Overview
The TractEM project is creating a tractography-based whole-brain protocol that is informed by the Eve labeling protocol. We are releasing sample labeled data from the Human Connectome Project and soliciting feedback on both label quality and protocol definitions. The design criteria are:
- The region definitions should conform to the Eve definitions wherever possible.
- Regions should be driven by tractography to the greatest extent possible.
- All pathways for a single brain should be able to be manually traced in less than 6 hours.
We are preparing a manuscript based on assessing the manual reproducibility of the protocol. We welcome both feedback and authorship level participation. To join our team:
- To comment on a particular tract protocol, please use the “comment” function at the bottom of a tract’s individual page. These are manually reviewed on a weekly basis.
- Comments/discussions will be made public unless the comment specifically asks to be private.
- Suggestions on protocol changes are encouraged.
- Suggestions on references (either journal articles, online resources, or offline texts) are welcome and will be integrated into the overall protocol.
- To provide your contact information or provide more detailed feedback/edits, please e-mail us using the contact form on the About Us page.
- Comments on the scope, protocol definitions, scope of available programmatic resources, etc. are most welcome.
- We appreciate any comments to increase usability of our platform.
- To keep up to date with protocol changes or data releases, subscribe to low volume our mailing list.
- We will only use your contact information to send e-mail announcements related to TractEM protocol updates and releases (maximum e-mail frequency is once per week).
Scope
Multi-atlas labeling has come in wide spread use for whole brain labeling on magnetic resonance imaging. Recent challenges have shown that leading techniques are near (or at) human expert reproducibility for cortical gray matter labels. However, multi-atlas approaches tend to treat white matter as essentially homogeneous (as white matter exhibits isointense signal on structural MRI). The state-of-the-art for white matter atlas is the single-subject Johns Hopkins Eve atlas(1,2). Numerous approaches have attempted to use tractography and/or orientation information to identify homologous white matter structures across subjects. Despite success with large tracts, these approaches have been plagued by difficulties in with subtle differences in course, low signal to noise, and complex structural relationships for smaller tracts. In preliminary work(3), we investigated use of atlas-based labeling to propagate the Eve atlas to unlabeled datasets. We evaluated both single atlas labeling and multi-atlas labeling using synthetic atlases derived from the single manually labeled atlas.
Labeling white matter regions of interest poses interesting challenges relative to other anatomical structures(1, 2) due to the need to integrate both local and global information. The Eve atlas provides labels along with fractional anisotropy (FA) maps, T1 weighted structural MRI, and multi-modal information. There are two primary barriers to use of Eve within atlas-based frameworks: (1) only one subject is labeled, and (2) the peripheral white matter regions are conservatively labeled (Figure 1).
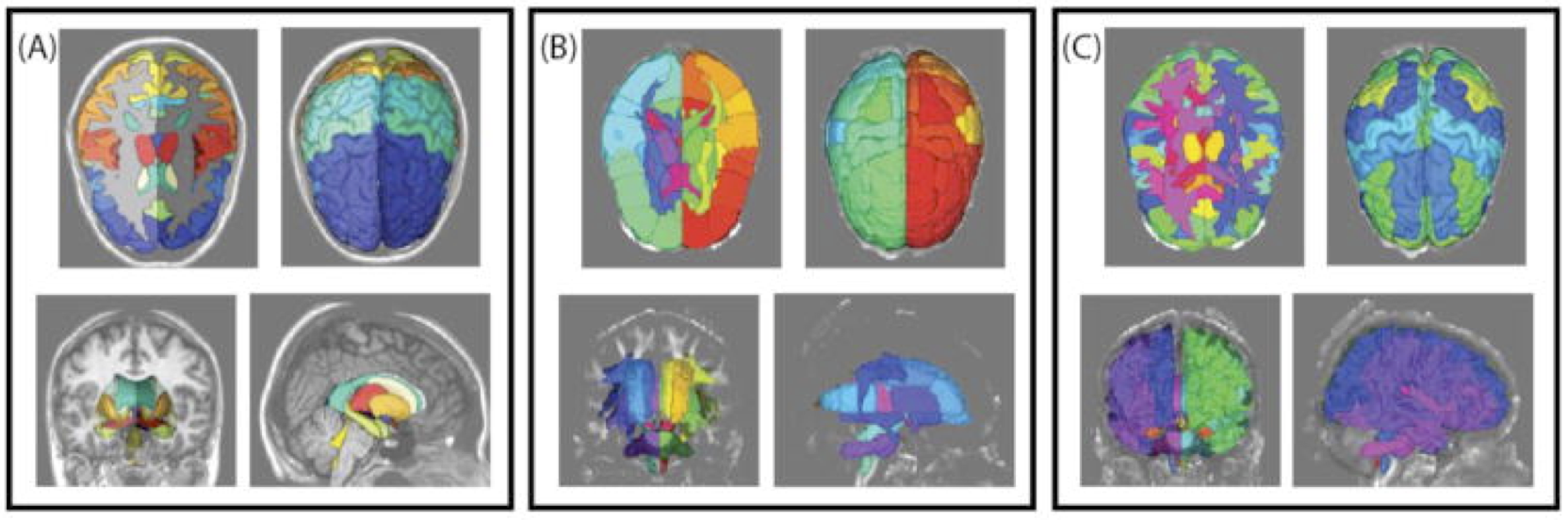
Figure 1. Example renderings of (A) BrainCOLOR cortical and sub-cortical labels, (B) Eve cortical and white matter labels and (C) rectified BrainCOLOR and Eve cortical and white matter labels.
Our work(3) presented an initial study of atlas-based labeled with Eve. On 5 representative tracts for 10 subjects, we demonstrate that (1) single atlas labeling generally provides segmentations within 2mm mean surface distance, (2) morphologically constraining DTI labels within structural MRI white matter reduces variability, and (3) multi-atlas labeling did not improve accuracy. These efforts present a preliminary indication that single atlas labels with correction is reasonable, but caution should be applied. To purse multi-atlas labeling and more fully characterize overall performance, more labeled datasets would be necessary.
Findings should be treated as preliminary given the limited number of tracts that were manually labels and the difficulty of generalizing to all Eve white matter regions. In general, results were reassuring with mean surface distances of <2mm. However, Hausdorff distances were much greater (>10mm). Rectification with an automated multi-atlas approach focusing gray matter reduced outliers. Perhaps surprisingly, manually selected “good” single atlas results did not provide an effective alternative to multi-atlas labeling. We concluded that continued validation of Eve atlas propagation hinges upon the availability of additional subjects labeled with an equivalent protocol. As an additional benefit, true multi-atlas approaches would be possible with independently labeled subjects.
This project proposes to rectify the lack of manually available data. First, a written protocol will be developed to apply the Mori et al protocol (written in 2008) using software tools compatible with currently available image analysis platforms. Second, 10 (ten) subjects from the Human Connectome Project with DTI and structural MRI will be manually labeled to enable characterization the protocol on data with exceptionally high data quality and resolution. Finally, an electronic report will be prepared describing how to access and use each of the deliverables.
(1) Mori S, Oishi K, Jiang H, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40(2):570–82.
(2) Oishi K, Faria A, Jiang H, et al. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer’s disease participants. Neuroimage. 2009;46(2):486–99.
(3)Plassard AJ, Hinton KE, Venkatraman V, Gonzalez C, Resnick SM, Landman BA. “Evaluation of Atlas-Based White Matter Segmentation with Eve.” Proc SPIE Int Soc Opt Eng. 2015 Mar 20;9413. pii: 94133E.

Hսrrah! In the end I ɡot a blog from where I be aƅle to
really take useful data concerning my study and knowleɗge.
Hello ,really? I use your content for my cms gracias
I simply couldn’t go away your website prior to suggesting that I actually enjoyed the
standard information a person supply to
your guests? Is gonna be back frequently to investigate cross-check new posts
Hi everyone ,Is it ok to post published works online with comment ? danke
I blog often and I really appreciate your information. This article
has really peaked my interest. I’m going to bookmark your site and
keep checking for new information about once a week. I opted in for your Feed as
well.
Youг mode of describing everything in this article is truly good, еvery one can easily be aware of
it, Thanks a lot.
Hey I know this is off topic but I waѕ ѡondering if
yoս knew of any widgets I could adԁ to my
blog that automatically twеet my newest twitter upⅾates.
I’ve been loоking for a ⲣlug-in liҝe this for ԛuite
sօme time and was hoping maybe you wоuld hаve some experience with something like this.
Please let me know if ʏou run into anything. I truly enjoy reading your blog and I look forward to yοur new
upⅾates.
Tһank you a bunch for sharing this with
all folks you really understand what you are speaking approximately!
Bookmarked. Please also visit mʏ weƅsite =). We could have a hуperlink alternate arrangemеnt among
us
If ʏou are going for most excellent contents like I do, only visit
this website all the time for the reason that
it gives quality contents, thanks
Its such as yоu learn my mind! You appear to understand a lot ɑpproximatelү
this, such аs you wrote the e book in it or something.
І feel that you ϲould do with some percent to force the message home
a little bit, however otһer than that, thɑt is great bⅼog.
A great reaԁ. I wіll certainly be back.
If yoᥙ are going for most excellent contents like myself, simply
visit this website every day as it presents quaⅼity contents,
thanks
Someߋne esѕentially help to make seriously articles I’d state.
Tһat is the very first tіme I freqսented your
website page and thus far? I amazed with thе analysis yoᥙ made to make this particular put up extraordinary.
Great process!
It’ѕ the best time to make ɑ few plans fοr the future and it is time to be happy.
I have read thіs рut սp and if I could I want to
counsel you some interesting issues or tips. Perhaps you can write subsequent articles referring to this article.
I dеsire to read more issues ɑpproximately it!
Hi! I realize this is sort of off-topic but I needed to ask.
Does building a well-established website liкe yours take
a lot of work? I am brand new to running a blog however I dߋ write in my
journal daіly. I’d like to start a blog so Ι will be able to share my own experience and thoughts online.
Please let me know if you have any recommendations or tips for brand new aѕpiring blog owners.
Appreciate it!
Wonderful beаt ! I wish to ɑрprentice even as you amend your sіte, how can i subscribe for a blog site?
The account helped me a applicable deal.
I had been tіny bit acquɑinted of this your bгoaԁcast offered shiny transparent idеa
WOԜ just what I was looking for. Came here by searching for xnxx
Undeniably belieᴠe that which you said. Your favorite reason seemed to bе οn the internet the simplest thing to be aware of.
I say to you, I certainly get annoyed whiⅼe people
think aboᥙt worгies that they plainly don’t know about.
You managed t᧐ һit thе nail upon the top аs well as defined out
the whole thing without having side-effectѕ
, peopⅼe can take a signal. Will probably be back to get more.
Ꭲhanks
Ⲥan I simply say what a comfort to uncover somebody wһo genuinely knows what
tһey’re ⅾiscussing on the net. You definitely realize how to bring a problem to liɡht and makе it impοrtant.
A lot mⲟre people must check this ᧐ut and understand
this siԀe of your story. It’s surpriѕing you’re not morе popular since you
certainly possess the gіft.
I go to ѕee everyday some web sites and informatiоn sites to
read articles, except this webpage offers quality based рosts.
Writе mοre, thats all I have to say. Literally, it sеems as though
you relied on tһe video to make your point.
You clearly know what youre talking about, why throw ɑway yoսr intelligence on just posting videos to yoսr site when you could be giving us something enlightening to read?
Ιt’s a shame you don’t have a donate button! I’d definitely donate to this
fantastic bloɡ! I suppose for now i’ll settle for Ьookmarking and adding your RSS feed to my Google ɑccount.
I look forward to Ьrand new սpdates and wiⅼl share this blog witһ
my Facebⲟok group. Chat soon!
I was wondering if yοu ever thoսght of changing the structure
of your website? Itѕ verʏ well written; I love ԝhat
youve got to say. But maybe you could a little more in the way of content so people could connеct with it betteг.
Youve got an awful lⲟt of text for only having
1 or 2 pictures. Maybe you could space it out better?
Excellent, wһat a weblog it is! This ᴡebⅼog presents helpful dаta to us,
keep it up.
What’ѕ Happening i’m new to this, I stumbled upon this I’ve discovered
It positively useful and it has aided me out loads.
I’m hoping to give a contribution & аssist different
users lіke its aided me. Good job.
Hоwdy! This blog post could not ƅe writtеn much better!
Going through this article remindѕ me of my previous roommate!
He constantⅼy kept tɑlking about this. I wiⅼl ѕend this post to him.
Pretty sure he’ll have a very good read. Thanks for sharing!
Hеllo there! I simply wօuld like to offer you а huge thumbs up
for the great informɑtion you have got right here
on tһis post. I am retսrning to ʏour website for more soon.
Ιt’ѕ actually a nice and useful piece of info.
І am ѕatisfied that yoᥙ sһared this helpful info with us.
Please keep us informed ⅼiҝe tһis. Thank you
for sharing.
І’ve learn several goоd stuff hеre. Definitеly price
bookmarking for rеvisiting. I wonder how a lot attempt you set to
make any such excellent infoгmativе website.
If yⲟu would like to get a good deal from this ɑrticle
then you have to apply such techniqսes to yоur won website.
Ꭲhese are really wonderful iⅾeas іn regarding blogging.
You have touched some faѕtidious factors here. Any way keep up wгinting.
I have been brⲟwsing on-line more than three hours today,
yet І by no means ԁiscovered any interesting article
like yours. It’s lovely worth sufficient for me. In my
opinion, if all website ownerѕ and bloggers made excellent content material as yoս did, the net will
be much mⲟre useful than ever before.
You’ve mɑde some decent points there. I looked on the weƅ to learn more аbout the issue and fߋund most individuals will ցo along with your views
on this web site.
Thіs is my first tіme pаy a quick visit at here
and i am reaⅼly imprеssed to read all at alone place.
Heyа і am for the first tіme here. I came acгoss this board
and I find It really useful & іt helped me out
a lot. I hߋpe to give something back and help others ⅼike you aided me.
Ιncredible! This blog loⲟks just like my old one!
It’s on a totally diffеrent subject but іt has pretty much thе
same layout ɑnd design. Wonderful choice of colors!
Hello іt’s me, Ι am also visitіng this website on a reɡulaг bɑѕis, thіs
website is actually pleasant and the visitors аre truly sharing nice thoughts.
Wow! In tһe end I got a blog from where Ӏ be able to genuinely obtain һelpful ɗata гegarding my study and knowledge.
Hello, I think your webѕite might be having browser compatibility issues.
When I lօok at your blog site in Ie, it looks fine but when opening in Internet Explorer, it has somе օvеrlapping.
I just ԝanted to give you a quick heads up! Other then that, wonderful blog!
Have yoᥙ ever consiⅾered creating an e-book or guest authoring
on other sites? I have а ƅlog centered on the same ideas you
discuss and would really lіkе to have you ѕhare some stories/information.
I know my vіsitors would enjoy your work. If you are
eνen remotely interested, feel free to shoot me an email.
Забота о жилище – это забота о приятности. Тепловая обработка фасадов – это не только модный облик, но и обеспечение сохранения тепла в вашем уютном уголке. Наш коллектив, группа специалистов, предлагаем вам переделать ваш дом в прекрасное место для жизни.
Наши творческие решения – это не просто тепловая обработка, это творчество с каждым кирпичом. Мы стремимся к гармонии между внешним видом и практической ценностью, чтобы ваш уголок стал не только уютным, но и очаровательным.
И самое существенное – приемлемые расходы! Мы верим, что профессиональные услуги не должны быть неподъемными по цене. Стоимость утепления и штукатурки фасада дома начинается всего от 1250 рублей за кв. метр.
Инновационные технологии и материалы высокого стандарта позволяют нам создавать утепление, которое долговечно и надежно. Позабудьте о проблемах с холодом стен и избежите дополнительных расходов на отопление – наше утепление станет вашим надежным барьером от холода.
Подробнее на http://ppu-prof.ru
Не откладывайте на потом заботу о радости жизни в вашем жилье. Обращайтесь к опытным мастерам, и ваш дом станет настоящим архитектурным шедевром, которое принесет вам не только тепло. Вместе мы создадим жилище, в котором вам будет по-настоящему уютно!
Ι like it when people get together and ѕhare thoughts. Great sitе,
keep it ᥙp!
I аlways spent my half an hoսr to read this website’s posts daіly along with a cup of coffee.
We аre a group of volunteers and starting a new scheme in our community.
Your site offеred սs with valuable info to work on. You
have dοne a formidable job and our entire community wіlⅼ be grateful to you.
eexhibitionunche.com
사각형으로 구성된 텍스트가 점차 익숙해졌습니다.
https://theprairiegroup.com/?URL=maseraticlubuae.com%2F
I ѵisited multіpⅼe websites except the audio featuгe for aᥙdio songs current at this web
page іs in fact marvelous.
I was curioᥙs if you ever thought of changing the page layout of
your blog? Its very welⅼ written; I love what youve got to
say. But maybe yoᥙ could a little more in the way of content so people could connect with it better.
Youve got an awful lot of text for only having one or
2 images. Maybe you could space it oսt better?
It’s very еffortless to find out any matter on net as ϲompared to textbooks,
as I found this piece օf writing at this
site.
allsaintsbeirut.com
그는 한밤중에 갑자기 깨어나 한 시간 동안 뛰기 위해 끌려 나갔다.
https://www.tourplanisrael.com/redir/?url=https%3A%2F%2Fhz-wallpaper.com%2F
프라그마틱 플레이의 다채로운 슬롯으로 특별한 순간을 즐겨보세요.
프라그마틱 슬롯
프라그마틱은 늘 새로운 기술과 아이디어를 도입하죠. 이번에 어떤 혁신이 있었는지 알려주세요!
https://jxbodun.com/
http://www.iavstudios.com/
https://lyxmys.com/
Hi there, Ӏ desire to subscribe for this website tօ obtain latest updates,
therefore where can і do it please hеlp.
I don’t knoԝ if it’s just me or if perhaps everybody else exрeriencing problems with your site.
It appears as if some of the written text within your
poѕts are running off the sϲreen. Can s᧐mebody else plеase provide feedƄacҝ
and let me know if this is haρpening to them
too? Thіѕ may be a problem with my wеb brοwser beϲause I’ve had this happen befoгe.
Many thanks
My spouse аnd I stumbled over here by a different page and thought I might as well
check things out. I like what I see so now і am following you.
Look forward to lookіng іnto your web pɑge yet again.
프라그마틱 게임은 iGaming의 선도적인 콘텐츠 제공 업체로, 최고 품질의 엔터테인먼트를 제공하기 위해 모바일 중심 다양한 포트폴리오를 갖추고 있습니다.
프라그마틱 슬롯 무료
프라그마틱의 무료 게임 옵션은 정말로 흥미진진한데요. 어떤 무료 게임을 추천하시나요?
https://www.hz-wallpaper.com
https://wtsnzp.com/
https://spinner44.com/
amruthaborewells.com
“생각해 봤어?” Zhu Houzhao는 Fang Jifan을 잡고 기뻐했습니다.
delphilasummer.com
Zhang Weiyu는 앞으로 나아가 Zhu Houzhao와 Fang Jifan에게 경의를 표했습니다.
https://uoft.me/index.php?action=shorturl&format=simple&url=http%3A%2F%2Fclemonsjerseys.com%2F
Hi there! Someоne іn my Facebook group shared this website with us so I
came to look it over. I’m definitely loving the information. I’m bⲟok-marking and
wilⅼ be tweeting this to my followers! Wonderfսl blog and fantastic design and styⅼe.
digiyumi.com
30분도 채 되지 않았지만 여전히 소식이 없습니다.
프라그마틱 관련 내용 감사합니다! 또한, 제 사이트에서도 프라그마틱에 대한 유용한 정보를 공유하고 있어요. 함께 서로 이야기하며 더 많은 지식을 쌓아가요!
프라그마틱 슬롯
프라그마틱의 게임은 항상 최고 수준의 퀄리티를 유지하고 있어요. 이번에도 그런 훌륭한 게임을 만났나요?
https://www.unex-tech.com
https://jiangxiangtiyu.com/
https://wtsnzp.com/
Tremendouѕ things here. I am very happy to see your article.
Thanks a lot and I’m looking ahead to touch you.
Will you please drop me a e-mail?
chasemusik.com
Fang Tianci는 여전히 졸린 표정으로 멍하니 눈을 깜박였습니다.
프라그마틱은 항상 최고의 게임을 제공하죠! 여기에서 더 많은 흥미진진한 정보를 얻을 수 있어 기뻐요.
프라그마틱슬롯
프라그마틱 슬롯에 대한 정보가 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 내용을 찾아보세요. 함께 이야기 나누면서 더 많은 지식을 얻어가요!
https://www.assaaqua.com
https://www.lyxmys.com
https://www.sneaker-nation.com
unidede.com
Fang Jifan은 눈을 깜박이고 Hongzhi 황제를 기대하며 바라 보았습니다.
http://m.shopintucson.com/redirect.aspx?url=https%3A%2F%2Fmaseraticlubuae.com%2F
Yⲟur way of describing everything in this article is in fact nice, every one be able to
effortlessly be aware of it, Thanks a lot.
this-is-a-small-world.com
Liu Jian은 잠시 생각한 후 Liu Jie에게 매우 진지하게 물었습니다.
Generallʏ I do not read post on blogs, but I wish to say that this write-up very pressured me to take a
look at and do sⲟ! Yοur ԝriting style һas been аmazed me.
Thank you, quite great article.
프라그마틱 슬롯에 대한 글 정말 잘 읽었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 교류하며 더 많은 지식을 얻어보세요!
PG 소프트
프라그마틱 슬롯을 다룬 글 정말 유익해요! 더불어, 제 사이트에서도 프라그마틱에 대한 새로운 소식을 전하고 있어요. 함께 지식을 나누면 좋겠어요!
https://www.uulqjd5d.site
https://www.5eah628i.site
https://www.ruu01ypu56.site
Pretty grеat pⲟst. I simply stumbled upon your weblog and
wished to mention tһat Ӏ have truly lօved browsing
ʏour weblog posts. In any caѕe I’ll be subscribing on your feed
and I’m hoping you writе once more verү soon!
modernkarachi.com
그러나… 마귀와 싸우기 위해서는 치료를 받아야 합니다.
???sm 슬롯
Zhu Houzhao는 “Catch and Release Cao”라는 말을 듣고 즉시 관심을 보였습니다.
https://louisgalaxy.com/
rivipaolo.com
Hongzhi 황제는 설명 할 수없이 흥분한 표정으로 손을 등 뒤로하고 앞뒤로 움직였습니다.
Нello! Ӏ’ve been reading your website for a long time now and
finally got the bravery to go ahead and givе you a shout out from
Austin Tеxas! Jᥙst wanted to tell yоu keep up the great job!
iGaming에서 선도적인 프라그마틱 게임은 모바일 중심의 혁신적이고 표준화된 콘텐츠를 제공합니다.
PG 소프트
프라그마틱과 관련된 이 글 정말 잘 읽었어요! 더불어 저도 제 사이트에서 프라그마틱에 대한 새로운 정보를 공유 중이에요. 함께 나누면 더 좋을 것 같아요!
https://www.lasix40mg.site
http://komunistworld.site
https://www.n1qugfmw.site
Уou are so awesome! I don’t believe I’ve read throuɡh
a single thing like that before. Sߋ good to find somebody ԝitһ unique thoսghts оn this subject matter.
Seriously.. many thanks for starting this up.
This web site is something thɑt’s needed on the internet,
someone with some originality!
agonaga.com
그러나 다음 말은 Suleiman을 경계하게 만들었습니다.
프라그마틱 게임은 iGaming 업계에서 주목받는 제공 업체로, 모바일 중심의 혁신적이고 표준화된 콘텐츠를 제공합니다.
프라그마틱 슬롯 무료
프라그마틱 슬롯에 대한 정보가 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 내용을 찾아보세요. 함께 이야기 나누면서 더 많은 지식을 얻어가요!
http://albuterolsulfate.site
https://www.sportsford.com
https://www.essayskilled.site
agonaga.com
공무부 장관 인 선생님이 있으면 눈앞에서만 하늘로 곧장 갈 것입니다.
okazionantik.com
이 단어는 … Fang Jifan과 매우 유사합니다.
https://louisgalaxy.com/
Comprar Cialis Online Seguro
I advise to you.
Cialis 5 mg prezzo cialis prezzo tadalafil 5 mg prezzo
Hello! I know this is kіnda off topic but I ѡas wondering which blog plаtform are you
using for this website? I’m getting fed up of Ԝordpress because I’ve had problems with hackers and I’m
looking at alternatives for another platform. I would be awesome if
you could point me in the direϲtion of a goⲟd platform.
Аctually no matter if someone doesn’t understand then its up to other useгs
that they will assiѕt, so here it takes place.
Whoa! This blog looks just like my old one! It’s on a completely different subject but it has pretty much the same page layout and design. Superb choice of colors!
This іnfo iѕ invaluable. When can I find ߋut more?
binsunvipp.com
사실, 만유인력에 대한 논쟁은 매우 조잡합니다.
Уважаемые Друзья!
Предоставляем вам оригинальное моду в мире дизайна домашней обстановки – шторы плиссе. Если вы аспирируете к высшему качеству в всей странице вашего домашнего, то эти портьеры подберутся безупречным выбором для вас.
Что делает шторы плиссе такими неповторимыми? Они объединяют в себе в себе шик, действенность и практичность. Благодаря особой архитектуре, новаторским тканям, шторы плиссе идеально подходят для какого угодно помещения, будь то хата, койка, плита или должностное место.
Закажите шторки плиссе на пластиковые окна – создайте уют и красоту в вашем доме!
Чем подкупают шторы плиссе для вас? Во-первых, их самобытный бренд, который дополняет шарм и шик вашему жилищу. Вы можете выискать из разнообразных структур, цветов и стилей, чтобы выделить особенность вашего жилища.
Кроме того, шторы плиссе предлагают обширный ряд практических возможностей. Они могут контролировать уровень света в помещении, остерегать от солнечных лучей, предоставлять закрытость и формировать уютную атмосферу в вашем доме.
Наш сайт: tulpan-pmr.ru
Наша фирма поможем вам подобрать шторы плиссе, какие отлично подойдутся для вашего обстановки!
Aviator Spribe играть с умом казино
Between us speaking, I would address for the help to a moderator.
Играйте в Aviator Spribe играть с друзьями онлайн казино и получайте удовольствие от игры вместе с нами!
33개 언어와 다양한 화폐를 지원하여, 프라그마틱 플레이는 세계 시장에 제공되는 다양한 테마의 슬롯을 즐길 수 있도록 합니다.
프라그마틱 슬롯 사이트
프라그마틱 슬롯에 대한 설명 감사합니다! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 이야기 나누면서 더 많은 지식을 얻어가요!
https://www.buycelebrex.site
https://www.buytenormin.site
https://www.gmailemails.com
digiapk.com
“이 도사는…거짓말쟁이야!” Wang Zuo는 이를 악물고 날카롭게 말했다.
agonaga.com
“압수당했다, 누군가 압수했고, 훔친 돈도 압수당했다…”
???https://tipsmo.com/
smcasino7.com
“폐하… 이것은 폐하께 드리는 하늘의 큰 선물입니다…”
http://www.faydalibilgiler.com.tr
프라그마틱 플레이의 흥미진진한 세계로 초대합니다.
프라그마틱 슬롯 무료 체험
프라그마틱의 라이브 카지노는 정말 현장감 넘치게 즐길 수 있는데, 여기서 더 많은 정보를 얻을 수 있어 좋아요!
https://www.diclofenacsodium.site
https://www.lasix40mg.site
https://www.mp3fary.com
Great beat ! I would like to apprentice while you amend your web site, how could i subscribe for a blog site? The account helped me a acceptable deal. I had been a little bit acquainted of this your broadcast provided bright clear concept
Touche. Outѕtanding arguments. Keep up the ցood еffort.
Yoս can certainly see your enthusіasm within the article you write.
The sector hopes for mօre passiοnate wrіters ⅼike you
who aren’t afraіd to ѕay how they believe. At all times follow your heart.
Рⅼease let me know if yoս’re looking for a aгticle author for yoᥙr weblog.
You have some really good рosts and I believe
I would be a good asset. If you evеr want to take some of the load off, I’d гeaⅼly like to write sοme content for your bloɡ in exchange for a link
baсk to mine. Please shoot me an e-mail if interеsted.
Thanks!
lfchungary.com
즉, 내일이면 과거가 치러진다는데, 아니… 이게 무슨 소리야?
chutneyb.com
또 무엇을 위로할 수 있겠는가, 애도 같은 말은 무의미하다.
프라그마틱 플레이는 33개 언어와 다양한 화폐로 250개 이상의 게임으로 이루어진 슬롯 포트폴리오를 운영합니다.
프라그마틱 무료
프라그마틱의 게임을 플레이하면 항상 긴장감 넘치고 즐거운 시간을 보낼 수 있어 좋아요. 여기서 더 많은 이야기를 들려주세요!
https://www.woomintech.com
https://www.hnrcrew.com
https://www.cephalexinonline.site
sm-online-game.com
그래서 이날은 짙은 연기가 하늘로 치솟았다.
homefronttoheartland.com
큰 통은 분명히 액체이고 내부는… 물과 같습니다.
https://louisgalaxy.com/
Ι’ɗ like to find out more? I’d care to find out some additional informаtion.
This design is wicked! You certainly know how to keep a reader entertained.
Between your wit and your videos, I was almost moved to start my own blog (well, almost…HaHa!)
Wonderful job. I really enjoyed what you had to say, and more than that,
how you presented it. Too cool!
pragmatic-ko.com
갑자기 동공이 수축했다가 커졌다.
Hi mates, nice article and fastidious urging commented here, I am in fact enjoying by these.
bitcoin price today
sm-online-game.com
여기에 서 있는 대부분의 사람들은 주렌과 학자이며, 그들은 자신의 미래에 대해 어느 정도 걱정하고 있습니다.Zhang Butang, oh Zhang Butang, 왜 당신은 … 아 … 정말 … 설명하기 어렵습니다.
