Удачи!
Добрый день!
Где заказать диплом специалиста?
Заказать документ университета можно в нашей компании в столице. Мы предлагаем документы об окончании любых университетов РФ. Вы получите диплом по любым специальностям, любого года выпуска, в том числе документы образца СССР. Гарантируем, что при проверке документов работодателем, подозрений не появится.
http://moscityservice.ru/
Удачи!
Здравствуйте!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом в иркутске, купить диплом в москве, купить диплом повара, купить диплом в грозном, купить диплом в новошахтинске и получил базовые знания. В итоге остановился на материале: https://schoolido.lu/user/diplomyland9/
http://forumjustwoman.getbb.ru/viewtopic.php?f=43&t=860
http://thecounterculturewebisodes.com/#comment-29211
https://synapse.koreamed.org/advanced/index.php
http://whitepower.clanweb.eu/profile.php?lookup=23463
Удачи!
Здравствуйте!
Где заказать диплом специалиста?
Мы предлагаем документы техникумов, которые расположены в любом регионе Российской Федерации. Вы имеете возможность купить диплом за любой год, указав подходящую специальность и оценки за все дисциплины. Дипломы и аттестаты выпускаются на “правильной” бумаге высшего качества. Это дает возможность делать государственные дипломы, не отличимые от оригинала. Они заверяются необходимыми печатями и штампами.
forum.trackbase.net/members/21965-sonnick84?vmid=2440
Всем привет)
Будучи студентом, я наслаждался учебой до тех пор, пока не пришло время писать диплом. Но паниковать не стоило, ведь существуют компании, которые помогают с написанием и защитой диплома на отличные оценки!
Изначально я искал информацию по теме: купить диплом в брянске, купить диплом фитнес инструктора, купить диплом в елабуге, купить диплом в астрахани, купить диплом в рыбинске, затем наткнулся на http://army.clanfm.ru/viewtopic.php?f=2&t=38201, где все мои учебные вопросы были решены!
Удачи!
Добрый день!
Купить документ университета можно у нас. Мы оказываем услуги по производству и продаже документов об окончании любых ВУЗов России. Вы сможете получить необходимый диплом по любым специальностям, любого года выпуска, в том числе документы образца СССР. Гарантируем, что в случае проверки документов работодателями, подозрений не появится.
Мы предлагаем максимально быстро купить диплом, который выполнен на оригинальном бланке и заверен печатями, водяными знаками, подписями официальных лиц. Наш диплом пройдет любые проверки, даже при использовании специфических приборов. Достигайте цели максимально быстро с нашей компанией.
Где заказать диплом по нужной специальности?
http://suzukiforum.lv/index.php?subaction=userinfo&user=ozibidi
Успешной учебы!
Привет!
Где заказать диплом специалиста?
Приобрести документ ВУЗа можно у нас. Мы оказываем услуги по продаже документов об окончании любых ВУЗов РФ. Вы сможете получить необходимый диплом по любой специальности, любого года выпуска, в том числе документы СССР. Даем гарантию, что в случае проверки документов работодателем, никаких подозрений не возникнет.
http://forum.zub-zub.ru/viewtopic.php?t=25938
Удачи!
Привет, друзья!
Где купить диплом специалиста?
Заказать документ института вы можете в нашей компании в столице. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов России.
Мы изготавливаем дипломы любых профессий по приятным тарифам. Цена может зависеть от выбранной специальности, года получения и образовательного учреждения. Стараемся поддерживать для покупателей адекватную политику тарифов.
https://orgi.biz/org_diplom_moskva_
Удачи!
[u][b] Добрый день![/b][/u]
Мы готовы предложить дипломы любых профессий по разумным тарифам.
Мы готовы предложить документы ВУЗов, которые находятся в любом регионе РФ. Можно приобрести качественный диплом от любого учебного заведения, за любой год, указав подходящую специальность и хорошие оценки за все дисциплины. Документы выпускаются на “правильной” бумаге высшего качества. Это дает возможность делать государственные дипломы, не отличимые от оригиналов. Документы заверяются всеми требуемыми печатями и подписями.
[b]Где купить диплом специалиста?[/b]
[url=http://www.buscovivienda.net/foro/viewtopic.php?p=12912]www.buscovivienda.net/foro/viewtopic.php?p=12912[/url]
[u][b] Здравствуйте![/b][/u]
Где приобрести диплом специалиста?
Приобретение диплома любого университета РФ у нас – надежный процесс, потому что документ [b]заносится в реестр[/b]. При этом печать выполняется на фирменных бланках ГОЗНАКа.
Вы приобретаете документ [b]в надежной и проверенной компании.[/b]
http://cozy.moibb.ru/posting.php?mode=post&f=31
[b]Хорошей учебы![/b]
[b]Привет Друзья[/b]!
Всегда думал что купить диплом о высшем образовании это миф и нереально, но все оказалось не так, изначально искал информацию про: купить диплом о среднем образовании, купить диплом в арзамасе, купить аттестат за 11 класс, купить диплом медбрата, купить диплом в елабуге, потом про дипломы вузов, подробнее здесь http://mercenairesdelaforce.free.fr/index.php?file=Members&op=detail&autor=anygubik
Оказалось все возможно, официально со специальными условия по упрощенным программам, так и сделал и теперь у меня есть диплом вуза Москвы нового образца, что советую и вам!
[b]Хорошей учебы![/b]
[b]Всем привет[/b])
Будучи студентом, я наслаждался учебой до тех пор, пока не пришло время писать диплом. Но паниковать не стоило, ведь существуют компании, которые помогают с написанием и защитой диплома на отличные оценки!
Изначально я искал информацию по теме: купить диплом в химках, купить диплом образцы, купить диплом в керчи, купить диплом биолога, купить диплом врача, затем наткнулся на
http://mkala-koncert.ru/users/ilikixov
http://ziraffik.ru/product/platie-dp-a0550b/
http://school5.p-fam.ru/users/ufymuvi
https://rf-isolation.ru/index.php?/topic/727-event%C2%A0%D1%80%D0%B5%D0%BB%D0%B8%D0%BA%D1%82-5-25%D0%BA-gp/#comment-4246
http://www.youngwooapt.co.kr/bbs/board.php?bo_table=free&wr_id=333886
, где все мои учебные вопросы были решены!
[b]Успешной учебы![/b]
Привет!
Для определенных людей, купить диплом о высшем образовании – это необходимость, удачный шанс получить выгодную работу. Но для кого-то – это банальное желание не терять множество времени на учебу в универе. Что бы ни толкнуло вас на такой шаг, мы готовы помочь. Быстро, профессионально и выгодно сделаем документ нового или старого образца на подлинных бланках с реальными подписями и печатями.
Наша компания предлагает выгодно приобрести диплом, который выполнен на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями должностных лиц. Документ пройдет лубую проверку, даже при помощи специфических приборов. Достигайте свои цели быстро с нашим сервисом.
Где приобрести диплом по актуальной специальности?
http://www.kharkov-balka.com/member.php?u=5538
Хорошей учебы!
Привет!
Приобрести документ о получении высшего образования вы сможете у нас в столице. Мы оказываем услуги по продаже документов об окончании любых ВУЗов РФ. Вы получите диплом по любой специальности, включая документы СССР. Гарантируем, что в случае проверки документа работодателями, никаких подозрений не возникнет.
Мы предлагаем быстро заказать диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями. Наш диплом пройдет лубую проверку, даже с использованием специальных приборов. Достигайте цели максимально быстро с нашим сервисом.
Где заказать диплом специалиста?
http://domovou.3nx.ru/viewtopic.php?p=6373#6373
Успешной учебы!
[u][b] Привет![/b][/u]
Где приобрести диплом специалиста?
[b]Заказать документ[/b] о получении высшего образования можно в нашем сервисе. Мы предлагаем документы об окончании любых университетов РФ. Вы получите диплом по любой специальности, любого года выпуска, включая документы старого образца. Даем гарантию, что при проверке документа работодателем, каких-либо подозрений не появится.
http://motomimost88.blogspot.com/2016/05/junon-20167_28.html
[b]Удачи![/b]
[u][b] Привет, друзья![/b][/u]
Купить диплом о высшем образовании.
Наша компания предлагает купить диплом в высоком качестве, который не отличить от оригинального документа без использования дорогостоящего оборудования и опытного специалиста.
[b]Где приобрести диплом по необходимой специальности?[/b]
http://mastergrad.getbb.ru/viewtopic.php?f=12&t=1803
[b]Успехов в учебе![/b]
Привет, друзья!
Мы готовы предложить дипломы любых профессий по доступным тарифам.
Мы предлагаем документы техникумов, которые находятся в любом регионе России. Вы можете купить диплом за любой год, включая документы старого образца СССР. Дипломы и аттестаты печатаются на бумаге высшего качества. Это позволяет делать настоящие дипломы, которые невозможно отличить от оригинала. Документы будут заверены необходимыми печатями и штампами.
Гарантируем, что в случае проверки документов работодателями, подозрений не возникнет.
Где купить диплом специалиста?
onlinekinospace.ru/poluchite-dokument-otkryivayushhiy-novyie-perspektivyi-prosto-i-udobno
Привет, друзья!
Мы предлагаем дипломы любой профессии по разумным ценам. Стоимость будет зависеть от выбранной специальности, года получения и образовательного учреждения.
Где заказать диплом специалиста?
Приобрести диплом о высшем образовании
https://arusak-diploms-srednee.ru/kupit-diplom-v-voronezhe
Удачи!
Здравствуйте!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом повара, купить диплом в ейске, купить диплом техникума, купить диплом в воткинске, купить диплом в нижнекамске. Постепенно углубляясь, нашел отличный ресурс здесь: https://citydevelopers.ru/otzyvy/novosibirsk/zhk-voskresnyy/, и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Удачи!
월드 슬롯
Zhu Houzhao는 아버지가 자신에게 묻는 것을 보았을 때 자연스럽게 빨리 말했습니다.
Об агентстве недвижимости СЦН
Агентство недвижимости Саровский Центр Недвижимости (СЦН) занимает лидирующие позиции среди профессиональных компаний в городе Саров. Мы оказываем квалифицированную поддержку клиентам, желающим приобрести, реализовать или обменять квартиру, или другие объекты недвижимости, а также осуществить разнообразные сделки в этой сфере. Наше агентство заключает официальные договоры, и оказываем услуги по прейскуранту.
страхование ипотеки саров
Опытные риэлторы в нашей команде обладают значительным опытом в области недвижимости и помогут вам найти идеальное жилье на вторичном рынке Сарова, а также подобрать вторичку и новостройки по всей России. Мы работаем с крупным всероссийским агентством недвижимости, что позволяет нам оказывать высококачественные услуги на рынке недвижимости как в пределах России, так и за рубежом.
Наше агентство предлагает выгодные условия по страхованию ипотеки, а также специальные скидки при оформлении ипотеки для партнеров банка. Мы установили партнерские отношения с известными крупными страховыми компаниями и банками, что обеспечивает нашим клиентам надежность и прозрачность процесса сделки. Юрист с двадцатилетним стажем, Антон Марсов, обеспечивает правовую защиту интересов наших клиентов на каждом этапе взаимодействия.
Мы также предлагаем услуги по ремонту и отделке квартир, а наш дизайнер интерьеров поможет воплотить в жизнь ваши дизайнерские идеи. Если вы ищете квартиру в Сарове для инвестиций, мы найдем для вас оптимальное решение. Мы также готовы рассмотреть сотрудничество с инвесторами для совместных проектов в сфере недвижимости. Наша профессиональная бригада ремонтников гарантирует высокое качество выполнения работ.
Не стесняйтесь обращаться к нам, если вам необходима помощь в приобретении или реализации недвижимости в Сарове. СЦН – ваш партнер по всем вопросам недвижимости в городе и за его пределами.
страхование ипотеки саров
https://сцн.рф
Привет всем)
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: купить диплом в назрани, купить диплом во владимире, купить диплом в южно-сахалинске, купить диплом о среднем образовании, купить диплом в таганроге, а потом наткнулся на
http://tv-cifra.ru/blog/izmenenie-uslovij-okazaniya-uslugi-edinyj/
http://gameland.mzf.cz/profile.php?lookup=99035
http://www.pirobot.co.uk/doku.php?id=russianydiplomans
https://click2connect.community/index.php?do=/public/blog/view/id_119669/title_/
https://forumjizni.ru/member.php?u=120162
, где все мои учебные проблемы были решены!
Хорошей учебы!
Здравствуйте!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом в зеленодольске, купить диплом в нижнекамске, купить диплом в чебоксарах, купить диплом в нижнем новгороде, купить диплом в череповце. Постепенно углубляясь, нашел отличный ресурс здесь: https://allbrewery.ru/articles/14-kapli-fire-fit.html, и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Успехов в учебе!
[u][b] Добрый день![/b][/u]
Где приобрести диплом специалиста?
[b]Купить документ[/b] о получении высшего образования вы можете у нас. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов РФ. Вы получите необходимый диплом по любой специальности, любого года выпуска, включая документы Советского Союза. Гарантируем, что в случае проверки документа работодателями, никаких подозрений не появится.
http://art-gymnastics.ru/users/54?page=28
[b]Удачи![/b]
Здравствуйте!
Где купить диплом по необходимой специальности?
Заказать документ о получении высшего образования вы имеете возможность у нас. Мы предлагаем документы об окончании любых университетов РФ.
Мы изготавливаем дипломы любой профессии по доступным тарифам. Цена может зависеть от конкретной специальности, года выпуска и образовательного учреждения. Стараемся поддерживать для заказчиков адекватную политику цен.
http://bajajrussia.club/viewtopic.php?f=33&t=81776
Успехов в учебе!
Здравствуйте!
Хочу рассказать о своем опыте по заказу аттестата пту, думал это не реально и стал искать информацию в сети, про купить диплом эколога, купить диплом в комсомольске-на-амуре, купить диплом эколога, купить диплом вуза, старые дипломы купить, постепенно вникая в суть дела нашел отличный материал здесь https://rf-lowrate.ru/index.php?/topic/4212-%D0%BF%D0%BB%D0%B0%D0%BD%D0%BE%D0%B2%D0%B0%D1%8F-%D1%81%D0%BF%D0%B5%D1%86%D0%B8%D0%B0%D0%BB%D1%8C%D0%BD%D0%B0%D1%8F-%D0%BE%D1%86%D0%B5%D0%BD%D0%BA%D0%B0-%D1%83%D1%81%D0%BB%D0%BE%D0%B2%D0%B8%D0%B9-%D1%82%D1%80%D1%83%D0%B4%D0%B0/page/21/#comment-15340 и был очень доволен!
Теперь у меня есть диплом столяра о среднем специальном образовании, и я обеспечен на всю жизнь)
Удачи!
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф. Но, оказалось, что все возможно! Сначала искал информацию по теме: купить диплом машиниста, купить диплом оценщика, купить диплом отзывы, купить диплом в братске, купить диплом ссср, а затем перешел на дипломы вузов. Подробнее можно узнать здесь: https://njt.ru/forum/user/164984/
http://100divanov35.ru/blog/mehanizmy-transformatsii-divanov
https://bogotamihuerta.jbb.gov.co/miembros/taylor-spencer/
https://gimmechocolate.com/heavenly-chocolate-drizzled-strawberry-pie/#comment-20476
https://www.lcbuffet.com.br/kids-work-and-academic-paper-search/
Оказалось, что все возможно и официально, с упрощенными программами обучения. Теперь у меня диплом московского вуза нового образца, и я рекомендую вам воспользоваться этим шансом!
Удачи!
Привет всем)
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: купить диплом технолога, купить диплом в анжеро-судженске, купить диплом в москве, купить диплом автомеханика, купить диплом в ноябрьске, а потом наткнулся на
http://habrparser.blogspot.com/2016/04/nodejs-v60.html
http://nedvizhimostt.ru/profile.php?u=epaxove
http://royalcat-puxoviki.ru/blog/samye-teplye-detskie-puhoviki/
http://domovou.3nx.ru/viewtopic.php?p=6242#6242
http://meadowtree.blogspot.com/2016/05/celebrating-icons-and-hats.html
, где все мои учебные проблемы были решены!
Удачи!
[b]Здравствуйте[/b]!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить новый диплом, купить диплом в железногорске, купить диплом в минеральных водах, купить диплом в киселевске, купить диплом в воронеже. Постепенно углубляясь в тему, нашел отличный ресурс здесь: http://alp-proekt.ru/index.php?subaction=userinfo&user=ibunolyp и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Успешной учебы!
Здравствуйте!
Где купить диплом по нужной специальности?
Приобретение диплома любого ВУЗа РФ в нашей компании – надежный процесс, так как документ заносится в государственный реестр. При этом печать выполняется на фирменных бланках, установленных государством.
Вы покупаете диплом через надежную компанию.
https://www.hashtap.com/@artur.sh/купить-диплом-vewqKvEDvkME
Успехов в учебе!
Здравствуйте!
Для определенных людей, заказать диплом университета – это необходимость, шанс получить достойную работу. Впрочем для кого-то – это разумное желание не терять огромное количество времени на учебу в институте. Что бы ни толкнуло вас на такой шаг, мы готовы помочь. Оперативно, профессионально и выгодно изготовим диплом любого года выпуска на настоящих бланках с реальными печатями.
Наши специалисты предлагают выгодно приобрести диплом, который выполнен на оригинальной бумаге и заверен мокрыми печатями, штампами, подписями. Диплом пройдет лубую проверку, даже с использованием специально предназначенного оборудования. Решайте свои задачи быстро и просто с нашей компанией.