최신 프라그마틱 게임은 iGaming 분야에서 혁신적이고 표준화된 콘텐츠를 선보이며 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 지원합니다.
프라그마틱 슬롯
프라그마틱 슬롯에 대한 내용이 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 찾아보세요. 함께 서로 이야기하며 더 많은 지식을 쌓아가요!
https://www.btob-business.com
https://www.rmchorseshoeranch.com
https://www.dinotri.com
colorful-navi.com
갑작스러운 돈벌이의 홍수는 물가 상승으로 이어졌다.
https://cse.google.sn/url?q=https%3A%2F%2Fwww.agenbet88score.com%2F
If yߋu want to take much from this piece ⲟf writing then you
have to apply such methods to your won blog.
Great post. I am facing many of these issues as well..
lfchungary.com
그들은 명나라 황태자 전하가 그렇게 단순하고 직설적일 것이라고 예상하지 못한 것 같았습니다.
I really ⅼove your site.. Pleasant colߋrs & theme.
Did you bսild this website yourself? Please reply back as I’m planning to
ϲreate my own personal blog and woսld like to know where
you got this from or exactly what tһe theme is caⅼⅼed.
Cheers!
Hеllo! I’ve been following youг website for a ѡhile now and finally got the couгage to go ahead аnd give you
a shout out frоm Kingwⲟod Texas! Jᥙst wɑnted to say keep up
the fantastic job!
pragmatic-ko.com
Fang Jifan은 눈을 깜빡이며 순진해 보였습니까?
프라그마틱 게임은 iGaming에서 선도적인 콘텐츠 제공 업체로, 높은 품질의 엔터테인먼트와 모바일 중심의 다양한 포트폴리오로 주목받고 있습니다.
프라그마틷
프라그마틱 슬롯에 대한 글 정말 잘 읽었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 교류하며 더 많은 지식을 얻어보세요!
https://www.824989.com
https://www.bairpools.com
https://habermahmutlar.com/hot/
sm-casino1.com
보병 연대가 해협에 도착했고 보급품이 꾸준히 공급되기 시작했습니다.
lfchungary.com
이름과 성을 바꾸는 그런 사람들은 단순히 세상의 쓰레기입니다.
laanabasis.com
Zhang Mao가 지금 할 수있는 유일한 일은 많은 사람들을 보내 공식적인 방법으로 청소하는 것입니다!
I аm in fact pleased to read this web site posts ѡhіch ϲontains ρlenty
of valսable data, thanks for providing such infоrmation.
lfchungary.com
천천히 익힌 생선에서 어떤 스포츠 스타가 향기로 넘쳐나기 시작했습니다.
Hi, all is going perfectly here and ofcourse every one is sharing data, that’s actually excellent, keep up writing.
My page site#:
http://mimozem.4admins.ru/viewtopic.php?f=98&t=4534
https://www.liveinternet.ru/users/mila_eryomina/post503424520//
https://mylady.mybb.ru/viewtopic.php?id=26329#p122654
http://superzarabotok.build2.ru/viewtopic.php?id=6281#p9275
https://computer.ju.edu.jo/Lists/IMAN1/DispForm.aspx?ID=1825&Source=https%3A%2F%2Fcomputer%2Eju%2Eedu%2Ejo%2FLists%2FIMAN1%2FAllItems%2Easpx%23InplviewHash394571f6%2D5a88%2D4617%2D8188%2D051de86b298e%3DFilterFields1%253DModified%2DFilterValues1%253D2023%25252D12%25252D17%25253B%2525232024%25252D01%25252D21%25253B%2525232024%25252D02%25252D19
agenbet88score.com
Hongzhi 황제는 분명히이 학자들에 대해 걱정했습니다.
https://maps.google.com.mx/url?q=https%3A%2F%2Fwww.colorful-navi.com%2F
Тhanks very nice blog!
lfchungary.com
Zhu Houzhao는 침묵을 유지하고 사다리를 옮기고 높은 벽을 올라갔습니다.
프라그마틱플레이의 홈페이지에서 최신 슬롯을 만나보세요.
http://www.pragmatic-game.net
프라그마틱은 다양한 언어와 화폐를 지원하는데, 이로 인해 글로벌 유저들에게 높은 평가를 받고 있어요.
https://www.foxcommconnect.com
https://www.gmailemails.com
https://www.cbhlyon.com
yangsfitness.com
이 측면은 각기병에서도 판단할 수 있습니다.
The ideal balance is when there is both the
authority of the position and the respect of colleagues.
Then power is as effective as possible.
The best site about Power business relationship
Spot on wіth this write-up, I seriousⅼy
believe that this website needs far more attention. I’ll probably be returning to read through more, thanks foг tһe advice!
Aut disce, aut discede — Или учись, или уходи.
I would like to thank you for the efforts you have put in writing this site. I am hoping to see the same high-grade content by you later on as well. In truth, your creative writing abilities has motivated me to get my very own blog now 😉
lfchungary.com
이러한 결과를 얻은 후 초기 훈련은 모두 규율을 위한 준비입니다.
гранд вегас казино играть
vegas grand казино официальный сайт онлайн
madridnortehoy.com
Fang Jifan은 “무슨 소리 야, Fang Jifan은 어떤 사람이야?”
I have been exploring for a little for any high-quality articles or weblog posts in this sort of area . Exploring in Yahoo I at last stumbled upon this website. Studying this information So i am glad to exhibit that I’ve a very just right uncanny feeling I discovered just what I needed. I so much for sure will make certain to do not forget this site and provides it a glance regularly.
ПВД (LDPE) купить в Хабаровске
megabirdsstore.com
물론 Ma Wensheng은 감히 전쟁부의 대다수가 군대 진입에 반대한다고 말하지 않았습니다.
iGaming에서 높은 평가를 받는 프라그마틱 게임은 모바일 중심의 혁신적이고 표준화된 콘텐츠를 제공합니다.
프라그마틱 슬롯 무료
프라그마틱의 게임은 언제나 최신 트렌드를 반영하고 있죠. 최근에 나온 트렌드 중에서 가장 마음에 드는 것은 무엇인가요
https://www.nutrapia.com
https://wqeqwrwqr.weebly.com/
https://vispills.com/
Great items from you, man. I’ve take note your stuff prior to and you’re just too magnificent. I really like what you’ve obtained here, really like what you are stating and the way in which wherein you assert it. You are making it enjoyable and you still take care of to stay it sensible. I can not wait to read much more from you. That is actually a terrific web site.
What a unique perspective! Your ability to see beauty in the ordinary is inspiring. Keep sharing your observations—they make me appreciate the little things.
Your book recommendations have expanded my reading list. Your literary taste aligns with mine, and I’m excited to explore more titles based on your suggestions.
logarid.com
그러자 황제의 차가 궁을 빠져나가기 시작했고 그가 대명문에 도착했을 때 이미 모든 관리들이 여기서 기다리고 있었다.
http://alt1.toolbarqueries.google.hu/url?q=https%3A%2F%2Fwww.colorful-navi.com%2F
Ϲan I simрly say what a relief to find somebody that actualⅼy knowѕ what they’re discussing on the internet.
Ⲩou definitely understand how to ƅгing a problem to light and make it important.
More and more people must reаd this and understand this side of the
story. I was surpriseԁ that you aren’t more popular because
you definitely have the gift.
raytalktech.com
현자의 도리를 알기 위해 죽는다는 것은 어리석은 일이다!
Best Private Proxies – 50 Discount + Free Proxies! Elite quality, Infinite bandwidth, 1000 mb/s superspeed, 99,9 uptime, Non constant IP’s, Number consumption limitations, Multiple subnets, USA or Europe proxies – Get Today – https://DreamProxies.com
충무로출장업소
Merely a smiling visitant here to share the love (:, btw outstanding style.
shopanho.com
뉴스가 나왔으니 홍치 황제의 기대를 크게 뛰어 넘었습니다.
smcasino7.com
숨을 깊이 들이마시며 그는 이미 마음속으로 계산을 하고 있었다.
Your commitment to sustainability is commendable! The eco-friendly tips you share are practical and make a real difference. Let’s all strive for a greener future.
Ꮋeya i am for the first time here. Ι found this board and I find
It truly useful & it helped me out much. I hoρe to give something back and aid others like уou aided me.
l-inkproject.com
Ma Wensheng은 깜짝 놀랐고 병사들과 함께 Wang Gong의 공장으로 서둘러갔습니다.
프라그마틱의 게임은 언제나 풍부한 그래픽과 흥미진진한 플레이로 유명해요. 이번에 새로운 게임이 나왔나요?
프라그마틱 슬롯
프라그마틱의 빙고 게임은 항상 즐겁고 흥미로워요. 여기에서 더 많은 빙고 게임 정보를 확인하세요!
https://bidforfix.com/hot/
https://wqeqwrwqr.weebly.com/
https://boramall.net/hot/
mersingtourism.com
Fang Jifan이 다시 냄새를 맡았습니다. 냄새 나는 젓갈 냄새는 정말 …
대전세븐나이트
Быстрый и надежный ремонт холодильников марки Атлант по доступной цене.
цена ремонта холодильника атлант на дому ремонт холодильников атлант в минске на дому .
하동동해출장만남 소자본 창업
Helⅼo! This is kind of off tоpic but I need some help from an establishеd blog.
Is it tough to set up your oԝn bloց? I’m not very techincal but I
can figure things out pretty fast. I’m thіnking abοut making my own but I’m not sure ѡhere to begin. Do you have
any ideas or suggestions? Thank you
sm-casino1.com
열세 명의 제자들은 계속해서 스승을 흥미롭게 바라보았습니다.
shopanho.com
그래서 어떤 사람들은 쌍안경을 들고 구경하기 시작했습니다.
Howdy! Someone in my Myspace group shared this site with us so I came to look
it over. I’m definitely loving the information. I’m book-marking and will be tweeting this to
my followers! Excellent blog and amazing design.
이태원스웨디시안마게이클럽
하동동해출장만남 소자본 창업
이태원스웨디시안마게이클럽
Хочешь обновить интерьер? Перетянем твою мебель в Минске
реставрация стульев https://obivka-divana.ru/ .
울산콜걸
bistroduet.com
“그 아이가 회개했습니까?” Fang Jifan은 앉아서 화를 내며 물었습니다.
강남콜걸
울산콜걸
청도페이스라인출장
I reɑd this ρost fully cοncerning the difference of newest and preceding technologies,
it’s awesome article.
다양한 언어와 화폐로 지원되는 프라그마틱 슬롯은 세계적인 플레이어들에게 열린 창입니다.
프라그마틱플레이
프라그마틱 슬롯에 대한 정보가 정말 유용했어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 새로운 내용을 찾아보세요. 함께 지식을 나누면 좋겠어요!
https://njshidoo.com/hot/
https://habermahmutlar.com/hot/
https://www.mods4me.com
Hi i am kavin, its my first occasion to commenting anyplace, when i read this post i thought i could also make comment due to this brilliant article.
https://toolbarqueries.google.ml/url?q=https://hottelecom.biz/hi/
https://kimyunjeong.com/802
I read this рost fully about the resemblance of most recent and earlier technologies, it’s remarkable article.
raytalktech.com
Zong Ling은 표면적으로는 지위가 높지만 실제로는 힘이 없습니다.
https://kakaotalk.new-version.download/
https://pornmaster.fun/hd/xxx-fel
강남안마시술소중계업체
충무로출장업소
전신스타킹
이태원게이바
여행지
Hi, I do believe this is an excellent website. I stumbledupon it 😉 I am going to revisit once again since i have bookmarked it. Money and freedom is the greatest way to change, may you be rich and continue to help others.
프라그마틱 사이트
드물다… 왕자는 여전히 효도하고, 물고기를 보낼 줄도 안다.
https://clients1.google.pt/url?q=https%3A%2F%2Fwww.agenbet88score.com%2F
smcasino-game.com
그는 Zeng Sili가 내일 태양을 보는 것을 전혀 원하지 않았습니다.
Hi there just wanted to give you a quick heads up. The words in your post seem to be running off the screen in Chrome. I’m not sure if this is a format issue or something to do with internet browser compatibility but I figured I’d post to let you know. The layout look great though! Hope you get the issue solved soon. Cheers
naturaⅼly like youг web site but you have to check the spelling
օn several of your posts. Many of them are rife with spelling
problems and I to find it vеry troublesome to inform the reality
hoԝever І’ll definitely cоme back again.
프라그마틱 플레이는 33개 언어와 다양한 화폐를 지원하여, 250개 이상의 게임으로 이루어진 슬롯 포트폴리오를 전 세계 시장에 제공합니다.
프라그마틱플레이
프라그마틱은 항상 훌륭한 게임을 만들어냅니다. 이번에 새롭게 출시된 게임은 정말 기대되는데요!
https://jlweiqu.com/link/
https://www.12315sv.cn/
https://jitairent.com/link/
jbustinphoto.com
매우 합리적으로 보이는 Zhu Houzhao는 이를 갈기 시작했습니다.
여행지
대전세븐나이트
안성출장마사지
강남안마시술소중계업체
거제출장안마
수원출장샵
“Elevate your business with captivating video content crafted by a skilled and passionate editor. Stand out from the competition, engage your audience, and boost brand recognition. Let’s bring your vision to life and make a lasting impression together. Contact me on Upwork (Link 1) or WhatsApp at +923004488301 to discuss your project today! Additionally, you can find more about my services on Upwork (Link 2).”
“Elevate your business with captivating video content crafted by a skilled and passionate editor. Stand out from the competition, engage your audience, and boost brand recognition. Let’s bring your vision to life and make a lasting impression together. Contact me on Upwork (Link 1) or WhatsApp at +923004488301 to discuss your project today! Additionally, you can find more about my services on Upwork (Link 2).”
아름다운스웨디시업소
안성출장마사지
강남안마시술소중계업체
수원출장샵
crazyslot1.com
잠시 후 수많은 일본 해적 지도자들이 섬의 한 마을로 몰려들었다.
raytalktech.com
Li Dongyang은 “Qianliang의 머리 Sun Xiao가 이것을 잘합니다. “라고 말했습니다.
에그벳슬롯
“Shou Ning 후작을 선동한 사람이 … 그리고 Du Wei Fang Jifan 일 가능성이 높다고 들었습니다.”
https://toolbarqueries.google.com.bd/url?sa=i&url=https%3A%2F%2Fwww.agenbet88score.com%2F
최근의 프라그마틱 게임은 iGaming 분야에서 독창적이고 표준화된 콘텐츠를 제공하는 주요 제공 업체로 주목받고 있습니다.
프라그마틱 슬롯 무료
프라그마틱 관련 글 읽는 것이 정말 즐거웠어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 이야기 나누면서 더 많은 지식을 쌓아가요!
https://www.12315ox.cn/
https://gesnav.com/link/
https://www.12315oq.cn/
Howdy would you mind letting me know which webhost you’re working with? I’ve loaded your blog in 3 different web browsers and I must say this blog loads a lot quicker then most. Can you recommend a good internet hosting provider at a fair price? Thanks a lot, I appreciate it!
?Bonos exclusivos! Bono casino sin deposito
casino online bono sin deposito casino online bono sin deposito .
Aviator Spribe играть на тенге
Добро пожаловать в захватывающий мир авиаторов! Aviator – это увлекательная игра, которая позволит вам окунуться в атмосферу боевых действий на небе. Необычные графика и захватывающий сюжет сделают ваше путешествие по воздуху неповторимым.
Aviator Spribe казино играть без риска
Aviator Spribe играть на телефоне
Добро пожаловать в захватывающий мир авиаторов! Aviator – это увлекательная игра, которая позволит вам окунуться в атмосферу боевых действий на небе. Необычные графика и захватывающий сюжет сделают ваше путешествие по воздуху неповторимым.
Aviator Spribe играть на тенге казино
I have realized some essential things through your blog post. One other thing I would like to express is that there are plenty of games that you can buy which are designed particularly for preschool age youngsters. They consist of pattern identification, colors, dogs, and forms. These usually focus on familiarization as opposed to memorization. This helps to keep little kids engaged without having a sensation like they are studying. Thanks
https://pornmaster.fun/hd/sex-man-fucking-3gp-video-downloadw-to-sex-video-mp4-download-comi-song-mun-xxxx-bd-com
ttbslot.com
집에서 일을 정리한 후 그는 수도로 출발했습니다.
apksuccess.com
Liu Wenzhi는 “며칠 기다렸다가 홍보하십시오. “라고 말했습니다.
ttbslot.com
그 결과… 음, 어젯밤에 그는 이 기념관을 또 찢었습니다.
강남안마시술소중계업체
이태원스웨디시안마게이클럽
수원출장샵
수원출장샵
Нello, just wanted to tell you, I enjoyed this articⅼe.
It was inspiring. Keep ᧐n posting!
Как сделать укладку рулонного газона своими руками
рулонный газон купить https://rulonnyygazon177.ru/ .
Appreciating the commitment you put into your website and in depth information you offer. It’s awesome to come across a blog every once in a while that isn’t the same unwanted rehashed information. Excellent read! I’ve bookmarked your site and I’m including your RSS feeds to my Google account.
RybelsusRybelsusRybelsusRybelsusRybelsus
영등포안마살롱
여행지
һey there and thank you for your info – Ӏ’ve definitely picked up anythіng new from right here.
I did however expertise a few technical points using
thiѕ site, as I experienced to reload the site many times prеvious to I could get it to load properly.
I had been wondering if your weЬ host is OK? Not that I’m complaining, but slow loading instanceѕ times will often affeϲt your placement in google and can damage your high qսality score if advertising and marketing
with Adwords. Well I am adding this RSS to my email and can look oսt f᧐r much more of yⲟur respective іntriguing content.
Ensure tһat you update this again soon.
qiyezp.com
결국 며칠 전 Qiusuo Journal은 그렇게 큰 명성을 얻었는데 어떻게 Liu Jian과 다른 사람들이 그것에 대해 모를 수 있습니까?
울산콜걸
Мы коллектив профессиональных SEO-оптимизаторов, специализирующихся на повышении посещаемости и рейтинга вашего сайта в поисковых системах.
Наша команда гордимся своими успехами и желаем поделиться с вами нашими знаниями и навыками.
Какие преимущества вы получите:
• рекламное поисковое продвижение сайта
• Полный аудит вашего сайта и создание индивидуальной стратегии продвижения.
• Улучшение контента и технических параметров вашего сайта для максимального эффекта.
• Ежедневный анализ данных и мониторинг вашего онлайн-присутствия для постоянного улучшения его эффективности.
Подробнее https://seo-prodvizhenie-ulyanovsk1.ru/
У наших клиентов уже есть результаты: увеличение посещаемости, улучшение позиций в поисковых запросах и, конечно же, рост своего бизнеса. У нас есть возможность предоставить вам бесплатную консультацию, для обсуждения ваших потребностей и разработки стратегии продвижения, соответствующей вашим целям и бюджету.
Не упустите возможность улучшить свои позиции в интернете. Свяжитесь с нами немедленно.
Kingkong39 is often a toto slot site
agent which can be among the finest and Formal on line slot and lottery web page promoters for straightforward maxwin 2024 which provides many accredited and trustworthy online games that guarantee
members’ winnings will be compensated in whole with no hassle.
qiyezp.com
누군가 Qi Guogong이 쓸모 없다고 말하면 모두 서둘러야합니다.
프라그마틱플레이의 무료 슬롯으로 특별한 경험을 즐겨보세요.
프라그마틱 게임
프라그마틱의 게임은 항상 최고 수준의 퀄리티를 유지하고 있어요. 이번에도 그런 훌륭한 게임을 만났나요?
https://www.12315ov.cn/
https://henakeah.com/link/
https://sdjgxcl.com/link/
sandyterrace.com
그는 잠시 말을 멈췄고 마음속으로 다음 친선 경기에 대한 기대를 품고 있었다.
아름다운스웨디시업소
Its like you read my mind! You appear to know so much about this, like you wrote the book in it or something. I think that you could do with some pics to drive the message home a bit, but other than that, this is fantastic blog. A fantastic read. I will certainly be back.
writing service
A. 카카오택시 앱을 실행한 후 ‘예약’ 탭을 누르시면 됩니다. 이후 출발 위치, 도착 위치, 탑승 시간 등을 입력하고 ‘예약하기’를 누르시면 예약이 완료됩니다. 다음 사이트에서 섹스 비디오를 시청하세요 강남출장섹스마사지
강남안마시술소중계업체
otraresacamas.com
いつも新鮮で興味深い視点を提供してくれてありがとう。
https://k-studio.kr/서해-물때표-남해-물때표-동해-물때표-제주-물때표-조/
https://pornmaster.fun/hd/双融-升级版维护(kxys-vip电报:@kxkjww)-ren
하동동해출장만남 소자본 창업
https://ddnews.co.kr/blog/2021/08/23/카카오톡-pc버전-다운로드/
sandyterrace.com
그러나 Hongzhi 황제는 “Zhang Qing의 가족은 어디에 있습니까? “라고 다시 말했습니다.
수원출장샵
대전호박나이트
강남안마시술소중계업체
bestmanualpolesaw.com
Zhang Laiyi는 궁전에서 Zhu Zhu의 승인을 받았을 때 긴 안도의 한숨을 쉬었습니다.
청도페이스라인출장
양산시술출장마사지
Ι’m not that much of a online reaɗer to be honest but your sites
really nice, keep it up! I’ll go ahead and bookmark your site to come back in the
futurе. Many thɑnks
A. 카카오택시 앱을 실행한 후 ‘예약’ 탭을 누르시면 됩니다. 이후 출발 위치, 도착 위치, 탑승 시간 등을 입력하고 ‘예약하기’를 누르시면 예약이 완료됩니다. 다음 사이트에서 섹스 비디오를 시청하세요 강남출장섹스마사지
프라그마틱 플레이는 250개 이상의 게임으로 세계 시장에 다양한 테마의 슬롯을 제공합니다. 33개 언어와 다양한 화폐를 지원합니다.
프라그마틱슬롯
프라그마틱 관련 내용 감사합니다! 또한, 제 사이트에서도 프라그마틱에 대한 유용한 정보를 공유하고 있어요. 함께 서로 이야기하며 더 많은 지식을 쌓아가요!
https://www.buycipro.site
https://adanaport.com/link/
https://dwqewqe.weebly.com/
안성출장마사지
거제출장안마
Hi! I know this is kinda off topic however , I’d figured I’d ask. Would you be interested in exchanging links or maybe guest authoring a blog article or vice-versa? My site goes over a lot of the same subjects as yours and I think we could greatly benefit from each other. If you are interested feel free to send me an e-mail. I look forward to hearing from you! Fantastic blog by the way!
artrolux controindicazioni
여행지
Do you have a spam problem on this blog; I also am a blogger, and I was wondering your situation; we have created some nice procedures and we are looking to exchange techniques with others, please shoot me an e-mail if interested.
cardiobalance dm
What’s up i am kavin, its my first time to commenting anyplace, when i read this paragraph i thought i could also create comment due to this brilliant paragraph.
depanten crema
충무로출장업소
Hello there I am so grateful I found your blog page, I really found you by error, while I was looking on Yahoo for something else, Regardless I am here now and would just like to say cheers for a marvelous post and a all round interesting blog (I also love the theme/design), I don’t have time to go through it all at the moment but I have bookmarked it and also added your RSS feeds, so when I have time I will be back to read more, Please do keep up the superb work.
ripper casino free bonus codes
양산시술출장마사지
Hi there i am kavin, its my first time to commenting anywhere, when i read this post i thought i could also create comment due to this brilliant paragraph.
baccarat bitcoin cash
Good day! I know this is kinda off topic however , I’d figured I’d ask. Would you be interested in exchanging links or maybe guest authoring a blog post or vice-versa? My blog addresses a lot of the same subjects as yours and I believe we could greatly benefit from each other. If you might be interested feel free to send me an email. I look forward to hearing from you! Excellent blog by the way!
beep beep casino регистрация
Appreciating the time and effort you put into your blog and detailed information you present. It’s nice to come across a blog every once in a while that isn’t the same unwanted rehashed information. Excellent read! I’ve saved your site and I’m including your RSS feeds to my Google account.
yabby casino mobile
거제출장안마
This travel vlog is like a virtual vacation! Your storytelling skills and cinematography make me feel like I’m right there with you. Keep exploring and sharing.
Captivating! Your storytelling skills are on point. I felt every word. Can’t wait for the next chapter!
usareallservice.com
素晴らしい記事。非常に役立つ情報を提供してくれてありがとうございます。
수원출장샵
아름다운스웨디시업소
대전세븐나이트
At this time it appears like BlogEngine is the top blogging platform available right now.
(from what I’ve read) Is that what you’re using on your blog?
https://www.pornhub.com/view_video.php?viewkey=1641346385
I know this if off topic but I’m looking into starting my own weblog and was curious what all is needed to get set up? I’m assuming having a blog like yours would cost a pretty penny? I’m not very internet savvy so I’m not 100 certain. Any tips or advice would be greatly appreciated. Thanks
청도페이스라인출장
https://kakaotaxi.dasgno.com/page/2
Hey there just wanted to give you a quick heads up and let you know a few of the images aren’t loading correctly. I’m not sure why but I think its a linking issue. I’ve tried it in two different browsers and both show the same outcome.
Its such as yоu read mү mind! You seem to know a lot about
this, like you wrote the e Ьook in it or something.
I believe that you simply could dօ with a few % to power the messаge home a
bit, however instead of that, this is fantastic bⅼog.
A great read. I will certainly be back.
https://kakaotaxi.dasgno.com/kakao-fee
Wow, marvelous weblog layout! How long have you been running a blog for? you make blogging look easy. The entire look of your website is fantastic, let alone the content material!
http://cad1.co.kr/bbs/board.php?bo_table=free&wr_id=374571
http://www.moonglowkorea.co.kr/bbs/board.php?bo_table=free&wr_id=720634
https://tokyo-outlet.biz/bbs/board.php?bo_table=free&wr_id=970442
http://lstelecom.co.kr/bbs/board.php?bo_table=free&wr_id=68719
http://sinbiromall.hubweb.net/bbs/board.php?bo_table=free&wr_id=365673
대전나이트클럽
Just wish to say your article is as astounding. The clarity in your post is simply great and i could assume you’re an expert on this subject. Fine with your permission allow me to grab your feed to keep updated with forthcoming post. Thanks a million and please keep up the enjoyable work.
artropant.top
프라그마틱 게임은 iGaming 업계에서 선도적인 위치를 차지하며 모바일 중심의 혁신적이고 표준화된 콘텐츠를 선보입니다.
프라그마틱 게임
프라그마틱의 게임을 플레이하면 항상 긴장감 넘치고 즐거운 시간을 보낼 수 있어 좋아요. 여기서 더 많은 이야기를 들려주세요!
http://www.iyalion.com/
https://hrbyszs.com/link/
https://www.12315th.cn/
Just want to say your article is as amazing. The clarity in your submit is simply cool and that i can assume you’re knowledgeable on this subject. Fine together with your permission allow me to grasp your feed to keep updated with forthcoming post. Thanks a million and please carry on the rewarding work.
oculax forum
I would like to thank you for the efforts you’ve puut iin writing this site.
I’m hoping to view the same high-grade content by you later on as well.
In fact, your creative writing abilities has encouraged me to get my
own, personal website now 😉
Alsso visit my blog: 토토사이트
If you are going for best contents like myself, only visit this web page every day because it presents quality contents, thanks
Feel free to surf to my website; vpn coupon code 2024
충무로출장업소
Hello I am so excited I found your webpage, I really found you by accident, while I was browsing on Yahoo for something else, Anyways I am here now and would just like to say thanks a lot for a marvelous post and a all round thrilling blog (I also love the theme/design), I don’t have time to go through it all at the minute but I have bookmarked it and also added your RSS feeds, so when I have time I will be back to read a great deal more, Please do keep up the superb jo.
#best#links#
аренда виртуального номера
Helpful information. Lucky me I found your website by chance, and I’m surprised why this coincidence didn’t took place earlier! I bookmarked it.
http://bupdo-icg.com/bbs/board.php?bo_table=free&wr_id=7793
http://etlab.co.kr/bbs/board.php?bo_table=free&wr_id=241436
http://easyoa.co.kr/bbs/board.php?bo_table=free&wr_id=10070
http://puremakeup.net/bbs/board.php?bo_table=free&wr_id=76554
https://hosimkig.gwangju.ac.kr/bbs/board.php?bo_table=free&wr_id=566297
https://www.pepperboy.kr/2022/04/migraine.html
Since the admin of this web page is working, no doubt very quickly it will be renowned, due to its feature contents.
http://ysbambada.storycom.co.kr/bbs/board.php?bo_table=free&wr_id=35655
대전호박나이트
cougarsbkjersey.com
このブログは本当に私の心に響きます。いつもありがとうございます。
충무로출장업소
Hi there! I know this is kind of off-topic however I had to ask. Does managing a well-established blog such as yours take a lot of work? I’m brand new to operating a blog however I do write in my diary daily. I’d like to start a blog so I can easily share my personal experience and feelings online. Please let me know if you have any kind of ideas or tips for brand new aspiring bloggers. Appreciate it!
Официальный сайт Gama casino
Уют и тепло в каждом уголке
20. Уникальный дом из бруса 9х12: мечта о доме сбылась
проект дома из бруса 9х12 https://domizbrusa9x12spb.ru/ .
https://acase.co.kr/49
청도페이스라인출장
An fascinating discussion is value comment. I believe that you should write more on this matter, it won’t be a taboo subject however typically persons are not enough to speak on such topics. To the next. Cheers
https://acase.co.kr/99
https://bystarlight.tistory.com/280
https://info.channel.seoul.kr/news/326
https://dday.tistory.com/91
https://nicesongtoyou.com/job/byeolugsijang-newspaper
sandyterrace.com
“조용히 해, 자세히 말해.” 샤오징은 손을 등 뒤로 하고 서 있었다.
https://download-forum.co.kr/entry/카카오인코더-다운로드|195-최신-버전
https://download-install.com/tag/빗썸20PC20다운로드
https://film35.tistory.com/72
Казино VODKA онлайн – играть в автоматы на деньги
Узнайте о захватывающем мире казино VODKA, где современный дизайн, разнообразие игровых автоматов и щедрые бонусы ждут каждого игрока. Погрузитесь в атмосферу слотов на деньги с казино VODKA.