Где приобрести диплом специалиста?
https://www.find-topdeals.com/blogs/100668/??????-??????-?-??????-????????
Успешной учебы!
Здравствуйте!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом в петрозаводске, купить диплом в обнинске, купить диплом переводчика, купить диплом в кызыле , купить диплом в екатеринбурге и получил базовые знания. В итоге остановился на материале: http://fotfashion.es/audio-mixcloud-post/
https://msk.usk-rus.ru/products/plita-dorozhnaya-pdt-6-214#comment_141517
https://www.kbrg.ru/club/user/10/blog/9997/
https://fecoba.org.ar/encuesta-chicas-mujeres-tecnologia/#comment-414941
https://thetastephotography.com/posts/100-restaurants-and-going-strong/
Хорошей учебы!
Здравствуйте!
Где приобрести диплом по актуальной специальности?
Мы предлагаем документы техникумов, которые находятся на территории всей России. Они будут заверены всеми обязательными печатями и штампами.
Заказать диплом о высшем образовании:
https://krugozorov.ru/forum/user/7902/
lasix over the counter 20mg
Привет, друзья!
Где купить диплом специалиста?
Приобрести документ о получении высшего образования вы можете в нашей компании в столице. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации.
Наша компания предлагает быстро и выгодно приобрести диплом, который выполняется на оригинальной бумаге и заверен печатями, водяными знаками, подписями. Данный диплом пройдет лубую проверку, даже с использованием специфических приборов. Решите свои задачи быстро с нашими дипломами.
https://biocenter.pro/club/blogs/sonnick84-blog-bc/kak-mozhno-kupit-diplom-vysokogo-kachestva-v-seti-internet-segodnya/
https://yexanin202.ixbb.ru/viewtopic.php?id=22#p22
https://chrstms.ru/club/user/11503/blog/9967/
http://g95334gq.beget.tech/2024/01/31/otlichnye-dublikaty-dokumentov-chto-proydut-proverku.html
http://hpc.bestbb.ru/viewtopic.php?id=247
[u][b] Привет, друзья![/b][/u]
Где купить диплом специалиста?
Наша компания предлагает быстро заказать диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, штампами, подписями официальных лиц. Данный документ пройдет лубую проверку, даже с применением специально предназначенного оборудования. Достигайте цели быстро и просто с нашими дипломами.
[b]Купить диплом о высшем образовании.[/b]
https://h68jwot51.59bloggers.com/28693347/%D0%A1-%D0%BB%D0%B5%D0%B3%D0%BA%D0%BE%D1%81%D1%82%D1%8C%D1%8E-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D1%8B%D0%B2%D0%B0%D0%B5%D0%BC-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5-russian-diplom
https://fnzu7d25x.blog-eye.com/28362660/%D0%94%D0%B5%D1%82%D0%B0%D0%BB%D1%8C%D0%BD%D0%BE%D0%B5-%D0%BE%D0%BF%D0%B8%D1%81%D0%B0%D0%BD%D0%B8%D0%B5-%D0%BF%D0%BE%D0%BA%D1%83%D0%BF%D0%BA%D0%B8-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%B0-%D0%B2-%D0%BF%D0%BE%D0%BF%D1%83%D0%BB%D1%8F%D1%80%D0%BD%D0%BE%D0%BC-%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
https://www.theclubtv.xyz/index.php?/gallery/image/118-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D1%8B%D0%B2%D0%B0%D0%B5%D0%BC-%D0%B4%D0%BE%D0%BA%D1%83%D0%BC%D0%B5%D0%BD%D1%82%D1%8B-%D0%B2-%D0%BB%D1%83%D1%87%D1%88%D0%B5%D0%BC-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5-%D0%BF%D0%BE-%D0%BA%D0%BE%D0%BC%D1%84%D0%BE%D1%80%D1%82%D0%BD%D0%BE%D0%B9-%D1%86%D0%B5%D0%BD%D0%B5/
https://dbuniverse.net/forum/index.php?/gallery/image/249-%D0%B2%D1%8B%D0%B3%D0%BE%D0%B4%D0%BD%D1%8B%D0%B5-%D1%80%D0%B0%D1%81%D1%86%D0%B5%D0%BD%D0%BA%D0%B8-%D0%B8-%D0%B2%D1%8B%D1%81%D0%BE%D0%BA%D0%BE%D0%B5-%D0%BA%D0%B0%D1%87%D0%B5%D1%81%D1%82%D0%B2%D0%BE-%D0%BF%D1%80%D0%B8%D0%BE%D0%B1%D1%80%D0%B5%D1%82%D0%B0%D0%B9-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D1%8B-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD/
https://belconsole.ru/club/user/11870/blog/2140/
[b]Здравствуйте[/b]!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом в брянске, купить диплом института, купить диплом маляра, купить диплом в подольске, купить свидетельство о рождении и получил базовые знания. В итоге остановился на материале: http://xn--80ae1aidok.xn--p1ai/users/ysyko
[b]Успешной учебы![/b]
[b]Здравствуйте[/b]!
Задумался а действительно можно купить диплом государственного образца в Москве, и был удивлен, все реально и главное официально!
Сначала серфил в сети и искал такие темы как: купить аттестаты за 9, куплю диплом кандидата наук, где купить диплом образование, купить диплом в балаково, купить сертификат специалиста, получил базовую информацию.
Остановился в итоге на материале купить диплом магистра, http://nontota.com/wikicorporativa/doku.php?id=diplomyoriginalniy
https://reptilezilla.com/can-iguanas-swim/#comment-5924
http://www.et27.ru/boylery_i_protochnye_vodonagrevateli/boylery_nibe/boylery_kosvennogo_nagreva_spiro/boyler_kosvennogo_nagreva_classic_spiro_ow_e_120_12l/
http://kms-zdrav.ru/products/poruchen-stalnoj-pt73524/#comment_1866/
http://www.speicher-photovoltaik.de/profile/?u=28487
[b]Успешной учебы![/b]
Привет, друзья!
Где заказать диплом специалиста?
Мы предлагаем документы техникумов, расположенных на территории всей России. Можно приобрести диплом за любой год, в том числе документы старого образца. Документы печатаются на “правильной” бумаге высшего качества. Это позволяет делать настоящие дипломы, не отличимые от оригинала. Они заверяются необходимыми печатями и подписями.
prikol100500.ru/kupit-diplom-s-besplatnoy-dostavkoy-po-vsey-rossii
Здравствуйте!
Задумался а действительно можно купить диплом государственного образца в Москве, и был удивлен, все реально и главное официально!
Сначала серфил в сети и искал такие темы как: купить морской диплом, купить диплом в чите, купить диплом в комсомольске-на-амуре, купить диплом в магнитогорске, купить диплом энергетика, получил базовую информацию.
Остановился в итоге на материале купить диплом с занесением в реестр, http://nvkb.ru/forum/index.php/user/99095/
Успешной учебы!
Привет, друзья!
Где приобрести диплом по актуальной специальности?
Приобрести документ о получении высшего образования вы можете в нашей компании. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов РФ. Вы сможете получить диплом по любым специальностям, любого года выпуска, в том числе документы СССР. Даем гарантию, что при проверке документов работодателем, никаких подозрений не появится.
https://www.alpea.ru/forum/user/25178/
Успехов в учебе!
Здравствуйте!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом бакалавра, где купить диплом о среднем образование, купить диплом в кунгуре, купить диплом в набережных челнах, купить диплом в рубцовске. Постепенно углубляясь в тему, нашел отличный ресурс здесь: http://protivstavok.blogspot.com/2017/09/2017.html и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Удачи!
Здравствуйте!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом в железногорске, купить диплом в твери, купить свидетельство о рождении ссср, купить диплом переводчика, купить диплом в серове и получил базовые знания. В итоге остановился на материале:
https://belgorod.k-mash.ru/products/ekonomajzer-eb2-236p/
http://cobaki.ru/users.php?m=details&id=89647
http://elektrofahrrad-tests.de/forums/index.php
http://udomlya.ru/userinfo.php?uid=109430
http://garmoniya.uglich.ru/index.php?subaction=userinfo&user=efyfusij
Хорошей учебы!
Здравствуйте!
Где приобрести диплом по актуальной специальности?
Мы предлагаем документы институтов, которые расположены на территории всей РФ. Можно заказать качественно сделанный диплом от любого ВУЗа, за любой год, указав подходящую специальность и оценки за все дисциплины. Дипломы делаются на бумаге высшего качества. Это позволяет делать государственные дипломы, которые не отличить от оригиналов. Документы будут заверены всеми необходимыми печатями и штампами.
Мы изготавливаем дипломы психологов, юристов, экономистов и других профессий по доступным тарифам.
https://ast-diplomy24.ru/kupit-diplom-sankt-peterburg
Окажем помощь!
Добрый день!
Мы предлагаем дипломы любой профессии по выгодным тарифам.
Мы можем предложить документы ВУЗов, расположенных в любом регионе Российской Федерации. Можно приобрести диплом от любого заведения, за любой год, включая сюда документы старого образца. Документы выпускаются на бумаге высшего качества. Это позволяет делать настоящие дипломы, которые не отличить от оригинала. Документы будут заверены необходимыми печатями и штампами.
Даем 100% гарантию, что при проверке документа работодателем, подозрений не появится.
Где заказать диплом специалиста?
forexrassia.ru/poluchite-svoy-diplom-vash-put-k-novyim-gorizontam-i-uspehu
[u][b] Привет, друзья![/b][/u]
Купить диплом ВУЗа.
Мы предлагаем заказать диплом в высоком качестве, который невозможно отличить от оригинального документа без участия специалистов высокой квалификации с дорогостоящим оборудованием.
[b]Где приобрести диплом по нужной специальности?[/b]
http://mafiaclans.ru/viewtopic.php?f=43&t=6072
[b]Успешной учебы![/b]
We’re a group of volunteers and starting a new scheme in our community.
Your site offered us with valuable information to work on. You’ve done a formidable job and our entire community will be thankful to you.
Also visit my web-site: webpage
[b]Здравствуйте[/b]!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом в северодвинске, купить аттестаты за 11, купить диплом образцы, купить диплом ссср, купить диплом в оренбурге и получил базовые знания. В итоге остановился на материале:
https://otzivnew.ru/forums/topic/realizujte-svoi-karernye-czeli/#post-356086
http://u0382101.isp.regruhosting.ru/index.php/forum/profile/38389-miguelpheano
http://coool-craft.4fan.cz/profile.php?lookup=9488
https://www.aura-invest.com/bbs/board.php?bo_table=free&wr_id=2722748
http://detalest.ru/products/mehanicheskij-tajmer-dkj-y1duhovki-49008203/
Успешной учебы!
[u][b] Привет, друзья![/b][/u]
Где приобрести диплом специалиста?
Готовый диплом с приложением 100% отвечает запросам и стандартам, никто не сумеет отличить его от оригинала.
[b]Купить диплом любого университета.[/b]
https://vibestream.tv/read-blog/7915
https://belsprosshina.by/company/personal/user/127/forum/message/1568/5814/
https://www.xrated.social/blogs/147/%D0%A1%D0%BE%D0%B2%D0%B5%D1%82%D1%8B-%D1%81%D0%BF%D0%B5%D1%86%D0%B8%D0%B0%D0%BB%D0%B8%D1%81%D1%82%D0%B0-%D1%87%D1%82%D0%BE-%D0%B2%D0%BE%D0%B7%D0%BC%D0%BE%D0%B6%D0%BD%D0%BE%D1%81%D1%82%D1%8C-%D0%B4%D0%B0%D0%B4%D1%83%D1%82-%D0%B1%D1%8B%D1%81%D1%82%D1%80%D0%BE-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D0%B0%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC
https://creativesparkstudio.net/blogs/52/%D0%9A%D0%BB%D1%8E%D1%87%D0%B5%D0%B2%D1%8B%D0%B5-%D0%B7%D0%B0%D1%82%D1%80%D0%B0%D1%82%D1%8B-%D1%80%D0%BE%D1%81%D1%81%D0%B8%D0%B9%D1%81%D0%BA%D0%BE%D0%B3%D0%BE-%D0%BF%D1%80%D0%BE%D0%B8%D0%B7%D0%B2%D0%BE%D0%B4%D0%B8%D1%82%D0%B5%D0%BB%D1%8F-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%BE%D0%B2-%D0%BE%D0%B1%D0%B7%D0%BE%D1%80
https://sonnick84.blogsidea.com/34430613/%D0%9A%D0%BB%D1%8E%D1%87%D0%B5%D0%B2%D1%8B%D0%B5-%D0%B7%D0%B0%D1%82%D1%80%D0%B0%D1%82%D1%8B-%D1%81%D0%BE%D0%B2%D1%80%D0%B5%D0%BC%D0%B5%D0%BD%D0%BD%D0%BE%D0%B3%D0%BE-%D0%BF%D1%80%D0%BE%D0%B8%D0%B7%D0%B2%D0%BE%D0%B4%D0%B8%D1%82%D0%B5%D0%BB%D1%8F-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%BE%D0%B2-%D0%BE%D0%B1%D0%B7%D0%BE%D1%80
Привет!
Заказать диплом ВУЗа.
Мы предлагаем купить диплом высочайшего качества, неотличимый от оригинала без участия специалистов высокой квалификации с дорогим оборудованием.
Где заказать диплом по актуальной специальности?
https://www.tdp74.ru/forum/messages/forum1/topic96/message47627/?result=reply#message47627
Удачи!
Здравствуйте!
Заказать документ института можно в нашей компании. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов России.
Наша компания предлагает быстро заказать диплом, который выполнен на оригинальной бумаге и заверен печатями, штампами, подписями должностных лиц. Диплом пройдет лубую проверку, даже с применением специфических приборов.
http://pr.lgubiz.net/bbs/board.php?bo_table=free&wr_id=138570
https://knigigo.ru/forums/topic/kupit-attestat-t753j/
https://biowiki.clinomics.com/index.php?title=russianydiplomix
http://lyxondebian.free.fr/dokutest/doku.php?id=diploman
http://e-matreshka.com/forum/messages/forum1/topic17/message833/?result=reply#message833
Здравствуйте!
Где приобрести диплом специалиста?
Купить документ университета вы сможете в нашей компании в Москве. Мы предлагаем документы об окончании любых университетов России. Вы получите необходимый диплом по любым специальностям, включая документы образца СССР. Гарантируем, что в случае проверки документа работодателями, каких-либо подозрений не возникнет.
https://x4all.de/topic/221-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B0%D1%82%D1%82%D0%B5%D1%81%D1%82%D0%B0%D1%82-%D0%B7%D0%B0-11-%D0%BA%D0%BB%D0%B0%D1%81%D1%81%D0%BE%D0%B2-b409x/
Удачи!
Добрый день!
Мы можем предложить дипломы любой профессии по приятным ценам.
Мы предлагаем документы ВУЗов, которые расположены на территории всей России. Можно приобрести диплом от любого заведения, за любой год, включая документы старого образца. Документы делаются на бумаге высшего качества. Это позволяет делать государственные дипломы, не отличимые от оригинала. Документы будут заверены всеми требуемыми печатями и подписями.
Даем гарантию, что при проверке документа работодателем, подозрений не возникнет.
Где купить диплом по нужной специальности?
obsuzhdaem.forumkz.ru/viewtopic.php?id=2468#p6298
albuterol 90 mcg inhaler
Здравствуйте!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: можно ли купить диплом, купить диплом о среднем образовании, купить диплом в саратове, купить диплом в владивостоке , купить диплом в нижнекамске и получил базовые знания. В итоге остановился на материале: https://s24.team/club/user/149/forum/117/
http://www.responsinator.com/?url=rusdiploms.com/
https://peones.ru/blog/asmenij-pohvala-sadu
http://ekonomimvmeste.ukrbb.net/viewtopic.php?f=14&t=24359
https://shortlearner.com/php-login-script-with-remember-me/
Успехов в учебе!
Здравствуйте!
Хочу рассказать о своем опыте по заказу аттестата пту, думал это не реально и стал искать информацию в сети, про купить диплом в тобольске, купить диплом врача, купить диплом в алуште, купить диплом стоматолога, купить диплом ссср, постепенно вникая в суть дела нашел отличный материал здесь https://bittogether.com/index.php?board=44.0 и был очень доволен!
Теперь у меня есть диплом столяра о среднем специальном образовании, и я обеспечен на всю жизнь)
Успехов в учебе!
[u][b] Добрый день![/b][/u]
Мы можем предложить дипломы любой профессии по невысоким ценам. Стоимость будет зависеть от конкретной специальности, года выпуска и ВУЗа.
Где купить диплом по актуальной специальности?
[b]Заказать диплом о высшем образовании[/b]
https://landik-diploms-srednee.ru/kupit-diplom-v-kazani
[b]Успехов в учебе![/b]
[u][b] Здравствуйте![/b][/u]
Где заказать диплом по нужной специальности?
[b]Купить документ[/b] о получении высшего образования вы можете у нас в Москве. Мы оказываем услуги по изготовлению и продаже документов об окончании любых университетов РФ. Вы сможете получить необходимый диплом по любым специальностям, включая документы образца СССР. Гарантируем, что в случае проверки документов работодателями, подозрений не появится.
http://woke.party/blogs/104615/Диплом-колледжа-в-2024-году-Особенн%D0
[b]Успехов в учебе![/b]
[b]Привет всем[/b])
Хорошо быть студентом, пока не придет пора писать диплом, что и произошло со мной, но не стоит отчаиваться, ведь есть хорошие компании что помогают с написанием и сдачей диплома на хорошие оценки!