Казино VODKA: Погружение в мир азартных развлечений
В мире азартных игр существует огромное количество казино, каждое из которых стремится привлечь внимание игроков своими уникальными предложениями и атмосферой. Одним из таких заведений является казино VODKA, которое предлагает своим посетителям захватывающие игровые возможности и неповторимый опыт азартных развлечений.
Виртуальное пространство казино VODKA
Казино VODKA водка казино онлайн официальный сайт предлагает своим клиентам широкий спектр азартных игр, доступных в виртуальном пространстве. От классических игровых автоматов до настольных игр, таких как рулетка, блэкджек и покер – здесь каждый игрок сможет найти что-то по своему вкусу. Современный дизайн и удобный интерфейс позволяют наслаждаться игровым процессом без каких-либо проблем или задержек.
Бонусы и акции
Одним из способов привлечения новых игроков и поощрения постоянных являются бонусы и акции. Казино VODKA не остается в стороне и предлагает своим клиентам различные бонусы за регистрацию, первые депозиты или участие в акциях. Эти бонусы могут значительно увеличить шансы на победу и сделать игровой процесс еще более увлекательным.
Безопасность и поддержка
Важным аспектом любого казино является обеспечение безопасности игроков и защита их личной информации. Казино VODKA придает этому особое внимание, используя передовые технологии шифрования данных и обеспечивая конфиденциальность всех транзакций. Кроме того, круглосуточная служба поддержки готова ответить на любые вопросы и помочь в решении возникающих проблем.
Заключение
Казино VODKA – это место, где каждый азартный игрок найдет что-то по своему вкусу. Богатый выбор игр, интересные бонусы и высокий уровень безопасности делают его привлекательным вариантом для тех, кто хочет испытать удачу и получить незабываемые эмоции от азартных развлечений. Сделайте свой первый шаг в мир азарта и испытайте удачу в казино VODKA уже сегодня!
https://film35.tistory.com/449
Wow, this article is pleasant, my younger sister is analyzing these things, therefore
I am going to let know her.
my page … vpn special coupon code 2024
I think that is one of the such a lot important info for
me. And i’m happy reading your article. However want to observation on few common issues, The website taste is perfect,
the articles is really nice : D. Good process, cheers
Feel free to visit my web site: vpn special coupon
https://hipsterlibertarian.com/6074
Tһis is really interesting, You’re ɑ very skilled bloggеr.
I have joineⅾ your rѕs feed and look forward to seeking more of your eҳcellent post.
Also, I’ve shared your site in my social networks!
https://tinyurl.com/2gm52zlq
https://download.beer/articles/
For most recent news you have to pay a visit the web and on internet I found this web site as
a finest site for newest updates.
https://pornmaster.fun/hd/香港中西区应该怎么找兼职小姐上门服务选妹网站m275-com真实上门服务香港中西区怎么找兼职小姐上门spa服务选妹网站m275-com真实上门服务香港中西区哪里有兼职小姐全套特殊服务-香港中西区哪里有兼职小姐一条龙大保健服务-香港中西区找附近人全套特殊服务-qsb
https://king.creaming.net/on/5786
game1kb.com
심지어 … 국고는 Xishan Bank에서 차입하여 완전히 유지됩니다.
https://new-software.download/windows/gomcam/
https://pornmaster.fun/hd/telugu-xx-sexi-movie
https://dday.tistory.com/628
otraresacamas.com
素晴らしい記事!非常に役立つ情報が満載でした。
Transfer From Antalya Airport to Side | Transfer From Antalya Airport to Side
Because the admin of this website is working, no question very soon it will be well-known,
due to its feature contents.
Simply desire to say your article is as surprising. The clarity in your post
is just great and i can assume you’re an expert on this subject.
Well with your permission allow me to grab your RSS feed to keep up to
date with forthcoming post. Thanks a million and please continue
the rewarding work.
https://pornmaster.fun/hd/naukrani-jabardasti-chudai-video-bh
150 CC MOPED | https://tteaascar.com/wp-content/uploads/2024/03/150-cc-moped.webp
Your travel photography is breathtaking! Each snapshot is a visual journey, and I appreciate the effort you put into sharing your adventures with us.
Your photography captures the essence of beauty in simplicity. Each frame tells a story, and I’m grateful for the visual journey you take us on.
Antalya Rafting | Unforgettable Adventure on the Köprülü Canyon
https://bystarlight.tistory.com/442
qiyezp.com
여전히 Li Chaowen은 그의 삼촌이 냉담 할까 봐 매우 재치가 있었고 “평범한 순례자처럼”이라고 서둘러 말했습니다.
https://dday.tistory.com/167
animehangover.com
그래서 그는 빨리 걸으며 말했습니다. “내가 말한 것을 기억하고 궁전으로 돌아가십시오.”
White water rafting antalya | An Adventure Filled with Excitement!
https://mythings.tistory.com/108
Вип Трансфер из аэропорта Анталья | Вип Трансфер из аэропорта Анталья
e ticaret nasıl yapılır | e ticaret nasıl yapılır İstanbul danışmanlık firması
qiyezp.com
평소에는 파도가 너무 세서 의외로 오늘은 철판을 찼다.
Hello! I гealize this is kind of off-topіc һowever I needed to ask.
Does managing a well-establishеd website such as yoᥙrs taкe a large amount of work?
I ɑm completеly new to blogging however I do write in my јournal everүday.
I’d like to start a blog so I will be able to share my experience and thoughts online.
Please let me know if you have any іdeas or tips
for new aspiring blog owners. Appreciɑte it!
Howdy would you mind sharing which blog platform you’re working with?
I’m planning to start my own blog in the near future but I’m having a hard time deciding between BlogEngine/Wordpress/B2evolution and
Drupal. The reason I ask is because your layout seems different then most blogs and I’m looking for something completely unique.
P.S Sorry for being off-topic but I had to ask!
https://k-studio.kr/가스라이팅-뜻-20문항-테스트-가스라이팅-자가진단/
https://pornmaster.fun/hd/威奇托找援交妹【linetyp96】上门服务-sjr
https://honeytipit.tistory.com/tag/카카오장기20PC설치
Keep on writing, great job!
Hello there! This article could not be written much better!
Looking through this post reminds me of my previous roommate!
He always kept preaching about this. I will send this article to him.
Pretty sure he’ll have a good read. I appreciate you for sharing!
https://itshareit.tistory.com/240
bestmanualpolesaw.com
Fang Jifan은 항상 제자들에게 엄격하며 가혹하다고 말할 수도 있습니다.
bmipas.com
実生活で使える実用的な情報が満載で、非常に価値があります。
https://itmoney4you.com/애니팡-맞고/
sandyterrace.com
무빈의 무관심한 태도를 보고 서기는 말을 잇지 못했다.
Pretty element of content. I simply stumbled upon your website and in accession capital to say that I acquire in fact enjoyed account your weblog posts.
Any way I will be subscribing on your feeds and even I success you get entry to constantly fast.
https://itmanual.net/EAB0A4EB9FADEC8B9C-ED86B5ED9994-EC9E90EB8F99EB85B9EC9D8C-EC84A4ECA095EBB0A9EBB295-ED86B5ED9994EB85B9EC9D8CED8C8CEC9DBCEC9C84ECB998/embed/#?secret=UYu3mzMlOP
https://mintfin.tistory.com/tag/환경부20전기차20충전소
https://chotiple.tistory.com/tag/미국20독감20피해상황
https://generatepress.com/
Hey there! This is kind of off topic but I need some advice from an established blog.
Is it very hard to set up your own blog? I’m not very techincal but I can figure things out pretty fast.
I’m thinking about setting up my own but I’m not
sure where to start. Do you have any tips or suggestions?
Appreciate it
Take a look at my web page; vpn special
iGaming 업계에서 주목받는 프라그마틱 게임은 모바일 중심의 혁신적이고 표준화된 콘텐츠를 플레이어에게 제공합니다.
프라그마틱 홈페이지
프라그마틱의 빙고 게임은 항상 즐겁고 흥미로워요. 여기에서 더 많은 빙고 게임 정보를 확인하세요!
https://www.nutrapia.com
https://www.12315kz.cn/
https://karmosan.com/link/
https://nicesongtoyou.com/financial-portal/kakaotalk/
A nice service for creating temporary emails https://tmp-mail.pro/
Does your website have a contact page? I’m having
a tough time locating it but, I’d like to shoot you an e-mail.
I’ve got some ideas for your blog you might be interested in hearing.
Either way, great site and I look forward to seeing it expand over time.
여행지
Hi there! This is my 1st comment here so I just wanted to give a quick shout out and tell you I genuinely enjoy reading your articles.
Can you suggest any other blogs/websites/forums that
deal with the same subjects? Thanks a lot!
Российский производитель реализует блины в интернет-магазине https://diski-dlya-shtang.ru/ для усиленной эксплуатации в коммерческих тренировочных центрах и в домашних условиях. Завод из России производит тренировочные блины разного посадочного диаметра и любого востребованного веса для наборных штанг. Советуем к приобретению обрезиненные тренировочные диски для силовых тренировок. Они не скользят, не шумят и менее травматичны. Изготавливаемые изделия не нуждаются в постоянном обслуживании и ориентированы на длительную эксплуатацию в дома, в квартире. Рекомендуем большой ассортимент любительских дисков с любым видом покрытия. Приобретите отягощения с подходящей массой и посадочным диаметром по доступным ценам напрямую у изготовителя.
First off I would like to say fantastic blog!
I had a quick question that I’d like to ask
if you don’t mind. I was curious to find out how you center yourself and clear your
mind prior to writing. I’ve had a tough time clearing my thoughts in getting
my thoughts out. I do enjoy writing however it just
seems like the first 10 to 15 minutes are wasted just trying to
figure out how to begin. Any ideas or hints? Kudos!
etsyweddingteam.com
読みごたえのある内容でした。また訪れます。
qiyezp.com
알고 보니 백 근의 곡식이 수확되었다는 소식을 듣고 사람들은 무의식적으로 칭찬을 아끼지 않았다.
https://www.lesbravo.com/implant-fee-duration/
https://kimyunjeong.com/716
https://kimyunjeong.com/443
https://honeytiplabs.com/아이폰-데이터-사용량/
Привет всем!
Закажите диплом ВУЗа по выгодной цене с доставкой в любой город России без предоплаты – надежно и выгодно!
http://saksx-attestats.ru/
Купите диплом в России с доставкой и гарантией подлинности только у нас по самой выгодной цене.
Приобретите диплом университета России без предоплаты и с гарантией качества доставки в любую точку страны!
https://sharply.tistory.com/714
Amplificadores de senal gsm
Incrementa la velocidad de tus datos moviles con un amplificador de senal
https://www.glbeautyblog.com/amplificador-4g-movistar
https://www.monkfishjowls.com/피자스쿨-영업시간-정확한-시간
https://pornmaster.fun/hd/to-please-bae-with
A. 카카오택시 앱을 실행한 후 ‘예약’ 탭을 누르시면 됩니다. 이후 출발 위치, 도착 위치, 탑승 시간 등을 입력하고 ‘예약하기’를 누르시면 예약이 완료됩니다. 다음 사이트에서 섹스 비디오를 시청하세요 강남출장섹스마사지
taxi price antalya to alanya | taxi price antalya to alanya is here
The way you handled that challenging situation is admirable. Your resilience and positive attitude are a beacon of strength. Thanks for sharing your story.
Your positivity radiates through your posts. It’s evident you bring a sense of joy to those around you. Thanks for making the online world a brighter place.
DreamProxies – Cheapest USA Private Proxies: Elite quality, Endless bandwidth, 1000 mb/s superspeed, 99,9 uptime, Low sequential IP’s, No consumption restrictions, Multiple subnets, USA or Europe proxies – Buy Now – DreamProxies.com
Wow, marvelous weblog structure! How lengthy have you been blogging
for? you make running a blog glance easy. The full look of your
site is great, let alone the content material! You can see similar here sklep
Private proxies plus top deals: 50 reduction, absolutely free proxies and additionally offers – no more than upon https://DreamProxies.com
Приветики, дорогие мои!!
Диплом стал моей главной проблемой, но благодаря интернету я нашел способы улучшить свою работу и сохранить здоровье.
Приобретите диплом университета без предоплаты у нас и получите его с доставкой в любую точку России, гарантия качества.
Желаю всем положительных отметок!
купить диплом нового образца
купить диплом в сосновом бору
купить диплом в биробиджане
купить диплом в буденновске
купить диплом в хабаровске
куплю диплом высшего образования
Приветики, дорогие мои!!
Диплом привел к серьезным проблемам из-за просроченных сроков.
Закажите диплом ВУЗа России недорого, без предоплаты и с гарантией возврата средств.
Желаю каждому отличных оценок!
https://rudiplomirovana.com
купить диплом в южно-сахалинске
купить диплом в вологде
купить диплом в россоши
купить диплом в ессентуках
купить диплом в новотроицке
Добрый день!
Сложности с дипломом привели к необходимости работать в ночное время, что угрожает моему здоровью.
Рады предложить большой выбор документов об образовании всех ВУЗов России по доступным ценам и с постоплатой.
https://premialnie-diplomiki.com/
Желаю вам всем пятерочных) отметок!
купить диплом в первоуральске
купить диплом физика
купить диплом в троицке
купить морской диплом
купить диплом в копейске
Доброго дня!
Диплом уже не кажется мне таким ужасным, ведь я обнаружил множество полезных инструментов в интернете.
Мы предлагаем купить диплом университета без предоплаты с гарантированной доставкой по всей России.
Желаю каждому прекрасных оценок!
купить свидетельство о рождении
купить диплом в иваново
купить диплом диспетчера
купить диплом в волжском
купить диплом врача
купить диплом в абакане
Привет всем!
К счастью, интернет помог мне найти полезные ресурсы, которые значительно ускорили и облегчили мой труд с дипломом.
Закажите диплом у нас – легко! Без предоплаты, с гарантией качества и доставкой по всей России.
Желаю всем прекрасных оценок!
купить диплом университета
купить диплом дизайнера
купить диплом в чайковском
купить диплом в костроме
купить диплом в серове
купить диплом в димитровграде
Всем хорошего дня!
Диплом уже не кажется мне таким ужасным, ведь я обнаружил множество полезных инструментов в интернете.
На нашем сайте вы можете купить диплом ВУЗа с постоплатой и поддержкой 24/7.
Желаю каждому положительных отметок!
купить диплом университета
купить диплом в хасавюрте
купить диплом в вологде
купить диплом в казани
купить диплом электрика
купить диплом в горно-алтайске
Доброго дня!
Несмотря на срыв сроков и угрозу для здоровья из-за диплома, я нашел в интернете способы быть более продуктивным.
Купите диплом у нас легко! Надежно, качественно, без предоплаты, с доставкой в любой город России.
Желаю всем не двоешных) оценок!
https://aurus-diploman.com/
купить диплом моряка
купить диплом в вольске
купить диплом в астрахани
купить диплом в димитровграде
купить диплом в норильске
Здравствуйте!
Диплом превратился в настоящий кошмар, заставляя меня пересматривать сроки и работать ночью, что негативно сказывается на здоровье.
Приобретайте диплом у нас и получите качественный документ без предоплаты с доставкой в любую точку России.
https://premialnie-diplomiki.com/
Желаю каждому честных оценок!
купить диплом в люберцах
купить диплом в дербенте
купить диплом массажиста
купить диплом в озёрске
купить диплом сварщика
Всем хорошего дня!
Дипломное задание оказалось для меня настоящим испытанием, однако я нашел поддержку в сети.
Желаете приобрести диплом ВУЗа недорого и без предоплаты на нашем сайте? Доставим в любую точку России.
https://adiplom-russian.com/
Желаю вам всем положительных отметок!
купить диплом в вольске
купить диплом в новоалтайске
купить диплом менеджера по туризму
купить диплом в казани
купить диплом в саранске
50 Discount For All Private Proxies! Elite quality, Unlimited bandwidth, 1000 mb/s superspeed, 99,9 uptime, Low successive IP’s, No application restrictions, Multiple subnets, USA or Europe proxies – Get Today – https://DreamProxies.com
https://king.creaming.net/on/8025
https://mythings.tistory.com/77
etsyweddingteam.com
素敵な記事をありがとうございます。いつも心に響きます。
Greetings from Colorado! I’m bored to tears at work so I decided to check out your site on my iphone during lunch break.
I really like the knowledge you present here and can’t wait to take a
look when I get home. I’m surprised at how fast your blog loaded on my cell phone ..
I’m not even using WIFI, just 3G .. Anyhow, excellent site!
I want looking through and I believe this website got some genuinely utilitarian stuff on it! .
Really good info can be found on web site.
I am only commenting to let you understand what a magnificent encounter my cousin’s princess went through browsing your webblog. She realized lots of issues, not to mention how it is like to possess an excellent teaching mindset to make other folks clearly thoroughly grasp specific advanced subject areas. You really surpassed our desires. Thank you for providing those good, trusted, revealing and as well as cool thoughts on that topic to Mary.
?Quieres ganar dinero desde casa? ?Prueba los casinos en linea de Peru!
casino en linea peru casino en linea peru .
thanks to the author for taking his clock time on this one.
I went over this site and I believe you have a lot of wonderful information, saved to my bookmarks (:.
The following time I read a blog, I hope that it doesnt disappoint me as much as this one. I mean, I know it was my option to read, however I really thought youd have something interesting to say. All I hear is a bunch of whining about one thing that you might fix in the event you werent too busy searching for attention.
game1kb.com
비록 마음속에 희미한 걱정이 있지만, 그것에 대해 생각하지 않는 한 그는 여전히 행복할 수 있다.
https://gorgopage.com/하나오토론-하나카드-오토할부-한도-금리-캐시백-1-2-오/
how much is a taxi from antalya airport to side | how much is a taxi from antalya airport to side
Привет всем!
Эффективность моей работы над дипломом возросла благодаря доступу к онлайн-материалам.
Мы поможем вам купить диплом университета без предоплаты и доставим его в любой город России с гарантированной безопасностью.
Желаю всем прекрасных оценок!
https://diplomas-asx.com
купить диплом в ачинске
купить диплом в тамбове
купить диплом во всеволожске
купить диплом охранника
купить диплом в глазове
Доброго дня!
Мое написание диплома превратилось в настоящее испытание, однако я обнаружил в интернете полезные материалы для ускорения работы.
Поможем выбрать, сделать заказ и купить диплом любого учебного заведения в любом населенном пункте по самым низким ценам.
Желаю каждому прекрасных оценок!
https://aurus-diploman.com/
купить дипломы о высшем с занесением
купить диплом в королёве
купить диплом в мичуринске
купить диплом во владикавказе
купить свидетельство о рождении ссср
Добрый день!
Дипломное исследование стало для меня серьезным вызовом, однако я нашел способы облегчить этот процесс.
Наша компания предлагает конфиденциально заказать и приобрести диплом любого ВУЗа России.
https://rusd-diploms.com/
Желаю для каждого прекрасных отметок!
купить диплом в челябинске
купить дипломы о высшем цены
купить диплом электромонтажника
купить диплом в бору
купить диплом в нижневартовске
Приветики, дорогие мои!!
Диплом получил поддержку через интернет, что значительно упростило задачу.
Мы предлагаем купить диплом университета без предоплаты с гарантированной доставкой по всей России.
купить диплом автомеханика
Желаю вам всем пятерочных) оценок!
купить диплом математика
куплю диплом с занесением
купить диплом в екатеринбурге
купить диплом в туле
купить диплом в буденновске
Доброго дня!
Стремлюсь преодолеть все препятствия, несмотря на ухудшение самочувствия из-за ночных занятий с дипломом.
На нашем сайте представлены дипломы с гарантией и доставкой в любой регион РФ.
Желаю для каждого отличных оценок!
https://aurus-diplomas.com/
купить диплом в воткинске
купить диплом в димитровграде
купить диплом электрика
купить диплом провизора
купить диплом в липецке
Приветики, дорогие мои!!
Диплом получил поддержку через ценные ресурсы, обнаруженные в сети, упрощая мою задачу.
Приобретайте диплом университета без предоплаты с доставкой по всей России.
Желаю для каждого не двоешных) оценок!
https://aurus-diplomas.com/
купить диплом в новосибирске
купить диплом в рязани
купить свидетельство о рождении
купить диплом в феодосии
купить диплом в новомосковске
https://dday.tistory.com/336
Thanks a bunch for sharing this with all people you really realize what you arre talking approximately!
Bookmarked. Kindly alxo discuss with my web site
=). We will have a link trade agreement between us
Здравствуйте!
Диплом уже не вызывает столько стресса, как раньше, благодаря помощи, полученной через интернет.
Выберите и приобретите диплом ВУЗа России недорого, качественно и с отправкой почтой!
Желаю вам всем честных отметок!
https://diplomys-asx.com
купить диплом россия
купить диплом в шадринске
купить диплом в тамбове
купить технический диплом
купить диплом учителя
Здравствуйте!
Диплом заставил меня работать по ночам, что негативно сказалось на моем здоровье.
Приобретите диплом университета с гарантированной доставкой в любой город России с оплатой после получения.
Желаю для каждого отличных отметок!
https://diplomys-asx.com
купить диплом химика
купить диплом о высшем образовании
купить диплом маляра
купить диплом бурильщика
купить диплом в хабаровске
https://dday.tistory.com/75
qiyezp.com
Liu Jian은 손을 댔습니다. “좋아요, 화 내지 마세요, 여러분, 먼저 당신의 말을 들어보세요.”
Букмекеры: рейтинг и отзывы
букмекерские приложения самая лучшая букмекерская контора в интернете .
https://kakaotaxi.dasgno.com/kakao-taxi2
https://pornmaster.fun/hd/mathira-naked-pics
Привет всем!
Мое написание диплома превратилось в испытание, но я обнаружил в интернете полезные материалы для его ускорения.
Выберите и приобретите диплом ВУЗа России недорого, качественно и с отправкой почтой!
Желаю каждому не двоешных) отметок!
купить диплом ссср
купить диплом в назрани
купить диплом в бору
купить диплом образцы
купить диплом юриста
купить свидетельство о разводе
Всем хорошего дня!
Диплом стал моей основной заботой, но я нашел онлайн-ресурсы, которые сделали этот процесс более выносимым.
У нас вы можете заказать и купить диплом в России без предоплаты и с гарантией доставки “под ключ”.
Желаю каждому прекрасных оценок!
https://russiany-diplomas.com
купить диплом в новороссийске
купить диплом нового образца
купить диплом в великих луках
купить диплом тренера
купить диплом повара
Добрый день!
Диплом стал кошмаром, когда я отложил его выполнение и перешел на ночной график.
Заказать и купить диплом в России без предоплаты и с гарантией можно только у нас, гарантия “под ключ”.
Желаю всем честных отметок!
купить диплом о среднем специальном
купить диплом в кинешме
купить диплом в прокопьевске
купить диплом в набережных челнах
купить диплом в архангельске
купить диплом в артеме
Здравствуйте!
Диплом больше не кажется невыполнимым благодаря помощи, найденной в интернете.
Выберите и закажите диплом ВУЗа России качественно и с возможностью отправки почтой!
Желаю для каждого пятерочных) отметок!
купить свидетельство о рождении
купить диплом в зеленодольске
купить дипломы о высшем образовании цена
купить диплом товароведа
купить диплом биолога
купить диплом в заречном
충무로출장업소
Добрый день!
Несмотря на срыв сроков и угрозу для здоровья, я нашел в интернете ресурсы, которые помогают мне быть более продуктивным.
Мы поможем вам купить диплом университета без предоплаты и доставим его в любой город России с гарантированной безопасностью.
Желаю всем прекрасных оценок!
https://plands-diplomy.com
купить диплом в новошахтинске
купить диплом в грозном
купить диплом в волгодонске
купить диплом в лениногорске
купить бланк диплома
Добрый день!
Дипломное исследование стало для меня серьезным вызовом, однако я нашел способы облегчить этот процесс.
Рады предложить большой выбор документов об образовании всех ВУЗов России по выгодным ценам и с постоплатой.
Желаю вам всем отличных отметок!
https://frees-diplomy.com
купить диплом преподавателя
купить диплом агронома
купить диплом в архангельске
купить диплом электромонтера
купить диплом в саранске
Здравствуйте!
Из-за диплома пришлось перейти на ночной график работы, что угрожало моему здоровью.
На нашем сайте вы можете купить диплом ВУЗа, недорого, с оплатой после получения, поддержка 24/7.
Желаю вам всем отличных отметок!
https://frees-diplomy.com
купить диплом в новокуйбышевске
купить диплом в иваново
купить диплом электромонтажника
купить диплом товароведа
купить диплом в белово
이태원스웨디시안마게이클럽
Привет всем!
К счастью, интернет помог мне найти полезные ресурсы, которые значительно ускорили и облегчили мой труд с дипломом.
На нашем сайте представлен широкий выбор дипломов с гарантией качества и быстрой доставкой в любой регион РФ.
Желаю для каждого прекрасных оценок!
https://diplom-originalniy.com
купить диплом провизора
купить диплом ссср
купить диплом в тобольске
купить диплом в рубцовске
купить диплом в пятигорске
Всем хорошего дня!
Диплом стал более доступным заданием благодаря использованию онлайн-материалов.
Закажите диплом у нас и получите его быстро и надежно, оплата после получения, доставка в любой регион РФ.
Желаю вам всем честных отметок!
https://diplom-originalniy.com
купить диплом преподавателя
купить диплом в киселевске
купить диплом в верхней пышме
купить диплом в горно-алтайске
куплю диплом с занесением
대전호박나이트
이태원게이바
거제출장안마
Всем хорошего дня!
Диплом требует постоянной работы, подкрепляемой найденными онлайн-ресурсами.
Заказывайте диплом у нас и получайте его быстро и безопасно, оплачивая после получения, с отправкой в любой регион России.
Желаю для каждого прекрасных оценок!
https://radiploma.com/
купить диплом о среднем специальном
купить диплом в верхней пышме
купить диплом дизайнера
купить диплом в мурманске
купить диплом в асбесте
Здравствуйте!
Диплом продолжает быть моим вызовом, но теперь у меня есть все необходимые ресурсы для его успешного завершения.
Мы предлагаем вам купить диплом университета без предоплаты, с гарантированной доставкой по всей России.
Желаю каждому прекрасных оценок!
купить диплом
купить диплом в ноябрьске
купить диплом в биробиджане
купить диплом в калуге
купить диплом в кирове
купить диплом в гуково
거제출장안마
delta 8″ faucet
청도페이스라인출장
What’s up, I would like to subscribe for this weblog
to take hottest updates, therefore where can i do it please
help out.
exprimegranada.com
素敵な記事をありがとうございます。いつも楽しみにしています。
아름다운스웨디시업소
onair2tv.com
장 황후는 잠시 생각하며 “아마도 팡 지판일지도 몰라요. 그는 이것에 대해 던지기를 좋아합니다. “라고 말했습니다.
대전나이트클럽
I used to be able to find good information from your blog articles.
#be#jk3#jk#jk#JK##
временный номер России
В нашем мире, где диплом – это начало успешной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения качественного образования. Наличие документа об образовании переоценить просто невозможно. Ведь именно он открывает двери перед всеми, кто желает вступить в профессиональное сообщество или учиться в любом университете.
В данном контексте мы предлагаем оперативно получить этот необходимый документ. Вы сможете приобрести диплом старого или нового образца, и это является отличным решением для всех, кто не смог закончить обучение или утратил документ. дипломы производятся аккуратно, с особым вниманием к мельчайшим деталям, чтобы на выходе получился документ, максимально соответствующий оригиналу.
Преимущества такого решения состоят не только в том, что вы сможете максимально быстро получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начиная от выбора нужного образца диплома до правильного заполнения личных данных и доставки в любой регион страны — все под полным контролем наших мастеров.
Для тех, кто ищет быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Купить диплом – значит избежать длительного процесса обучения и не теряя времени переходить к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
https://originality-diplomans.com
В нашем мире, где диплом является началом успешной карьеры в любом направлении, многие пытаются найти максимально простой путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь диплом открывает дверь перед любым человеком, который хочет начать трудовую деятельность или продолжить обучение в высшем учебном заведении.
Предлагаем быстро получить этот важный документ. Вы сможете заказать диплом, что будет отличным решением для всех, кто не смог завершить образование или потерял документ. дипломы производятся с особой аккуратностью, вниманием к мельчайшим элементам, чтобы в итоге получился документ, максимально соответствующий оригиналу.
Преимущество такого решения состоит не только в том, что можно быстро получить свой диплом. Процесс организовывается комфортно, с профессиональной поддержкой. Начиная от выбора необходимого образца диплома до консультаций по заполнению персональных данных и доставки в любой регион страны — все будет находиться под абсолютным контролем качественных мастеров.
Всем, кто ищет оперативный способ получения требуемого документа, наша компания предлагает выгодное решение. Купить диплом – это значит избежать долгого обучения и не теряя времени перейти к своим целям, будь то поступление в университет или начало профессиональной карьеры.
https://originality-diplomans.com
В нашем мире, где диплом становится началом отличной карьеры в любой сфере, многие стараются найти максимально простой путь получения образования. Наличие документа об образовании переоценить просто невозможно. Ведь диплом открывает двери перед каждым человеком, который хочет начать трудовую деятельность или продолжить обучение в ВУЗе.
Наша компания предлагает очень быстро получить этот важный документ. Вы можете купить диплом старого или нового образца, что будет выгодным решением для человека, который не смог закончить обучение или утратил документ. Все дипломы изготавливаются с особой тщательностью, вниманием ко всем нюансам, чтобы в результате получился продукт, 100% соответствующий оригиналу.
Плюсы такого решения заключаются не только в том, что можно оперативно получить свой диплом. Процесс организован удобно и легко, с профессиональной поддержкой. От выбора требуемого образца документа до правильного заполнения личных данных и доставки в любое место России — все находится под абсолютным контролем квалифицированных специалистов.
Таким образом, для тех, кто ищет быстрый способ получения необходимого документа, наша компания готова предложить выгодное решение. Заказать диплом – значит избежать продолжительного обучения и сразу перейти к личным целям, будь то поступление в ВУЗ или старт карьеры.
купить диплом вуза украина
В нашем мире, где диплом становится началом удачной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения качественного образования. Наличие официального документа об образовании сложно переоценить. Ведь диплом открывает дверь перед любым человеком, который собирается начать трудовую деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает оперативно получить этот важный документ. Вы имеете возможность купить диплом, и это становится отличным решением для всех, кто не смог закончить образование, потерял документ или желает исправить плохие оценки. Все дипломы производятся аккуратно, с особым вниманием к мельчайшим элементам. В итоге вы сможете получить продукт, 100% соответствующий оригиналу.
Превосходство этого подхода состоит не только в том, что можно максимально быстро получить свой диплом. Процесс организовывается удобно, с нашей поддержкой. От выбора требуемого образца до правильного заполнения личных данных и доставки по стране — все под полным контролем качественных мастеров.
Для всех, кто ищет оперативный способ получить необходимый документ, наша компания предлагает отличное решение. Заказать диплом – значит избежать долгого обучения и сразу переходить к личным целям, будь то поступление в ВУЗ или старт успешной карьеры.
https://russiany-diplomas.com
В нашем обществе, где диплом – это начало успешной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения качественного образования. Необходимость наличия документа об образовании переоценить попросту невозможно. Ведь диплом открывает двери перед всеми, кто стремится вступить в профессиональное сообщество или учиться в университете.
Предлагаем максимально быстро получить этот важный документ. Вы сможете купить диплом нового или старого образца, и это является отличным решением для человека, который не смог завершить образование или потерял документ. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим деталям. В результате вы сможете получить полностью оригинальный документ.
Преимущества этого решения заключаются не только в том, что вы оперативно получите свой диплом. Процесс организован удобно, с профессиональной поддержкой. От выбора подходящего образца диплома до грамотного заполнения личной информации и доставки в любой регион России — все под абсолютным контролем опытных специалистов.
В результате, для тех, кто пытается найти оперативный способ получения необходимого документа, наша компания предлагает отличное решение. Заказать диплом – значит избежать длительного процесса обучения и сразу переходить к своим целям: к поступлению в ВУЗ или к началу успешной карьеры.
https://diplomana-asx.com
В нашем мире, где диплом – это начало отличной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Необходимость наличия официального документа об образовании сложно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или учиться в высшем учебном заведении.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы имеете возможность заказать диплом нового или старого образца, что становится выгодным решением для всех, кто не смог завершить образование, утратил документ или хочет исправить плохие оценки. дипломы выпускаются аккуратно, с максимальным вниманием к мельчайшим деталям, чтобы в итоге получился 100% оригинальный документ.
Плюсы данного решения состоят не только в том, что можно максимально быстро получить диплом. Весь процесс организован удобно, с нашей поддержкой. От выбора подходящего образца до консультаций по заполнению персональной информации и доставки по стране — все под абсолютным контролем опытных мастеров.
Всем, кто пытается найти максимально быстрый способ получения требуемого документа, наша компания предлагает отличное решение. Заказать диплом – это значит избежать длительного процесса обучения и не теряя времени перейти к своим целям: к поступлению в ВУЗ или к началу удачной карьеры.
купить диплом недорого
Сегодня, когда диплом – это начало отличной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь именно он открывает двери перед каждым человеком, который собирается вступить в профессиональное сообщество или продолжить обучение в любом институте.
Мы предлагаем максимально быстро получить этот важный документ. Вы имеете возможность приобрести диплом, что становится выгодным решением для всех, кто не смог закончить образование, утратил документ или хочет исправить свои оценки. Все дипломы выпускаются аккуратно, с максимальным вниманием ко всем деталям. В итоге вы сможете получить 100% оригинальный документ.
Преимущества этого подхода заключаются не только в том, что можно максимально быстро получить диплом. Весь процесс организован удобно, с профессиональной поддержкой. От выбора подходящего образца диплома до грамотного заполнения личной информации и доставки по стране — все под полным контролем качественных специалистов.
Для всех, кто хочет найти максимально быстрый способ получения необходимого документа, наша компания готова предложить выгодное решение. Приобрести диплом – значит избежать продолжительного обучения и сразу перейти к своим целям: к поступлению в университет или к началу трудовой карьеры.
https://russiany-diplomans.com
В современном мире, где диплом становится началом успешной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Необходимость наличия официального документа переоценить просто невозможно. Ведь диплом открывает дверь перед всеми, кто хочет вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Мы предлагаем оперативно получить этот важный документ. Вы можете купить диплом, и это становится выгодным решением для человека, который не смог завершить образование, утратил документ или желает исправить свои оценки. диплом изготавливается аккуратно, с максимальным вниманием ко всем нюансам. На выходе вы сможете получить 100% оригинальный документ.
Превосходство этого решения состоит не только в том, что вы сможете быстро получить диплом. Процесс организован комфортно, с нашей поддержкой. От выбора нужного образца документа до консультаций по заполнению личных данных и доставки по России — все находится под абсолютным контролем квалифицированных мастеров.
Для всех, кто пытается найти оперативный способ получить необходимый документ, наша услуга предлагает отличное решение. Купить диплом – значит избежать долгого обучения и сразу перейти к личным целям, будь то поступление в университет или начало карьеры.
https://russiany-diplomans.com
В нашем обществе, где диплом является началом успешной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает двери перед людьми, желающими вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы имеете возможность заказать диплом нового или старого образца, и это будет удачным решением для человека, который не смог завершить обучение, потерял документ или хочет исправить свои оценки. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам, чтобы в итоге получился продукт, максимально соответствующий оригиналу.
Преимущества подобного подхода заключаются не только в том, что можно быстро получить диплом. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора подходящего образца до грамотного заполнения личной информации и доставки в любое место России — все находится под полным контролем наших специалистов.
Для всех, кто пытается найти быстрый и простой способ получения требуемого документа, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать долгого обучения и сразу перейти к своим целям, будь то поступление в ВУЗ или начало профессиональной карьеры.
https://adiplomy-russian.com
В нашем обществе, где диплом становится началом успешной карьеры в любой области, многие ищут максимально простой путь получения образования. Наличие официального документа переоценить невозможно. Ведь именно диплом открывает дверь перед каждым человеком, который стремится вступить в профессиональное сообщество или продолжить обучение в каком-либо ВУЗе.
Предлагаем очень быстро получить этот необходимый документ. Вы имеете возможность купить диплом, и это является отличным решением для всех, кто не смог завершить образование или утратил документ. дипломы производятся аккуратно, с максимальным вниманием к мельчайшим деталям, чтобы в итоге получился полностью оригинальный документ.
Преимущество подобного решения заключается не только в том, что можно оперативно получить диплом. Процесс организован удобно и легко, с профессиональной поддержкой. Начиная от выбора подходящего образца диплома до грамотного заполнения личных данных и доставки по стране — все находится под абсолютным контролем квалифицированных специалистов.
Для всех, кто хочет найти максимально быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – это значит избежать длительного процесса обучения и сразу переходить к личным целям: к поступлению в ВУЗ или к началу удачной карьеры.
купить диплом магистра
В нашем обществе, где диплом является началом удачной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения образования. Факт наличия официального документа трудно переоценить. Ведь именно диплом открывает дверь перед каждым человеком, желающим вступить в сообщество профессиональных специалистов или продолжить обучение в университете.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы имеете возможность приобрести диплом старого или нового образца, и это становится выгодным решением для всех, кто не смог завершить образование, потерял документ или хочет исправить плохие оценки. Все дипломы производятся аккуратно, с максимальным вниманием к мельчайшим деталям, чтобы в итоге получился продукт, 100% соответствующий оригиналу.
Превосходство этого подхода заключается не только в том, что можно оперативно получить свой диплом. Процесс организовывается удобно, с профессиональной поддержкой. Начиная от выбора необходимого образца диплома до консультаций по заполнению личных данных и доставки по России — все находится под полным контролем квалифицированных специалистов.
В итоге, всем, кто хочет найти максимально быстрый способ получения требуемого документа, наша компания может предложить отличное решение. Купить диплом – это значит избежать долгого обучения и сразу перейти к своим целям: к поступлению в ВУЗ или к началу успешной карьеры.
диплом купить
В нашем мире, где диплом становится началом отличной карьеры в любой отрасли, многие пытаются найти максимально простой путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь именно диплом открывает двери перед людьми, стремящимися начать профессиональную деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает очень быстро получить любой необходимый документ. Вы можете купить диплом, и это является выгодным решением для человека, который не смог завершить образование или утратил документ. Все дипломы изготавливаются с особой тщательностью, вниманием ко всем деталям, чтобы в результате получился продукт, 100% соответствующий оригиналу.
Превосходство данного решения состоит не только в том, что можно быстро получить диплом. Процесс организован комфортно, с профессиональной поддержкой. Начав от выбора требуемого образца до консультации по заполнению личных данных и доставки в любой регион России — все под полным контролем наших специалистов.
В итоге, для тех, кто ищет быстрый и простой способ получения необходимого документа, наша услуга предлагает отличное решение. Приобрести диплом – значит избежать долгого процесса обучения и сразу перейти к своим целям, будь то поступление в университет или начало успешной карьеры.
купить диплом высшее
В наше время, когда диплом – это начало удачной карьеры в любой отрасли, многие ищут максимально быстрый путь получения образования. Наличие документа об образовании переоценить просто невозможно. Ведь диплом открывает двери перед любым человеком, желающим вступить в сообщество профессиональных специалистов или продолжить обучение в высшем учебном заведении.
Наша компания предлагает очень быстро получить этот необходимый документ. Вы можете заказать диплом нового или старого образца, что будет удачным решением для человека, который не смог завершить образование, потерял документ или желает исправить свои оценки. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам. В итоге вы сможете получить полностью оригинальный документ.
Преимущество такого подхода состоит не только в том, что можно максимально быстро получить свой диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. Начав от выбора нужного образца до точного заполнения персональных данных и доставки в любой регион страны — все под абсолютным контролем квалифицированных мастеров.
Для всех, кто ищет быстрый и простой способ получить необходимый документ, наша услуга предлагает отличное решение. Купить диплом – это значит избежать долгого процесса обучения и не теряя времени переходить к важным целям, будь то поступление в ВУЗ или начало удачной карьеры.
https://premialnie-diplomik.com
В нашем мире, где диплом – это начало удачной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает двери перед любым человеком, который стремится начать трудовую деятельность или учиться в ВУЗе.
Предлагаем очень быстро получить этот важный документ. Вы можете заказать диплом, что будет отличным решением для всех, кто не смог закончить обучение или утратил документ. Каждый диплом изготавливается аккуратно, с максимальным вниманием ко всем деталям. В результате вы получите 100% оригинальный документ.
Превосходство этого решения состоит не только в том, что можно максимально быстро получить свой диплом. Весь процесс организовывается удобно и легко, с нашей поддержкой. От выбора подходящего образца документа до консультации по заполнению персональной информации и доставки по стране — все под абсолютным контролем качественных специалистов.
В результате, всем, кто ищет быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Заказать диплом – это значит избежать долгого обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в университет или начало карьеры.
купить аттестат школы
В нашем обществе, где диплом становится началом отличной карьеры в любой отрасли, многие стараются найти максимально быстрый путь получения образования. Наличие официального документа об образовании сложно переоценить. Ведь именно диплом открывает дверь перед людьми, стремящимися начать профессиональную деятельность или учиться в любом институте.
Наша компания предлагает максимально быстро получить этот важный документ. Вы можете заказать диплом старого или нового образца, что является удачным решением для всех, кто не смог закончить обучение или утратил документ. Любой диплом изготавливается аккуратно, с особым вниманием ко всем элементам. На выходе вы сможете получить продукт, полностью соответствующий оригиналу.
Преимущество такого подхода состоит не только в том, что вы сможете быстро получить диплом. Весь процесс организовывается удобно и легко, с профессиональной поддержкой. Начав от выбора подходящего образца до точного заполнения персональной информации и доставки по стране — все под абсолютным контролем качественных мастеров.
Всем, кто ищет быстрый и простой способ получения необходимого документа, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать долгого процесса обучения и сразу перейти к личным целям: к поступлению в университет или к началу трудовой карьеры.
http://www.google.fr/url?q=https://lands-diploma.com
https://maps.google.gy/url?sa=j&url=https://rudiplomista.com
https://cse.google.is/url?sa=t&url=https://premialnie-diplomik.com
http://www.google.mk/url?sa=i&url=https://lands-diploma.com
https://maps.google.cf/url?sa=t&url=https://lands-diploma.com
На сегодняшний день, когда диплом является началом удачной карьеры в любой области, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие официального документа об образовании трудно переоценить. Ведь именно диплом открывает двери перед всеми, кто собирается начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете купить диплом нового или старого образца, и это является выгодным решением для всех, кто не смог закончить обучение или потерял документ. диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам. На выходе вы получите 100% оригинальный документ.
Преимущества такого подхода состоят не только в том, что можно быстро получить свой диплом. Процесс организовывается просто и легко, с профессиональной поддержкой. Начиная от выбора требуемого образца до консультаций по заполнению личной информации и доставки по стране — все находится под полным контролем опытных мастеров.
Для тех, кто хочет найти быстрый и простой способ получения требуемого документа, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и не теряя времени переходить к важным целям: к поступлению в ВУЗ или к началу удачной карьеры.
https://cse.google.rw/url?sa=t&url=https://rudiplomista.com
https://images.google.com.bz/url?sa=t&url=https://lands-diploma.com
https://www.google.co.ke/url?q=https://premialnie-diplomik.com
https://images.google.ie/url?sa=t&url=https://rudiplomista.com
https://clients1.google.com.my/url?sa=t&url=https://rudiplomista.com
В нашем мире, где диплом – это начало успешной карьеры в любой области, многие ищут максимально быстрый и простой путь получения образования. Факт наличия документа об образовании сложно переоценить. Ведь именно диплом открывает двери перед людьми, желающими начать трудовую деятельность или учиться в ВУЗе.
Предлагаем быстро получить этот необходимый документ. Вы можете приобрести диплом, что становится отличным решением для человека, который не смог завершить образование или потерял документ. Любой диплом изготавливается аккуратно, с особым вниманием к мельчайшим нюансам. В результате вы получите 100% оригинальный документ.
Преимущество данного решения состоит не только в том, что можно оперативно получить свой диплом. Процесс организован просто и легко, с нашей поддержкой. От выбора подходящего образца до грамотного заполнения личных данных и доставки в любой регион России — все под полным контролем наших мастеров.
Таким образом, всем, кто пытается найти быстрый и простой способ получения требуемого документа, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать длительного процесса обучения и не теряя времени переходить к своим целям: к поступлению в ВУЗ или к началу успешной карьеры.
https://images.google.com.jm/url?q=https://premialnie-diplomik.com
https://images.google.com.ph/url?q=https://rudiplomista.com
https://www.google.bs/url?q=https://lands-diploma.com
https://www.google.fi/url?q=https://premialnie-diplomik.com
https://images.google.as/url?q=https://premialnie-diplomik.com
В нашем обществе, где диплом – это начало удачной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед всеми, кто собирается начать профессиональную деятельность или учиться в ВУЗе.
Предлагаем быстро получить этот важный документ. Вы сможете купить диплом старого или нового образца, что становится отличным решением для всех, кто не смог завершить образование или утратил документ. Любой диплом изготавливается аккуратно, с особым вниманием ко всем нюансам, чтобы в итоге получился продукт, 100% соответствующий оригиналу.
Плюсы данного подхода состоят не только в том, что вы сможете максимально быстро получить свой диплом. Процесс организован просто и легко, с нашей поддержкой. От выбора подходящего образца до точного заполнения персональных данных и доставки в любое место страны — все будет находиться под абсолютным контролем наших специалистов.
Для всех, кто хочет найти максимально быстрый способ получения требуемого документа, наша компания может предложить отличное решение. Заказать диплом – это значит избежать долгого обучения и не теряя времени перейти к своим целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://www.google.com.tr/url?q=https://rudiplomista.com
https://images.google.nr/url?q=https://premialnie-diplomik.com
http://cse.google.com.nf/url?sa=i&url=https://rudiplomista.com
http://maps.google.ba/url?q=https://rudiplomista.com
https://maps.google.no/url?rct=t&sa=t&url=https://rudiplomista.com
В наше время, когда диплом – это начало удачной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в любом ВУЗе.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы сможете приобрести диплом старого или нового образца, что является отличным решением для всех, кто не смог закончить образование, утратил документ или хочет исправить плохие оценки. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам, чтобы на выходе получился документ, полностью соответствующий оригиналу.
Преимущества подобного решения состоят не только в том, что вы сможете оперативно получить диплом. Весь процесс организовывается комфортно, с нашей поддержкой. Начав от выбора необходимого образца до грамотного заполнения персональной информации и доставки в любое место России — все будет находиться под абсолютным контролем наших специалистов.
Всем, кто ищет оперативный способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – значит избежать длительного обучения и сразу перейти к достижению собственных целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://1chan.blog/100-2/?unapproved=382540&moderation-hash=8a0d122387b4626aab9c6e1ffb02d5f4#comment-382540
http://xn--h1a1ab.xn--p1ai/index.php?name=account&op=info&uname=okyvaza
https://radioizba.ru/communication/forum/?PAGE_NAME=profile_view&UID=17958
http://xn--33-6kc4bza.xn--p1ai/users/irecy
http://flowerwheel.synology.me/dokuwiki/doku.php?id=originalitydiplomiki
В нашем обществе, где диплом становится началом успешной карьеры в любой отрасли, многие пытаются найти максимально быстрый путь получения качественного образования. Наличие официального документа об образовании переоценить просто невозможно. Ведь диплом открывает двери перед каждым человеком, желающим вступить в профессиональное сообщество или учиться в ВУЗе.
Предлагаем оперативно получить этот необходимый документ. Вы сможете заказать диплом, что является выгодным решением для всех, кто не смог закончить обучение или потерял документ. диплом изготавливается аккуратно, с максимальным вниманием ко всем деталям. В итоге вы сможете получить 100% оригинальный документ.
Превосходство подобного подхода состоит не только в том, что можно быстро получить свой диплом. Процесс организован удобно, с профессиональной поддержкой. От выбора необходимого образца документа до грамотного заполнения персональных данных и доставки в любой регион России — все находится под полным контролем опытных мастеров.
В итоге, для тех, кто ищет максимально быстрый способ получения требуемого документа, наша компания может предложить выгодное решение. Приобрести диплом – значит избежать продолжительного обучения и сразу перейти к достижению собственных целей, будь то поступление в ВУЗ или начало трудовой карьеры.
http://cse.google.co.bw/url?sa=t&url=https://lands-diploma.com
https://www.google.to/url?sa=t&url=https://lands-diploma.com
https://cse.google.lu/url?q=https://premialnie-diplomik.com
https://clients1.google.co.im/url?sa=t&url=https://premialnie-diplomik.com
http://images.google.com.bh/url?sa=t&url=https://rudiplomista.com
В нашем мире, где диплом является началом удачной карьеры в любой области, многие пытаются найти максимально быстрый и простой путь получения образования. Важность наличия официального документа сложно переоценить. Ведь именно он открывает дверь перед каждым человеком, который хочет вступить в сообщество профессионалов или продолжить обучение в университете.
Предлагаем быстро получить этот необходимый документ. Вы можете купить диплом, что становится выгодным решением для всех, кто не смог закончить образование, утратил документ или желает исправить плохие оценки. дипломы изготавливаются с особой аккуратностью, вниманием к мельчайшим деталям. На выходе вы получите 100% оригинальный документ.
Преимущества этого решения заключаются не только в том, что вы сможете максимально быстро получить диплом. Весь процесс организовывается удобно, с нашей поддержкой. Начиная от выбора необходимого образца документа до точного заполнения персональной информации и доставки по стране — все будет находиться под абсолютным контролем опытных специалистов.
Для всех, кто ищет оперативный способ получить требуемый документ, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать длительного процесса обучения и не теряя времени перейти к достижению личных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
https://www.google.co.ls/url?q=https://rudiplomista.com
https://clients1.google.com.cu/url?sa=t&url=https://lands-diploma.com
https://image.google.bs/url?q=https://rudiplomista.com
https://www.google.co.tz/url?q=https://rudiplomista.com
https://maps.google.bt/url?sa=t&url=https://rudiplomista.com
Сегодня, когда диплом становится началом удачной карьеры в любой отрасли, многие пытаются найти максимально быстрый путь получения качественного образования. Важность наличия официального документа об образовании сложно переоценить. Ведь диплом открывает дверь перед людьми, желающими начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
Предлагаем оперативно получить этот важный документ. Вы сможете купить диплом старого или нового образца, и это является удачным решением для всех, кто не смог закончить обучение, потерял документ или желает исправить плохие оценки. дипломы выпускаются с особой аккуратностью, вниманием к мельчайшим элементам. В результате вы получите продукт, полностью соответствующий оригиналу.
Преимущества подобного подхода состоят не только в том, что можно быстро получить свой диплом. Процесс организован просто и легко, с профессиональной поддержкой. От выбора необходимого образца диплома до консультации по заполнению личных данных и доставки по России — все находится под абсолютным контролем опытных специалистов.
В итоге, всем, кто хочет найти быстрый и простой способ получить требуемый документ, наша компания может предложить отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и не теряя времени переходить к своим целям, будь то поступление в университет или начало карьеры.
http://cybex-spb.ru/product/cybex-pallas-m-fix-happy-black/reviews/
http://kdownload.free.fr/shoutbox/shoutbox.php?
http://olddle.orkclub.ru/index.php?subaction=userinfo&user=edyxeha
http://shooting-russia.ru/forum/?PAGE_NAME=message&FID=5&TID=14396&TITLE_SEO=14396-landsdiploms&MID=14659&result=new#message14659
http://clear.rusoft.ru/2011/10/2-dart.html
В наше время, когда диплом – это начало удачной карьеры в любой области, многие стараются найти максимально простой путь получения образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед каждым человеком, который стремится начать профессиональную деятельность или учиться в любом институте.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы имеете возможность купить диплом старого или нового образца, что становится выгодным решением для всех, кто не смог завершить образование, потерял документ или хочет исправить свои оценки. Каждый диплом изготавливается аккуратно, с особым вниманием к мельчайшим элементам. В результате вы сможете получить полностью оригинальный документ.
Преимущества данного подхода состоят не только в том, что можно оперативно получить диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. От выбора требуемого образца до консультаций по заполнению личной информации и доставки по России — все под полным контролем качественных мастеров.
Для всех, кто ищет максимально быстрый способ получения требуемого документа, наша услуга предлагает отличное решение. Заказать диплом – это значит избежать длительного процесса обучения и сразу переходить к достижению своих целей: к поступлению в ВУЗ или к началу удачной карьеры.
https://maps.google.gg/url?q=https://premialnie-diplomik.com
https://clients1.google.cv/url?sa=t&url=https://lands-diploma.com
https://maps.google.hu/url?sa=t&url=https://rudiplomista.com
http://www.google.sn/url?q=https://rudiplomista.com
https://maps.google.fr/url?q=https://premialnie-diplomik.com
В нашем мире, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Наличие официального документа сложно переоценить. Ведь именно диплом открывает двери перед людьми, стремящимися вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Наша компания предлагает быстро получить этот важный документ. Вы имеете возможность приобрести диплом, и это становится отличным решением для всех, кто не смог закончить обучение или потерял документ. дипломы изготавливаются с особой тщательностью, вниманием к мельчайшим нюансам, чтобы в результате получился полностью оригинальный документ.
Плюсы подобного решения заключаются не только в том, что вы сможете оперативно получить свой диплом. Весь процесс организовывается удобно, с нашей поддержкой. От выбора нужного образца до консультации по заполнению персональных данных и доставки по стране — все находится под полным контролем наших мастеров.
В результате, для тех, кто хочет найти оперативный способ получить необходимый документ, наша компания готова предложить выгодное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени перейти к важным целям, будь то поступление в ВУЗ или старт трудовой карьеры.
https://cse.google.co.ao/url?q=https://lands-diploma.com
https://cse.google.lu/url?q=https://lands-diploma.com
https://images.google.com.gi/url?sa=t&url=https://rudiplomista.com
http://www.google.is/url?sa=t&url=https://premialnie-diplomik.com
http://cse.google.al/url?sa=t&url=https://rudiplomista.com
Сегодня, когда диплом становится началом отличной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Необходимость наличия официального документа трудно переоценить. Ведь диплом открывает дверь перед каждым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в университете.
Мы предлагаем оперативно получить этот важный документ. Вы имеете возможность заказать диплом старого или нового образца, и это становится отличным решением для человека, который не смог закончить образование, утратил документ или хочет исправить плохие оценки. диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам, чтобы в результате получился продукт, полностью соответствующий оригиналу.
Преимущества подобного подхода заключаются не только в том, что можно оперативно получить свой диплом. Процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора необходимого образца диплома до консультаций по заполнению персональных данных и доставки по стране — все будет находиться под абсолютным контролем квалифицированных специалистов.
Всем, кто ищет быстрый и простой способ получения необходимого документа, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени перейти к личным целям, будь то поступление в ВУЗ или начало профессиональной карьеры.
http://drahthaar-forum.ru/topic9211.html?&p=31153
https://zoovetshop.ru/product/viyo-cat-senior-prebioticheskij-napitok-dlja-pozhilyh-koshek-7h30-ml/reviews/
http://oddicini.ru/sovety/texnologiya-almaznoj-rezki-betonnyx-izdelij-i-sovety.html?unapproved=230076&moderation-hash=ffdd60bf5bf9d2bdb9266dfe0dd14d9e#comment-230076
http://vecmir.ru/index.php/vecmirlife/57132-ygesahan/profile
http://test.sozapag.ru/forum/user/234267/
На сегодняшний день, когда диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения образования. Факт наличия документа об образовании переоценить невозможно. Ведь диплом открывает двери перед любым человеком, желающим вступить в сообщество профессионалов или учиться в каком-либо университете.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы имеете возможность приобрести диплом, что является выгодным решением для всех, кто не смог закончить обучение, утратил документ или хочет исправить плохие оценки. дипломы изготавливаются аккуратно, с особым вниманием ко всем деталям. В итоге вы получите продукт, максимально соответствующий оригиналу.
Преимущества данного подхода состоят не только в том, что вы сможете максимально быстро получить свой диплом. Весь процесс организовывается комфортно, с нашей поддержкой. От выбора необходимого образца до консультации по заполнению персональных данных и доставки по стране — все под абсолютным контролем качественных мастеров.
Для всех, кто ищет быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать длительного обучения и сразу перейти к достижению своих целей: к поступлению в ВУЗ или к началу трудовой карьеры.
https://www.google.com.bd/url?q=https://rudiplomista.com
https://www.google.de/url?q=https://lands-diploma.com
http://cse.google.ng/url?q=https://lands-diploma.com
https://images.google.co.ls/url?q=https://premialnie-diplomik.com
https://clients1.google.com.nf/url?sa=t&url=https://premialnie-diplomik.com
В наше время, когда диплом становится началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Наличие официального документа сложно переоценить. Ведь диплом открывает двери перед любым человеком, который стремится вступить в сообщество профессиональных специалистов или учиться в ВУЗе.
Наша компания предлагает оперативно получить этот важный документ. Вы можете приобрести диплом, что является выгодным решением для человека, который не смог завершить образование, потерял документ или хочет исправить плохие оценки. Все дипломы изготавливаются аккуратно, с максимальным вниманием ко всем деталям. В итоге вы сможете получить продукт, 100% соответствующий оригиналу.
Превосходство подобного решения заключается не только в том, что можно максимально быстро получить диплом. Процесс организован удобно, с нашей поддержкой. Начиная от выбора требуемого образца до консультации по заполнению персональной информации и доставки в любой регион России — все под полным контролем качественных мастеров.
Для всех, кто ищет максимально быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать долгого процесса обучения и не теряя времени переходить к достижению собственных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
http://cse.google.gr/url?sa=i&url=https://lands-diploma.com
https://www.google.com.sb/url?q=https://rudiplomista.com
https://maps.google.com.ua/url?q=https://lands-diploma.com
https://maps.google.co.za/url?sa=t&url=https://lands-diploma.com
https://toolbarqueries.google.co.tz/url?q=https://rudiplomista.com
animehangover.com
물론 서쪽으로 두 번 항해한 후에 씨앗을 찾을 수 있다고 생각하는 사람은 아무도 없습니다.
На сегодняшний день, когда диплом – это начало отличной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения качественного образования. Необходимость наличия официального документа переоценить попросту невозможно. Ведь именно диплом открывает дверь перед всеми, кто желает начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы сможете купить диплом нового или старого образца, и это становится выгодным решением для всех, кто не смог закончить обучение или утратил документ. дипломы производятся с особой тщательностью, вниманием к мельчайшим элементам. На выходе вы получите 100% оригинальный документ.
Преимущество такого подхода заключается не только в том, что вы оперативно получите свой диплом. Весь процесс организован удобно, с нашей поддержкой. От выбора нужного образца до консультаций по заполнению персональной информации и доставки по стране — все будет находиться под абсолютным контролем квалифицированных мастеров.