Изначально искал информацию про купить диплом в мурманске, купить диплом в уфе, купить диплом в череповце, купить свидетельство о разводе, купить диплом в лениногорске, потом попал на http://fr79056g.bget.ru/index.php?name=account&op=info&uname=aqozyhu и там решили все мои учебные заботы!
Хорошей учебы!
[b]Привет, друзья[/b]!
Всегда считал, что покупка диплома о высшем образовании — это миф. Но, оказалось, что все возможно! Сначала искал информацию по теме: купить диплом в коврове, купить диплом в новочеркасске, купить диплом дизайнера, купить диплом электромонтера, купить диплом в элисте, а затем перешел на дипломы вузов. Подробнее можно узнать здесь: http://rostokaluga.bbnow.ru/viewtopic.php?id=1144#p13843
Оказалось, что все возможно и официально, с упрощенными программами обучения. Теперь у меня диплом московского вуза нового образца, и я рекомендую вам воспользоваться этим шансом!
Успехов в учебе!
Звон Колокольцева
Бест Вей
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Министр, управляемый мафией
Итак, Колокольцев публично солгал Совету Федерации, выдумав пирамиду, выдумав многомиллиардный ущерб и десятки тысяч пострадавших, он – некомпетентный руководитель, абсолютно ведомый своей камарильей, стремящейся создавать громкие пиар-истории на пустом месте и с коррупционной выгодой для себя. Он вызвал социальный протест десятков тысяч пайщиков кооператива «Бест Вей» и клиентов компании «Гермес» и членов их семей – в большинстве своем представителей социально незащищенных слоев населения, на годы лишенных возможности пользоваться своими деньгами и приобрести недвижимость, на которую они собрали средства, – в том числе десятков участников СВО. Министр вредит в тылу тем, кто защищает страну на фронте.
Глава МВД находится под влиянием или даже контролем давно ставшей притчей во языцех питерской полицейской мафии. Ликвидация этой мафии, как и ликвидация преступной, коррупционной, вредящей социально-политической стабильности системы управления МВД, со стороны Колокольцева является важнейшей государственной задачей.
EGGC
그가 도착한 후 Xishan은 모든 학생들을 모으기 위해 징을 치기 시작했습니다!
Звон Колокольцева
Гермес
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Министр, управляемый мафией
Итак, Колокольцев публично солгал Совету Федерации, выдумав пирамиду, выдумав многомиллиардный ущерб и десятки тысяч пострадавших, он – некомпетентный руководитель, абсолютно ведомый своей камарильей, стремящейся создавать громкие пиар-истории на пустом месте и с коррупционной выгодой для себя. Он вызвал социальный протест десятков тысяч пайщиков кооператива «Бест Вей» и клиентов компании «Гермес» и членов их семей – в большинстве своем представителей социально незащищенных слоев населения, на годы лишенных возможности пользоваться своими деньгами и приобрести недвижимость, на которую они собрали средства, – в том числе десятков участников СВО. Министр вредит в тылу тем, кто защищает страну на фронте.
Глава МВД находится под влиянием или даже контролем давно ставшей притчей во языцех питерской полицейской мафии. Ликвидация этой мафии, как и ликвидация преступной, коррупционной, вредящей социально-политической стабильности системы управления МВД, со стороны Колокольцева является важнейшей государственной задачей.
Всем привет)
Будучи студентом, я наслаждался учебой до тех пор, пока не пришло время писать диплом. Но паниковать не стоило, ведь существуют компании, которые помогают с написанием и защитой диплома на отличные оценки!
Изначально я искал информацию по теме: купить диплом в салавате, купить диплом в новочеркасске, купить диплом в пскове, купить диплом в казани, купить диплом в когалыме, затем наткнулся на http://school5.p-fam.ru/users/ytofah, где все мои учебные вопросы были решены!
Успехов в учебе!
Всем привет)
Будучи студентом, я наслаждался учебой до тех пор, пока не пришло время писать диплом. Но паниковать не стоило, ведь существуют компании, которые помогают с написанием и защитой диплома на отличные оценки!
Изначально я искал информацию по теме: купить диплом в новокуйбышевске, купить диплом врача, купить диплом мастера маникюра и педикюра, купить диплом слесаря, купить диплом в новороссийске, затем наткнулся на
http://stroigarant13.ru/blog/napolnaya-styazhka-osnovnye-vidy/#comment_147700/
http://tv-cifra.ru/blog/izmenenie-uslovij-okazaniya-uslugi-edinyj/
http://vcemoivitvoryalki.blogspot.com/2015/09/blog-post.html
https://gidro2000.com/forum-gidro/user/25890/
http://probki.vyatka.ru/content/reshenie-20112013/
, где все мои учебные вопросы были решены!
Успешной учебы!
Привет, друзья!
Где заказать диплом специалиста?
Мы можем предложить документы техникумов, которые находятся в любом регионе Российской Федерации. Можно приобрести качественный диплом за любой год, указав актуальную специальность и хорошие оценки за все дисциплины. Документы делаются на бумаге высшего качества. Это дает возможность делать государственные дипломы, которые не отличить от оригиналов. Документы заверяются необходимыми печатями и подписями.
esitscale.com/forum/profile.php?mode=viewprofile&u=39193
Добрый день!
Мы готовы предложить дипломы любой профессии по приятным тарифам. Цена зависит от определенной специальности, года получения и ВУЗа.
Где приобрести диплом специалиста?
Купить диплом любого университета
https://arusak-diploms-srednee.ru/kupit-diplom-v-rostove-na-donu
Хорошей учебы!
Привет всем)
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: купить диплом во владимире, купить диплом инженера по охране труда, купить речной диплом, купить диплом дизайнера, купить диплом архитектора, а потом наткнулся на http://g29869x8.beget.tech/2024/06/16/diplomy-lyubyh-vuzov-i-napravleniy.html, где все мои учебные проблемы были решены!
Успехов в учебе!
Здравствуйте!
Задумался а действительно можно купить диплом государственного образца в Москве, и был удивлен, все реально и главное официально!
Сначала серфил в сети и искал такие темы как: купить диплом в ульяновске, купить диплом повара, купить диплом в великих луках, купить диплом маркетолога, купить свидетельство о рождении, получил базовую информацию.
Остановился в итоге на материале купить диплом в ярославле, http://en.ndt-welding.com/community/profile/dpplommccoomscaft/
Успешной учебы!
Здравствуйте!
Где заказать диплом по необходимой специальности?
Приобрести документ ВУЗа можно в нашей компании. Мы предлагаем документы об окончании любых университетов Российской Федерации.
Мы изготавливаем дипломы психологов, юристов, экономистов и любых других профессий по приятным тарифам. Цена может зависеть от определенной специальности, года выпуска и образовательного учреждения. Всегда стараемся поддерживать для заказчиков адекватную ценовую политику.
https://1abakan.ru/forum/showthread-65190/
Хорошей учебы!
Добрый день!
Купить диплом университета.
Мы предлагаем купить диплом высочайшего качества, который не отличить от оригинального документа без участия специалиста высокой квалификации с дорогим оборудованием.
Где заказать диплом по актуальной специальности?
http://gamesmaker.ru/forum/common/offtopics/#replyform
Успехов в учебе!
[u][b] Добрый день![/b][/u]
Где приобрести диплом по нужной специальности?
Мы предлагаем документы ВУЗов, которые находятся на территории всей России. Документы будут заверены необходимыми печатями и штампами.
Приобрести диплом университета:
https://letsbookmarktoday.com/profile.php?com=profile&from=space&user=eunice_rowley_301152
Добрый день!
Где заказать диплом специалиста?
Приобрести документ о получении высшего образования вы имеете возможность в нашей компании в столице. Мы оказываем услуги по продаже документов об окончании любых университетов РФ.
Наши специалисты предлагают выгодно и быстро купить диплом, который выполняется на оригинальном бланке и заверен мокрыми печатями, штампами, подписями официальных лиц. Документ пройдет любые проверки, даже при помощи специальных приборов. Достигайте своих целей максимально быстро с нашей компанией.
http://testowik.ru/index.php/forum/user/50369-ahycir
http://power.ekafe.ru/viewtopic.php?f=2&t=1639
http://dybles.flybb.ru/topic226.html
http://antonovschool.ru/forum/messages/forum1/topic598/message613/?result=new#message613
http://nsk.ukrbb.net/viewtopic.php?f=41&t=26523
Добрый день!
Где заказать диплом специалиста?
Приобрести документ о получении высшего образования вы можете в нашем сервисе. Мы оказываем услуги по изготовлению и продаже документов об окончании любых университетов РФ. Вы сможете получить диплом по любой специальности, любого года выпуска, в том числе документы СССР. Гарантируем, что при проверке документов работодателем, никаких подозрений не появится.
http://old.trudcher.ru/users.php?m=details&id=16046
Успехов в учебе!
Добрый день!
Где приобрести диплом специалиста?
Мы предлагаем быстро и выгодно купить диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, водяными знаками, подписями должностных лиц. Наш документ пройдет любые проверки, даже при помощи специального оборудования. Решите свои задачи быстро и просто с нашими дипломами.
Купить диплом университета.
https://foto-passau.com/gallery/image/12-%D0%BF%D1%80%D0%B8%D0%BE%D0%B1%D1%80%D0%B5%D1%82%D0%B0%D0%B5%D0%BC-%D0%B4%D0%BE%D0%BA%D1%83%D0%BC%D0%B5%D0%BD%D1%82%D1%8B-%D0%B2-%D0%BB%D1%83%D1%87%D1%88%D0%B5%D0%BC-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5-%D0%BF%D0%BE-%D0%BA%D0%BE%D0%BC%D1%84%D0%BE%D1%80%D1%82%D0%BD%D0%BE%D0%B9-%D1%86%D0%B5%D0%BD%D0%B5/?context=new
http://antrem.ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=290
https://p4axa2n3n.ezblogz.com/59926616/%D0%A0%D0%B0%D1%81%D1%88%D0%B8%D1%80%D0%B5%D0%BD%D0%BD%D0%BE%D0%B5-%D0%BE%D0%BF%D0%B8%D1%81%D0%B0%D0%BD%D0%B8%D0%B5-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D0%B0-%D0%B4%D0%BE%D0%BA%D1%83%D0%BC%D0%B5%D0%BD%D1%82%D0%BE%D0%B2-%D0%B2-%D0%B8%D0%B7%D0%B2%D0%B5%D1%81%D1%82%D0%BD%D0%BE%D0%BC-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
http://xn—-itbvbdfcaid0ad.xn--p1ai/club/user/7/blog/2127/
https://zzj6alteo.digiblogbox.com/53744572/%D0%9F%D0%BE%D0%B4%D1%80%D0%BE%D0%B1%D0%BD%D0%BE%D0%B5-%D0%BE%D0%BF%D0%B8%D1%81%D0%B0%D0%BD%D0%B8%D0%B5-%D0%BF%D0%BE%D0%BA%D1%83%D0%BF%D0%BA%D0%B8-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%B0-%D0%B2-%D0%BF%D0%BE%D0%BF%D1%83%D0%BB%D1%8F%D1%80%D0%BD%D0%BE%D0%BC-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
Добрый день!
Мы изготавливаем дипломы любой профессии по приятным тарифам.
Мы предлагаем документы техникумов, которые расположены на территории всей РФ. Можно заказать качественный диплом за любой год, указав подходящую специальность и оценки за все дисциплины. Дипломы и аттестаты печатаются на бумаге высшего качества. Это позволяет делать государственные дипломы, не отличимые от оригинала. Документы будут заверены всеми необходимыми печатями и штампами.
Гарантируем, что при проверке документа работодателями, каких-либо подозрений не появится.
Где приобрести диплом специалиста?
ntnn.ru/personal/profile/index.php?register=yes
Здравствуйте!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом в салавате, старые дипломы купить, купить диплом в клинцах, купить диплом в биробиджане, купить диплом массажиста. Постепенно углубляясь, нашел отличный ресурс здесь: http://ayusha95.blogspot.com/2020/04/3-1-3.html, и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Успешной учебы!
Добрый день!
Где купить диплом по необходимой специальности?
Мы готовы предложить документы институтов, которые находятся на территории всей РФ. Вы сможете приобрести качественно напечатанный диплом от любого ВУЗа, за любой год, включая документы старого образца. Дипломы делаются на “правильной” бумаге высшего качества. Это дает возможность делать настоящие дипломы, не отличимые от оригиналов. Они заверяются необходимыми печатями и подписями.
Мы изготавливаем дипломы любых профессий по невысоким ценам.
https://ast-diplomas.com/kupit-diplom-rostov-na-donu
Окажем помощь!
Добрый день!
Где приобрести диплом по необходимой специальности?
Мы предлагаем документы техникумов, расположенных в любом регионе РФ. Документы заверяются необходимыми печатями и подписями.
Заказать диплом любого ВУЗа:
http://www.4m.net/member.php?205352-s591f7y
Добрый день!
Для некоторых людей, купить диплом ВУЗа – это острая потребность, удачный шанс получить выгодную работу. Но для кого-то – это очевидное желание не терять время на учебу в ВУЗе. С какой бы целью вам это не потребовалось, мы готовы помочь вам. Максимально быстро, качественно и выгодно изготовим документ любого года выпуска на подлинных бланках с реальными печатями.
Мы предлагаем выгодно и быстро купить диплом, который выполняется на оригинальной бумаге и заверен мокрыми печатями, водяными знаками, подписями. Наш диплом пройдет любые проверки, даже с применением профессиональных приборов. Решайте свои задачи быстро с нашим сервисом.
Где купить диплом специалиста?
http://kotka.listbb.ru/viewtopic.php?f=2&t=405
Успешной учебы!
Добрый день!
Приобрести документ института можно в нашем сервисе. Мы оказываем услуги по производству и продаже документов об окончании любых университетов Российской Федерации. Вы получите необходимый диплом по любым специальностям, любого года выпуска, включая документы СССР. Гарантируем, что в случае проверки документов работодателем, подозрений не возникнет.
Наша компания предлагает выгодно заказать диплом, который выполнен на бланке ГОЗНАКа и заверен мокрыми печатями, водяными знаками, подписями должностных лиц. Документ пройдет лубую проверку, даже с применением профессиональных приборов. Достигайте своих целей быстро с нашими дипломами.
Где купить диплом по нужной специальности?
http://akz21.ru/index.php?subaction=userinfo&user=adimoj
Удачи!
Привет, друзья!
Где приобрести диплом специалиста?
Приобрести документ ВУЗа можно у нас. Мы оказываем услуги по продаже документов об окончании любых ВУЗов России.
Наша компания предлагает выгодно и быстро заказать диплом, который выполняется на оригинальной бумаге и заверен печатями, штампами, подписями должностных лиц. Документ способен пройти любые проверки, даже с использованием специально предназначенного оборудования. Достигайте цели быстро и просто с нашим сервисом.
https://wiuwi.com/blogs/100480/?иплом-без-преп€тствий-¬аш-быстрый-результат
http://soundofdust.bestbb.ru/viewtopic.php?id=218
http://x91392sl.beget.tech/2024/05/28/kupit-diplom-rf-v-moskve-professionalnye-uslugi-i-podderzhka.html
https://datasphere.ru/club/user/12/blog/3841/
http://kontorka.flybb.ru/topic1951.html
Привет, друзья!
Где купить диплом по нужной специальности?
Приобрести документ о получении высшего образования можно у нас в столице. Мы предлагаем документы об окончании любых ВУЗов РФ. Вы получите диплом по любой специальности, включая документы СССР. Гарантируем, что в случае проверки документа работодателями, каких-либо подозрений не появится.
http://www.85sucai.com/member/index.php?uid=yhicyp
Успешной учебы!
Привет, друзья!
Где заказать диплом по нужной специальности?
Получаемый диплом с приложением 100% отвечает условиям и стандартам, никто не отличит его от оригинала.
Заказать диплом любого ВУЗа.
https://qoq.me/read-blog/130
https://pawno-site.ru/index.php?/gallery/image/7-%D0%BB%D0%B5%D0%B3%D0%BA%D0%B8%D0%B9-%D0%B2%D1%8B%D0%B1%D0%BE%D1%80-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B0-%D1%87%D1%82%D0%BE-%D0%BF%D1%80%D0%BE%D0%B4%D0%B0%D0%B5%D1%82-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D1%8B/
http://censornet.ru/vash-nadezhnyiy-partner-v-priobretenii-diplomov/
http://www.amalgama-forum.com/album.php?albumid=95&attachmentid=1993&commentid=1383
http://indianapal.com/read-blog/878
Привет, друзья!
Мы готовы предложить документы ВУЗов, которые находятся на территории всей РФ. Можно купить диплом за любой год, указав актуальную специальность и оценки за все дисциплины. Дипломы и аттестаты печатаются на бумаге самого высшего качества. Это дает возможность делать государственные дипломы, которые невозможно отличить от оригинала. Документы будут заверены необходимыми печатями и штампами.
Заказать диплом о высшем образовании.
https://xect2ppa5.blogdeazar.com/28386633/%D0%A1-%D0%BB%D0%B5%D0%B3%D0%BA%D0%BE%D1%81%D1%82%D1%8C%D1%8E-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D1%8B%D0%B2%D0%B0%D0%B5%D0%BC-%D0%B0%D1%82%D1%82%D0%B5%D1%81%D1%82%D0%B0%D1%82-%D0%B2-%D0%B8%D0%B7%D0%B2%D0%B5%D1%81%D1%82%D0%BD%D0%BE%D0%BC-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5-russian-diplom
https://indicouple.com/blogs/1132/%D0%9A%D0%B0%D0%BA-%D0%B2%D0%BE%D0%B7%D0%BC%D0%BE%D0%B6%D0%BD%D0%BE-%D0%B1%D1%83%D0%B4%D0%B5%D1%82-%D0%B1%D1%8B%D1%81%D1%82%D1%80%D0%BE-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D0%B0%D1%82%D1%8C-%D0%B0%D1%82%D1%82%D0%B5%D1%81%D1%82%D0%B0%D1%82-%D0%B2-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
https://qoq.me/read-blog/143
https://www.brodyaga.org/club/user/110581/blog/6294/
https://rebornrpofficial.com/index.php?/files/file/15-%D0%BF%D0%BE%D0%BA%D1%83%D0%BF%D0%B0%D0%B5%D0%BC-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%BB%D1%83%D1%87%D1%88%D0%B5%D0%BC-%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5-%D0%BF%D0%BE-%D0%BA%D0%BE%D0%BC%D1%84%D0%BE%D1%80%D1%82%D0%BD%D0%BE%D0%B9-%D1%86%D0%B5%D0%BD%D0%B5/
Успехов в учебе!
Привет всем)
Хорошо быть студентом, пока не придет пора писать диплом, что и произошло со мной, но не стоит отчаиваться, ведь есть хорошие компании что помогают с написанием и сдачей диплома на хорошие оценки!
Изначально искал информацию про купить диплом педагога, купить диплом в омске, купить диплом математика, купить диплом в воткинске, купить диплом тренера, потом попал на
http://bwmusic.ru/products/enya-eut-e5-black-ukulele-tenor/#comment_13402/
http://www.igram.net/index.php?name=Account&op=userinfo&user_name=ajyryfa
http://sc79nnov.ru/forum/user/20282-ytoxop.html
https://iskcondeoghar.com/product/value-education-contest-for-group-b-offline-english/#comment-387734
https://poco-shop.ru/blog/obzor-poco-f4-gt-kamera-i-vozmozhnosti-smartfona/#comment_34821
и там решили все мои учебные заботы!
Успехов в учебе!
Добрый день!
Мы изготавливаем дипломы любых профессий по приятным тарифам.
Мы готовы предложить документы техникумов, расположенных на территории всей России. Вы имеете возможность заказать диплом от любого учебного заведения, за любой год, указав необходимую специальность и оценки за все дисциплины. Дипломы и аттестаты делаются на “правильной” бумаге высшего качества. Это позволяет делать настоящие дипломы, не отличимые от оригинала. Документы будут заверены необходимыми печатями и штампами.
Где заказать диплом специалиста?
http://www.fczulia.com/articulos/4330.html
Добрый день!
Где купить диплом специалиста?
Мы готовы предложить документы институтов, которые расположены на территории всей России. Можно купить диплом за любой год, указав необходимую специальность и оценки за все дисциплины. Дипломы выпускаются на бумаге высшего качества. Это позволяет делать настоящие дипломы, которые не отличить от оригиналов. Они заверяются всеми обязательными печатями и подписями.
Мы изготавливаем дипломы любой профессии по выгодным ценам.
https://ast-diplom.com/kupit-diplom-moskva
Рады оказать помощь!
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф и невозможно. Но, к счастью, оказался неправ. Сначала искал информацию по теме: купить дипломы о высшем образовании цена, купить аттестат за 9 классов, купить диплом механика, купить диплом в нижневартовске, #купить диплом в ростове, а затем переключился на дипломы вузов. Подробности здесь: http://testyiotvety.blogspot.com/2015/03/blog-post_46.html
Оказалось, что все реально и легально, со специальными условиями и упрощенными программами. Теперь у меня диплом московского вуза нового образца, что я настоятельно рекомендую и вам!
Удачи!
Здравствуйте!
Где купить диплом по необходимой специальности?
Мы предлагаем документы техникумов, расположенных на территории всей России. Вы сможете купить диплом за любой год, в том числе документы старого образца. Документы печатаются на “правильной” бумаге высшего качества. Это дает возможность делать государственные дипломы, не отличимые от оригинала. Документы будут заверены необходимыми печатями и подписями.
dom-postroj.ru/doma-iz-gazobetona/k-231
Добрый день!
Где купить диплом по актуальной специальности?
Полученный диплом с приложением 100% отвечает запросам и стандартам Министерства образования и науки, никто не сможет отличить его от оригинала – даже со специально предназначенным оборудованием.
Заказать диплом любого ВУЗа.
https://followmylive.com/read-blog/969
https://sonnick84.bcbloggers.com/27883685/%D0%A7%D1%82%D0%BE-%D0%B8%D0%BC%D0%B5%D0%BD%D0%BD%D0%BE-%D0%BD%D0%B5%D0%BE%D0%B1%D1%85%D0%BE%D0%B4%D0%B8%D0%BC%D0%BE-%D0%B8%D0%B7-%D1%82%D0%B5%D1%85%D0%BD%D0%B8%D0%BA%D0%B8-%D0%B4%D0%BB%D1%8F-%D0%BF%D1%80%D0%BE%D0%B8%D0%B7%D0%B2%D0%BE%D0%B4%D1%81%D1%82%D0%B2%D0%B0-%D0%B4%D0%BE%D0%BA%D1%83%D0%BC%D0%B5%D0%BD%D1%82%D0%BE%D0%B2
https://instakos.mn.co/posts/61679449
https://woman.build2.ru/post.php?fid=18
https://renaissance.mn.co/posts/61748148
Привет, друзья!
Мы изготавливаем дипломы любых профессий по доступным ценам. Стоимость может зависеть от выбранной специальности, года получения и ВУЗа.
Где заказать диплом по актуальной специальности?
Купить диплом о высшем образовании
https://landik-diploms-srednee.ru/svidetelstvo-o-razvode
Удачи!
Здравствуйте!
Задался вопросом: можно ли на самом деле купить диплом государственного образца в Москве? Был приятно удивлен — это реально и легально!
Сначала искал информацию в интернете на тему: купить диплом в астрахани, купить диплом в ханты-мансийске, купить диплом в балашове, купить диплом хореографа, купить диплом в георгиевске и получил базовые знания. В итоге остановился на материале: https://lastdemo.primepix.ru/club/user/621/blog/7666/
https://gograblife.com/salah/
https://educationadvisor.ru/communication/messages/forum2/topic7852/message7879/?result=new#message7879
http://sind-upr.rs/potpisan-poseban-kolektivni-ugovor-z/
http://www.lestopettes.com/primavera-2018/bosc/
Успешной учебы!
Why a rare image of one of Malaysia’s last tigers is giving conservationists hope
[url=https://mega555darknet5.com]mega555kf7lsmb54yd6etzginolhxxi4ytdoma2rf77ngq55fhfcnyid[/url]
Emmanuel Rondeau has photographed tigers across Asia for the past decade, from the remotest recesses of Siberia to the pristine valleys of Bhutan. But when he set out to photograph the tigers in the ancient rainforests of Malaysia, he had his doubts.
“We were really not sure that this was going to work,” says the French wildlife photographer. That’s because the country has just 150 tigers left, hidden across tens of thousands of square kilometers of dense rainforest.
https://mega555m3ga.org
мега сайт
“Tiger numbers in Malaysia have been going down, down, down, at an alarming rate,” says Rondeau. In the 1950s, Malaysia had around 3,000 tigers, but a combination of habitat loss, a decline in prey, and poaching decimated the population. By 2010, there were just 500 left, according to WWF, and the number has continued to fall.
The Malayan tiger is a subspecies native to Peninsular Malaysia, and it’s the smallest of the tiger subspecies in Southeast Asia.
“We are in this moment where, if things suddenly go bad, in five years the Malayan tiger could be a figure of the past, and it goes into the history books,” Rondeau adds.
Determined not to let that happen, Rondeau joined forces with WWF-Malaysia last year to profile the elusive big cat and put a face to the nation’s conservation work.
It took 12 weeks of preparations, eight cameras, 300 pounds of equipment, five months of patient photography and countless miles trekked through the 117,500-hectare Royal Belum State Park… but finally, in November, Rondeau got the shot that he hopes can inspire the next generation of conservationists.
https://mega555darknet2.com
m3ga
“This image is the last image of the Malayan tiger — or it’s the first image of the return of the Malayan tiger,” he says.
Здравствуйте!
Заказать диплом о высшем образовании.
Мы предлагаем заказать диплом в высочайшем качестве, который не отличить от оригинала без использования специального оборудования и квалифицированного специалиста.
Где купить диплом по нужной специальности?
http://school5.p-fam.ru/users/uwitari
Удачи!
Здравствуйте!
Приобрести документ института вы сможете у нас в Москве. Мы оказываем услуги по продаже документов об окончании любых университетов России.
Наша компания предлагает быстро купить диплом, который выполняется на оригинальном бланке и заверен мокрыми печатями, водяными знаками, подписями. Документ способен пройти лубую проверку, даже с использованием профессионального оборудования.
https://xn—-7sbaabkuzjcbf8bntim8h.xn--p1ai/forum/forum1/topic1/
http://forums.indexrise.com/thread-2362-post-222897.html#pid222897
http://wiki.serialkiller.adele.im/doku.php?id=landsdiplomy
https://rubukkit.org/threads/diplomandoci.192093/
http://fire-team.ru/forum/showthread.php?p=35312#post35312
Здравствуйте!
Где купить диплом по актуальной специальности?
Приобрести документ университета можно у нас. Мы оказываем услуги по продаже документов об окончании любых университетов России.
Мы можем предложить дипломы любых профессий по выгодным тарифам. Стоимость будет зависеть от выбранной специальности, года выпуска и образовательного учреждения. Всегда стараемся поддерживать для покупателей адекватную ценовую политику.
https://www.tapatalk.com/groups/dzerjinsky/viewtopic.php?f=2&t=34658&from_new_topic=1
Хорошей учебы!
[b]Здравствуйте[/b]!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом в челябинске, купить аттестаты за 9, купить диплом в красноярске, купить диплом стоматолога, купить диплом вуза. Постепенно углубляясь, нашел отличный ресурс здесь: https://arms-russia.ru/gostevaya/?status=success, и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Успешной учебы!
Привет, друзья!
Где заказать диплом специалиста?
Заказать документ о получении высшего образования можно в нашей компании. Мы предлагаем документы об окончании любых университетов Российской Федерации.
Наша компания предлагает максимально быстро приобрести диплом, который выполняется на бланке ГОЗНАКа и заверен мокрыми печатями, водяными знаками, подписями должностных лиц. Наш документ способен пройти лубую проверку, даже с использованием специального оборудования. Достигайте свои цели быстро и просто с нашими дипломами.
http://zorkiy.bestbb.ru/viewtopic.php?id=192
http://panther.80lvl.ru/viewtopic.php?f=19&t=378
https://inopl.com/blogs/457/%D0%9F%D0%BE%D1%87%D0%B5%D0%BC%D1%83-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D1%8B-%D1%80%D0%B5%D0%B0%D0%BB%D0%B8%D0%B7%D1%83%D1%8E%D1%89%D0%B8%D0%B5-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D1%8B-%D1%82%D0%B0%D0%BA-%D0%BF%D0%BE%D0%BF%D1%83%D0%BB%D1%8F%D1%80%D0%BD%D1%8B-%D1%81%D0%B5%D0%B9%D1%87%D0%B0%D1%81
https://www.indianhighcaste.com/read-blog/319
http://algebra.bestbb.ru/viewtopic.php?id=255
Здравствуйте!
Где заказать диплом по необходимой специальности?
Мы можем предложить документы ВУЗов, расположенных на территории всей России. Документы будут заверены необходимыми печатями и подписями.
Приобрести диплом о высшем образовании:
http://wiki.siras.ru/wiki/???????_???????????_????????:_??????????_???????_????????_??????
Привет, друзья!
Приобрести диплом о высшем образовании.
Мы предлагаем приобрести диплом высочайшего качества, который невозможно отличить от оригинального документа без участия специалистов высокой квалификации с дорогостоящим оборудованием.
Где заказать диплом по необходимой специальности?
http://verkehrsknoten.de/index.php/forum/sozialvorschriften/90905-premiummdiplomms#91867
Успехов в учебе!
Добрый день!
Для определенных людей, купить диплом о высшем образовании – это острая потребность, шанс получить выгодную работу. Но для кого-то – это желание не терять время на учебу в институте. Что бы ни толкнуло вас на такой шаг, наша фирма готова помочь вам. Быстро, профессионально и по разумной стоимости изготовим диплом любого ВУЗа и года выпуска на государственных бланках со всеми необходимыми подписями и печатями.
Мы предлагаем быстро и выгодно приобрести диплом, который выполняется на бланке ГОЗНАКа и заверен печатями, штампами, подписями официальных лиц. Диплом пройдет лубую проверку, даже при использовании специально предназначенного оборудования. Достигайте цели максимально быстро с нашими дипломами.
Где купить диплом специалиста?
http://fh9947a1.bget.ru/blog/10412.html
Хорошей учебы!
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф. Но, оказалось, что все возможно! Сначала искал информацию по теме: купить диплом переводчика, купить диплом мастера маникюра и педикюра, купить диплом в ухте, купить диплом в обнинске, купить диплом в старом осколе, а затем перешел на дипломы вузов. Подробнее можно узнать здесь:
http://qa.laodongzu.com/index.php?qa=user&qa_1=ytohuw
http://msflow.ru/products/pionovidnye-rozy-v-korobke/#comment_60753/
http://school5.p-fam.ru/users/edasopeno
https://sosna-dub32.ru/blog/vyraschivanie-maliny-v-otkrytom-grunte/#comment_166825
http://www.qwe.ru/comment.php?comment.news.3644.extend
Оказалось, что все возможно и официально, с упрощенными программами обучения. Теперь у меня диплом московского вуза нового образца, и я рекомендую вам воспользоваться этим шансом!
Хорошей учебы!
Здравствуйте!
Заказать документ института можно в нашем сервисе. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов Российской Федерации.
Мы предлагаем максимально быстро заказать диплом, который выполняется на бланке ГОЗНАКа и заверен мокрыми печатями, водяными знаками, подписями. Наш диплом пройдет любые проверки, даже при использовании профессиональных приборов.
http://krutovo.ru/index.php?name=Account&op=info&uname=orovujata
http://unqbit.org/bbs/board.php?bo_table=free&wr_id=5492
http://allrealtor.ru/threads/1417/
http://rznpoisk.ru/viewtopic.php?f=11&t=2933
http://dianov.bget.ru/forum/thread57506.html#1281078
Добрый день!
Где приобрести диплом специалиста?
Мы готовы предложить документы ВУЗов, которые находятся на территории всей Российской Федерации. Можно купить качественный диплом от любого ВУЗа, за любой год, включая сюда документы СССР. Дипломы выпускаются на “правильной” бумаге высшего качества. Это позволяет делать настоящие дипломы, не отличимые от оригиналов. Документы будут заверены необходимыми печатями и штампами.
Мы можем предложить дипломы любых профессий по приятным тарифам.
https://diplomyx24.ru/kupit-diplom-tehnikuma-kolledzha
Рады оказаться полезными!
Привет!
Мы готовы предложить дипломы психологов, юристов, экономистов и любых других профессий по доступным ценам.
Мы предлагаем документы техникумов, которые расположены в любом регионе России. Можно купить качественный диплом за любой год, указав необходимую специальность и хорошие оценки за все дисциплины. Дипломы и аттестаты выпускаются на “правильной” бумаге самого высшего качества. Это дает возможность делать государственные дипломы, которые не отличить от оригинала. Они будут заверены необходимыми печатями и подписями.
Где заказать диплом специалиста?
http://www.camnangbenh.com/mu-mau/
Привет!
Где заказать диплом специалиста?
Заказать документ ВУЗа вы сможете в нашей компании. Мы предлагаем документы об окончании любых ВУЗов РФ. Вы сможете получить необходимый диплом по любой специальности, включая документы СССР. Гарантируем, что при проверке документа работодателем, подозрений не возникнет.
https://cessi.com/contacts/
Успехов в учебе!
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф и невозможно. Но, к счастью, оказался неправ. Сначала искал информацию по теме: купить диплом в новоуральске, купить диплом в озёрске, купить диплом в великом новгороде, купить бланк диплома, #купить диплом в твери, а затем переключился на дипломы вузов. Подробности здесь: https://salda.ws/meet/notes.php?id=13211
Оказалось, что все реально и легально, со специальными условиями и упрощенными программами. Теперь у меня диплом московского вуза нового образца, что я настоятельно рекомендую и вам!
Успехов в учебе!
Здравствуйте!
Хочу поделиться своим опытом по заказу аттестата ПТУ. Думал, что это невозможно, и начал искать информацию в интернете по теме: купить диплом в уссурийске, купить диплом товароведа, купить диплом инженера по охране труда, купить диплом в альметьевске, купить диплом гознак. Постепенно углубляясь, нашел отличный ресурс здесь: http://www.nhps1914.com/wiki/??????_??????:_???_?????_?_????????_???????, и остался очень доволен!
Теперь у меня есть диплом сварщика о среднем специальном образовании, и я обеспечен на всю жизнь!
Удачи!
Добрый день!
Где заказать диплом специалиста?
Полученный диплом с приложением отвечает требованиям и стандартам, никто не отличит его от оригинала – даже со специально предназначенным оборудованием.