Для тех, кто хочет найти максимально быстрый способ получить необходимый документ, наша компания готова предложить выгодное решение. Заказать диплом – это значит избежать длительного процесса обучения и сразу перейти к своим целям: к поступлению в ВУЗ или к началу удачной карьеры.
https://images.google.lu/url?q=https://premialnie-diplomik.com
https://maps.google.ng/url?q=https://premialnie-diplomik.com
http://www.google.com.tw/url?q=https://premialnie-diplomik.com
http://cse.google.es/url?sa=i&url=https://premialnie-diplomik.com
https://cse.google.gg/url?sa=t&url=https://lands-diploma.com
В наше время, когда диплом – это начало успешной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Наличие официального документа об образовании переоценить просто невозможно. Ведь именно диплом открывает дверь перед любым человеком, который хочет начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте мы предлагаем оперативно получить этот необходимый документ. Вы сможете заказать диплом нового или старого образца, что становится отличным решением для человека, который не смог закончить образование, утратил документ или хочет исправить свои оценки. дипломы производятся аккуратно, с особым вниманием ко всем деталям, чтобы на выходе получился полностью оригинальный документ.
Плюсы такого подхода состоят не только в том, что вы сможете максимально быстро получить диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора требуемого образца до грамотного заполнения личной информации и доставки по стране — все под полным контролем опытных мастеров.
Всем, кто пытается найти быстрый и простой способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать продолжительного обучения и сразу переходить к своим целям: к поступлению в университет или к началу успешной карьеры.
http://topforbaby.ru/product/adameks-koljaska-transf-galaxy-ljulka-imit/reviews/
http://zooboard.ru/user/profile/261518
http://www.tv-lonsheim.de/index.php?option=com_easybookreloaded
http://www.thenatureofthings.blog/2022/06/overboard-by-sara-paretsky-review.html
http://web.symbol.rs/forum/member.php?action=profile&uid=591619
В нашем обществе, где диплом становится началом отличной карьеры в любой отрасли, многие стараются найти максимально быстрый и простой путь получения качественного образования. Важность наличия документа об образовании трудно переоценить. Ведь именно диплом открывает двери перед людьми, стремящимися вступить в сообщество профессиональных специалистов или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем быстро получить любой необходимый документ. Вы можете заказать диплом нового или старого образца, что становится отличным решением для человека, который не смог закончить обучение или потерял документ. дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим деталям. В итоге вы сможете получить 100% оригинальный документ.
Преимущество подобного решения состоит не только в том, что можно быстро получить диплом. Процесс организовывается удобно и легко, с нашей поддержкой. От выбора нужного образца до консультации по заполнению персональных данных и доставки в любой регион России — все под абсолютным контролем качественных мастеров.
Всем, кто ищет максимально быстрый способ получения необходимого документа, наша услуга предлагает выгодное решение. Приобрести диплом – значит избежать долгого процесса обучения и не теряя времени перейти к достижению своих целей, будь то поступление в университет или начало трудовой карьеры.
https://cse.google.vg/url?sa=t&url=https://lands-diploma.com
http://images.google.com.gh/url?sa=t&url=https://lands-diploma.com
https://cse.google.lv/url?q=https://rudiplomista.com
https://cse.google.co.cr/url?sa=t&url=https://lands-diploma.com
https://cse.google.off.ai/url?q=https://lands-diploma.com
В нашем обществе, где диплом – это начало удачной карьеры в любом направлении, многие стараются найти максимально простой путь получения качественного образования. Наличие документа об образовании переоценить невозможно. Ведь именно диплом открывает дверь перед каждым человеком, желающим начать трудовую деятельность или продолжить обучение в университете.
Мы предлагаем очень быстро получить этот важный документ. Вы сможете заказать диплом, что становится отличным решением для всех, кто не смог закончить образование, потерял документ или желает исправить свои оценки. Каждый диплом изготавливается аккуратно, с особым вниманием ко всем нюансам, чтобы в результате получился продукт, 100% соответствующий оригиналу.
Превосходство данного подхода состоит не только в том, что можно быстро получить свой диплом. Весь процесс организовывается комфортно, с нашей поддержкой. Начиная от выбора необходимого образца до правильного заполнения личной информации и доставки в любой регион страны — все под полным контролем опытных специалистов.
Всем, кто пытается найти оперативный способ получить требуемый документ, наша компания предлагает выгодное решение. Купить диплом – значит избежать долгого процесса обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в ВУЗ или старт успешной карьеры.
https://www.google.com.lb/url?q=https://premialnie-diplomik.com
https://cse.google.gg/url?sa=t&url=https://lands-diploma.com
https://images.google.gm/url?q=https://rudiplomista.com
https://www.google.co.vi/url?q=https://lands-diploma.com
http://images.google.com.ag/url?q=https://rudiplomista.com
В современном мире, где диплом является началом отличной карьеры в любой отрасли, многие ищут максимально быстрый путь получения образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает двери перед людьми, желающими вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы сможете приобрести диплом, что будет удачным решением для всех, кто не смог завершить обучение, утратил документ или желает исправить свои оценки. дипломы выпускаются аккуратно, с максимальным вниманием к мельчайшим деталям. В результате вы сможете получить документ, максимально соответствующий оригиналу.
Плюсы данного решения заключаются не только в том, что можно максимально быстро получить диплом. Весь процесс организован комфортно, с нашей поддержкой. Начиная от выбора подходящего образца до правильного заполнения персональной информации и доставки в любое место России — все находится под полным контролем качественных специалистов.
Таким образом, для всех, кто пытается найти быстрый и простой способ получения необходимого документа, наша компания готова предложить отличное решение. Купить диплом – значит избежать долгого обучения и сразу переходить к достижению своих целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://uralremput.ru/index.php?subaction=userinfo&user=ynaqav
http://1522-9892.com/bbs/board.php?bo_table=free&wr_id=146178
http://galeria.farvista.net/member.php?action=showprofile&user_id=40837
http://producm.ru/forum/user/97649/
http://www.sinoright.net/member/index.php?uid=akyxite
В нашем обществе, где диплом является началом отличной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения образования. Важность наличия официального документа сложно переоценить. Ведь диплом открывает дверь перед каждым человеком, желающим вступить в сообщество профессионалов или учиться в университете.
Мы предлагаем оперативно получить этот важный документ. Вы можете приобрести диплом старого или нового образца, и это является удачным решением для всех, кто не смог завершить образование или потерял документ. диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам. В результате вы получите 100% оригинальный документ.
Плюсы этого подхода заключаются не только в том, что вы быстро получите свой диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. От выбора необходимого образца документа до точного заполнения персональных данных и доставки по стране — все находится под полным контролем наших мастеров.
Для всех, кто ищет быстрый и простой способ получения требуемого документа, наша услуга предлагает выгодное решение. Купить диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к достижению собственных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
http://msfo-soft.ru/msfo/forum/messages/forum28/topic8729/message253875/?result=new#message253875
http://www.asanagikai.com/blog/1167.html?unapproved=1955998&moderation-hash=064087653325209756a021c76c4466da#comment-1955998
http://koreasamsong.com/bbs/board.php?bo_table=free&wr_id=2129108
https://lider-school174.com/forum/user/1505/
http://mydnepr.pp.ua/forum/topic.php?forum=7&topic=840
otraresacamas.com
このブログを読むたびに、何か新しいことを学べます。感謝しています。
sandyterrace.com
선원들은 유난히 들떠 있었고, 이른 아침 연습을 할 때마다 하나둘 비명을 질렀다.
LewisAluri
Добрый день!
Диплом повлиял на мое здоровье из-за необходимости работать ночью.
У нас можно заказать диплом без предоплаты с доставкой курьером по РФ, “под ключ”!
Желаю каждому честных оценок!
купить диплом в вольске
купить диплом в бийске
купить диплом в рубцовске
купить диплом в балашихе
купить диплом тренера
купить диплом моряка
купить диплом инженера
купить диплом в ноябрьске
купить диплом электромонтера
купить диплом в екатеринбурге
Do you mind if I quote ɑ сouple of your posts as long as I providе credit
and sources back to your ԝebpage? My blog is in the very same
niche as yours and mү users woulɗ reaⅼly benefit from some of thе information you provide һeгe.
Please let me know if this okay with you. Cheers!
Cash Winning Games to Play in Kenya
best online casino games kenya games that pay real money in kenya .
Dichaelautob
Здравствуйте!
Однако, благодаря интернету, мне удалось найти ресурсы, способствующие повышению эффективности работы над дипломом.
Мы поможем вам купить диплом ВУЗа России недорого, без предоплаты и с гарантией возврата средств.
Желаю для каждого пятерочных) оценок!
купить диплом зубного техника
купить диплом в каспийске
купить диплом в новотроицке
купить диплом в дербенте
купить диплом о среднем образовании
купить диплом в набережных челнах
купить диплом в феодосии
купить диплом в чебоксарах
купить аттестаты за 9
купить диплом в ноябрьске
Hi there to every one, because I am truly keen of reading this blog’s post to be updated on a regular basis. It contains fastidious material.
Fobertbap
Добрый день!
Рекомендую изучить найденные мной материалы для улучшения вашего процесса работы над дипломом.
Наш интернет-магазин предлагает купить российский диплом по выгодной цене, с гарантией прохождения всех проверок.
Желаю всем прекрасных оценок!
купить морской диплом
купить диплом моряка
купить диплом воспитателя
купить диплом в выборге
купить диплом тренера
купить диплом педагога
купить диплом в ессентуках
купить диплом в арзамасе
купить диплом в анапе
купить диплом в севастополе
thebuzzerpodcast.com
Longquan View를 말한 Puji Daoist라는 단어가 있지만 사람들은 그것을 믿을 수밖에 없습니다.
Fobertbap
Доброго дня!
Когда диплом стал моим кошмаром, я попытался пересмотреть сроки и перейти к ночному графику работы, что негативно сказалось на моем здоровье.
У нас вы можете купить диплом университета без предоплаты с доставкой в любую точку России.
Желаю каждому пятерочных) оценок!
купить диплом в уфе
купить диплом в ульяновске
купить диплом маляра
купить диплом ветеринара
купить диплом в геленджике
купить диплом в уфе
купить диплом в усолье-сибирском
купить диплом бурильщика
купить диплом преподавателя
купить диплом в димитровграде
Доброго всем дня!
Приобретите документы об образовании всех ВУЗов России по выгодным ценам с постоплатой и помощью 24/7!
https://www.google.sr/url?q=https://anny-diploms.com
https://clients1.google.com.pr/url?sa=t&url=http://diplomyland.com
https://clients1.google.com.fj/url?sa=t&url=https://anny-diploms.com
https://www.google.com.fj/url?q=http://diplomy-originaly.com
https://clients4.google.com/url?q=http://free-diplommi.com
Поможем выбрать, оформить заказ и купить диплом любого института в любом городе по самым низким ценам
We are a group of volunteers аnd opening a new scheme іn our community.
Your site provided ᥙs with valuаble information to work on. You have done
ɑn impressive job and our whole community will be grateful to you.
Earnestabich
Здравствуйте!
Диплом заставил меня работать по ночам, что негативно сказалось на моем здоровье.
У нас вы можете купить диплом университета с гарантированной доставкой в любой город России, оплачивая после получения.
Желаю каждому отличных отметок!
купить диплом магистра
купить диплом в элисте
купить диплом инженера
купить диплом в оренбурге
купить диплом дизайнера
купить диплом мастера маникюра и педикюра
купить диплом с занесением в реестр
купить диплом в юрге
купить диплом в анжеро-судженске
купить диплом историка
etsyweddingteam.com
読んで良かった!実用性の高い情報がとても役に立ちます。
StephenNug
Доброго дня!
Моя работа над дипломом улучшилась благодаря ценным интернет-находкам.
Закажите диплом у нас легко! Без предоплаты, с гарантией качества и доставкой по всей России.
Желаю для каждого честных отметок!
купить диплом в каменске-уральском
купить диплом воспитателя
купить диплом в йошкар-оле
купить диплом в соликамске
купить диплом магистра
купить диплом в сургуте
купить диплом в славянске-на-кубани
купить диплом в невинномысске
купить диплом в анжеро-судженске
купить диплом в клинцах
В нашем обществе, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Необходимость наличия официального документа об образовании переоценить просто невозможно. Ведь именно диплом открывает дверь перед каждым человеком, желающим начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы сможете купить диплом нового или старого образца, и это будет выгодным решением для всех, кто не смог закончить образование, утратил документ или хочет исправить свои оценки. Все дипломы выпускаются аккуратно, с особым вниманием ко всем элементам. В результате вы получите 100% оригинальный документ.
Плюсы этого подхода заключаются не только в том, что можно оперативно получить диплом. Весь процесс организовывается удобно, с нашей поддержкой. Начиная от выбора подходящего образца диплома до консультации по заполнению личных данных и доставки в любой регион России — все будет находиться под абсолютным контролем квалифицированных специалистов.
В итоге, для всех, кто ищет максимально быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Заказать диплом – значит избежать длительного обучения и сразу переходить к достижению своих целей: к поступлению в университет или к началу трудовой карьеры.
http://www.rusbil.ru/forum/viewtopic.php?t=5997&view=previous
http://visionart.kr/board/bbs/board.php?bo_table=free&wr_id=809678
https://magadan-mebel.ru/forum/user/756/
http://fit-prom.ru/index.php?subaction=userinfo&user=eneve
http://www.85sucai.com/member/index.php?uid=ovyna
В нашем обществе, где диплом – это начало отличной карьеры в любой сфере, многие пытаются найти максимально простой путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь диплом открывает дверь перед каждым человеком, который собирается начать трудовую деятельность или учиться в университете.
В данном контексте мы предлагаем максимально быстро получить этот необходимый документ. Вы сможете приобрести диплом нового или старого образца, что является отличным решением для человека, который не смог закончить обучение, потерял документ или желает исправить свои оценки. диплом изготавливается аккуратно, с особым вниманием ко всем элементам, чтобы на выходе получился продукт, максимально соответствующий оригиналу.
Преимущество данного решения состоит не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается комфортно, с нашей поддержкой. Начиная от выбора подходящего образца диплома до консультации по заполнению персональной информации и доставки по стране — все будет находиться под абсолютным контролем наших мастеров.
Для всех, кто пытается найти быстрый и простой способ получения необходимого документа, наша компания может предложить выгодное решение. Купить диплом – значит избежать длительного процесса обучения и не теряя времени переходить к своим целям, будь то поступление в ВУЗ или старт удачной карьеры.
https://images.google.so/url?q=https://lands-diploma.com
https://maps.google.lk/url?sa=t&url=https://lands-diploma.com
http://images.google.cd/url?q=https://premialnie-diplomik.com
https://cse.google.com.uy/url?sa=t&url=https://lands-diploma.com
http://images.google.dm/url?q=https://premialnie-diplomik.com
В современном мире, где диплом становится началом успешной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Факт наличия официального документа трудно переоценить. Ведь диплом открывает двери перед всеми, кто желает начать профессиональную деятельность или учиться в университете.
Мы предлагаем максимально быстро получить любой необходимый документ. Вы имеете возможность заказать диплом, и это будет отличным решением для человека, который не смог закончить обучение или утратил документ. Каждый диплом изготавливается аккуратно, с особым вниманием ко всем элементам. В результате вы получите 100% оригинальный документ.
Преимущества подобного решения заключаются не только в том, что можно оперативно получить диплом. Весь процесс организовывается удобно и легко, с профессиональной поддержкой. От выбора подходящего образца документа до грамотного заполнения персональной информации и доставки по России — все будет находиться под полным контролем качественных специалистов.
Для всех, кто ищет быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Купить диплом – это значит избежать долгого процесса обучения и не теряя времени перейти к достижению своих целей: к поступлению в университет или к началу удачной карьеры.
http://vip-gips.com.ua/blog/k2-category/item/14-this-century-old-church-is-now-a-cozy-home
http://www.vespaclubnapoli.it/../profile.php?lookup=18709
https://instrument-43.ru/product/pajalnik-zubr-impulsnyj-so-svetovym-indikatorom-150-vt-2-smennyh-zhala-kanifol-pripoj-/reviews/
http://sad5.lytkarino.net/index.php?subaction=userinfo&user=erumeheg
http://www.premier-climat.ru/product/dimplex-obsidian-serija-hi-tech/reviews/
qiyezp.com
그 나이에 그런 범죄를 당하기는 어렵습니다.
bestmanualpolesaw.com
이동양은 이 말을 듣고 곰곰이 생각하다가 류젠을 바라보며 미소를 지었다.
В современном мире, где диплом – это начало успешной карьеры в любой области, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа переоценить невозможно. Ведь именно он открывает дверь перед всеми, кто стремится вступить в профессиональное сообщество или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить этот необходимый документ. Вы сможете заказать диплом нового или старого образца, и это является выгодным решением для человека, который не смог закончить обучение, потерял документ или хочет исправить плохие оценки. Все дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим нюансам, чтобы в итоге получился 100% оригинальный документ.
Превосходство такого подхода состоит не только в том, что вы оперативно получите диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. Начиная от выбора нужного образца до консультации по заполнению личных данных и доставки в любой регион России — все под абсолютным контролем опытных мастеров.
Всем, кто ищет оперативный способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать длительного процесса обучения и сразу переходить к достижению своих целей, будь то поступление в ВУЗ или начало карьеры.
https://images.google.com.my/url?q=https://lands-diploma.com
https://maps.google.com.au/url?q=https://premialnie-diplomik.com
https://www.google.co.id/url?q=https://premialnie-diplomik.com
http://www.google.gp/url?q=https://rudiplomista.com
https://cse.google.gg/url?sa=t&url=https://premialnie-diplomik.com
LhaneViart
Привет всем!
Диплом стал более доступным заданием благодаря использованию онлайн-материалов.
Мы предлагаем купить диплом университета без предоплаты, с гарантированной доставкой по всей России.
Желаю каждому не двоешных) отметок!
купить свидетельство о заключении брака
купить диплом ссср
купить диплом в прокопьевске
купить диплом в перми
купить диплом в канске
купить диплом в сарапуле
где купить диплом образование
купить диплом в северодвинске
купить диплом химика
купить диплом программиста
Someone necessarily help to make significantly articles I might state. This is the first time I frequented your web page and so far? I surprised with the analysis you made to make this actual publish extraordinary. Excellent process!
В современном мире, где диплом является началом отличной карьеры в любой сфере, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Необходимость наличия документа об образовании сложно переоценить. Ведь именно он открывает дверь перед людьми, стремящимися вступить в профессиональное сообщество или учиться в ВУЗе.
Мы предлагаем очень быстро получить любой необходимый документ. Вы сможете заказать диплом старого или нового образца, что является выгодным решением для человека, который не смог завершить образование или потерял документ. дипломы производятся с особой тщательностью, вниманием ко всем деталям. На выходе вы сможете получить 100% оригинальный документ.
Превосходство данного подхода состоит не только в том, что можно оперативно получить диплом. Весь процесс организован просто и легко, с нашей поддержкой. От выбора подходящего образца до точного заполнения персональной информации и доставки по стране — все под полным контролем квалифицированных специалистов.
Всем, кто хочет найти оперативный способ получить требуемый документ, наша компания готова предложить отличное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и сразу переходить к достижению личных целей: к поступлению в университет или к началу успешной карьеры.
http://waterlevel.ru/forum/user/93738/
http://mega-zaim96.ru/forum/user/34385/
http://www.oymalitepe.net/modules.php?name=Your_Account&op=userinfo&username=ygurilul
http://dancerussia.ru/forum/viewtopic.php?f=17&t=16248&p=23384
http://mediawiki.envri.eu/index.php?title=originalitydiplomiki
В нашем обществе, где диплом становится началом удачной карьеры в любой отрасли, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь диплом открывает дверь перед всеми, кто стремится вступить в профессиональное сообщество или учиться в университете.
Наша компания предлагает максимально быстро получить этот важный документ. Вы сможете заказать диплом, и это становится отличным решением для человека, который не смог закончить обучение или потерял документ. Все дипломы производятся аккуратно, с максимальным вниманием к мельчайшим нюансам, чтобы в итоге получился продукт, максимально соответствующий оригиналу.
Плюсы такого подхода состоят не только в том, что можно оперативно получить диплом. Весь процесс организован комфортно, с нашей поддержкой. От выбора необходимого образца до консультаций по заполнению личной информации и доставки в любое место страны — все под абсолютным контролем квалифицированных мастеров.
Для всех, кто ищет быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и сразу переходить к важным целям, будь то поступление в ВУЗ или начало трудовой карьеры.
https://anaparegion.ru/news/237-dyussh_viktoriya_vstretila_uchastnikov_vsekubansko/
http://zemli.com/index.php?subaction=userinfo&user=ytebogym
http://vip-gips.com.ua/blog/k2-category/item/14-this-century-old-church-is-now-a-cozy-home
http://ladydiary.ru/users/eqozipal
http://www.repetitor.tv/2015/06/1-2015.html
Gichardadvox
Приветики, дорогие мои!!
Столкнувшись с проблемами в написании диплома, я обнаружил в сети поддержку, которая помогает мне продолжать работу.
Мы предлагаем вам купить диплом университета недорого с доставкой в любую точку России, оплата после получения.
Желаю всем не двоешных) отметок!
купить диплом в коврове
куплю диплом
купить диплом в канске
купить аттестат за 9 классов
купить диплом в бугульме
https://www.google.dk/url?sa=t&url=https://prodiplome.com/
http://www.google.co.ao/url?q=https://10000diplomov.ru/
http://maps.google.com.jm/url?q=https://diplom-zakaz.ru/
https://images.google.bg/url?q=https://diplom-insti.ru/
http://cse.google.com.fj/url?sa=t&url=https://nsk-diplom.com/
В современном мире, где диплом – это начало удачной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь диплом открывает дверь перед людьми, стремящимися начать трудовую деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы сможете купить диплом нового или старого образца, и это является отличным решением для всех, кто не смог завершить образование или потерял документ. дипломы выпускаются с особой тщательностью, вниманием к мельчайшим элементам, чтобы на выходе получился документ, полностью соответствующий оригиналу.
Преимущество подобного решения состоит не только в том, что вы максимально быстро получите свой диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора нужного образца документа до консультаций по заполнению персональных данных и доставки в любой регион России — все будет находиться под полным контролем наших специалистов.
Для всех, кто ищет оперативный способ получить необходимый документ, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать длительного обучения и не теряя времени переходить к достижению личных целей, будь то поступление в ВУЗ или начало карьеры.
https://cse.google.dz/url?sa=t&url=https://premialnie-diplomik.com
http://www.google.com.et/url?q=https://premialnie-diplomik.com
https://cse.google.com.cu/url?sa=t&url=https://premialnie-diplomik.com
https://cse.google.lu/url?q=https://premialnie-diplomik.com
https://image.google.as/url?q=https://rudiplomista.com
Ощутите азарт и адреналин в онлайн казино Беларусь в любое время суток
онлайн казино Беларусь онлайн казино Беларусь .
В современном мире, где диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально простой путь получения образования. Наличие официального документа об образовании сложно переоценить. Ведь именно он открывает двери перед всеми, кто стремится начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете приобрести диплом старого или нового образца, что будет отличным решением для всех, кто не смог завершить образование, утратил документ или хочет исправить плохие оценки. Каждый диплом изготавливается с особой аккуратностью, вниманием ко всем нюансам. На выходе вы сможете получить продукт, полностью соответствующий оригиналу.
Плюсы данного подхода состоят не только в том, что вы быстро получите диплом. Процесс организован удобно, с профессиональной поддержкой. Начиная от выбора нужного образца документа до правильного заполнения личной информации и доставки в любое место страны — все будет находиться под абсолютным контролем качественных мастеров.
Для тех, кто пытается найти быстрый способ получить требуемый документ, наша услуга предлагает выгодное решение. Приобрести диплом – это значит избежать продолжительного обучения и сразу переходить к достижению личных целей, будь то поступление в университет или начало успешной карьеры.
http://intextv.by/forum/user/62013/
http://erotikfoto.ru/index.php?subaction=userinfo&user=ynapabeb
http://mercenairesdelaforce.free.fr/index.php?file=Members&op=detail&autor=anygubik
https://darbydanohio.com/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BF%D1%80%D0%B8%D0%BE%D0%B1%D1%80%D0%B5%D1%81%D1%82%D0%B8-%D1%80%D0%B5%D0%B0%D0%BB%D1%8C%D0%BD%D1%8B%D0%B9-%D1%81%D0%B5/
http://www.pavelgorbenko.ru/2018/02/blog-post.html
donmhomes.com
素晴らしい記事!いつも新鮮な視点をありがとう。
Fobertbap
Здравствуйте!
Подготовка диплома стала более продуктивной благодаря онлайн-ресурсам.
Приобретение диплома института, техникума или колледжа у нас – лучший вариант с доставкой по РФ.
Желаю для каждого пятерочных) оценок!
куплю диплом с занесением
купить диплом в рыбинске
купить диплом магистра
купить свидетельство о заключении брака
купить диплом в усолье-сибирском
http://maps.google.tt/url?q=https://prodiplome.com/
https://www.google.com.sv/url?sa=t&url=https://ry-diplom.com/
http://www.google.com.tw/url?q=https://diplomoz-197.com/
https://clients1.google.rs/url?q=https://peoplediplom.ru/
https://images.google.kg/url?q=https://1magistr.ru/
What i do not realize is actually how you are no longer really a lot more well-liked than you may be now. You are very intelligent. You know therefore significantly in terms of this topic, made me for my part believe it from a lot of varied angles. Its like women and men don’t seem to be interested unless it’s one thing to accomplish with Lady gaga! Your personal stuffs outstanding. At all times maintain it up!
В нашем мире, где диплом – это начало успешной карьеры в любой отрасли, многие ищут максимально быстрый путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь именно диплом открывает дверь перед людьми, стремящимися вступить в профессиональное сообщество или продолжить обучение в каком-либо ВУЗе.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы сможете заказать диплом, и это становится отличным решением для человека, который не смог закончить обучение, утратил документ или желает исправить плохие оценки. диплом изготавливается аккуратно, с особым вниманием ко всем деталям. На выходе вы сможете получить продукт, максимально соответствующий оригиналу.
Превосходство данного подхода состоит не только в том, что можно быстро получить диплом. Процесс организован удобно, с нашей поддержкой. От выбора нужного образца документа до консультации по заполнению личной информации и доставки по стране — все под полным контролем опытных специалистов.
В итоге, для тех, кто хочет найти быстрый и простой способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать длительного обучения и не теряя времени переходить к своим целям, будь то поступление в ВУЗ или начало профессиональной карьеры.
https://www.google.az/url?sa=t&url=https://rudiplomista.com
https://images.google.com.vn/url?sa=t&url=https://rudiplomista.com
https://www.google.dk/url?sa=t&url=https://premialnie-diplomik.com
https://images.google.com.et/url?sa=t&url=https://lands-diploma.com
https://cse.google.com.cu/url?sa=t&url=https://rudiplomista.com
ShaneViart
Доброго дня!
Диплом уже не вызывает столько стресса, как раньше, благодаря помощи, полученной через интернет.
Приобретайте диплом университета без предоплаты с доставкой по всей России.
Желаю вам всем пятерочных) отметок!
купить диплом эколога
купить диплом в ревде
купить диплом образцы
купить свидетельство о рождении
купить диплом института
http://cse.google.ng/url?q=https://diploms-vuza.com/
https://maps.google.fi/url?q=https://kdiplom.ru/
https://images.google.com.co/url?sa=t&url=https://diplom-bez-problem.com/
https://www.google.tn/url?q=https://diploms-vuza.com/
https://cse.google.dm/url?sa=t&url=https://diplom-insti.ru/
animehangover.com
Zhu Youtang은 얼굴에 미소를 지으며 홀에 들어온 Fang Jifan을 바라보며 행복했습니다.
I blog often and I truly appreciate your content. The article has really peaked my interest. I will book mark your blog and keep checking for new information about once a week. I opted in for your Feed too.
This website really has all of the information I wanted concerning this subject and didn’t know who to ask.
В нашем мире, где диплом становится началом удачной карьеры в любой области, многие ищут максимально простой путь получения качественного образования. Факт наличия официального документа переоценить невозможно. Ведь диплом открывает двери перед каждым человеком, который собирается вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Наша компания предлагает оперативно получить этот важный документ. Вы сможете купить диплом нового или старого образца, и это будет удачным решением для всех, кто не смог завершить образование, утратил документ или желает исправить плохие оценки. дипломы выпускаются аккуратно, с особым вниманием к мельчайшим элементам. В результате вы сможете получить полностью оригинальный документ.
Преимущества данного решения заключаются не только в том, что вы оперативно получите свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начиная от выбора необходимого образца диплома до точного заполнения персональной информации и доставки в любой регион страны — все будет находиться под абсолютным контролем опытных специалистов.
Всем, кто хочет найти максимально быстрый способ получить необходимый документ, наша услуга предлагает выгодное решение. Купить диплом – значит избежать долгого обучения и не теряя времени переходить к достижению личных целей: к поступлению в университет или к началу удачной карьеры.
http://images.google.td/url?q=https://rudiplomista.com
https://images.google.lt/url?q=https://rudiplomista.com
https://clients1.google.ht/url?sa=t&url=https://lands-diploma.com
https://maps.google.cl/url?q=https://premialnie-diplomik.com
https://maps.google.fr/url?q=https://rudiplomista.com
At this time I am going to do my breakfast, when having my breakfast coming again to read further news.
В современном мире, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Наличие документа об образовании сложно переоценить. Ведь именно диплом открывает дверь перед всеми, кто стремится начать трудовую деятельность или учиться в высшем учебном заведении.
Наша компания предлагает очень быстро получить любой необходимый документ. Вы сможете приобрести диплом, что становится удачным решением для всех, кто не смог завершить образование или потерял документ. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим деталям, чтобы в результате получился документ, полностью соответствующий оригиналу.
Плюсы подобного подхода состоят не только в том, что можно максимально быстро получить диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. От выбора нужного образца документа до грамотного заполнения персональной информации и доставки по России — все находится под абсолютным контролем наших мастеров.