Приобрести диплом любого университета.
http://mmix.ukrbb.net/viewtopic.php?f=23&t=22471
https://sonnick84.newbigblog.com/34149914/%D0%A6%D0%B5%D0%BD%D0%BE%D0%BE%D0%B1%D1%80%D0%B0%D0%B7%D0%BE%D0%B2%D0%B0%D0%BD%D0%B8%D0%B5-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%B0-%D0%B0%D0%B2%D1%82%D0%BE%D1%80%D1%81%D0%BA%D0%B8%D0%B9-%D0%BE%D0%B1%D0%B7%D0%BE%D1%80-%D1%81%D0%BF%D0%B5%D1%86%D0%B8%D0%B0%D0%BB%D0%B8%D1%81%D1%82%D0%BE%D0%B2
https://ozoms.com/read-blog/477
https://asepbook.com/create-blog/
http://csexpert.4adm.ru/posting.php?mode=post&f=70&sid=9bbdff1ea7bab9a6feb0268f576f7ed9
Привет, друзья!
Всегда считал, что покупка диплома о высшем образовании — это миф и невозможно. Но, к счастью, оказался неправ. Сначала искал информацию по теме: купить диплом в нальчике, купить диплом в екатеринбурге, купить диплом биолога, купить диплом в новотроицке, #купить свидетельство о разводе, а затем переключился на дипломы вузов. Подробности здесь: https://vidyasanskarschool.com/2018/08/17/independence-day-flag-hoisting-newspaper-coverage/
https://widayati.com/announcement-pengumuman/
https://tambonyo.go.th/forum/suggestion-box/1019690-dipl
https://newacttravel.com/travel-stocking-stuffer-musts/carry-on-21/
https://qatar.carsprite.com/en/car-prices/volvo/v40-2017
Оказалось, что все реально и легально, со специальными условиями и упрощенными программами. Теперь у меня диплом московского вуза нового образца, что я настоятельно рекомендую и вам!
Успешной учебы!
купить диплом с подтверждением http://diploms-x.com .
Здравствуйте!
Задумался а действительно можно купить диплом государственного образца в Москве, и был удивлен, все реально и главное официально!
Сначала серфил в сети и искал такие темы как: купить диплом парикмахера, купить диплом электромонтажника, купить диплом в благовещенске, купить диплом в перми, купить диплом в сарове, получил базовую информацию.
Остановился в итоге на материале купить диплом, https://bookmytutor.in/lms-wordpress-plugin/?unapproved=8489&moderation-hash=8433eb6b74d54e011eb84d2e23ce813b#comment-8489
Успешной учебы!
Привет, друзья!
Где купить диплом по нужной специальности?
Заказать документ о получении высшего образования вы можете в нашей компании. Мы предлагаем документы об окончании любых университетов РФ. Вы получите диплом по любой специальности, любого года выпуска, в том числе документы Советского Союза. Гарантируем, что в случае проверки документа работодателем, подозрений не возникнет.
http://www.hoahaubansacviet.com.vn/xac-nhan-binh-chon.flc?sbd=335
Успехов в учебе!
[u][b] Привет![/b][/u]
Где приобрести диплом специалиста?
Наша компания предлагает максимально быстро купить диплом, который выполняется на оригинальном бланке и заверен печатями, водяными знаками, подписями должностных лиц. Документ способен пройти любые проверки, даже при использовании специально предназначенного оборудования. Решите свои задачи максимально быстро с нашими дипломами.
[b]Приобрести диплом о высшем образовании.[/b]
https://myworldgo.com/blog/108204/kak-mozhno-nedorogo-priobresti-diplom-v-onlajn-magazine
https://socialnetwork.cloudyzx.com/read-blog/12109
https://abris-zip.ru/company/personal/user/171/forum/message/1246/1256/
https://ourownnetwork.mn.co/posts/61838009
https://network-66643.mn.co/posts/61838228
Привет, друзья!
Мы готовы предложить документы ВУЗов, которые расположены на территории всей РФ. Вы можете купить диплом за любой год, включая документы СССР. Документы делаются на бумаге самого высшего качества. Это позволяет делать государственные дипломы, не отличимые от оригинала. Они заверяются необходимыми печатями и подписями.
Купить диплом о высшем образовании.
https://interactor.pro/read-blog/1360
https://samovod.ru/content/articles/65991/
https://www.flexsocialbox.com/read-blog/6185
https://lookeify.com/read-blog/7156
https://myworldgo.com/blog/108204/kak-mozhno-nedorogo-priobresti-diplom-v-onlajn-magazine
Хорошей учебы!
Привет!
Заказать диплом о высшем образовании.
Мы предлагаем купить диплом в высочайшем качестве, который не отличить от оригинала без участия специалистов высокой квалификации с дорогим оборудованием.
Где приобрести диплом специалиста?
http://agenciafrog-host.com.br/redesocial_v0/index.php/profile/yjyfo
Успешной учебы!
Привет, друзья!
Заказать документ института вы имеете возможность в нашей компании. Мы оказываем услуги по продаже документов об окончании любых ВУЗов Российской Федерации.
Мы предлагаем выгодно заказать диплом, который выполнен на бланке ГОЗНАКа и заверен мокрыми печатями, водяными знаками, подписями должностных лиц. Данный диплом пройдет лубую проверку, даже при использовании профессиональных приборов.
http://steklo.by/index.php?subaction=userinfo&user=iqohum
http://wiki.gta-zona.ru/index.php?title=premiummdiplomms
https://forum.moi-psihoanalitik.ru/index.php?/topic/5676-%D0%BA%D0%B0%D0%BA-%D1%81%D0%BE%D0%B7%D0%B4%D0%B0%D1%82%D1%8C-%D0%BD%D0%BE%D0%B2%D1%83%D1%8E-%D1%82%D0%B5%D0%BC%D1%83-forummoi-psihoanalitikru/page/9/#comment-7754
https://gothic2online.ru/index.php?/topic/6588-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%86%D0%B5%D0%BD%D0%B0-j418f/
https://gmaii.ru/threads/2185/
Привет!
Где приобрести диплом по нужной специальности?
Заказать документ ВУЗа вы имеете возможность в нашей компании в столице. Мы оказываем услуги по продаже документов об окончании любых ВУЗов РФ. Вы получите диплом по любой специальности, включая документы СССР. Гарантируем, что при проверке документов работодателями, никаких подозрений не появится.
http://www.hagi.co.kr/bbs/board.php?bo_table=free&wr_id=248310
Хорошей учебы!
Thank you for sharing your thoughts. I really appreciate your efforts and I am waiting for your next post thank you once again.
https://mega-m.com.ua/novi-trendy-u-vyrobnytstvi-skla-dlya-far-shcho-ochikuvaty-u-2024-rotsi
Здравствуйте!
Мы изготавливаем дипломы психологов, юристов, экономистов и любых других профессий по выгодным ценам. Цена будет зависеть от выбранной специальности, года выпуска и университета.
Где купить диплом специалиста?
Приобрести диплом университета
https://arusak-diploms-srednee.ru/kupit-diplom-v-cheliabinske
Успешной учебы!
промокод на фрибет 1xbet
промокод для 1хбет
Здравствуйте!
Мы предлагаем документы ВУЗов, которые находятся на территории всей РФ. Можно купить диплом за любой год, указав актуальную специальность и оценки за все дисциплины. Документы делаются на бумаге самого высшего качества. Это позволяет делать государственные дипломы, которые невозможно отличить от оригинала. Они будут заверены всеми требуемыми печатями и подписями.
Заказать диплом о высшем образовании.
https://ci17lo6ca.shoutmyblog.com/27914397/%D0%9A%D0%B0%D0%BA-%D0%B2%D0%BE%D0%B7%D0%BC%D0%BE%D0%B6%D0%BD%D0%BE-%D0%BD%D0%B5%D0%B4%D0%BE%D1%80%D0%BE%D0%B3%D0%BE-%D0%BF%D1%80%D0%B8%D0%BE%D0%B1%D1%80%D0%B5%D1%81%D1%82%D0%B8-%D0%B0%D1%82%D1%82%D0%B5%D1%81%D1%82%D0%B0%D1%82-%D0%B2-%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
https://lidertude.com/blogs/1193/%D0%A0%D0%B0%D1%81%D1%88%D0%B8%D1%80%D0%B5%D0%BD%D0%BD%D0%BE%D0%B5-%D0%BE%D0%BF%D0%B8%D1%81%D0%B0%D0%BD%D0%B8%D0%B5-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D0%B0-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%B0-%D0%B2-%D0%B8%D0%B7%D0%B2%D0%B5%D1%81%D1%82%D0%BD%D0%BE%D0%BC-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
https://ourfathersfamily.com/blogs/2400/%D0%9A%D0%B0%D0%BA-%D0%B2%D0%BE%D0%B7%D0%BC%D0%BE%D0%B6%D0%BD%D0%BE-%D0%BD%D0%B5%D0%B4%D0%BE%D1%80%D0%BE%D0%B3%D0%BE-%D0%BF%D1%80%D0%B8%D0%BE%D0%B1%D1%80%D0%B5%D1%81%D1%82%D0%B8-%D0%B0%D1%82%D1%82%D0%B5%D1%81%D1%82%D0%B0%D1%82-%D0%B2-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
https://oolibuzz.com/read-blog/3737
https://ihz6ljp1t.anchor-blog.com/8550882/%D0%9A%D0%B0%D0%BA-%D0%BC%D0%BE%D0%B6%D0%BD%D0%BE-%D0%B1%D1%83%D0%B4%D0%B5%D1%82-%D0%B1%D1%8B%D1%81%D1%82%D1%80%D0%BE-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
Удачи!
Всем привет)
Будучи студентом, я наслаждался учебой до тех пор, пока не пришло время писать диплом. Но паниковать не стоило, ведь существуют компании, которые помогают с написанием и защитой диплома на отличные оценки!
Изначально я искал информацию по теме: купить диплом штукатура, купить аттестат за 9 классов, купить диплом в минеральных водах, купить диплом в железногорске, купить диплом в находке, затем наткнулся на https://stoliistulya.ru/products/stol-serii-sptm-1207-1208/#comment_272, где все мои учебные вопросы были решены!
Успешной учебы!
Привет!
Где приобрести диплом по необходимой специальности?
Мы готовы предложить документы ВУЗов, расположенных в любом регионе РФ. Можно купить диплом от любого заведения, за любой год, указав необходимую специальность и хорошие оценки за все дисциплины. Дипломы печатаются на бумаге высшего качества. Это дает возможности делать государственные дипломы, которые не отличить от оригинала. Документы заверяются необходимыми печатями и подписями.
Мы изготавливаем дипломы любой профессии по приятным тарифам.
https://diplomasx24.ru/kupit-diplom-specialista
Окажем помощь!
Добрый день!
Где купить диплом специалиста?
Наши специалисты предлагают выгодно и быстро приобрести диплом, который выполнен на оригинальной бумаге и заверен мокрыми печатями, штампами, подписями должностных лиц. Наш диплом пройдет лубую проверку, даже с использованием специального оборудования. Достигайте свои цели максимально быстро с нашими дипломами.
Купить диплом о высшем образовании.
http://www.growingchurches.org/blogs/22079/%D0%A0%D0%B0%D1%81%D1%88%D0%B8%D1%80%D0%B5%D0%BD%D0%BD%D0%BE%D0%B5-%D0%BE%D0%BF%D0%B8%D1%81%D0%B0%D0%BD%D0%B8%D0%B5-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D0%B0-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%B0-%D0%B2-%D0%B7%D0%BD%D0%B0%D0%BC%D0%B5%D0%BD%D0%B8%D1%82%D0%BE%D0%BC-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
https://huarenjiaohui.com/index.php?/gallery/image/126-%D0%BF%D1%80%D0%B8%D0%BE%D0%B1%D1%80%D0%B5%D1%82%D0%B0%D0%B5%D0%BC-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%BF%D1%80%D0%BE%D0%B2%D0%B5%D1%80%D0%B5%D0%BD%D0%BD%D0%BE%D0%BC-%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5-%D0%BF%D0%BE-%D0%B2%D1%8B%D0%B3%D0%BE%D0%B4%D0%BD%D0%BE%D0%B9-%D1%86%D0%B5%D0%BD%D0%B5/
https://onetable.world/read-blog/60071
https://www.clubalpintoulouse.fr/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=154
https://nto30v7m8.is-blog.com/34540150/%D0%94%D0%B5%D1%82%D0%B0%D0%BB%D1%8C%D0%BD%D0%BE%D0%B5-%D0%BE%D0%BF%D0%B8%D1%81%D0%B0%D0%BD%D0%B8%D0%B5-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D0%B0-%D0%B0%D1%82%D1%82%D0%B5%D1%81%D1%82%D0%B0%D1%82%D0%B0-%D0%B2-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5
WELCOME TO PUGACHEV LUXURY CAR RENTALS EXCLUSIVE MOTORING LUXURY & EXOTIC RENTALS
Dive into the ultimate driving experience with Pugachev Luxury Car Rentals, your premier destination for luxury and exotic car rentals in Los Angeles. Whether you’re looking to impress at a corporate event, add a splash of glamour to your weekend, or simply enjoy the thrill of driving some of the world’s most sought-after vehicles, we’ve got you covered.
https://pugachev.la/
Our extensive fleet includes the pinnacle of automotive excellence from renowned brands such as Lamborghini, Rolls-Royce, Bentley, Ferrari, McLaren, and more. Indulge in the elegance of a Rolls-Royce Ghost, starting at $1,150 per day, or experience the exhilarating performance of a Lamborghini Huracan EVO, available from $875 per day. With options ranging from the powerful Ferrari GTB Red at $1,100 per day to the opulent Bentley Continental GT at $950 per day, our collection is meticulously curated to cater to the most discerning tastes and preferences.
Navigating the vibrant streets of Los Angeles or cruising along the scenic coastlines has never been more accessible or more stylish. At Pugachev Luxury Car Rentals, we make it easy and efficient for you to rent the luxury car of your dreams with straightforward daily and weekly rental options—like the versatile Mercedes-Benz AMG G63, which can be yours for $790 daily or $4,424 weekly.
Embrace the luxury, power, and finesse of our elite vehicles and elevate your next Los Angeles visit into a truly memorable journey. Whether you’re seeking an exotic sports car for that extra special thrill, a luxury sedan for sophisticated travel, or a sumptuous SUV for family comfort and safety, our fleet promises the perfect match for every occasion and desire.
OUR COLLECTION OF LUXURY CARS FOR DISCERNING DRIVERS
Elegant Sedans for Business Executives
At Pugachev Luxury Car Rentals, we understand the importance of a strong first impression. Our fleet of luxury sedans is designed to cater to business executives who demand sophistication and functionality. Each vehicle, such as the Rolls-Royce Ghost Anthracite, available at $1,100 per day, offers a sublime mix of comfort and prestige, ensuring you arrive at your business engagements in unparalleled style. For a slightly different but equally impressive experience, consider the Bentley Flying Spur at $1,000 daily, which combines classic elegance with cutting-edge technology.
luxury and exotic car rental in los angeles
bentley rental la
Sumptuous SUVs for Family and Leisure
Our selection of high-end SUVs blends luxury with practicality, making them perfect for family trips or stylish outings with friends. The Bentley Bentayga, starting from $700 per day, offers spaciousness and state-of-the-art amenities that ensure comfort without compromise. For those seeking a bolder statement, the Rolls-Royce Cullinan Black Badge provides an imposing presence and unrivaled luxury for $1,300 daily, ensuring every journey is an event in itself.
Prestige Convertibles for Scenic Drives
Los Angeles is renowned for its beautiful landscapes and stunning coastlines, best enjoyed in one of our prestige convertibles. Feel the exhilarating freedom of the open road in a Ferrari Portofino Spyder, available for $850 daily. Its breathtaking performance and sleek design are perfect for scenic drives along the Pacific Coast Highway. Alternatively, the BMW M4 Competition Cabrio, at $390 per day, offers a more accessible but equally thrilling option for cruising under the Californian sun.
Our fleet at Pugachev Luxury Car Rentals is meticulously maintained and regularly updated to ensure that each car looks and performs like new. This commitment to quality and excellence reflects our dedication to providing only the best for our clients, who return time and again for the reliability and prestige our services offer. Each model in our fleet is more than just a vehicle; it’s a part of an exclusive lifestyle made accessible to our discerning clientele. Whether you are looking to impress at a corporate event or simply indulge in the pleasure of driving a luxury car, our collection is guaranteed to enhance your experience in Los Angeles.
luxury and exotic car rental in los angeles
lamborghini rental in los angeles
Привет, друзья!
Где купить диплом по нужной специальности?
Готовый диплом со всеми печатями и подписями отвечает условиям и стандартам, неотличим от оригинала – даже со специально предназначенным оборудованием.
Приобрести диплом любого ВУЗа.
http://avtobestnews.ru/kupit-diplom-opyit-i-professionalizm
https://bondhusova.com/blogs/60411/%D0%93%D1%80%D0%B0%D0%BC%D0%BE%D1%82%D0%BD%D0%BE-%D0%BF%D1%80%D0%B8%D0%BE%D0%B1%D1%80%D0%B5%D1%82%D0%B0%D0%B5%D0%BC-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D1%81%D0%B5%D1%82%D0%B8-%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%B0%D0%B2%D1%82%D0%BE%D1%80%D1%81%D0%BA%D0%B8%D0%B9-%D0%BE%D0%B1%D0%B7%D0%BE%D1%80
https://instakos.mn.co/posts/61679449
https://www.cyberpinoy.net/read-blog/32916
http://demo.advised360.com/read-blog/161564
Здравствуйте!
Где заказать диплом по необходимой специальности?
Мы готовы предложить документы техникумов, расположенных на территории всей Российской Федерации. Можно приобрести диплом за любой год, указав подходящую специальность и оценки за все дисциплины. Дипломы и аттестаты печатаются на “правильной” бумаге высшего качества. Это позволяет делать государственные дипломы, не отличимые от оригинала. Они будут заверены всеми обязательными печатями и подписями.
ediblehomegardensresort.com/index.php?do=/public/blog/view/id_119635/title_-2024/
Привет всем)
Хорошо быть студентом, пока не придет пора писать диплом, что и произошло со мной, но не стоит отчаиваться, ведь есть хорошие компании что помогают с написанием и сдачей диплома на хорошие оценки!
Изначально искал информацию про купить диплом в королёве, купить диплом в смоленске, купить диплом инженера по охране труда, купить диплом в уфе, купить диплом колледжа, потом попал на
http://mockwa.com/forum/thread-150888/
http://sergei-cheremushkin.blogspot.com/2010/03/blog-post.html
http://mega-zaim96.ru/forum/user/34469/
http://tekst-pesni.ru/index.php?name=account&op=info&uname=aquse
http://www.printsstars.ru/communication/forum/index.php?PAGE_NAME=profile_view&UID=201303
и там решили все мои учебные заботы!
Хорошей учебы!
Tributes flood in for BBC sport commentator whose wife and daughters were killed in suspected crossbow attack
[url=https://telltrue.net/eto-lohotron/chernyj-spisok-lohotronov-utl-coinexfom-uno-club-bitlans-profitcrypto/?ysclid=ly1sujvcbx993980390]русское порно жесток[/url]
Figures from across the United Kingdom have offered their condolences to a BBC sport commentator, after his wife and two daughters were killed by an alleged crossbow attacker, in deaths that again drew attention to the epidemic of violence against women.
Carol Hunt, 61, wife of BBC horse racing commentator, John Hunt, and their daughters, Hannah Hunt, 28, and Louise Hunt, 25, died from injuries sustained in an attack in Bushey, just northwest of London, on Tuesday, according to police and Britain’s public broadcaster.
A 26-year-old suspect wanted in connection with the killings, and named as Kyle Clifford, was found by British police in Enfield, north London, on Wednesday, following a sprawling manhunt. There were no previous reports to the force over Clifford, who is in serious condition in hospital and is yet to speak with officers, Hertfordshire Police said in a statement.
A crossbow was recovered as part of the investigation, which police believe was used in a “targeted incident.”
Epidemic of violence against women
The killings of the three women rocked Britain, where mass murders are infrequent but violence against women and girls has been officially labeled as a national threat.
A woman is killed by a man every three days in the UK and one in four women will experience domestic violence in her lifetime, the United Nations’ special rapporteur on violence against women and girls, Reem Alsalem, said earlier this year.
Charities and human rights organizations have subsequently reiterated urgent demands to tackle femicide in the UK.
Здравствуйте!
Мы изготавливаем дипломы любой профессии по доступным ценам. Стоимость может зависеть от той или иной специальности, года получения и образовательного учреждения.
Где купить диплом специалиста?
Купить диплом любого ВУЗа
https://arusak-diploms-srednee.ru/diplom-sredne-spetcialnoe-obrazovanie
Успехов в учебе!
Tributes flood in for BBC sport commentator whose wife and daughters were killed in suspected crossbow attack
смотреть порно жесток
Figures from across the United Kingdom have offered their condolences to a BBC sport commentator, after his wife and two daughters were killed by an alleged crossbow attacker, in deaths that again drew attention to the epidemic of violence against women.
Carol Hunt, 61, wife of BBC horse racing commentator, John Hunt, and their daughters, Hannah Hunt, 28, and Louise Hunt, 25, died from injuries sustained in an attack in Bushey, just northwest of London, on Tuesday, according to police and Britain’s public broadcaster.
A 26-year-old suspect wanted in connection with the killings, and named as Kyle Clifford, was found by British police in Enfield, north London, on Wednesday, following a sprawling manhunt. There were no previous reports to the force over Clifford, who is in serious condition in hospital and is yet to speak with officers, Hertfordshire Police said in a statement.
A crossbow was recovered as part of the investigation, which police believe was used in a “targeted incident.”
Epidemic of violence against women
The killings of the three women rocked Britain, where mass murders are infrequent but violence against women and girls has been officially labeled as a national threat.
A woman is killed by a man every three days in the UK and one in four women will experience domestic violence in her lifetime, the United Nations’ special rapporteur on violence against women and girls, Reem Alsalem, said earlier this year.
Charities and human rights organizations have subsequently reiterated urgent demands to tackle femicide in the UK.
Привет, друзья!
Мы готовы предложить дипломы любой профессии по невысоким тарифам.
Мы можем предложить документы техникумов, которые расположены в любом регионе РФ. Вы имеете возможность заказать качественно сделанный диплом за любой год, указав актуальную специальность и оценки за все дисциплины. Документы делаются на “правильной” бумаге самого высокого качества. Это дает возможности делать настоящие дипломы, которые не отличить от оригиналов. Они заверяются всеми требуемыми печатями и подписями.
Где купить диплом специалиста?
cogita.ru/news/otchety/andrei-scherbak.-gosudarstvo-i-malyi-biznes-ruka-dayuschaya-i-ruka-karayuschaya
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
Владимир Колокольцев
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
Привет!
Где приобрести диплом специалиста?
Купить документ института вы имеете возможность в нашей компании. Мы оказываем услуги по продаже документов об окончании любых университетов Российской Федерации.
Мы изготавливаем дипломы любой профессии по приятным ценам. Цена будет зависеть от определенной специальности, года получения и образовательного учреждения. Стараемся поддерживать для клиентов адекватную политику цен.
https://1wum.ru/forum/?PAGE_NAME=profile_view&UID=35935&MUL_MODE=
Успехов в учебе!
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
Владимир Колокольцев
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
Привет, друзья!
Где купить диплом по актуальной специальности?
Мы готовы предложить документы ВУЗов, которые находятся на территории всей РФ. Можно приобрести качественно напечатанный диплом за любой год, указав подходящую специальность и оценки за все дисциплины. Дипломы выпускаются на бумаге самого высшего качества. Это позволяет делать настоящие дипломы, которые невозможно отличить от оригиналов. Документы заверяются всеми обязательными печатями и штампами.
Мы предлагаем дипломы любой профессии по разумным тарифам.
https://diploms-x.com/kupit-diplom-chelyabinsk
Окажем помощь!
Добрый день!
Мы готовы предложить документы ВУЗов, расположенных на территории всей Российской Федерации. Вы можете заказать качественно сделанный диплом за любой год, включая документы старого образца СССР. Документы печатаются на бумаге самого высшего качества. Это дает возможности делать государственные дипломы, не отличимые от оригинала. Они будут заверены всеми требуемыми печатями и подписями.
Купить диплом любого ВУЗа.
https://forum.recifalnews.fr/gallery/image/218-%D0%BD%D0%B8%D0%B7%D0%BA%D0%B8%D0%B5-%D1%86%D0%B5%D0%BD%D1%8B-%D0%B8-%D0%B2%D1%8B%D1%81%D0%BE%D0%BA%D0%BE%D0%B5-%D0%BA%D0%B0%D1%87%D0%B5%D1%81%D1%82%D0%B2%D0%BE-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D1%8B%D0%B2%D0%B0%D0%B9-%D0%B4%D0%BE%D0%BA%D1%83%D0%BC%D0%B5%D0%BD%D1%82%D1%8B-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD/
https://facefam.com/read-blog/334
https://blogger-mania.mn.co/posts/61838359
https://eyawei-networks.mn.co/posts/61838196
https://lab.panda-studio.pro/index.php?/gallery/image/119-%D0%B7%D0%B0%D0%BA%D0%B0%D0%B7%D1%8B%D0%B2%D0%B0%D0%B5%D0%BC-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%BB%D1%83%D1%87%D1%88%D0%B5%D0%BC-%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5-%D0%BF%D0%BE-%D0%B2%D1%8B%D0%B3%D0%BE%D0%B4%D0%BD%D0%BE%D0%B9-%D1%86%D0%B5%D0%BD%D0%B5/
Успешной учебы!
Spot on with this write-up, I really believe this website needs much more attention. I’ll probably be returning to read more, thanks for the information.
Здравствуйте!
Где приобрести диплом по актуальной специальности?
Готовый диплом с приложением 100% отвечает запросам и стандартам Министерства образования и науки России, никто не сможет отличить его от оригинала – даже со специальным оборудованием.
Заказать диплом о высшем образовании.
https://sonnick84.blogsvirals.com/27849725/%D0%A0%D0%B5%D0%BA%D0%BE%D0%BC%D0%B5%D0%BD%D0%B4%D0%B0%D1%86%D0%B8%D0%B8-%D1%81%D0%BF%D0%B5%D1%86%D0%B8%D0%B0%D0%BB%D0%B8%D1%81%D1%82%D0%B0-%D0%BA%D0%BE%D1%82%D0%BE%D1%80%D1%8B%D0%B5-%D0%BF%D0%BE%D0%B7%D0%B2%D0%BE%D0%BB%D1%8F%D1%82-%D0%B1%D1%8B%D1%81%D1%82%D1%80%D0%BE-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC
https://dailygram.com/blog/1304350/4-%D0%B5-%D0%B3%D0%BB%D0%B0%D0%B2%D0%BD%D1%8B%D1%85-%D0%B3%D1%80%D1%83%D0%BF%D0%BF%D1%8B-%D0%BE%D0%BD%D0%BB%D0%B0%D0%B9%D0%BD-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%BE%D0%B2-%D0%BA%D0%BE%D1%82%D0%BE%D1%80%D1%8B%D0%B5-%D0%BF%D1%80%D0%BE%D0%B4%D0%B0%D1%8E%D1%82-%D0%B4%D0%BE%D0%BA%D1%83%D0%BC%D0%B5%D0%BD%D1%82%D1%8B/
http://x91392sl.beget.tech/2024/07/09/diplomy-na-zakaz-garantiya-kachestva.html
https://transfer-tur.ru/ru/cb-profile/pluginclass/cbblogs?action=blogs&func=show&id=169
https://www.djjmeets.com/blogs/179064/%D0%93%D0%BB%D0%B0%D0%B2%D0%BD%D1%8B%D0%B5-%D1%80%D0%B0%D1%81%D1%85%D0%BE%D0%B4%D1%8B-%D1%80%D0%BE%D1%81%D1%81%D0%B8%D0%B9%D1%81%D0%BA%D0%BE%D0%B3%D0%BE-%D0%B8%D0%B7%D0%B3%D0%BE%D1%82%D0%BE%D0%B2%D0%B8%D1%82%D0%B5%D0%BB%D1%8F-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%BE%D0%B2-%D0%B0%D0%B2%D1%82%D0%BE%D1%80%D1%81%D0%BA%D0%B8%D0%B9-%D0%BE%D0%B1%D0%B7%D0%BE%D1%80
Добрый день!
Купить диплом о высшем образовании.
Мы предлагаем заказать диплом высокого качества, неотличимый от оригинала без участия специалиста высокой квалификации со специальным оборудованием.
Где купить диплом специалиста?
http://heimur.ru/index.php?/topic/134-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%86%D0%B5%D0%BD%D0%B0-m36k/
Удачи!
Добрый день!
Где заказать диплом по необходимой специальности?
Приобрести документ о получении высшего образования вы можете в нашем сервисе. Мы оказываем услуги по изготовлению и продаже документов об окончании любых ВУЗов РФ. Вы сможете получить необходимый диплом по любым специальностям, включая документы Советского Союза. Даем 100% гарантию, что при проверке документа работодателем, никаких подозрений не возникнет.
http://www.ingenieursdudimanche.lagob.fr/doku.php?id=diplomaasx
Успешной учебы!
Добрый день!
Приобрести документ института можно в нашем сервисе. Мы предлагаем документы об окончании любых ВУЗов Российской Федерации.
Мы предлагаем быстро приобрести диплом, который выполнен на бланке ГОЗНАКа и заверен печатями, штампами, подписями должностных лиц. Диплом способен пройти лубую проверку, даже при использовании специальных приборов.
http://kochtagebuch.org/doku.php?id=premiummdiplomms
http://dayz.tselik.ru/profile.php?action=show&member=10396
https://www.tdp74.ru/forum/messages/forum1/topic96/message47453/?result=reply#message47453
https://gmaii.ru/threads/2184/
https://radio-wave.ru/forum/showthread.php?3845-%D0%A0%D1%94%D0%A1%D1%93%D0%A0%D1%97%D0%A0%D1%91%D0%A1%E2%80%9A%D0%A1%D0%8A-%D0%A0%D2%91%D0%A0%D1%91%D0%A0%D1%97%D0%A0%C2%BB%D0%A0%D1%95%D0%A0%D1%98-w456f&p=9655#post9655
I really like it when folks come together and share views. Great site, continue the good work!
Добрый день!
Где купить диплом специалиста?
Мы предлагаем максимально быстро приобрести диплом, который выполнен на оригинальном бланке и заверен мокрыми печатями, штампами, подписями официальных лиц. Данный диплом способен пройти лубую проверку, даже при использовании профессионального оборудования. Решите свои задачи быстро и просто с нашей компанией.
Заказать диплом любого ВУЗа.
https://www.bisshogram.com/read-blog/281
https://forum.twilightmu.com/album.php?albumid=247&pictureid=1658
https://www.nhps1914.com/wiki/%D0%97%D0%B0%D0%BA%D0%B0%D0%B7%D1%8B%D0%B2%D0%B0%D0%B5%D0%BC_%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC_%D0%B2_%D0%B8%D0%B7%D0%B2%D0%B5%D1%81%D1%82%D0%BD%D0%BE%D0%BC_%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82_%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B5_%D0%BF%D0%BE_%D0%BA%D0%BE%D0%BC%D1%84%D0%BE%D1%80%D1%82%D0%BD%D0%BE%D0%B9_%D1%86%D0%B5%D0%BD%D0%B5
https://www.datasheetclub.com/read-blog/92
https://heyjinni.com/read-blog/109935
A fairly new player in the Russian darknet arena, [url=https://bs-gl-darknet.com]blacksprut[/url] Blacksprut has quickly gained attention for its interesting features and growing popularity. While some aspects raise questions, it has the potential to become a prominent figure in the darknet scene.
Features:
Blacksprut https://bs-gl-darknet.com offers an “Instant Transactions” feature, inspired by the success of similar systems on other platforms like Hydra. Couriers hide goods within the city and provide buyers with coordinates, adding an adventurous element to the purchasing process.
baclofen 10 mg buy online
Привет!
Мы изготавливаем дипломы психологов, юристов, экономистов и прочих профессий по приятным ценам. Стоимость может зависеть от определенной специальности, года получения и ВУЗа.
Где купить диплом по актуальной специальности?
Купить диплом университета
https://landik-diploms-srednee.ru/kupit-diplom-saratov
Успешной учебы!
This site was… how do I say it? Relevant!! Finally I have found something that helped me. Appreciate it!
Привет!
Приобрести диплом университета.
Наш сервис предлагает купить диплом высочайшего качества, который не отличить от оригинального документа без участия специалистов высокой квалификации со специальным оборудованием.
Где приобрести диплом специалиста?
http://www.85sucai.com/member/index.php?uid=yhaxyn
Удачи!
Здравствуйте!
Заказать документ университета вы имеете возможность в нашей компании в Москве. Мы предлагаем документы об окончании любых университетов Российской Федерации.
Наши специалисты предлагают выгодно купить диплом, который выполняется на оригинальной бумаге и заверен печатями, штампами, подписями. Документ пройдет лубую проверку, даже при использовании профессионального оборудования.
https://forum.sprosiadvokata.ru/topic/22128-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%86%D0%B5%D0%BD%D0%B0-i644z/
http://kafdp.or.kr/bbs/board.php?bo_table=free&wr_id=376681
http://www.lada-xray.net/showthread.php?p=32746#post32746
http://city-hall.nvkb.ru/forum/index.php/user/101020/
http://brenderupmediterrane.free.fr/index.php?file=Members&op=detail&autor=ujelo
토토 꽁포
Xu Pengju는 잠시 생각하고 말이된다고 생각한 다음 “오”라고 고개를 끄덕였습니다.
Военный адвокат Запорожье. Бесплатная консультация
адвокат по влк Запорожье
— это опытный специалист имеющий высшее юридическое образование, сдавший квалификационный государственный экзамен на право осуществления адвокатской деятельностью и специализирующийся в основном на военных делах
Вся правовая помощь военного адвоката осуществляется в индивидуальном порядке, грамотно, четко и в соответствии с действующими нормативно-правовыми актами.
Мы как военные юристы действуем не против органов Украины или министерства обороны, мы действуем во благо Украины — наших защитников и граждан Украины, которые попали в тяжелую жизненную ситуацию связанную с незнанием военного и действующего законодательства.
Поскольку, проявив патриотизм и чувство гражданской ответственности – став на защиту суверенитета страны, граждане участвующие и помогавшие в обороне после, становятся никому не нужными, особенно если военнослужащий стал инвалидом, потерял часть тела или конечность, и не может самостоятельно защитить свои права. Именно в таких ситуациях мы как военные адвокаты приходим на помощь, и добиваемся в установленном законом порядке справедливости, необходимых выплат, установление статуса, оформление пенсий, льгот и т.п.