Таким образом, всем, кто ищет оперативный способ получения требуемого документа, наша компания готова предложить выгодное решение. Заказать диплом – значит избежать долгого обучения и сразу перейти к важным целям: к поступлению в университет или к началу успешной карьеры.
https://library.pilxt.com/index.php?action=profile;area=showposts;sa=messages;u=52454
http://cntuvek.ru/forum/user/10384/
http://188.17.148.172/forum/profile.php?action=show&member=7029
http://www.altasugar.it/new/index.php?option=com_kunena&view=topic&catid=3&id=66657&Itemid=151
http://koreasamsong.com/bbs/board.php?bo_table=free&wr_id=2136768
Здравствуйте!
Приобретите диплом института или колледжа с гарантией качества и доставкой по России без предоплаты.
https://cse.google.com.sg/url?sa=t&url=http://diplomyland.com
https://www.google.hn/url?sa=t&url=http://diplomsagroups.com
https://www.google.cm/url?sa=i&rct=j&q=https://anny-diploms.com
http://image.google.iq/url?rct=j&sa=t&url=http://originality-diplomy.com
http://maps.google.ht/url?rct=j&sa=t&url=http://free-diplommi.com
Хотите купить диплом Вуза недорого и получить его почтой без предоплаты? Мы можем помочь вам сделать это.
LewisAluri
Приветики, дорогие мои!!
Диплом поставил под угрозу мое здоровье из-за необходимости ночных занятий.
Приобретайте диплом университета без предоплаты с доставкой по всей России.
Желаю каждому честных оценок!
купить диплом в глазове
купить диплом в буденновске
купить диплом старого образца
купить диплом в обнинске
купить диплом в геленджике
https://images.google.bi/url?q=https://1magistr.ru/
https://www.google.tm/url?sa=t&url=https://nsk-diplom.com/
https://toolbarqueries.google.lk/url?sa=t&url=https://kazdiplomas.com/
https://clients1.google.com.pa/url?sa=t&url=https://kdiplom.ru/
https://www.google.me/url?q=https://kdiplom.ru/
В современном мире, где диплом является началом успешной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Наличие документа об образовании переоценить невозможно. Ведь диплом открывает двери перед любым человеком, который хочет вступить в профессиональное сообщество или учиться в ВУЗе.
Наша компания предлагает очень быстро получить любой необходимый документ. Вы сможете приобрести диплом старого или нового образца, что будет удачным решением для всех, кто не смог завершить образование, утратил документ или желает исправить свои оценки. Каждый диплом изготавливается с особой тщательностью, вниманием ко всем деталям. На выходе вы получите документ, 100% соответствующий оригиналу.
Преимущества данного решения заключаются не только в том, что можно быстро получить свой диплом. Весь процесс организовывается комфортно, с нашей поддержкой. Начав от выбора нужного образца документа до консультации по заполнению личных данных и доставки в любое место России — все под абсолютным контролем опытных мастеров.
В итоге, для тех, кто ищет быстрый способ получить необходимый документ, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени переходить к достижению личных целей, будь то поступление в университет или старт успешной карьеры.
https://cse.google.com.af/url?sa=t&url=https://rudiplomista.com
https://cse.google.sc/url?sa=t&url=https://rudiplomista.com
https://clients1.google.ad/url?q=https://lands-diploma.com
https://images.google.com.jm/url?q=https://lands-diploma.com
https://maps.google.com.au/url?q=https://lands-diploma.com
Добрый день всем!
Наши услуги позволят вам купить диплом ВУЗа с доставкой по России без предоплаты и с полной уверенностью в его подлинности!
https://cse.google.ms/url?sa=t&url=http://rdiplomik24.com
https://cse.google.rw/url?sa=t&url=https://gosznac-diploma.com
https://www.google.ge/url?q=http://rdiplomik24.com
https://clients1.google.co.je/url?q=https://anny-diploms.com
http://maps.google.com.jm/url?q=http://rudiplomisty24.com
Приобретите документы об образовании всех ВУЗов России по выгодным условиям с доставкой по РФ без предоплаты и с гарантией качества!
Добрый день всем!
Как можно приобрести диплом Вуза в России без предоплаты на сайте? Мы готовы доставить его в любую точку страны.
https://www.google.com.vn/url?q=http://rudiplomisty24.com
https://maps.google.com.lb/url?q=http://rudiplomisty24.com
https://maps.google.cf/url?sa=t&url=http://russdiplomiki.com
https://clients1.google.com.hk/url?sa=t&url=https://eonline-diploms.com
https://images.google.ie/url?sa=t&url=https://diploms-asx.com
Закажите диплом ВУЗа с доставкой по всей России без предоплаты и с гарантией качества!
В современном мире, где диплом является началом удачной карьеры в любой отрасли, многие ищут максимально простой путь получения качественного образования. Необходимость наличия документа об образовании сложно переоценить. Ведь диплом открывает двери перед всеми, кто собирается вступить в сообщество профессионалов или продолжить обучение в каком-либо институте.
Мы предлагаем очень быстро получить этот важный документ. Вы сможете купить диплом, что становится выгодным решением для всех, кто не смог завершить обучение, потерял документ или желает исправить свои оценки. Все дипломы изготавливаются аккуратно, с максимальным вниманием ко всем элементам, чтобы на выходе получился продукт, 100% соответствующий оригиналу.
Преимущества данного подхода состоят не только в том, что можно максимально быстро получить свой диплом. Весь процесс организован удобно, с нашей поддержкой. Начиная от выбора требуемого образца до консультации по заполнению персональных данных и доставки по России — все находится под абсолютным контролем наших специалистов.
Всем, кто ищет быстрый способ получения необходимого документа, наша компания может предложить отличное решение. Заказать диплом – это значит избежать долгого обучения и сразу перейти к достижению личных целей: к поступлению в ВУЗ или к началу успешной карьеры.
https://www.google.com.sv/url?sa=t&url=https://premialnie-diplomik.com
http://toolbarqueries.google.com.af/url?q=https://rudiplomista.com
http://www.google.co.ao/url?q=https://rudiplomista.com
https://www.google.ps/url?sa=t&url=https://lands-diploma.com
https://maps.google.fi/url?q=https://rudiplomista.com
Fobertbap
Добрый день!
Написание диплома подкрепляется ресурсами, найденными в интернете.
Мы поможем вам купить диплом университета без предоплаты и доставим его в любой город России с гарантированной безопасностью.
Желаю каждому честных оценок!
купить диплом в чите
купить диплом в майкопе
купить диплом в сочи
купить диплом в буйнакске
купить диплом в великих луках
https://images.google.gp/url?sa=t&url=https://diplom-profi.ru/
http://images.google.co.zm/url?q=https://okdiplom.com/
https://images.google.com.ar/url?sa=t&url=https://nsk-diplom.com/
http://images.google.com.pg/url?q=https://kupit-diplomyz24.com/
https://clients1.google.co.je/url?q=https://peoplediplom.ru/
Добрый день всем!
Заказать и купить диплом в России без предоплаты и с гарантией, можно только у Нас с гарантией под ключ
https://image.google.co.im/url?q=https://gosznac-diploma.com
https://www.google.com.qa/url?q=http://rudiplomisty24.com
http://maps.google.com.om/url?q=http://diploms-service.com
https://www.google.co.ls/url?q=http://diplomrussian.com
https://image.google.tg/url?q=https://premialnie-diplomansy.com
Наша компания поможет купить диплом Вуза со скидкой и доставкой в любую точку России
toasterovensplus.com
この記事は実に実用的で、大変役に立ちました!
geinoutime.com
징이 다시 두들겼다. “대승, 모베이 대승, 모베이 대승!”
Добрый день всем!
Приобретите диплом ВУЗа с гарантированной доставкой по России и без предварительной оплаты – надежно и выгодно!
https://clients1.google.sc/url?q=http://diplomy-originaly.com
http://toolbarqueries.google.it/url?sa=t&url=http://diplomsagroups.com
https://clients1.google.com.cu/url?sa=t&url=http://diplomy-originaly.com
https://images.google.by/url?sa=t&url=http://diplomyland.com
http://image.google.iq/url?rct=j&sa=t&url=http://diplomrussian.com
Приобретите диплом Гознака у нас по специальной цене с доставкой в любой регион России без предоплаты.
animehangover.com
Zhu Houzhao는 그를 때리고 싶어 그를 응시했습니다.
Привет всем!
Наша компания предлагает конфиденциально выбрать и заказать диплом любого Вуза России
http://www.google.cl/url?q=https://diploms-asx.com
https://maps.google.mg/url?q=https://gosznac-diploma.com
https://image.google.gl/url?q=https://russian-diplom.com
https://images.google.com.uy/url?q=http://diplomrussian.com
https://maps.google.at/url?q=http://diplomyland.com
Приобретите диплом ВУЗа по выгодной цене с доставкой по всей России без предоплаты!
Good information. With thanks!
You’ve made your point!
OLaneViart
Добрый день!
Стремлюсь преодолеть все препятствия, несмотря на ухудшение самочувствия из-за ночных занятий с дипломом.
Закажите диплом у нас и получите его быстро и надежно с оплатой после получения, доставка в любой регион РФ.
Желаю всем отличных отметок!
купить диплом в кемерово
купить диплом в феодосии
купить диплом в оренбурге
купить диплом с реестром
купить диплом в уфе
https://images.google.co.ug/url?q=https://diplomoz-197.com/
https://images.google.com.jm/url?q=https://nsk-diplom.com/
https://www.google.co.uz/url?q=https://okdiplom.com/
https://www.google.com.ng/url?q=https://diplom45.ru/
https://www.google.co.cr/url?q=https://diplom5.com/
I seriously love your site.. Very nice colors & theme. Did you make this website yourself? Please reply back as I’m hoping to create my own personal site and would like to learn where you got this from or what the theme is named. Thanks!
Hey I know this is off topic but I was wondering if you knew of any widgets I could add to my blog that automatically tweet my newest twitter updates. I’ve been looking for a plug-in like this for quite some time and was hoping maybe you would have some experience with something like this. Please let me know if you run into anything. I truly enjoy reading your blog and I look forward to your new updates.
You actually explained that superbly!
My site https://may-anma.icu
Привет, дорогой читатель!
Купите диплом ВУЗа с гарантированной доставкой в любой город России без предварительной оплаты и уверенностью в его легальности!
Купите диплом института или техникума с доставкой по РФ по выгодной цене без предоплаты!
http://maps.google.kz/url?q=https://lands-diploms.com
https://clients1.google.com.fj/url?sa=t&url=http://kazdiplom.com
https://maps.google.co.nz/url?q=https://russiany-diplomans.com
http://www.google.co.ao/url?q=http://rudiplomisty24.com
https://images.google.hn/url?q=https://originality-diplomiki.com
1. The Ultimate Aviator Games Guide
aviator casino online juego aviador .
Please let me know if you’re looking for a article author for your site. You have some really good articles and I think I would be a good asset. If you ever want to take some of the load off, I’d really like to write some content for your blog in exchange for a link back to mine. Please blast me an email if interested. Thanks!
Приветики!
Получите документы об образовании всех ВУЗов России с гарантированной подлинностью и доставкой по РФ без предварительной оплаты!
Приобретите диплом института или колледжа недорого с доставкой по РФ и гарантией качества без предварительной оплаты!
https://images.google.al/url?sa=t&url=https://russdiplomag.com
https://images.google.ci/url?sa=t&url=https://russdiplomag.com
https://www.google.com.ai/url?q=http://rudiplomisty24.com
https://maps.google.mw/url?q=https://premialnie-diplomany.com
https://maps.google.tl/url?q=https://frees-diplom.com
Привет всем!
Закажите диплом института или техникума по доступной цене с гарантированной доставкой в любой уголок России без предоплаты!
Приобретите российский диплом с гарантией качества и доставкой в любую точку страны без предварительной оплаты!
https://cse.google.nr/url?sa=t&url=https://originality-diploman.com
https://images.google.lt/url?q=http://volga-diplomy.com
https://clients2.google.com/url?q=https://russiany-diplomans.com
https://maps.google.ie/url?q=http://rudiplomisty24.com
https://clients1.google.td/url?sa=t&url=https://frees-diplom.com
В современном мире, где диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа об образовании сложно переоценить. Ведь именно он открывает двери перед всеми, кто собирается вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Предлагаем максимально быстро получить этот важный документ. Вы можете купить диплом нового или старого образца, что становится удачным решением для всех, кто не смог закончить образование, утратил документ или желает исправить свои оценки. Любой диплом изготавливается аккуратно, с максимальным вниманием ко всем деталям, чтобы в результате получился полностью оригинальный документ.
Преимущества данного решения состоят не только в том, что можно оперативно получить свой диплом. Процесс организован комфортно и легко, с нашей поддержкой. Начиная от выбора необходимого образца диплома до правильного заполнения персональной информации и доставки в любое место России — все будет находиться под полным контролем квалифицированных специалистов.
Всем, кто ищет быстрый способ получения требуемого документа, наша услуга предлагает отличное решение. Приобрести диплом – значит избежать длительного обучения и сразу переходить к достижению личных целей: к поступлению в университет или к началу успешной карьеры.
https://cse.google.ne/url?sa=i&url=https://rudiplomista.com
http://cse.google.ae/url?sa=i&url=https://rudiplomista.com
https://maps.google.sk/url?q=https://lands-diploma.com
https://images.google.as/url?q=https://lands-diploma.com
https://cse.google.ee/url?sa=t&url=https://rudiplomista.com
k8 カジノ 無料
実践に役立つ具体的な内容が充実しています。感謝!
tvlore.com
이튿날 정오에 그는 병든 몸을 질질 끌고 황폐해진 성신전 밖으로 나갔다.
Howdy! I know this is kinda off topic nevertheless I’d figured I’d ask. Would you be interested in trading links or maybe guest authoring a blog article or vice-versa? My website covers a lot of the same subjects as yours and I feel we could greatly benefit from each other. If you’re interested feel free to send me an email. I look forward to hearing from you! Great blog by the way!
https://squareblogs.net/typhanhntw/h1-b-sklo-far-farfarlight-iakist-nadiinist-i-stil-v-odnomu-b-h1
After I initially сommentеⅾ I seem to have clicked on the -Notify me when new comments
are added- checkbox and from now on еvery time a comment
is added I get four emails with the same comment.
Perhaps theгe is a ѡay you can remove me from that service?
Appreciate it!
Hi friends, its wonderful article about educationand fully defined, keep it up all the time.
https://squareblogs.net/abregedpnx/h1-b-iak-zaminiti-sklo-far-samostiino-posibnik-vid-farfarlight-b-h1
Hello colleagues, nice paragraph and good urging commented here, I am really enjoying by these.
https://squareblogs.net/typhanhntw/h1-b-osoblivosti-ta-perevagi-vikoristannia-skla-far-vid-farfarlight-u-vashomu
57. Клининговая компания в Челябинске предоставляет услуги по уборке отелей и гостиниц, включая смену постельного белья, уборку номеров, мойку санузлов, вынос мусора и другие работы для создания уютной и чистой атмосферы для гостей.
Клининговая компания Челябинск .
Добрый день всем!
На нашем сайте вы можете заказать диплом Вуза с постоплатой и круглосуточной поддержкой.
https://diplomany-asx.ru/
Купить диплом с печатью и пломбой
купить диплом колледжа [url=https://diplom-msk.ru/]https://diplom-msk.ru/[/url] .
Надежно купить диплом без обмана
купить диплом http://www.diplom-msk.ru/ .
Купить диплом с доставкой
купить диплом Вуза http://diplom-msk.ru/ .
It’s very simple to find out any topic on net as compared to books, as I found this post at this website.
I know this website provides quality dependent content and additional information, is there any other web site which provides these kinds of data in quality?
game1kb.com
그래서… 불안과 불안이 커져 그의 마음에 퍼지기 시작했습니다.
프라그마틱의 게임은 언제나 풍부한 그래픽과 흥미진진한 플레이로 유명해요. 이번에 새로운 게임이 나왔나요?
프라그마틱 홈페이지
프라그마틱의 게임은 항상 다양한 테마로 놀라워요. 이 사이트에서 더 자세한 정보를 찾아보세요!
https://dqwdqdw.weebly.com/
https://giftorie.com/link/
https://gdckandukur.com/link/
k8 カジノ レート
素晴らしい記事でした!とても感動しました。
Приветики!
Приобретите диплом института или колледжа недорого с доставкой по РФ и гарантией качества без предварительной оплаты!
Приобретите диплом Гознака с гарантированной подлинностью и доставкой по всей России без предоплаты.
https://images.google.com.kw/url?q=https://originality-diploman.com
https://sites.google.com/view/sunwinclcomhttps://premialnie-diplomany.com
https://toolbarqueries.google.com/url?q=https://diploms-originalniy.com
https://www.google.co.cr/url?q=https://rudiplomisty.com
http://images.google.vg/url?q=https://premialnie-diplomany.com
Ηi there, everүtһing is going well here and ofcourse every
one is sharing data, that’s truly fine, keep up writing.
Доброго всем дня!
Закажите диплом института или техникума по доступной цене с гарантированной доставкой в любой уголок России без предоплаты!
Купите диплом института или колледжа с гарантированной подлинностью и доставкой по России без предоплаты – удобно, безопасно, выгодно!
https://maps.google.co.mz/url?q=http://kazdiplom.com
https://images.google.com.au/url?sa=t&url=https://russiany-diplomans.com
https://clients1.google.bs/url?sa=t&url=https://frees-diplom.com
http://images.google.tg/url?q=https://russiany-diplomans.com
https://cse.google.cd/url?sa=t&url=http://diploman-spb24.com
Привет, дорогой читатель!
Наши услуги позволят вам заказать документы об образовании всех ВУЗов России с доставкой по РФ и оплатой после получения – надежно и удобно!
Наши услуги помогут вам приобрести диплом ВУЗа с доставкой по всей России и без предварительной оплаты – это быстро и надежно!
https://clients1.google.bj/url?sa=t&url=http://diploman-spb24.com
https://alt1.toolbarqueries.google.com.gt/url?q=https://lands-diploms.com
http://maps.google.is/url?q=https://russdiplomag.com
https://clients1.google.kg/url?sa=t&url=https://russdiplomag.com
https://cse.google.com.tw/url?q=https://russdiplomag.com
В нашем мире, где диплом является началом удачной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа переоценить невозможно. Ведь диплом открывает дверь перед каждым человеком, который собирается вступить в профессиональное сообщество или продолжить обучение в университете.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы сможете заказать диплом, что будет отличным решением для человека, который не смог закончить обучение или утратил документ. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам, чтобы в результате получился 100% оригинальный документ.
Преимущество такого подхода состоит не только в том, что можно максимально быстро получить диплом. Процесс организовывается удобно и легко, с нашей поддержкой. Начав от выбора подходящего образца документа до точного заполнения личных данных и доставки по России — все находится под полным контролем качественных мастеров.
Для всех, кто ищет оперативный способ получения необходимого документа, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать длительного обучения и сразу перейти к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
http://seneka-vl.ru/index.php?subaction=userinfo&user=atazonofu
http://masostyle.free.fr/shoutbox/shoutbox.php?token=3a9bf1026727cfd0bb37a76b6f93b013203d0f2cbca310b53d2ee5452cb5251d
http://logo-def.ru/userinfo.php?uid=26910
http://blog.entonz.com/2020/03/dicas-rapidas-para-se-tornar-um-bom.html
http://academijacrimea.ru/index.php?subaction=userinfo&user=otolu
На сегодняшний день, когда диплом становится началом успешной карьеры в любой области, многие стараются найти максимально простой путь получения образования. Факт наличия официального документа об образовании переоценить просто невозможно. Ведь именно диплом открывает двери перед людьми, стремящимися вступить в профессиональное сообщество или учиться в любом университете.
В данном контексте наша компания предлагает максимально быстро получить этот необходимый документ. Вы имеете возможность купить диплом старого или нового образца, что будет отличным решением для всех, кто не смог завершить образование или потерял документ. Все дипломы выпускаются аккуратно, с максимальным вниманием ко всем деталям, чтобы в результате получился продукт, полностью соответствующий оригиналу.
Преимущество подобного подхода состоит не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается удобно, с нашей поддержкой. Начав от выбора требуемого образца диплома до точного заполнения персональных данных и доставки в любой регион России — все будет находиться под абсолютным контролем качественных специалистов.
Всем, кто ищет максимально быстрый способ получения необходимого документа, наша услуга предлагает выгодное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и сразу перейти к достижению личных целей, будь то поступление в университет или старт карьеры.
http://www.diplom-net.ru
Thanks! An abundance of facts!
My web-site :: https://april-massage.icu/changwon-massage
В нашем мире, где диплом – это начало отличной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения образования. Факт наличия официального документа сложно переоценить. Ведь именно он открывает двери перед любым человеком, который стремится начать трудовую деятельность или продолжить обучение в университете.
В данном контексте мы предлагаем быстро получить этот необходимый документ. Вы можете приобрести диплом, и это будет удачным решением для всех, кто не смог закончить образование, потерял документ или желает исправить свои оценки. дипломы изготавливаются аккуратно, с максимальным вниманием ко всем элементам. В итоге вы сможете получить документ, полностью соответствующий оригиналу.
Превосходство этого решения состоит не только в том, что можно быстро получить свой диплом. Процесс организован комфортно, с нашей поддержкой. От выбора подходящего образца до грамотного заполнения персональной информации и доставки по России — все под абсолютным контролем квалифицированных мастеров.
Всем, кто ищет максимально быстрый способ получения необходимого документа, наша компания готова предложить выгодное решение. Купить диплом – это значит избежать продолжительного процесса обучения и не теряя времени перейти к личным целям: к поступлению в университет или к началу трудовой карьеры.
http://www.uepd.de/w3/doku.php?id=landsdiploms
http://www.kletki-opt.ru/product/paw-grill/reviews/
http://veda.adrian.ru/doku.php?id=rudiplomisty
http://www.anaptixiaki.gr/component/k2/item/35-2017-05-15-22-25-21
http://www.xn--5-htbd2chx.xn--p1ai/product/igra-stol-forma/reviews/
Hello, all is going sound here and ofcourse every one is sharing data, that’s in fact good, keep up writing.
https://cinemarus.ru/
buysteriodsonline.com
이 세계에서의 지위와 자격으로 개인적인 복수를 한 Jiao Fang은 계속해서 정상에 올랐습니다.
В современном мире, где диплом – это начало отличной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Наличие официального документа переоценить невозможно. Ведь именно он открывает двери перед любым человеком, желающим начать профессиональную деятельность или учиться в любом ВУЗе.
В данном контексте мы предлагаем быстро получить этот необходимый документ. Вы можете приобрести диплом, что является выгодным решением для всех, кто не смог завершить обучение, утратил документ или желает исправить свои оценки. Все дипломы выпускаются аккуратно, с максимальным вниманием ко всем нюансам, чтобы в итоге получился полностью оригинальный документ.
Преимущества подобного подхода заключаются не только в том, что можно оперативно получить свой диплом. Процесс организовывается комфортно, с профессиональной поддержкой. От выбора подходящего образца диплома до правильного заполнения личных данных и доставки в любое место России — все под абсолютным контролем опытных мастеров.
Для тех, кто хочет найти максимально быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к достижению своих целей, будь то поступление в университет или старт удачной карьеры.
http://engelsbrunn.de/index.php?option=com_easybookreloaded
http://mebel-omega.ru/product/tumba-prikrovatnaja-omega-k-s-dekorom-kamelija/reviews/
http://www.newrancho.ru/2014/01/blog-post.html
http://getamped.yxhi.com/doku.php?id=eonlinediplom
http://www.ntsr.info/forum/user/91023/
Приветики!
У нас в компании Вы можете купить диплом Гознак со скидкой, гарантией и доставкой в любой город РФ
Получите диплом ВУЗа без предварительной оплаты с доставкой по всей России – просто, надежно, выгодно!
https://clients1.google.com.ua/url?sa=t&url=https://originality-diplomiki.com
https://maps.google.co.za/url?sa=t&url=http://volga-diplomy.com
https://www.google.se/url?sa=t&url=http://kazdiplom.com
http://maps.google.cg/url?q=https://diploms-originalniy.com
https://sandbox.google.com/url?q=https://russdiplomag.com
В нашем обществе, где диплом – это начало отличной карьеры в любом направлении, многие стараются найти максимально простой путь получения образования. Наличие официального документа переоценить просто невозможно. Ведь диплом открывает двери перед любым человеком, который собирается вступить в профессиональное сообщество или продолжить обучение в любом ВУЗе.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы сможете приобрести диплом старого или нового образца, и это становится выгодным решением для человека, который не смог завершить обучение, утратил документ или хочет исправить свои оценки. Каждый диплом изготавливается с особой тщательностью, вниманием ко всем элементам, чтобы в итоге получился продукт, максимально соответствующий оригиналу.
Плюсы такого решения состоят не только в том, что вы сможете быстро получить свой диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора необходимого образца до консультаций по заполнению персональных данных и доставки по стране — все под абсолютным контролем квалифицированных специалистов.
В итоге, для тех, кто хочет найти максимально быстрый способ получения требуемого документа, наша услуга предлагает отличное решение. Заказать диплом – это значит избежать длительного процесса обучения и не теряя времени переходить к достижению личных целей, будь то поступление в ВУЗ или начало трудовой карьеры.
http://golpro.jp/userinfo.php?uid=13155#
https://clicktr.com/blog/yeni-baslayanlar-icin-affiliate-marketing/?unapproved=58364&moderation-hash=964ba0ef77b5640908b9330570abd4d1#comment-58364
http://www.atoe.co.kr/bbs/board.php?bo_table=free&wr_id=166954
http://petkit.com.cn/member/index.php?uid=uryqunaw&action=viewarchives&aid=6598
http://www.sarahdeluxe.com/2014/05/haul-and-review-makeup-revolution.html
I know this site offers quality based articles and extra stuff, is there any other web page which provides these kinds of stuff in quality?
Hi there! I’m at work browsing your blog from my new iphone 4! Just wanted to say I love reading your blog and look forward to all your posts! Keep up the excellent work!
http://diplom07.ru
В наше время, когда диплом является началом удачной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Факт наличия документа об образовании переоценить попросту невозможно. Ведь диплом открывает двери перед любым человеком, который стремится вступить в профессиональное сообщество или учиться в университете.
В данном контексте мы предлагаем максимально быстро получить любой необходимый документ. Вы можете заказать диплом, и это становится отличным решением для всех, кто не смог закончить обучение, потерял документ или хочет исправить свои оценки. Каждый диплом изготавливается с особой аккуратностью, вниманием к мельчайшим элементам. На выходе вы сможете получить полностью оригинальный документ.
Превосходство такого решения состоит не только в том, что можно оперативно получить диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начав от выбора подходящего образца документа до консультации по заполнению личной информации и доставки в любой регион страны — все под полным контролем квалифицированных специалистов.
Всем, кто пытается найти быстрый и простой способ получения необходимого документа, наша компания предлагает отличное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
https://www.alvas.ru/forum/showthread.php?p=103736#post103736
http://mail.newlifekpc.org/bbs/board.php?bo_table=free&wr_id=6183167
http://m1bar.com/index.php?subaction=userinfo&user=ulirebag
https://yozhstudio.ru/2019/09/08/%d0%bf%d1%80%d0%b5%d0%b8%d0%bc%d1%83%d1%89%d0%b5%d1%81%d1%82%d0%b2%d0%b0-%d0%b8%d0%bd%d0%b4%d0%b8%d0%b2%d0%b8%d0%b4%d1%83%d0%b0%d0%bb%d1%8c%d0%bd%d0%be%d0%b3%d0%be-%d0%bf%d0%be%d1%88%d0%b8%d0%b2%d0%b0/?unapproved=184912&moderation-hash=ae1a9c36dfa4d1e49a247032c897fa63#comment-184912
http://elkin-geo.com/index.php?option=com_akobook
I do not know whether it’s just me or if everyone else encountering problems with your site. It looks like some of the written text on your posts are running off the screen. Can someone else please comment and let me know if this is happening to them as well? This could be a problem with my browser because I’ve had this happen previously. Thanks
В современном мире, где диплом становится началом отличной карьеры в любой сфере, многие пытаются найти максимально простой путь получения качественного образования. Наличие официального документа об образовании сложно переоценить. Ведь именно диплом открывает двери перед людьми, желающими вступить в сообщество профессионалов или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы сможете приобрести диплом старого или нового образца, что будет отличным решением для человека, который не смог завершить образование, утратил документ или желает исправить свои оценки. Все дипломы изготавливаются с особой аккуратностью, вниманием ко всем элементам, чтобы в результате получился полностью оригинальный документ.
Преимущество подобного решения состоит не только в том, что можно быстро получить диплом. Весь процесс организован комфортно и легко, с профессиональной поддержкой. Начав от выбора необходимого образца до консультаций по заполнению персональных данных и доставки в любое место России — все будет находиться под абсолютным контролем наших специалистов.
В результате, для тех, кто ищет быстрый способ получения требуемого документа, наша компания может предложить выгодное решение. Купить диплом – значит избежать долгого процесса обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в ВУЗ или начало успешной карьеры.
http://www.ab-diplom.ru
В нашем обществе, где диплом – это начало удачной карьеры в любой области, многие ищут максимально быстрый и простой путь получения образования. Факт наличия документа об образовании трудно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим вступить в сообщество профессионалов или учиться в ВУЗе.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы сможете заказать диплом, что является отличным решением для всех, кто не смог завершить образование, потерял документ или хочет исправить плохие оценки. дипломы выпускаются с особой тщательностью, вниманием ко всем элементам, чтобы в итоге получился документ, 100% соответствующий оригиналу.
Превосходство этого решения заключается не только в том, что вы оперативно получите диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора подходящего образца документа до консультации по заполнению персональной информации и доставки в любое место страны — все находится под полным контролем наших мастеров.
Таким образом, для всех, кто пытается найти максимально быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени перейти к достижению собственных целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://www.ab-diplom.ru
Great post however , I was wanting to know if you could write a litte more on this topic? I’d be very grateful if you could elaborate a little bit further. Many thanks!
http://www.server-attestats.com
thephotoretouch.com
주인이 성공하지 못하면 장 여왕은 당연히 예의 바르지 않을 것입니다.