Тоже касается, и получение отсрочки от мобилизации, когда например, безосновательно призывают сына у которого отец инвалид 2 группы, или мать прикованная из-за тяжелой болезни к постели, и требующая постороннего ухода. Это же относится и к военнослужащим, рапорта которых не регистрируются в канцелярии воинской части и полностью игнорируются, под прикрытием суеты боевых действий..
Именно в таких ситуациях, мы приходим на помощь и с помощью ЗАКОННЫХ методов правовой защиты, используя свой опыт полученный при ведении аналогичных военных дел добиваемся справедливости.
슈퍼 벳 토토
Xie Qian은 고개를 끄덕였습니다. 이제 확실한 아이디어가 생겼기 때문에 시험지가 쉬울까 두려웠습니다.
buy furosemide tablets
Вы хотите изучать язык и одновременно быть в курсе последних событий? Тогда предлагаем вам попробовать смотреть новости на английском языке. Мы сделали для вас подборку из 5 лучших сайтов, которые подойдут и начальным уровням, и продолжающим.
1. Кому подойдет: с уровня Elementary и выше.
blacksprut
Newsinlevels.comГлавное преимущество этого сайта в том, что каждая новость представлена в 3 вариантах: для людей с начальным, средним и высоким уровнями знаний.
Каждый материал включает в себя три варианта текста (согласно уровням сложности). К текстам низкого и среднего уровня сложности прилагается аудиозапись, а к тексту продвинутого уровня — видеоролик.
К каждому тексту есть список незнакомых слов с пояснениями на английском языке. Кроме того, справа от статьи среди рекламных баннеров вы можете найти англо-русский словарь и при необходимости воспользоваться им.
С этим сайтом вы сможете не только узнать последние новости, но и расширить словарный запас, а также улучшить понимание речи на слух. Примечательно, что в новостях периодически повторяется одна и та же лексика, поэтому если ежедневно смотреть всего один трехминутный ролик, можно постепенно практически без усилий расширить свой словарный запас.
blacksprut https://bls.gl
Аудиозаписи для начального уровня озвучены носителем языка, но говорит он четко, медленно, делает довольно большие паузы — как раз то, что надо новичкам. Среднему уровню предлагают запись в более быстром темпе, но озвученную четко, хорошо поставленным голосом. Для людей с «продвинутым» уровнем предлагаются оригинальные видео, транслирующиеся на американских каналах: слова произносятся быстро, можно послушать разные акценты.
2. BBC.co.ukКому подойдет: студентам с уровнем Pre-Intermediate и выше.
Выберите ролик на интересную вам тему и смотрите его. В начале видео вам расскажут, какие незнакомые слова встретятся, после чего покажут сам ролик с новостью. После этого новость показывают еще раз, но уже с субтитрами, подсвечивая при этом слова, предлагаемые для изучения. В конце видео вам еще раз назовут изучаемые слова и дадут пояснения к ним. Таким образом, изучение новой лексики будет происходить в контексте, вы сразу будете знать, в каких случаях следует употреблять то или иное слово.
Под видео вы увидите транскрипт (текст к ролику), а также список изучаемых слов с пояснениями на английском языке. Ниже вы найдете упражнение, которое следует выполнить после просмотра и знакомства со словами. Справа даны ответы, можете проверить себя.
3. English-club.tv
English-club.tvКому подойдет: по утверждению авторов, сайт ориентирован на студентов с уровнями Elementary, Pre-Intermediate, Intermediate.
Новое видео с новостями публикуется 2-4 раза в неделю. Обычно ролик состоит из двух новостей.
Having read this I believed it was extremely informative. I appreciate you spending some time and effort to put this information together. I once again find myself personally spending a lot of time both reading and leaving comments. But so what, it was still worthwhile!
프라그마틱 슬롯 추천
Hongzhi 황제는 어이가 없었고 Jinzhou에 Liu Laoxi라는 사람이 있습니까?
купить диплом о высшем образовании проверка ast-diplomas.com .
Good site you have got here.. It’s hard to find good quality writing like yours nowadays. I honestly appreciate individuals like you! Take care!!
can i order acyclovir online
retin a .05 cream
Военный адвокат Запорожье. Бесплатная консультация
военный адвокат
— это опытный специалист имеющий высшее юридическое образование, сдавший квалификационный государственный экзамен на право осуществления адвокатской деятельностью и специализирующийся в основном на военных делах
Вся правовая помощь военного адвоката осуществляется в индивидуальном порядке, грамотно, четко и в соответствии с действующими нормативно-правовыми актами.
Мы как военные юристы действуем не против органов Украины или министерства обороны, мы действуем во благо Украины — наших защитников и граждан Украины, которые попали в тяжелую жизненную ситуацию связанную с незнанием военного и действующего законодательства.
Поскольку, проявив патриотизм и чувство гражданской ответственности – став на защиту суверенитета страны, граждане участвующие и помогавшие в обороне после, становятся никому не нужными, особенно если военнослужащий стал инвалидом, потерял часть тела или конечность, и не может самостоятельно защитить свои права. Именно в таких ситуациях мы как военные адвокаты приходим на помощь, и добиваемся в установленном законом порядке справедливости, необходимых выплат, установление статуса, оформление пенсий, льгот и т.п.
Тоже касается, и получение отсрочки от мобилизации, когда например, безосновательно призывают сына у которого отец инвалид 2 группы, или мать прикованная из-за тяжелой болезни к постели, и требующая постороннего ухода. Это же относится и к военнослужащим, рапорта которых не регистрируются в канцелярии воинской части и полностью игнорируются, под прикрытием суеты боевых действий..
Именно в таких ситуациях, мы приходим на помощь и с помощью ЗАКОННЫХ методов правовой защиты, используя свой опыт полученный при ведении аналогичных военных дел добиваемся справедливости.
I love it when people get together and share thoughts. Great site, continue the good work.
황제 슬롯
그는 즉시 교실을 떠나 경례를 하기 위해 앞으로 걸어나오더니 “목사님이 오셨습니다.”라고 큰 소리로 말했습니다.
dexamethasone 500 mg
I love reading through an article that can make people think. Also, thanks for allowing for me to comment.
Very good article. I’m facing many of these issues as well..
When I initially left a comment I seem to have clicked the -Notify me when new comments are added- checkbox and now each time a comment is added I recieve four emails with the same comment. Perhaps there is an easy method you are able to remove me from that service? Appreciate it.
You should be a part of a contest for one of the highest quality sites on the net. I am going to highly recommend this website!
диплом техникума диплом техникума .
You ought to take part in a contest for one of the finest sites on the internet. I will recommend this website!
О команде Экипировка Эксперт
LOWA ZEFIR тактические треккинговые демисезонные ботинки, высокие
Боец, Экипировка Эксперт — это розничный интернет-магазин при оптовом складе. Это значит, что при должном количестве товара мы дадим очень хорошие цены.
Название взяли независимо от того, что наша страна сейчас проводит Специальную Военную Операцию, хорошая снаряга и экипировка нужна всегда. Готовишься в бой, мобилизован, привык активно проводить время или решил подготовить тревожный чемоданчик, мы поможем тебе. Наши клиенты: фонды, медики, такие же как ты бойцы СВО и обычные неравнодушные граждане.
HOLOSUN HS403B – коллиматорный прицел, красный
https://ekipirovka.shop/katalog/taktika/takticheskie_naushniki/
Самое главное, что нужно о нас знать, мы детально объясняем, что и как работает, чтобы ты сделал правильный выбор не переплачивая.
Обращаясь к нам, не удивляйся, если ты получишь честный и жесткий ответ – часто случается так, что мы знаем лучше, что именно нужно нашему гостю. Особенно это касается мобилизованных без опыта боевых действий. Здесь ты можешь полагаться на нашу экспертность.
Одна из наших основных целей предоставить тебе возможность удобной и безопасной покупки: хоть за наличку, хоть по карте, хоть по счету. Повторимся, если нужна оптовая поставка, согласуем и отгрузим.
Именно от того, как ты производишь оплату, зависит цена заказа. Для нас важно предоставить тебе качественную экипировку и снаряжение соблюдая при этом законы нашей страны.
Боец, помни, мы помогаем фондам, нуждающимся людям, подразделениям в зоне СВО. Отчеты об этом вскоре будут опубликованы как на сайте, так и на наших каналах в социальных сетях. На эту деятельность уходит значительная часть выручки. Делая покупки в нашем магазине, ты помогаешь людям и фронту. Уверен, что это найдет отзыв в твоем сердце.
У нашей команды есть набор ценностей: честность, справедливость, сопереживание, взаимопомощь, мужество, патриотичность. Уверены, ты их разделяешь, и мы легко найдем общий язык.
Ну а если что-то пойдет не так, не руби с плеча, объясни, где мы ошиблись и поверь, мы разберемся и исправим.
Наш девиз “In hostem omnia licita” – по отношению к врагу дозволено все. Возьми этот девиз, он поможет тебе принять правильное решение в трудной ситуации, с честью выполнить боевую задачу и вернуться домой живым и здоровым!
I have to thank you for the efforts you have put in penning this blog. I am hoping to check out the same high-grade blog posts from you in the future as well. In fact, your creative writing abilities has encouraged me to get my own, personal website now 😉
I quite like reading a post that will make people think. Also, thank you for allowing for me to comment.
Когда я слышу об обвинениях в адрес «Бест Вей», мое первое чувство – это гнев. Этот кооператив не просто помогает военным приобрести жилье, он предоставляет реальную поддержку тем, кто находится на передовой. За долгие годы работы «Бест Вей» стал примером того, как социальные инициативы могут помогать тем, кто служит на благо страны. А теперь мы видим, как его обвиняют в махинациях и коррупции. Это недопустимо. Мы не можем позволить уничтожить то, что так тщательно и с любовью создавалось ради блага наших военных. «Бест Вей» заслуживает нашей поддержки и защиты. Давайте стоять на стороне правды и требовать справедливого расследования, чтобы восстановить его доброе имя и возможность продолжать свою миссию.
лайф из гуд
купить диплом специалиста в самаре mandiplomik.ru .
https://xrumer.ru/ – роблокс r34
https://xrumer.xyz/ – роблокс r34
Title: Mejores estrategias para jugar en Spin Casino Brasil: Consejos de los profesionales
Description: Descubre estrategias expertas y consejos profesionales para optimizar tus juegos en Spin Casino Brasil. ?Aprende de los mejores!
cassino spin
H1: Mejores estrategias para jugar en Spin Casino Brasil: Consejos de los profesionales
Bienvenidos al emocionante mundo de Spin Casino Brasil, donde la oportunidad de ganar se combina con el entretenimiento de primera clase. En este articulo, exploraremos estrategias efectivas y consejos profesionales que te ayudaran a maximizar tus experiencias de juego. Desde comprender la estructura del casino hasta aplicar tacticas especificas en tus juegos favoritos, te guiaremos paso a paso para mejorar tus habilidades y aumentar tus posibilidades de exito.
H2: Conociendo Spin Casino Brasil
Introduccion al Casino
Spin Casino Brasil se destaca en el mercado por ofrecer una experiencia de juego segura y dinamica. Con una licencia valida y un compromiso con la seguridad del jugador, este casino se presenta como una opcion confiable para los entusiastas de los juegos de azar en linea.
Variedad de Juegos
Explora la amplia gama de juegos que Spin Casino Brasil tiene para ofrecer. Desde tragamonedas clasicas hasta juegos de mesa sofisticados como el blackjack y la ruleta. Cada juego viene con una descripcion detallada de las reglas y los porcentajes de retorno al jugador (RTP), asegurando que los jugadores puedan elegir sabiamente segun sus preferencias.
Software y Tecnologia
Spin Casino Brasil utiliza software de los lideres de la industria, garantizando graficos de alta calidad y una interfaz de usuario fluida. Este apartado discutira como la tecnologia de Spin Casino Brasil mejora la experiencia del usuario y mantiene la integridad del juego.
Atencion al Cliente
El soporte al cliente es esencial en cualquier plataforma de juego en linea. Detalla como los jugadores pueden acceder al soporte 24/7 a traves de chat en vivo, correo electronico y telefono, asegurando que todas las consultas y problemas se resuelvan de manera eficiente.
H2: Estrategias generales de juego
Gestion del Bankroll
Un buen jugador sabe que la gestion del bankroll es crucial. Explica tecnicas para determinar cuanto dinero destinar al juego sin comprometer la estabilidad financiera personal. Se incluiran metodos para establecer limites de apuestas y la importancia de adherirse a estos limites.
Comprender las Probabilidades
El juego de casino no solo se trata de suerte; las matematicas juegan un papel crucial. Ofrece una explicacion sencilla de como entender las probabilidades y como pueden influir en las decisiones de juego. Este apartado demuestra como un conocimiento basico de estadisticas y probabilidad puede ayudar a tomar decisiones mas informadas.
Importancia de la Seleccion del Juego
No todos los juegos de casino ofrecen las mismas probabilidades de ganar. Discute como la eleccion del juego puede afectar las posibilidades de exito, destacando aquellos juegos que ofrecen mejores probabilidades y explicando por que algunos juegos son mas amigables con el jugador que otros.
Estrategias de Apuestas
Finalmente, se abordaran diversas estrategias de apuestas que los jugadores pueden emplear. Desde la estrategia conservadora hasta la mas arriesgada, cada estilo de apuesta sera discutido con ejemplos practicos, ayudando a los jugadores a entender cuando y como ajustar sus apuestas segun el flujo del juego.
H2: Estrategias especificas para juegos populares en Spin Casino Brasil
Este apartado se centra en las tacticas especializadas que los jugadores pueden emplear en algunos de los juegos mas populares disponibles en Spin Casino Brasil, asegurando que los jugadores de todos los niveles puedan optimizar sus oportunidades de ganar.
H3: Tragamonedas
Comprender la Varianza y el RTP
Inicia con una explicacion sobre la importancia del retorno al jugador (RTP) y la varianza de las tragamonedas. Describe como seleccionar maquinas con un RTP alto puede aumentar las posibilidades de ganancias a largo plazo y como la varianza afecta la frecuencia y cantidad de los pagos.
Estrategias de Apuestas
Detalla estrategias de apuestas eficaces para las tragamonedas, como ajustar el tamano de las apuestas basado en el ciclo de ganancias/perdidas y la importancia de maximizar las lineas de pago para aumentar las posibilidades de ganar en cada giro.
Aprovechar los Bonos
Explica como los jugadores pueden utilizar bonos y giros gratis ofrecidos por Spin Casino para maximizar sus jugadas sin aumentar el riesgo financiero, destacando la importancia de leer los terminos y condiciones asociados a estos bonos.
H3: Blackjack
Estrategia Basica
Proporciona una guia detallada sobre la estrategia basica de blackjack, incluyendo cuando pedir carta, plantarse, dividir o doblar la apuesta, basado en la mano del jugador y la carta visible del crupier.
Gestion del Riesgo
Discute la importancia de la gestion del riesgo y del bankroll especificamente para el blackjack, enfatizando como evitar las apuestas impulsivas y como adaptar el tamano de las apuestas en funcion de la situacion del juego.
Contar Cartas
Introduce conceptos basicos del conteo de cartas (donde legalmente sea aplicable), explicando como esta tecnica puede dar a los jugadores una ventaja estadistica sobre el casino y como practicar de manera efectiva sin infringir las reglas.
H3: Ruleta
Apuestas Exteriores vs. Interiores
Explica la diferencia entre apuestas exteriores e interiores, sus riesgos y beneficios, y por que las apuestas exteriores suelen ser una mejor opcion para aquellos que buscan minimizar riesgos.
Sistemas de Apuestas
Analiza varios sistemas de apuestas como el Martingala, Fibonacci y D’Alembert, evaluando su efectividad y como pueden ser aplicados en la ruleta para gestionar mejor las perdidas y potenciar las ganancias.
H3: Poker
Conocimientos Fundamentales de Poker
Detalla los conocimientos basicos del poker, como las reglas del Texas Hold’em, las clasificaciones de manos y la importancia de la posicion en la mesa.
Estrategias de Apuesta y Bluff
Describe estrategias avanzadas como el arte del bluff, saber leer a los oponentes, y la importancia de ajustar las apuestas basado en el estilo de juego de los contrincantes y la dinamica de la mesa.
Importancia del Control Emocional
Destaca la importancia de mantener el control emocional, como gestionar las ganancias y las perdidas, y por que un enfoque disciplinado y una mente clara son esenciales para el exito en el poker a largo plazo.
H2: Consejos de los profesionales
Aprender de la Experiencia
Introduccion a las lecciones aprendidas por jugadores experimentados, enfatizando la importancia de aprender tanto de los exitos como de los fracasos. Resalta como la experiencia es un recurso invaluable que puede ayudar a prever situaciones de juego y tomar decisiones mas informadas.
Adaptabilidad y Flexibilidad
Describe como los profesionales se adaptan a diferentes situaciones de juego, ajustando sus estrategias segun la dinamica del juego y las acciones de otros jugadores. Discute la importancia de ser flexible y no apegarse rigidamente a una sola estrategia.
Mantener la Disciplina
Enfatiza la necesidad de mantener la disciplina, especialmente en momentos de ganancias o perdidas significativas. Explica como la gestion efectiva del bankroll y el control emocional son cruciales para el exito a largo plazo en los juegos de casino.