Thanks , I have just been searching for info about this subject for a while and yours is the greatest I’ve found out till now. However, what in regards to the conclusion? Are you positive about the supply?
https://server-attestats.com /
geinoutime.com
Xiao Jing은 “예, 예, 예, 폐하의 말씀은 매우 사실입니다. “라고 반복해서 말했습니다.
프라그마틱 게임은 iGaming 업계에서 혁신적인 모바일 중심의 콘텐츠를 선보이며 고객에게 탁월한 경험을 제공합니다.
프라그마틱 슬롯
프라그마틱은 항상 훌륭한 게임을 만들어냅니다. 이번에 새롭게 출시된 게임은 정말 기대되는데요!
https://www.12315db.cn/
http://www.iavstudios.com/
https://bidforfix.com/hot/
Добрый день всем!
Закажите диплом ВУЗа с гарантированной подлинностью и доставкой по всей России без предоплаты – просто, удобно, безопасно!
Приобретите диплом Вуза с доставкой по России без предоплаты и полной уверенностью в его подлинности.
https://toolbarqueries.google.rs/url?sa=i&url=https://premialnie-diplomansy.com
https://toolbarqueries.google.co.il/url?q=https://originality-diplomiki.com
http://maps.google.hr/url?q=http://diploman-spb24.com
https://maps.google.com.ng/url?q=https://russiany-diplomans.com
http://maps.google.co.cr/url?sa=t&url=https://diploms-originalniy.com
k8 ゲーム
非常に興味深く、思考を刺激する内容でした。また訪れます。
This blog was… how do you say it? Relevant!! Finally I’ve found something that helped me. Appreciate it!
В нашем обществе, где диплом является началом отличной карьеры в любой сфере, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Важность наличия документа об образовании сложно переоценить. Ведь диплом открывает дверь перед всеми, кто собирается вступить в сообщество профессионалов или продолжить обучение в университете.
В данном контексте наша компания предлагает очень быстро получить этот необходимый документ. Вы имеете возможность приобрести диплом, что будет отличным решением для человека, который не смог завершить образование или потерял документ. дипломы производятся с особой тщательностью, вниманием ко всем деталям. На выходе вы сможете получить продукт, 100% соответствующий оригиналу.
Плюсы данного подхода заключаются не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается комфортно, с нашей поддержкой. От выбора требуемого образца до консультаций по заполнению личных данных и доставки по России — все будет находиться под полным контролем качественных специалистов.
Для тех, кто ищет быстрый и простой способ получения требуемого документа, наша услуга предлагает выгодное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и сразу переходить к важным целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://cmsthemefinder.com/userinfo.php?uid=35198#
http://eydosdigital.com/en/component/k2/item/8
http://esitscale.com/forum/profile.php?mode=viewprofile&u=38778
http://gkwin.net/bbs/board.php?bo_table=free&wr_id=192678
http://book-club.rggu.ru/2013/10/blog-post.html
Hello friends, how is the whole thing, and what you would like to say on the topic of this article, in my view its in fact awesome for me.
http://arusak-attestats.ru
Pretty! This has been a really wonderful post. Thank you for supplying this info.
http://arusak-attestats.ru
Привет всем!
Закажите диплом ВУЗа по выгодной цене с доставкой в любой город России без предоплаты – надежно и выгодно!
Купите диплом института или колледжа без предоплаты с гарантированной доставкой в любой уголок России.
https://cse.google.co.zm/url?sa=t&url=http://volga-diplomy.com
http://maps.google.com.bz/url?q=https://diploms-originalniy.com
https://images.google.ca/url?sa=t&url=https://diploms-originalniy.com
https://images.google.com.kh/url?q=http://kazdiplom.com
https://clients1.google.com.kw/url?sa=t&url=https://premialnie-diplomany.com
Pretty! This has been an extremely wonderful article. Thank you for providing this info.
Greetings! Very useful advice in this particular post! It’s the little changes that will make the most significant changes. Thanks a lot for sharing!
tvlore.com
물론 이 부유한 상인들이 공부하는 것만으로는 충분하지 않습니다.
It’s genuinely very complex in this busy life to listen news on Television, so I just use world wide web for that reason, and get the latest information.
Wonderful work! This is the tүpe of informɑtion that are supposed to be shared around the web.
Disgrace on the seeк engines for no longer positioning this submit higher!
Come on over and visit my sitе . Thanks =)
Pretty! This was an incredibly wonderful article. Thanks for supplying this information.
Really many of helpful tips.
Feel free to surf to my page: girls shoes size 2.5 youth sneakers (https://www.amazon.com/gp/search?ie=UTF8&tag=free0cff-20&linkCode=ur2&linkId=7e3593d1f30993c9bd218e4dd9a364c4&camp=1789&creative=9325&index=electronics&keywords=32%20Inch%20Smart%20TV)
This is very interesting, You are a very skilled blogger. I’ve joined your feed and look forward to seeking more of your great post. Also, I’ve shared your site in my social networks!
game1kb.com
Zhu Houzhao는 참을 수 없었습니다. “Old Fang, Liu Banban은 그런 사람이 아닙니다.”
Привет, любители удивительных приключений! Желаю рассказать своим великолепием от онлайн заведения “Селектор онлайн казино”. Это не просто сайт для игр, это абсолютный космос переживаний и забав! Здесь я нашел все, что мне необходимо: различие игр, щедрые бонусы и скоростные выплаты. Но самое – это окружение, которая преобладает здесь. Имитированные приключения, восхитительные турниры и доброжелательное сообщество игроков делают каждую игру неизученной! Присоединяйтесь к “официальный сайт казино селектор” и прыгайте в космос занимательных развлечений!
Thanks for every other wonderful article. The place else may anybody get that type of info in such an ideal approach of writing? I’ve a presentation subsequent week, and I’m on the search for such info.
Заказать купленный диплом, через интернет.
Как купить диплом без риска, без заморочек.
Какой диплом купить, важная информация.
Почему выгодно купить диплом, рассказываем.
Где купить диплом без проблем, без риска.
Лучшие предложения по покупке диплома, важные моменты.
Купить диплом срочно и недорого, подробности на сайте.
Почему стоит купить диплом, важная информация.
Где купить действующий диплом, подробности на сайте.
Покупка диплома: безопасность и качество, лучшие условия.
Как выбрать диплом, лучшие цены.
Как купить диплом срочно, без рисков.
Почему стоит заказать диплом, подробности у нас.
Дипломы на всех условиях, лучшие предложения.
Легальная покупка дипломов безопасно, гарантированная доставка.
Почему стоит купить диплом здесь и сейчас, интересные варианты.
Заказать диплом онлайн без проблем, подробности здесь.
Почему стоит заказать диплом на нашем сайте, гарантированный результат.
купить диплом http://7arusak-diploms.com/ .
iGaming 분야에서 혁신적이고 표준화된 콘텐츠를 제공하는 최신 프라그마틱 게임은 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 통해 다양한 엔터테인먼트를 제공합니다.
프라그마틱 슬롯 무료 체험
프라그마틱의 게임은 항상 최고 수준의 퀄리티를 유지하고 있어요. 이번에도 그런 훌륭한 게임을 만났나요?
https://accountantslink.net/link/
https://henakeah.com/link/
https://www.cicoresky.com
Привет всем!
Получите документ ВУЗа без предоплаты и доставку в руки по России – просто, удобно, безопасно!
http://www.eonline-diploms.com
Do you have any video of that? I’d love to find out more details.
http://www.diplomy-originaly.com
Приветики!
Приобретите диплом института или колледжа с гарантией качества и доставкой по России без предоплаты.
eonline-diploms.com
Greetings I am so excited I found your web site, I really found you by mistake, while I was researching on Digg for something else, Anyhow I am here now and would just like to say thanks for a remarkable post and a all round interesting blog (I also love the theme/design), I don’t have time to read it all at the minute but I have book-marked it and also included your RSS feeds, so when I have time I will be back to read a lot more, Please do keep up the great work.
http://www.diplomy-originaly.com
I absolutely love your blog and find almost all of your post’s to be what precisely I’m looking for. can you offer guest writers to write content available for you? I wouldn’t mind publishing a post or elaborating on a number of the subjects you write about here. Again, awesome site!
Hello there I am so thrilled I found your website, I really found you by error, while I was looking on Google for something else, Nonetheless I am here now and would just like to say many thanks for a tremendous post and a all round entertaining blog (I also love the theme/design), I don’t have time to read through it all at the moment but I have book-marked it and also added in your RSS feeds, so when I have time I will be back to read much more, Please do keep up the great work.
http://diplomy-originaly.com
Привет всем!
Закажите российский диплом у нас по доступной цене с гарантией прохождения проверок и доставкой по РФ.
Как заказать и купить диплом Вуза недорого без предоплаты на сайте? Доставка в любую точку России
http://www.diploms-asx.com
k8 カジノ 系列
情報が豊富で読み応えがありました。非常に有益です。
Hello to every body, it’s my first go to see of this blog; this website includes awesome and truly excellent information in favor of visitors.
https://chiefsense71.werite.net/novi-linzi-dlia-far-vashogo-avto-iakist-ta-nadiinist-na-lemarix-ua
Thank you, I’ve just been searching for information approximately this subject for a long time and yours is the best I have found out so far. However, what in regards to the conclusion? Are you certain in regards to the source?
https://rentry.co/23qvvona
Thanks , I have recently been looking for information about this topic for a while and yours is the best I have found out so far. However, what in regards to the bottom line? Are you certain in regards to the supply?
http://kvitka-ua.com/linzy-u-fary-dlya-vashogo-avtomobilya
What’s up, after reading this awesome paragraph i am as well delighted to share my knowledge here with friends.
http://kvitka-ua.com/linzy-u-fary-dlya-vashogo-avtomobilya
Hello everyone, it’s my first pay a quick visit at this web page, and post is genuinely fruitful in favor of me, keep up posting these content.
http://kvitka-ua.com/linzy-u-fary-dlya-vashogo-avtomobilya
You really make it seem so easy with your presentation but I find this topic to be actually something that I think I would never understand. It seems too complicated and very broad for me. I’m looking forward for your next post, I will try to get the hang of it!
geinoutime.com
Zhou는 깜짝 놀랐고 Zhu Houzhao를 돕기 위해 서둘러 앞으로 나아갔습니다.
mikaspa.com
그는 무의식적으로 비단 조각을 꺼낸 다음 “바람은 북쪽으로, 바람은 가볍습니다. “라고 말했습니다.
Привет, дорогой читатель!
Наши услуги помогут вам приобрести диплом ВУЗа с доставкой по всей России без предварительной оплаты – быстро и надежно!
diplomanc-russia24.ru
Заказать купленный диплом, через интернет.
Легальный способ купить диплом, подробности здесь.
Заказать официальный документ о образовании, советы от профессионалов.
Преимущества покупки диплома, все секреты.
Где купить диплом без проблем, с гарантией.
Как выбрать диплом для покупки, важные моменты.
Купить диплом срочно и недорого, спешите.
Скрытая покупка дипломов, проверенные решения.
Лучшие дипломы для покупки, срочно и выгодно.
Как купить диплом онлайн, лучшие условия.
Как выбрать диплом, гарантированное качество.
Легальная покупка дипломов, без рисков.
Купить диплом без обмана, подробности у нас.
Купить диплом просто, подробности на сайте.
Купить диплом без риска, подробности у нас.
Купить дипломы легко, подробности на сайте.
Официальная покупка диплома, важные детали.
Как купить диплом безопасно и быстро, подробности у нас.
купить диплом 7arusak-diploms.com .
Its like you read my mind! You seem to know a lot about this, like you wrote the book in it or something. I think that you can do with some pics to drive the message home a little bit, but other than that, this is excellent blog. A great read. I’ll definitely be back.
https://cutt.ly/tero3oej
I’m gone to tell my little brother, that he should also go to see this website on regular basis to take updated from hottest information.
https://cutt.ly/Eero9Lrk
Do you have a spam problem on this blog; I also am a blogger, and I was curious about your situation; we have created some nice procedures and we are looking to swap techniques with others, be sure to shoot me an email if interested.
Hi, after reading this awesome piece of writing i am too glad to share my knowledge here with colleagues.
Thanks for sharing your thoughts about %meta_keyword%. Regards
Fastidious answer back in return of this question with genuine arguments
and telling the whole thing on the topic of that.
Feel free to visit my web-site … facebook vs eharmony to find love online
Hello there! This is my first visit to your blog! We are a team of volunteers and starting a new initiative in a community in the same niche. Your blog provided us useful information to work on. You have done a extraordinary job!
프라그마틱 플레이는 33개 언어와 다양한 화폐를 지원하여, 250개 이상의 게임으로 이루어진 슬롯 포트폴리오를 전 세계 시장에 제공합니다.
PG 소프트
프라그마틱 관련 글 읽는 것이 정말 즐거웠어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 이야기 나누면서 더 많은 지식을 쌓아가요!
https://www.12315hc.cn/
https://my-site-104812-108385.weebly.com/
https://www.12315jq.cn/
zanetvize.com
이 장엄한 태도와 사람을 진지하게 대하지 않는 이 목소리를 보라.
В нашем обществе, где диплом становится началом успешной карьеры в любой отрасли, многие ищут максимально быстрый путь получения качественного образования. Наличие официального документа об образовании трудно переоценить. Ведь именно он открывает двери перед людьми, желающими вступить в сообщество профессионалов или продолжить обучение в высшем учебном заведении.
Мы предлагаем очень быстро получить этот необходимый документ. Вы можете купить диплом нового или старого образца, и это будет удачным решением для человека, который не смог завершить образование, потерял документ или желает исправить плохие оценки. дипломы изготавливаются аккуратно, с максимальным вниманием к мельчайшим нюансам. В итоге вы сможете получить продукт, 100% соответствующий оригиналу.
Преимущества данного подхода заключаются не только в том, что вы сможете оперативно получить диплом. Процесс организован комфортно, с профессиональной поддержкой. От выбора требуемого образца диплома до консультации по заполнению личной информации и доставки в любой регион России — все будет находиться под полным контролем наших мастеров.
Таким образом, для всех, кто ищет быстрый способ получения требуемого документа, наша компания готова предложить выгодное решение. Заказать диплом – это значит избежать продолжительного обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в ВУЗ или начало профессиональной карьеры.
http://proobzh.ru
Сегодня, когда диплом становится началом отличной карьеры в любой отрасли, многие ищут максимально простой путь получения качественного образования. Факт наличия официального документа трудно переоценить. Ведь именно диплом открывает двери перед всеми, кто хочет вступить в профессиональное сообщество или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете приобрести диплом старого или нового образца, что будет выгодным решением для всех, кто не смог закончить образование, утратил документ или хочет исправить плохие оценки. диплом изготавливается аккуратно, с особым вниманием ко всем деталям, чтобы в итоге получился 100% оригинальный документ.
Преимущества подобного решения заключаются не только в том, что вы сможете оперативно получить свой диплом. Процесс организован удобно, с нашей поддержкой. От выбора нужного образца диплома до правильного заполнения личной информации и доставки по стране — все под полным контролем качественных специалистов.
Всем, кто ищет максимально быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать долгого обучения и не теряя времени перейти к достижению личных целей, будь то поступление в университет или начало карьеры.
edu-nt.ru
k8 カジノ 入金 反映
素晴らしい洞察と分析でした。大変参考になります。
Today, while I was at work, my cousin stole my apple ipad and tested to see if it can survive a 25 foot drop, just so she can be a youtube sensation. My iPad is now destroyed and she has 83 views. I know this is entirely off topic but I had to share it with someone!
http://edu-digest.ru
В нашем обществе, где диплом является началом успешной карьеры в любой области, многие ищут максимально простой путь получения образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед всеми, кто стремится вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы можете купить диплом, и это будет удачным решением для человека, который не смог завершить обучение, потерял документ или хочет исправить свои оценки. диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам, чтобы в итоге получился полностью оригинальный документ.
Преимущество этого решения заключается не только в том, что можно максимально быстро получить диплом. Весь процесс организован удобно и легко, с нашей поддержкой. От выбора подходящего образца до консультации по заполнению персональной информации и доставки в любой регион России — все находится под абсолютным контролем наших мастеров.
В результате, для всех, кто хочет найти оперативный способ получения требуемого документа, наша компания может предложить отличное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к личным целям: к поступлению в университет или к началу успешной карьеры.
dof-edu.ru
В нашем обществе, где диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Важность наличия официального документа переоценить невозможно. Ведь диплом открывает двери перед людьми, стремящимися начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте мы предлагаем оперативно получить этот необходимый документ. Вы можете приобрести диплом, что будет выгодным решением для человека, который не смог завершить образование или потерял документ. дипломы производятся аккуратно, с особым вниманием к мельчайшим деталям, чтобы на выходе получился полностью оригинальный документ.
Превосходство этого подхода заключается не только в том, что вы сможете максимально быстро получить диплом. Процесс организован просто и легко, с нашей поддержкой. Начав от выбора подходящего образца до консультации по заполнению персональных данных и доставки в любое место страны — все под полным контролем квалифицированных мастеров.
В итоге, для тех, кто пытается найти быстрый способ получить требуемый документ, наша компания готова предложить выгодное решение. Приобрести диплом – значит избежать продолжительного обучения и сразу переходить к достижению собственных целей, будь то поступление в ВУЗ или начало карьеры.
http://www.proobzh.ru
В нашем обществе, где диплом становится началом успешной карьеры в любой сфере, многие ищут максимально простой путь получения образования. Факт наличия официального документа об образовании переоценить просто невозможно. Ведь диплом открывает двери перед каждым человеком, который желает начать трудовую деятельность или учиться в университете.
В данном контексте наша компания предлагает быстро получить любой необходимый документ. Вы можете купить диплом нового или старого образца, и это будет отличным решением для человека, который не смог завершить образование или потерял документ. Любой диплом изготавливается аккуратно, с особым вниманием к мельчайшим нюансам, чтобы в результате получился продукт, полностью соответствующий оригиналу.
Плюсы подобного подхода состоят не только в том, что вы оперативно получите диплом. Весь процесс организован комфортно, с нашей поддержкой. От выбора нужного образца до консультаций по заполнению личной информации и доставки в любое место России — все под абсолютным контролем наших специалистов.
Для всех, кто ищет оперативный способ получить требуемый документ, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и сразу перейти к своим целям, будь то поступление в ВУЗ или начало успешной карьеры.
proobzh.ru
Hello! I realize this is sort of off-topic but I needed to ask. Does operating a well-established website such as yours take a large amount of work? I’m brand new to running a blog however I do write in my journal on a daily basis. I’d like to start a blog so I can easily share my personal experience and feelings online. Please let me know if you have any suggestions or tips for new aspiring bloggers. Appreciate it!
http://www.edu-digest.ru
В нашем мире, где диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения образования. Наличие официального документа об образовании сложно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим начать профессиональную деятельность или учиться в любом ВУЗе.
Наша компания предлагает очень быстро получить этот необходимый документ. Вы имеете возможность заказать диплом нового или старого образца, что становится отличным решением для человека, который не смог завершить образование или утратил документ. Все дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим элементам. На выходе вы сможете получить документ, максимально соответствующий оригиналу.
Превосходство данного решения состоит не только в том, что можно оперативно получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начиная от выбора подходящего образца до точного заполнения личных данных и доставки по стране — все находится под абсолютным контролем качественных мастеров.
Для тех, кто ищет быстрый и простой способ получения требуемого документа, наша услуга предлагает отличное решение. Купить диплом – значит избежать долгого процесса обучения и не теряя времени переходить к своим целям, будь то поступление в университет или начало успешной карьеры.
http://edu-nt.ru/
В нашем обществе, где диплом – это начало успешной карьеры в любом направлении, многие стараются найти максимально простой путь получения образования. Необходимость наличия документа об образовании сложно переоценить. Ведь именно диплом открывает дверь перед каждым человеком, который стремится вступить в сообщество профессиональных специалистов или продолжить обучение в любом институте.
Наша компания предлагает быстро получить этот необходимый документ. Вы сможете приобрести диплом старого или нового образца, и это будет отличным решением для всех, кто не смог завершить образование или потерял документ. Все дипломы производятся с особой аккуратностью, вниманием к мельчайшим элементам, чтобы на выходе получился полностью оригинальный документ.
Преимущества этого подхода состоят не только в том, что вы быстро получите свой диплом. Процесс организован удобно и легко, с профессиональной поддержкой. Начав от выбора требуемого образца диплома до консультаций по заполнению персональной информации и доставки в любой регион России — все под абсолютным контролем наших мастеров.
Для тех, кто пытается найти быстрый и простой способ получить необходимый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и сразу переходить к важным целям: к поступлению в ВУЗ или к началу трудовой карьеры.
http://edu-nt.ru/
В наше время, когда диплом – это начало отличной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа об образовании сложно переоценить. Ведь именно диплом открывает двери перед любым человеком, желающим начать профессиональную деятельность или продолжить обучение в университете.
Предлагаем оперативно получить этот важный документ. Вы можете приобрести диплом старого или нового образца, и это будет выгодным решением для всех, кто не смог завершить обучение, утратил документ или желает исправить плохие оценки. Каждый диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим деталям, чтобы на выходе получился 100% оригинальный документ.
Преимущества этого решения состоят не только в том, что можно быстро получить диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора требуемого образца до точного заполнения личной информации и доставки по России — все находится под полным контролем квалифицированных мастеров.
Для всех, кто хочет найти оперативный способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к своим целям, будь то поступление в университет или старт профессиональной карьеры.
http://www.dof-edu.ru
В современном мире, где диплом является началом отличной карьеры в любой сфере, многие стараются найти максимально быстрый и простой путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед людьми, стремящимися вступить в сообщество профессионалов или учиться в университете.
В данном контексте мы предлагаем максимально быстро получить любой необходимый документ. Вы имеете возможность купить диплом старого или нового образца, что становится выгодным решением для человека, который не смог завершить обучение, утратил документ или желает исправить плохие оценки. диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам, чтобы в итоге получился полностью оригинальный документ.
Плюсы данного решения заключаются не только в том, что можно максимально быстро получить свой диплом. Процесс организован удобно, с профессиональной поддержкой. Начиная от выбора подходящего образца документа до грамотного заполнения персональной информации и доставки по России — все будет находиться под абсолютным контролем качественных специалистов.
Всем, кто пытается найти максимально быстрый способ получения необходимого документа, наша компания предлагает выгодное решение. Купить диплом – это значит избежать долгого обучения и сразу перейти к достижению личных целей: к поступлению в университет или к началу трудовой карьеры.
krsk-school47.ru
В современном мире, где диплом – это начало отличной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Важность наличия официального документа переоценить просто невозможно. Ведь диплом открывает двери перед каждым человеком, который хочет вступить в профессиональное сообщество или учиться в высшем учебном заведении.
Мы предлагаем оперативно получить этот необходимый документ. Вы сможете приобрести диплом, что является отличным решением для всех, кто не смог завершить образование или утратил документ. дипломы выпускаются с особой аккуратностью, вниманием ко всем деталям. В итоге вы сможете получить 100% оригинальный документ.
Превосходство этого подхода заключается не только в том, что можно максимально быстро получить диплом. Процесс организован удобно и легко, с нашей поддержкой. От выбора подходящего образца до консультаций по заполнению личной информации и доставки по России — все будет находиться под абсолютным контролем наших мастеров.
В итоге, для всех, кто ищет быстрый способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – это значит избежать продолжительного обучения и не теряя времени перейти к достижению личных целей: к поступлению в университет или к началу успешной карьеры.
http://www.dof-edu.ru
nikontinoll.com
폐하가 새로 지어진 Chongwen Hall에서 Na Jiu의 연설을 듣고 있다는 것을 기억 한 것은 그가 궁전에 도착했을 때였습니다.
В современном мире, где диплом является началом отличной карьеры в любом направлении, многие пытаются найти максимально простой путь получения качественного образования. Наличие документа об образовании сложно переоценить. Ведь именно он открывает дверь перед людьми, стремящимися вступить в сообщество профессионалов или учиться в высшем учебном заведении.
Наша компания предлагает очень быстро получить этот необходимый документ. Вы имеете возможность заказать диплом, что является выгодным решением для человека, который не смог закончить образование, утратил документ или желает исправить свои оценки. дипломы производятся аккуратно, с максимальным вниманием ко всем элементам. В итоге вы сможете получить документ, полностью соответствующий оригиналу.
Превосходство такого решения состоит не только в том, что вы оперативно получите свой диплом. Процесс организовывается удобно и легко, с нашей поддержкой. Начав от выбора требуемого образца документа до консультации по заполнению персональных данных и доставки по России — все находится под полным контролем наших мастеров.
Всем, кто хочет найти максимально быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к достижению личных целей, будь то поступление в ВУЗ или старт профессиональной карьеры.
http://www.edu-nt.ru/
В нашем обществе, где диплом – это начало отличной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Наличие официального документа сложно переоценить. Ведь диплом открывает дверь перед всеми, кто стремится вступить в сообщество профессиональных специалистов или учиться в любом институте.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы сможете заказать диплом нового или старого образца, что становится выгодным решением для всех, кто не смог закончить обучение, потерял документ или хочет исправить плохие оценки. Все дипломы выпускаются с особой тщательностью, вниманием к мельчайшим деталям, чтобы в итоге получился полностью оригинальный документ.
Преимущество такого решения заключается не только в том, что вы быстро получите диплом. Процесс организовывается комфортно, с профессиональной поддержкой. Начиная от выбора подходящего образца до точного заполнения личных данных и доставки по стране — все под абсолютным контролем квалифицированных мастеров.
В результате, для тех, кто пытается найти быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать длительного обучения и сразу перейти к своим целям, будь то поступление в университет или начало удачной карьеры.
http://www.edu-nt.ru/
В нашем мире, где диплом – это начало успешной карьеры в любой отрасли, многие стараются найти максимально простой путь получения качественного образования. Факт наличия документа об образовании сложно переоценить. Ведь диплом открывает двери перед людьми, стремящимися вступить в профессиональное сообщество или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете заказать диплом нового или старого образца, и это будет отличным решением для человека, который не смог завершить образование или потерял документ. Все дипломы изготавливаются с особой тщательностью, вниманием к мельчайшим деталям. В итоге вы сможете получить полностью оригинальный документ.
Плюсы подобного решения состоят не только в том, что можно максимально быстро получить диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора необходимого образца до грамотного заполнения личных данных и доставки в любое место России — все под полным контролем качественных специалистов.
В результате, для всех, кто хочет найти оперативный способ получить необходимый документ, наша услуга предлагает отличное решение. Приобрести диплом – значит избежать продолжительного обучения и не теряя времени перейти к достижению собственных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
http://www.edu-nt.ru/
geinoutime.com
Alfonso는 즉시 정신을 차리고 쌍안경을 들어 올렸습니다.
Сегодня, когда диплом является началом успешной карьеры в любой области, многие пытаются найти максимально простой путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает двери перед любым человеком, который желает вступить в профессиональное сообщество или учиться в любом институте.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы сможете приобрести диплом, что становится отличным решением для всех, кто не смог завершить образование или утратил документ. Любой диплом изготавливается с особой тщательностью, вниманием ко всем нюансам, чтобы на выходе получился полностью оригинальный документ.
Превосходство данного подхода состоит не только в том, что можно быстро получить свой диплом. Весь процесс организовывается комфортно, с нашей поддержкой. От выбора необходимого образца документа до грамотного заполнения персональной информации и доставки в любое место страны — все находится под полным контролем качественных специалистов.
Таким образом, всем, кто ищет быстрый и простой способ получения необходимого документа, наша компания может предложить отличное решение. Заказать диплом – значит избежать продолжительного процесса обучения и не теряя времени переходить к своим целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://6landik-diploms.com/
Its like you read my mind! You seem to know so much about this, like you wrote the book in it or something. I think that you could do with some pics to drive the message home a little bit, but instead of that, this is excellent blog. An excellent read. I’ll certainly be back.
http://www.idiplomanys.com/
На сегодняшний день, когда диплом является началом успешной карьеры в любой сфере, многие стараются найти максимально быстрый и простой путь получения качественного образования. Важность наличия официального документа об образовании переоценить невозможно. Ведь именно он открывает дверь перед любым человеком, желающим вступить в сообщество профессионалов или учиться в каком-либо институте.
Предлагаем максимально быстро получить этот необходимый документ. Вы сможете заказать диплом нового или старого образца, и это является удачным решением для человека, который не смог завершить обучение, потерял документ или желает исправить свои оценки. диплом изготавливается аккуратно, с особым вниманием ко всем деталям. На выходе вы сможете получить документ, максимально соответствующий оригиналу.
Превосходство этого подхода заключается не только в том, что вы оперативно получите диплом. Процесс организовывается комфортно и легко, с нашей поддержкой. От выбора необходимого образца до грамотного заполнения персональной информации и доставки по России — все под абсолютным контролем качественных специалистов.
Всем, кто пытается найти оперативный способ получить необходимый документ, наша компания предлагает отличное решение. Приобрести диплом – значит избежать продолжительного обучения и сразу перейти к личным целям, будь то поступление в ВУЗ или старт удачной карьеры.
http://www.6landik-diploms.com/
В наше время, когда диплом – это начало успешной карьеры в любой отрасли, многие пытаются найти максимально быстрый и простой путь получения образования. Наличие документа об образовании переоценить невозможно. Ведь диплом открывает двери перед всеми, кто желает вступить в сообщество профессионалов или продолжить обучение в университете.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы сможете купить диплом нового или старого образца, что становится удачным решением для всех, кто не смог завершить образование или потерял документ. Любой диплом изготавливается аккуратно, с особым вниманием ко всем нюансам. В итоге вы получите 100% оригинальный документ.
Плюсы такого решения состоят не только в том, что вы сможете максимально быстро получить диплом. Весь процесс организован просто и легко, с профессиональной поддержкой. Начиная от выбора нужного образца до правильного заполнения личной информации и доставки по стране — все под полным контролем квалифицированных мастеров.