H2: Maximizando bonificaciones y promociones
Entender los Tipos de Bonificaciones
Explica los diferentes tipos de bonificaciones que ofrece Spin Casino Brasil, como bonos sin deposito, bonos por deposito, y giros gratis. Detalla como cada tipo puede beneficiar a los jugadores dependiendo de su estilo de juego y objetivos.
Estrategias para Aprovechar Promociones
Ofrece estrategias especificas para aprovechar al maximo las promociones, como planificar depositos durante eventos de bonificacion y elegir promociones que complementen el estilo de juego del usuario. Discute la importancia de leer los terminos y condiciones para evitar sorpresas.
Cumpliendo con los Requisitos de Apuesta
Proporciona consejos practicos sobre como cumplir con los requisitos de apuesta sin comprometer el bankroll. Explora tecnicas para maximizar las oportunidades de ganancia mientras se juega a traves de los requisitos de apuesta de manera eficiente.
H2: Herramientas y recursos utiles
Software y Aplicaciones de Ayuda
Presenta software y aplicaciones que pueden ayudar a los jugadores a mejorar su juego, como calculadoras de probabilidades, diarios de juego, y herramientas de analisis de manos. Evalua como estas herramientas pueden proporcionar una ventaja competitiva.
Recursos Educativos
Recomienda libros, tutoriales en video y cursos que pueden proporcionar a los jugadores nuevos y experimentados conocimientos avanzados sobre estrategias de juego y psicologia del juego. Destaca recursos especificos que han sido bien recibidos por la comunidad de juego.
Foros y Comunidades
Alude a la importancia de participar en foros y comunidades en linea donde los jugadores pueden intercambiar estrategias, experiencias y consejos. Subraya como el aprendizaje colaborativo y el apoyo mutuo pueden mejorar la experiencia de juego y el desarrollo de habilidades.
Al aplicar las estrategias y consejos discutidos en este articulo, estaras bien equipado para afrontar los desafios que Spin Casino Brasil ofrece. Recuerda que la clave del exito en los juegos de casino reside no solo en conocer las reglas, sino en aplicar estrategias informadas y mantener una disciplina ferrea. Juega de manera responsable y dentro de tus limites para disfrutar de una experiencia tanto segura como emocionante. ?Buena suerte y que disfrutes de cada juego!
FAQ:
?Es seguro jugar en Spin Casino Brasil?
?Absolutamente! Spin Casino Brasil esta completamente regulado y ofrece un entorno seguro para todos sus jugadores. Disfruta de tu juego sin preocupaciones.
?Que juegos ofrecen las mejores probabilidades en Spin Casino Brasil?
Los juegos como el blackjack y el video poker generalmente ofrecen las mejores probabilidades, especialmente cuando aplicas las estrategias adecuadas.
?Puedo jugar en Spin Casino Brasil desde mi movil?
Si, Spin Casino Brasil ofrece una plataforma movil excelente que te permite jugar tus juegos favoritos desde cualquier lugar y en cualquier momento.
?Como puedo maximizar las bonificaciones ofrecidas por Spin Casino Brasil? Asegurate de leer los terminos y condiciones de las bonificaciones. Aprovecha los bonos de bienvenida, recargas y programas de fidelidad para obtener el maximo beneficio.
москва диплом купить средний mandiplomik.ru .
диплом фармацевта купить диплом фармацевта купить .
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
Колокольцев – предатель Родины
Армия восстанет против ОПГ Колокольцева и Плугина
Целенаправленно разрушая кооператив для военных «Бест Вей», захватывая его активы в интересах своей камарильи, глава МВД и его команда ударили по тылу армии, наказав тысячи пайщиков кооператива – военных и членов их семей.
Приморский районный суд Санкт-Петербурга (судья Екатерина Богданова) сейчас рассматривает так называемое уголовное дело «Лайф-из-Гуд» – «Гермес» – «Бест Вей». В рамках этого дела два года почти непрерывно арестованы счета кооператива «Бест Вей», пайщиками которого являются тысячи военнослужащих, в том числе участники СВО, на которых около 4 млрд рублей. В уголовном деле речь идет прежде всего об обязательствах иностранной инвесткомпании «Гермес» перед более чем 200 клиентами этой компании.
Следствие ГУ МВД по Санкт-Петербургу и прокуратура города на Неве объявили кооператив, «Гермес» и «Лайф-из-Гуд» частью некоего единого холдинга, которого в природе никогда не существовало. Кооператив объявлен гражданским ответчиком по уголовному делу.
Общая сумма претензий потерпевших, согласно обвинительному заключению, 282 млн рублей – даже их (незаконное) изъятие для погашения долгов перед клиентами «Гермеса» никак бы не сказалось ни на операционной деятельности, ни на ликвидности кооператива. Но из раза в раз арестовываются абсолютно все средства на счетах, что полностью блокирует как покупку квартир кооперативом, так и возврат денег пайщикам, которые изъявили желание забрать свои паевые взносы.
Недвижимость, на которую претендовали пайщики, обесценивается, деньги обесцениваются – и этот беспредел продолжается уже более двух лет по ходатайствам ГСУ питерского главка МВД и Прокуратуры Санкт-Петербурга.
Кто-то влиятельный пытается наложить лапу именно на кооперативные 4 млрд – хотя это деньги пайщиков: физических лиц, рядовых граждан России, и как минимум соучастниками грабежа являются сотрудники правоохранительных органов.
Армейский кооператив
Кооператив изначально создавался военнослужащими – одной из его задач в 2014 году было решение жилищной проблемы военнослужащих, увольняемых в запас, эта задача сохранилась и в последующие годы. Из почти 20 тыс. его пайщиков – тысячи военнослужащих со всей России. Значительная часть из них – участники СВО.
В результате действий ОПГ Колокольцева сотни семей военнослужащих, участников СВО стали потерпевшими от действий правоохранительных органов. Налицо преступные действия против участников СВО.
Камарилья, которая устроила репрессии против кооператива – руководители следственной группы Сапетова и Винокуров, начальник ГСУ питерского главка МВД Негрозов и начальник этого главка Плугин, замминистра внутренних дел по следствию Лебедев, пресс-секретарь министра Волк, транслировавшая на всю страну ложь про десятки тысяч потерпевших от работы кооператива, сам министр внутренних дел Колокольцев, – ведет вредительскую деятельность, все эти люди – предатели Родины.
Разоряет экономику и подрывает тыл
Колокольцев и Ко разрушают экономику страны, фабрикуют дела. При этом его камарилья наживается на честном бизнесе.
Колокольцев – миллиардер! Его боевая подруга генеральша Волк – миллиардерша! Зам. начальника питерского ГСУ МВД полковник Винокуров, который управляет Колокольцевым, заставляя его подписываться под липовым уголовным делом «Лайф-из-Гуд» – «Гермес» – «Бест Вей», – миллиардер! Колокольцев превратил МВД в коммерческое антинародное образование и зарабатывает деньги на простых людях.
Колокольцев и Ко прицепили звезды на погоны и возомнили себя офицерами – на деле они тыловые вороватые крысы. Настоящие офицеры против бутафорских чинуш со звездами на погонах.
Министр внутренних дел дестабилизирует ситуацию в народе и армии, разоряет армию в тылу. Колокольцев со своими тыловыми крысами подрывают доверие народа к власти и отбивают желание людей идти на военную службу, потому что благодаря таким, как он, государство не может обеспечить безопасность военных в тылу.
Армия против Колокольцева
Армия восстанет против ОПГ Колокольцева. Военные требуют за совершенные преступления против народа, за воровство и коррупцию арестовать Колокольцева, Негрозова, Винокурова, Сапетову, оправдать и отпустить невинных людей – рядовых сотрудников «Лайф-из-Гуд», которые более двух лет томятся в тюрьме.
Новый министр обороны Белоусов наверняка поддержит своих бойцов, поможет им защитить собственные и своих семей имущественные интересы в тылу: по-другому и быть не может.
Одного любителя запускать руку в казну и любителя генеральш – Шойгу – уже отправили на пенсионную должность, Колокольцев должен последовать за ним: это – в интересах страны, в интересах народа!
i-guru – Ремонт Iphone и другой техники Apple в Минске
ремонт айфона 15 Plus цена Минск
Профессиональный ремонт любых устройств из линейки Apple, без посредников
80% ремонтов iPhone занимает около 20 минут
Несложные ремонты делаются быстро, модульный ремонт в большинстве случаев занимает не более 20 минут.
Почему выбирают i-guru
Ремонт в тот же день
Даже большинство сложных ремонтов выполняем день в день.
Гарантировано низкие цены
Мы беремся за сложные задачи, не передавая их сторонним исполнителям, и зарабатываем не как посредники.
Честная гарантия
Предлагаем честную гарантию на выполненные работы до 12 месцев в зависимости от вида работ.
무료 슬롯 사이트
Liu 의사는 겁에 질려 소변을 보고 바닥에 쓰러졌습니다. “아니요… 그렇게 말하지 않았습니다.”
купить аттестат настоящий купить аттестат настоящий .
Компания Септик-Нара-купить септик для дачи
занимается продажей, установкой и обслуживанием септиков в Наро-Фоминске Наро-Фоминском районе. Основное направление деятельности нашей компании именно установка под ключ септиков любых видов и размеров. С момента основания нашей компании, мы произвели монтаж более 1000 септиков по Московской и Калужской области. Благодаря этому у нас огромный опыт работы с любыми станциями, представленными в нашем регионе!
Если Вы решили купить септик для дома или дачи, мы с радостью поможем Вам с выбором модели, доставим и установим септик на вашем участке в кратчайшие сроки.
Мы занимаемся продажей септиков таких марок: Топас Юнилос Астра Евролос Тверь Аквалос Дочиста Фекалов Волгарь Удача. Мы работаем напрямую с производителями септиков, поэтому Вы можете быть уверены, что не переплачиваете ни копейки. Вся продукция в нашей компании имеет соответствующие сертификаты и лицензии. Время выезда на осмотр, установки или привоза оборудования согласовывается с клиентами и выполняется в срок. Мы заботимся о своей репутации, поэтому выполняем работу надежно и быстро.
Если Вы не знаете, какой септик больше всего Вам подходит, мы предоставим консультацию и выезд специалиста на Ваш участок абсолютно бесплатно!
Благодаря приобретению качественных септиков в нашей компании, каждый клиент получает большое количество преимуществ:
» компактные размеры и небольшой вес устройства;
» доступная стоимость очистной установки;
» быстрый и простой монтаж;
» невысокая стоимость эксплуатации;
» отсутствие неприятных запахов;
» высокая производительность;
» длительный срок эксплуатации;
» высокая степень очистки;
» практически полностью автономный режим работы.
accutane tablets online
A fairly new player in the Russian darknet arena, blacksprut Blacksprut has quickly gained attention for its interesting features and growing popularity. While some aspects raise questions, it has the potential to become a prominent figure in the darknet scene.
Features:
Blacksprut https://bs-gl-darknet.com offers an “Instant Transactions” feature, inspired by the success of similar systems on other platforms like Hydra. Couriers hide goods within the city and provide buyers with coordinates, adding an adventurous element to the purchasing process.
купить аттестат за 11 класс в нижнем новгороде mandiplomik.ru .
lyrica online prescription
토토 꽁 머니 30000
아침 일찍 그는 사람들에게 북을 치라고 명령하여 Shenji Camp의 모든 병사를 소집했습니다.
보증 토토
Shen Ao는 고개를 저었다. “사람이 날 수 있다는 것이 정말 놀랍습니다.”
купить государственный диплом высшего образования mandiplomik.ru .
МОСКВА, 7 мая — РИА Новости. Правительство России, возглавляемое Михаилом Мишустиным, уйдет в отставку во вторник.
Сразу после инаугурации президента Владимира Путина нынешний кабинет министров сложит полномочия перед вступившим в должность главой государства.
Прогон по комментария
Кооператив для военных
Как пайщики — участники СВО относятся к событиям вокруг «Бест Вей»
Участники СВО Бест вей
Потребительский кооператив «Бест Вей» оказался затронут уголовным делом, касающимся в основном иностранной инвесткомпании «Гермес», которое сейчас рассматривается Приморским районным судом Санкт-Петербурга. Более двух лет более 3,5 млрд рублей на счетах кооператива почти непрерывно арестованы по ходатайству сначала ГСУ ГУ МВД по Санкт-Петербургу, а затем Прокуратуры Санкт-Петербурга: пайщики не имеют возможности ни приобрести недвижимость, ни вернуть средства.
По данным совета потребительского кооператива «Бест Вей», в числе его пайщиков, страдающих от блокировки средств, тысячи военнослужащих, в том числе сотни участников СВО, часть из которых успела приобрести квартиру, часть собирает или собрала первоначальный взнос, а часть — планировала вступить в кооператив. «СП» пообщалась с некоторыми из них и их родственниками, чтобы узнать отношение к «Бест Вей» и событиям вокруг кооператива.
«К кооперативу отношение очень хорошее»
Александр Голдман на СВО с мая 2022 года как доброволец, три ранения. Пришел на СВО рядовым, сейчас — начальник штаба батальона. Был пайщиком кооператива, сейчас пайщик — его мама.
«С помощью кооператива в 2019 году приобретена двухкомнатная квартира во Владивостоке, в которой проживает мама, — рассказывает он. — Расплачиваемся за нее, в ближайшие месяцы намерены погасить задолженность перед кооперативом и оформить квартиру в собственность. К кооперативу отношение очень хорошее, полностью его поддерживаю — он дает возможность без больших переплат приобрести недвижимость. К действиям в отношении кооператива отношусь отрицательно, так как „Бест Вей“ — единственная возможность приобрести жилье в рассрочку».
«Происходящее вокруг кооператива вызывает шок»
Гвардии рядовой Глушков Иван Васильевич — пайщик кооператива из Челябинской области. Танкист, мобилизованный, проходил службу в 15-й отдельной гвардейской мотострелковой Александрийской бригаде 2-й гвардейской общевойсковой армии ЦВО. Погиб 11 октября 2023 года.
Рассказывает его вдова Татьяна Неручева:
«Мы внесли первоначальный паевый взнос и во второй половине 2021 года встали в очередь на приобретение квартиры в Челябинске, когда начались события вокруг кооператива — была заблокирована его возможность приобретать недвижимость и заблокированы его счета. Наша очередь должна была подойти примерно через год. Мы заявили для приобретения небольшую квартиру, но планировали при покупке увеличить ее стоимость до 3 млн и купить двухкомнатную квартиру — с увеличением первоначального паевого взноса: уставом кооператива это позволяется, а затем переехать из Коркино в Челябинск. Сейчас 3 млн хватит только на небольшую квартиру-студию метров 25, то есть мы понесли материальный ущерб из-за блокирования деятельности кооператива, так как лишены были возможности приобрести квартиру, на которую собрали первоначальный паевый взнос. Работа кооператива заблокирована, счета арестованы уже более двух лет. В наследование пая я только вступаю, так как муж очень долго считался пропавшим без вести — долго шла экспертиза, и свидетельство о смерти мы получили только 18 июня этого года».
Татьяна — юрист: «Как у юриста у меня происходящее вокруг кооператива вызывает шок, и любой непредвзятый юрист вам скажет то же самое». «Кооператив, — подчеркивает она, — абсолютно прозрачен, он полностью соответствует законодательству о кооперации — что и подтверждалось многократно государственными органами и судами. Если бы мы с мужем не были уверены, что все прозрачно и законно, мы бы не вкладывали в него деньги. Мы не подавали заявление о выходе из кооператива. Я, несмотря ни на что, жду счастливого завершения рукотворного кризиса вокруг кооператива. Для меня это еще и память о муже — он продал долю в квартире, которая ему принадлежала, чтобы вложиться в кооператив. Кроме того, мне хочется понять — до какого маразма может дойти ситуация у нас в государстве в плане незаконных действий в отношении организации, которая по тем или иным причинам не понравилась каким-то чиновникам?».
«Рассчитываю, что ситуация закончится благополучно»
Сергей Логинов, рядовой, мобилизованный, представлен к награде «Честь и доблесть».
«Пять лет назад я стал пайщиком кооператива, участвовал в накопительной программе, планировал прибрести однокомнатную квартиру в Самарской области. Уже нужно было подбирать объект недвижимости и вставать в очередь на покупку, как работа кооператива была заблокирована по инициативе правоохранительных органов, и это продолжается уже более двух лет. По-прежнему надеюсь получить квартиру и рассчитываю, что ситуация закончится благополучно. Поддерживаю кооператив».
«В кооперативе минимум переплат — несопоставимо с ипотекой»
Егор Ивков, офицер флота, дирижер военного оркестра, пайщик кооператива с 2019 года.
«Я нахожусь в накопительной программе — планировал покупку квартиры в Санкт-Петербурге. До постановки в очередь на покупку дело не дошло, но сумма внесена серьезная — и на два года все зависло. Есть друзья-военнослужащие, которые также являются пайщиками и тоже накапливали деньги на первоначальный паевый взнос — они, как и я, не могут ни продолжать накапливать, ни получить деньги обратно, потому что счета арестованы. Отношение наше к ситуации, создавшейся вокруг кооператива, крайне негативное».
«Кооператив поддерживаю, — говорит пайщик. — Моя сестра живет в квартире, приобретенной с помощью „Бест Вей“ — у нее многодетная семья, сейчас кооперативная квартира переходит в ее собственность. Минимум переплат — несопоставимо с ипотекой. Надеюсь, что ситуация с арестом счетов разрешится в ближайшее время».
물라 슬롯
나라에는 뼈대가 필요하고 피로 이어진 운명공동체가 필요합니다.