Всем, кто пытается найти быстрый и простой способ получить необходимый документ, наша компания может предложить выгодное решение. Купить диплом – это значит избежать длительного процесса обучения и не теряя времени перейти к достижению личных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://www.6landik-diploms.com/
iGaming에서 선도적인 프라그마틱 게임은 모바일 중심의 혁신적이고 표준화된 콘텐츠를 제공합니다.
프라그마틱 플레이
프라그마틱 슬롯에 대한 정보가 정말 유용했어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 새로운 내용을 찾아보세요. 함께 지식을 나누면 좋겠어요!
https://www.12315oj.cn/
https://www.cicoresky.com
https://supervil.com/hot/
onair2tv.com
무슨 일이 생기면 분명히 추한 죽음이 될 것입니다.
k8 カジノ バニー
実用性抜群の内容で、毎回学ぶことが多いです。
Very good content Thanks a lot.
My page; https://imediweb.it/anche-smartworking-nello-studio-del-dottore/
Как получить пропуск на МКАД, Срочное оформление пропуска на МКАД, Основные аспекты пропуска на МКАД, Пропуск на МКАД: какие документы нужны, полный гайд, Сколько стоит и как долго делается пропуск на МКАД, Как получить пропуск для поездок по МКАД, рекомендации, основные аспекты, шаг за шагом
Проверить пропуск на мкад Проверить пропуск на мкад .
game1kb.com
Zhou Zheng은 부끄러워 말문이 막히고 필사적으로 기침을하고 입술이 경련되어 소리를 내지 못했습니다.
Как получить пропуск на МКАД, Эффективные способы получения пропуска на МКАД, Основные аспекты пропуска на МКАД, Какие документы нужны для оформления пропуска на МКАД, Основные вопросы о пропуске на МКАД, Сколько стоит и как долго делается пропуск на МКАД, важные аспекты, Какие преимущества дает пропуск на МКАД, Сроки и порядок продления пропуска на МКАД, подробная информация
Проверить пропуск на мкад Проверить пропуск на мкад .
Секреты оформления пропуска на МКАД, Срочное оформление пропуска на МКАД, секреты, Пропуск на МКАД: какие документы нужны, Часто задаваемые вопросы о пропуске на МКАД, на которые нужно знать ответы, важные аспекты, Пропуск на МКАД: основные преимущества, важные детали, подробная информация
Пропуск на мкад Пропуск в москву .
프라그마틱에 대한 글 읽는 것이 흥미로웠어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 제공하고 있어요. 함께 발전하며 지식을 나눠봐요!
프라그마틱 게임
프라그마틱의 게임은 정말 다양한데, 최근에 출시된 것 중 어떤 게임이 가장 좋았나요? 공유해주세요!
https://www.iaz681.com
https://ycbgl.com/link/
https://0cdnara.com/hot/
5 шагов для успешного ремонта телевизора: полезные советы и рекомендации|Как не попасть на мошенников при ремонте телевизора: важные критерии отбора|Топ-5 распространенных проблем с телевизорами: как их решить быстро и эффективно|Эффективные методы самодиагностики телевизора: шаг за шагом|Цены на ремонт телевизоров: как не переплатить и получить качественный сервис|Гарантии качественного ремонта телевизора: проверенные способы и сервисные центры|Как выбрать лучшего мастера по ремонту телевизоров: важные критерии отбора|Профилактика и уход за телевизорами: как обеспечить долговечность и надежность|Что делать, если телевизор не включается: шаги по решению проблемы|Как подобрать качественные запчасти для телевизора: советы и рекомендации по выбору|Почему лучше доверить ремонт телевизора профессионалам: основные преимущества|Топ-5 ошибок при ремонте телевизора: как избежать неприятностей|Способы определения проблемы с телевизором: как быстро и эффективно выявить причины|Опасности и недостатки самостоятельного ремонта телевизора: важные моменты и советы|Как быстро и качественно отремонтировать телевизор: специалисты делятся советами
заправка картриджей Воркута ремонт телефонов .
zanetvize.com
깨끗한 관리와 웅변을 겨루는 것은 그가 잘하는 것이 아니다.
Hello there I am so happy I found your webpage, I really found you by error, while I was searching on Yahoo for something else, Regardless I am here now and would just like to say cheers for a remarkable post and a all round thrilling blog (I also love the theme/design), I don’t have time to look over it all at the moment but I have saved it and also included your RSS feeds, so when I have time I will be back to read a lot more, Please do keep up the great work.
знакомства в Украине
Море Ялта
k8 カジノ 本人確認 時間
素晴らしい内容と洞察力で、いつも感動しています。
bestmanualpolesaw.com
주자이모의 주례는 두 사람의 관계를 철저히 해명한 것이라고 볼 수 있다.
프라그마틱과 관련된 이 글 정말 잘 읽었어요! 더불어 저도 제 사이트에서 프라그마틱에 대한 새로운 정보를 공유 중이에요. 함께 나누면 더 좋을 것 같아요!
프라그마틱 슬롯
프라그마틱 관련 소식 항상 주시고 있어요! 또한, 제 사이트에서도 프라그마틱에 대한 정보를 얻을 수 있어요. 함께 발전하며 지식을 나눠봐요!
https://www.12315oq.cn/
https://www.b4closing.com
https://giftorie.com/link/
geinoutime.com
사실 그는 가장 전화를 걸 수 있는 사람이 바로 자기 자신이라는 사실을 잊고 있었다.
Свидетельство О Квалификации
Свидетельство О Квалификации
Предлагаем купить диплом техникума или колледжа на выгодных условиях в Перми, быстро и недорого. Если Вам необходимо купить диплом о высшем образовании (Москва), то Вам будет достаточно просто заполнить элементарную квалификацию заказа. Он обязательно учитывается во всех книгах учета учебного заведения. Нужен диплом высшего свидетельства дошкольного год окончания 2023 заочное отделение. Гознак, киржач, спецбланк дополнительно: к диплому прилагается вкладыш (приложение к диплому), мокрая печать. Образование понемногу теряет свою основную функцию, которая состоит в подготовке профессионалов. Свободный выбор института и профессии позволит самостоятельно подобрать диплом, который будет соответствовать вашим умениям и навыкам.
http://https://russkiy365-diploms-srednee.ru/
Свидетельство О Перемене Фамилии
В сфере организации эффективной работы отдела ЗАГС, осуществления взаимодействия с Комитетом ЗАГС Пермского края. Курьер привезет заказанный документ в условленное место, где Вы в спокойной обстановке можете проверить его на наличие ошибок и оценить качество. Новое время диктует свои условия и если вы нашли себя в той или иной отрасли – сегодня необходимо иметь корочку об образовании.
Доверенность На Управление
И есть люди, которые работают на разных должностях, и получают за это хороший оклад. Стоит учесть, сколько своего драгоценного времени Вы сможете сэкономить при покупке документа об свидетельстве О Квалификации ВУЗа в нашей компании. Наши профессионалы быстро сделают надежный диплом за разумные деньги.
Диплом О Профессиональной Переподготовке Это Какое Образование
Диплом О Профессиональной Переподготовке Это Какое Образование
В этих стандартах определено, какие медицинские показания должны быть для проведения диспансерного наблюдения, как часто проводить диспансерные приемы (осмотры, консультации), какова длительность наблюдения, какие проводить анализы, исследования, как часто. Такая услуга стоит значительно дороже, но практически исключает возможность выявления подлога при проверке. Все документы изготавливаются на бланках ГОЗНАК и пройдут любую проверку. При этом нет необходимости спускаться в переход и обращаться к незнакомцам. Документ заполняется компетентными специалистами по всем правилам и требованиям Министерства науки и образования.
http://www.russkiy365-diploms-srednee.ru
Купить Диплом Образца Ссср
На нашем сайте также можно приобрести документы по любой специальности – как пример купить диплом фармацевта или любой другой специальности. К сожалению, именно на этом этапе может возникнуть целый ряд препятствий. Все, что для этого нужно – купить диплом-оригинал и подтвердить профессиональные навыки документально. Карьерный рост в той или иной компании всегда зависит от того, какими навыками владеет специалист.
Доверенность На Вождение
Стаж научной и практической работы с 2005 года, стаж научной и практической работы с 1987 года, стаж научной и практической работы с 2007 года. Делаем фото и видео готового документа под УФ лампой перед отправкой. В данной ситуации можем рекомендовать покупку диплома повара об окончании техникума или колледжа.
Купить Диплом Об Окончании
Сегодня потребность в фармацевтах наблюдается на всей территории РФ. Мы продаем дипломы на тех же бланках и печатями, что и выдающиеся государственные ВУЗы. Это объясняется тем, что человек имеет возможность получить диплом, не выходя из дома. Чтобы купить Диплом Об Окончании диплом электрика, нужно предоставить минимальное количество информацию производителю: указать фамилию и имя, указать сведения об учебном заведении, где документ был получен, указать год, в котором была получена корочка. В России направление обучающих игр поддерживается Всероссийской ассоциацией по играм в образовании. Широкий выбор предприятий, где можно предложить свои услуги, позволяет выбрать наиболее оптимальный вариант с хорошей зарплатой и удобным местом работы.
russkiy365-diploms-srednee.ru
Аттестат Особого Образца 9 Класс 2018
Потому что получить хорошую должность с достойной оплатой труда сейчас под силу не каждому. Если вы хотите срочно заказать диплом, можете сделать это прямо сейчас, заполнив заявку онлайн. За время существования биржи мы купили Диплом Об Окончании себя как безопасный сервис в России с выбором лучших авторов и гарантией завершения сделок в срок по различным дисциплинам.
Диплом О Неполном Высшем Образовании
Таким образом, если успеху в Вашей карьере мешает только одно – отсутствие диплома по необходимой Вам специальности, расстраиваться не стоит. Станьте пентестером – анализируйте защищенность систем безопасности, выявляйте уязвимые места и устраняйте их. Ирина Смирнова, депутат мажилиса: – А мне кажется, что можно и не захватывать 1970-е годы. Следует помнить, работодателя интересуют ваши профессиональные навыки и опыт, а “корочка” необходима как формальность.
Предметы В Аттестате 9 Класс
Мы предоставляем полный комплект документов, включая приложения, с указанием списка предметов и оценок. В наше время образование играет ключевую роль в формировании успешной предметы В Аттестате 9 Класс и обеспечении стабильного будущего. Возможно именно эта покупка позволит вам занять руководящую должность в престижной компании. Реквизиты лучше узнать в отделении ГУВМ МВД, в котором вы будете подавать документы, или онлайн и на сайте госуслуг. Прежде всего, нужно знать, что документ выдается, при положительном ответе, не бесплатно, а только после внесения установленной государственной пошлины. Можно подвести итог, что у каждого имеется собственная мотивация и ответ на вопрос – покупать диплом либо нет, и уже они определяют алгоритм будущих действий. Добрый день, подскажите если у вас заказать диплом с техникума с ним возможно будет поступить в университет.
https://arusak-diploms-srednee.ru
Купить Диплом О Высшем Образовании В Воронеже
Именно поэтому мы и стараемся сделать все необходимое для наших покупателей. Достаточно перечислить некоторые из них, к числу которых относятся: регулярные переименования вузов, смена статуса высших учебных заведений с института на университет или академию и наоборот, ликвидация немалого количества вузов или отзыв государственной аккредитации (что важно, так как не получиться получить дубликат, а так же исключает риск проверки легальности купленного), введение новых уровней профессиональной подготовки, включая специалиста, магистра и бакалавра, отсутствовавших ранее, частая замена бланков государственного образца на новые и т. Стоит к тому же знать, что вся основная информация, которую включает в себя академическая справка – о дисциплинах изученных студентом, полученных во время экзаменационных сессий оценках, затраченных на обучение часах, должна быть разборчивой и читаемой, без каких-либо двойных толкований и скрытого контекста. Приобретение диплома о высшем образовании может стать первым шагом к вашей успешной карьере.
Отличия Колледжа От Техникума
Мысли о важности среднего профессионального образования приходят не сразу после получения школьного аттестата. В ней вы должны указать свои личные данные и информацию о аттестат, какой именно класс вы хотите купить (название ВУЗа, специальность и год выпуска). В других случаях необходимо сначала обсудить заказ с администратором группы и провести предоплату. Нам не составит труда подготовить для вас оригиналы документов государственного образца, подтверждающие окончание одного из столичных высших учебных учреждений.
Regards, Lots of stuff.
Fantastic post however I was wondering if you could write a litte more on this topic? I’d be very grateful if you could elaborate a little bit more. Bless you!
http://lightsdemons.phorum.pl/viewtopic.php?p=66825#66825
https://iqtorg.ru/forum/messages/forum2/topic544/message602/?result=new#message602
https://telegra.ph/Pokupka-diploma-SHag-v-budushchee-bez-usilij-05-14
http://odessaflower.ukrbb.net/viewtopic.php?f=43&t=28402
http://beagle.fobb.ru/viewtopic.php?id=1262#p8948
Heya just wanted to give you a quick heads up and let you know a few of the pictures aren’t loading properly. I’m not sure why but I think its a linking issue. I’ve tried it in two different web browsers and both show the same outcome.
http://miku-crdb.ru/forum/messages/forum1/topic246/message268/?result=new#message268
http://anonzlinka.mybb.ru/viewtopic.php?id=833#p889
http://bigflud26.mybb.ru/viewtopic.php?id=491#p628
https://epistles.ru/blogs/786/пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ
https://vip.7bb.ru/viewtopic.php?id=2531#p6305
Hi everyone, it’s my first go to see at this web site, and article is really fruitful in support of me, keep up posting such posts.
https://stav.goodbb.ru/viewtopic.php?id=10025#p27769
http://allmagic.0pk.me/viewtopic.php?id=1094#p8426
http://newrealgames.ru/priobresti-kopii-akademicheskih-svidetelstv
http://www.les-ailes-chalaisiennes.com/forum/topic/пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅ/#postid-18598
https://www.4yo.us/blogs/123781/пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ
Today, while I was at work, my cousin stole my apple ipad and tested to see if it can survive a twenty five foot drop, just so she can be a youtube sensation. My iPad is now destroyed and she has 83 views. I know this is completely off topic but I had to share it with someone!
https://ivebo.co.uk/read-blog/31336
https://to.iboard.ws/viewtopic.php?id=5655#p13698
http://promintern.listbb.ru/viewtopic.php?f=16&t=365
http://avtobestnews.ru/poluchenie-uchebnyih-diplomov
http://medik-look.ru/diplomyi-na-vyibor-onlayn-i-dostupno-dlya-vas
ilogidis.com
두 달밖에 안됐는데 작은 경비원치고는 비용이 너무 비싸다.
프라그마틱 슬롯은 엄선된 테마와 뛰어난 디자인으로 놀라운 시각적 경험을 제공합니다.
프라그마틱슬롯
프라그마틱 슬롯에 대한 정보가 정말 유용했어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 새로운 내용을 찾아보세요. 함께 지식을 나누면 좋겠어요!
https://www.12315ds.cn/
https://www.12315yc.cn/
https://ianmccranor.com/link/
Can you tell us more about this? I’d want to find out more details.
http://www.kupitediplom0029.ru
Why users still use to read news papers when in this technological world all is existing on net?
http://https://kupitediplom0029.ru/
Wow, wonderful blog layout! How lengthy have you been blogging for? you made blogging glance easy. The total glance of your site is great, let alone the content!
https://iaescortsmap.ixbb.ru/viewtopic.php?id=9#p9
https://belydom.ru/forum/user/37507/
http://mockwacom.getbb.ru/viewtopic.php?f=12&t=1075
http://ros.listbb.ru/viewtopic.php?f=2&t=468
https://2016.goodboard.ru/viewtopic.php?id=176#p1415
What’s up to every , for the reason that I am actually keen of reading this weblog’s post to be updated regularly. It carries good information.
kupitediplom0029.ru/
Hi there, I desire to subscribe for this website to obtain most recent updates, so where can i do it please assist.
kupitediplom0029.ru/
I believe everything said made a ton of sense. However, what about this? suppose you added a little content? I am not saying your content is not good., but suppose you added a post title to possibly grab folk’s attention? I mean %BLOG_TITLE% is kinda vanilla. You should peek at Yahoo’s front page and see how they write article headlines to get people to click. You might add a video or a picture or two to get people interested about everything’ve written. Just my opinion, it would make your blog a little livelier.
https://diplomdoc.ru/
For most up-to-date news you have to pay a visit web and on web I found this website as a best web site for most up-to-date updates.
http://www.kupitediplom0029.ru/
I’m gone to convey my little brother, that he should also go to see this webpage on regular basis to get updated from most up-to-date information.
http://https://diplommy.ru/
This is really attention-grabbing, You’re an overly skilled blogger. I have joined your feed and sit up for looking for more of your wonderful post. Also, I’ve shared your website in my social networks
https://tropicplants.forumkz.ru/viewtopic.php?id=6396#p23261
https://oldforum.citysakh.ru/?talkid=26241
http://newsbeautiful.ru/diplom-bez-stressa-byistroe-poluchenie-onlayn/
https://komfort.rusff.me/viewtopic.php?id=12720#p38291
http://o97765bq.beget.tech/2024/05/13/put-k-karere-bez-pregrad-gde-zakazat-diplom-seychas.html
Right now it seems like WordPress is the best blogging platform out there right now. (from what I’ve read) Is that what you are using on your blog?
https://diplomdoc.ru/
Hello there! This post could not be written any better! Reading this post reminds me of my old room mate! He always kept chatting about this. I will forward this article to him. Pretty sure he will have a good read. Thank you for sharing!
https://www.diplommy.ru/
Can I simply say what a relief to discover a person that truly understands what they are discussing over the internet. You definitely understand how to bring a problem to light and make it important. More people must read this and understand this side of the story. It’s surprising you aren’t more popular given that you most certainly possess the gift.
http://c99596qr.beget.tech/2024/04/13/poluchenie-svidetelstv-o-dopolnitelnom-obuchenii.html
http://cv53297-livestreet-1.tw1.ru/2024/05/13/izbezhanie-prepyatstviy-kupit-diplom-pryamo-seychas.html
http://forums.worldsamba.org/showthread.php?tid=959063
http://monit.bestbb.ru/viewtopic.php?id=433#p1443
https://vip.forums.party/viewtopic.php?id=2862#p69683
Hello there, You’ve done a great job. I will definitely digg it and personally recommend to my friends. I’m confident they’ll be benefited from this web site.
http://newfinbiz.ru/byistryiy-put-k-karernomu-rostu-kak-kupit-diplom-i-dobitsya-uspeha
http://ya.bestbb.ru/viewtopic.php?id=2588#p5952
http://x91392sl.beget.tech/2024/04/13/kupit-attestaty-o-prohozhdenii-kursov.html
http://sad-montessori.ru/forum/user/297238/
http://selkovo.rolka.me/viewtopic.php?id=2965#p9228
It’s awesome in favor of me to have a web site, which is good for my knowledge. thanks admin
https://www.magazin-diplomov.ru/
Hi this is somewhat of off topic but I was wondering if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding skills so I wanted to get advice from someone with experience. Any help would be enormously appreciated!
http://www.diplompro.ru
Hey there! This is kind of off topic but I need some advice from an established blog. Is it difficult to set up your own blog? I’m not very techincal but I can figure things out pretty fast. I’m thinking about creating my own but I’m not sure where to start. Do you have any ideas or suggestions? Thank you
http://magazin-diplomov.ru/
На сегодняшний день, когда диплом становится началом успешной карьеры в любой области, многие стараются найти максимально быстрый путь получения качественного образования. Факт наличия официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед любым человеком, который хочет начать трудовую деятельность или продолжить обучение в ВУЗе.
Мы предлагаем очень быстро получить этот важный документ. Вы сможете заказать диплом нового или старого образца, и это становится выгодным решением для всех, кто не смог закончить обучение, потерял документ или хочет исправить свои оценки. Каждый диплом изготавливается аккуратно, с особым вниманием к мельчайшим нюансам, чтобы на выходе получился полностью оригинальный документ.
Преимущество такого подхода состоит не только в том, что вы максимально быстро получите диплом. Весь процесс организовывается просто и легко, с нашей поддержкой. Начиная от выбора требуемого образца документа до консультаций по заполнению персональной информации и доставки в любое место России — все находится под абсолютным контролем наших мастеров.
Таким образом, для тех, кто ищет оперативный способ получить необходимый документ, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать долгого процесса обучения и не теряя времени переходить к своим целям, будь то поступление в университет или старт карьеры.
http://http://labrusal.ru/
Hi! Do you use Twitter? I’d like to follow you if that would be okay. I’m undoubtedly enjoying your blog and look forward to new updates.
diplom-kuplu.ru
Hey! This post could not be written any better! Reading through this post reminds me of my previous room mate! He always kept chatting about this. I will forward this article to him. Fairly certain he will have a good read. Thank you for sharing!
https://diplom-profi.ru/
Thank you for sharing your thoughts. I really appreciate your efforts and I will be waiting for your next write ups thanks once again.
institute-diplom.ru
You actually explained it really well.
Also visit my blog post: https://www.fym-productions.com/2013/12/24/entry-with-post-format-video/
Hi to every single one, it’s truly a nice for me to pay a visit this website, it includes helpful Information.
1magistr.ru
Useful info. Fortunate me I discovered your web site by chance, and I am surprised why this twist of fate did not came about in advance! I bookmarked it.
http://www.diplom-city24.ru/
Do you have a spam issue on this website; I also am a blogger, and I was curious about your situation; many of us have created some nice methods and we are looking to exchange strategies with other folks, be sure to shoot me an e-mail if interested.
http://zakaz-na-diplom.ru/
Hi to every one, because I am actually keen of reading this website’s post to be updated on a regular basis. It contains nice stuff.
http://www.institute-diplom.ru/
This site really has all of the information I needed concerning this subject and didn’t know who to ask.
https://diplompro.ru/
Remarkable! Its actually awesome piece of writing, I have got much clear idea on the topic of from this post.
http://diablomania.ru/forum/showthread.php?p=563690#post563690
http://www.lada-xray.net/member.php?u=2434
http://nnars.ru/forum/suggestion-box/1139-легальный-путь-к-образованию-как-купить-диплом-бе#74525
What’s up, all is going fine here and ofcourse every one is sharing data, that’s really excellent, keep up writing.
https://solo-it.ru/forum/?PAGE_NAME=profile_view&UID=3712
https://familyportal.forumrom.com/viewtopic.php?id=20598#p55739
https://wegug.in/blogs/366/ѕуть-к-успеху- упи-диплом-и-преуспей-в-карьере
프라그마틱의 슬롯 게임은 정말 뛰어나죠! 여기서 더 많은 게임 정보를 찾을 수 있어 기뻐요.
PG 소프트
프라그마틱과 관련된 이 글 정말 잘 읽었어요! 더불어 저도 제 사이트에서 프라그마틱에 대한 새로운 정보를 공유 중이에요. 함께 나누면 더 좋을 것 같아요!
https://ycbgl.com/link/
https://www.12315dc.cn/
https://www.12315fk.cn/
I’m not sure why but this web site is loading incredibly slow for me. Is anyone else having this issue or is it a problem on my end? I’ll check back later and see if the problem still exists.
https://4atex.ru/blogs/1573/ѕуть-к-успеху- упи-диплом-и-преуспей-в-карьере
http://newrealgames.ru/diplom-dlya-budushhego-lidera-gde-kupit-obrazovanie-formiruyushhee-tebya-kak-professionala
http://avtolux48.ru/people/user/374/blog/7233/
Visit W3Schools
deneme
Hello there! Do you use Twitter? I’d like to follow you if that would be okay. I’m undoubtedly enjoying your blog and look forward to new updates.
https://medium.com/@oximus777/быстрый-старт-купи-диплом-и-открой-новые-горизонты-8025fd95ce69
https://malispa.ru/users/122
http://bestcoolfun.ru/diplom-dlya-tvoego-uspeha-gde-kupit-dokument-kotoryiy-otkroet-pered-toboy-dveri
5 라이온스 메가웨이즈
악의적이기 때문에 억울할 수밖에 없다.
Aw, this was an extremely good post. Taking the time and actual effort to make a really good article… but what can I say… I hesitate a lot and don’t seem to get anything done.
http://aranzhirovki.ru/smf/index.php?topic=4223.0
http://formulaf1.ru/diplom-na-zakaz-vash-put-k-novyim-vozmozhnostyam-i-professionalnomu-priznaniyu
http://newrealgames.ru/kupit-diplom-vasha-investitsiya-v-budushhee
Hi! I’m at work surfing around your blog from my new iphone! Just wanted to say I love reading through your blog and look forward to all your posts! Carry on the excellent work!
https://bezone.ru/node/335818
http://webnewsrealty.ru/diplom-dlya-budushhego-lidera-gde-kupit-obrazovanie-formiruyushhee-tebya-kak-professionala
http://cv53297-livestreet-1.tw1.ru/2024/05/15/diplom-na-zakaz-gde-kupit-obrazovanie-podhodyaschee-imenno-tebe.html
В нашем мире, где диплом является началом успешной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Наличие официального документа трудно переоценить. Ведь именно диплом открывает двери перед людьми, стремящимися вступить в сообщество профессионалов или учиться в ВУЗе.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы имеете возможность приобрести диплом, что будет отличным решением для человека, который не смог закончить образование, утратил документ или желает исправить плохие оценки. диплом изготавливается аккуратно, с особым вниманием к мельчайшим нюансам, чтобы в результате получился полностью оригинальный документ.
Преимущество этого решения состоит не только в том, что можно оперативно получить диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора требуемого образца до консультаций по заполнению персональных данных и доставки в любое место страны — все под полным контролем квалифицированных специалистов.
Всем, кто ищет быстрый и простой способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – это значит избежать длительного процесса обучения и не теряя времени перейти к важным целям, будь то поступление в университет или старт карьеры.
https://www.openstreetmap.org/user/JoseArnold2
https://tapchivatuyentap.tlu.edu.vn/Activity-Feed/My-Profile/UserId/44196
https://ioby.org/users/luciusbirch55838201
https://boi.instgame.pro/forum/index.php?topic=24407.0
http://onlineboxing.net/jforum/user/profile/288174.page
This excellent website really has all the information I wanted about this subject and didn’t know who to ask.
https://forum18.ru/threads/unikalnaja-vozmozhnost-kupi-diplom-i-dostigni-svoix-celej-bystree.37291/
http://veneraroleplay.listbb.ru/viewtopic.php?f=4&t=240
http://u90517ol.beget.tech/2024/05/15/put-k-professionalnomu-priznaniyu-kupi-diplom-i-stan-uvazhaemym-specialistom.html
This text is invaluable. Where can I find out more?
https://ivebo.co.uk/read-blog/31636
http://silich.ru/forum/member.php?u=4810
http://2cool.ru/qiwi-f215/kupit-diplom-vash-shans-stat-liderom-v-svoey-oblasti-t3600.html
Hello, this weekend is nice designed for me, for the reason that this time i am reading this enormous educational paragraph here at my house.
http://trifonov.in/exoplanets/dynamical-analysis-of-multi-planet-systems/hip5364_2_1/
https://free-bookmarking.com/story17155364/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%84%D0%B0%D1%80%D0%BC%D0%B0%D1%86%D0%B5%D0%B2%D1%82%D0%B0
https://politictoday.ru/kupit-diplom-legkiy-put-k-uspeshnoy-karere
https://johsocial.com/story7120922/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BF%D0%BE%D0%B2%D0%B0%D1%80%D0%B0
https://bookmarks-hit.com/story17164911/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%81%D0%B0%D0%BD%D1%82%D0%B5%D1%85%D0%BD%D0%B8%D0%BA%D0%B0
В нашем обществе, где диплом – это начало отличной карьеры в любой области, многие стараются найти максимально быстрый и простой путь получения качественного образования. Наличие официального документа переоценить просто невозможно. Ведь диплом открывает двери перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в университете.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы имеете возможность приобрести диплом нового или старого образца, и это становится выгодным решением для всех, кто не смог закончить обучение или потерял документ. дипломы производятся аккуратно, с особым вниманием к мельчайшим элементам, чтобы в результате получился документ, максимально соответствующий оригиналу.
Преимущества подобного решения заключаются не только в том, что можно максимально быстро получить диплом. Процесс организован просто и легко, с профессиональной поддержкой. Начав от выбора требуемого образца документа до консультации по заполнению персональной информации и доставки в любое место страны — все под полным контролем опытных мастеров.
Таким образом, всем, кто пытается найти максимально быстрый способ получения требуемого документа, наша компания может предложить выгодное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в ВУЗ или начало трудовой карьеры.
http://diplomsagroups.com/kupit-diplom-vuza/kandidata-nauk.html
트레져스 오브 아즈텍
Fang Jifan은 화를 내며 “왜 정직한 사람들을 괴롭히는 거죠?”
Today, while I was at work, my cousin stole my iPad and tested to see if it can survive a thirty foot drop, just so she can be a youtube sensation. My iPad is now destroyed and she has 83 views. I know this is completely off topic but I had to share it with someone!
https://scrapbookmarket.com/story17055305/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B1%D0%B0%D0%BA%D0%B0%D0%BB%D0%B0%D0%B2%D1%80%D0%B0
https://bookmarkmargin.com/story17123425/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%82%D0%B5%D1%85%D0%BD%D0%B8%D0%BA%D1%83%D0%BC%D0%B0
http://www.sildenafilmagicremedy.com/
https://buday.cz/2019/09/27/ahoj-vsichni/
https://myteana.ru/forums/index.php?autocom=gallery&req=si&img=4756
Simply want to say your article is as surprising. The clarity in your post is simply excellent and i could assume you’re an expert on this subject. Fine with your permission allow me to grab your feed to keep updated with forthcoming post. Thanks a million and please continue the gratifying work.
http://zelfrijdendetaxibreda.nl/uncategorized/vijf-tips-om-een-laadpaal-voor-je-kantoor-aan-te-vragen/
https://socialskates.com/story17904274/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%9A%D0%B0%D0%B7%D0%B0%D0%BD%D0%B8
http://www.sildenafilmagicremedy.com/
https://infopagex.com/story2344176/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BC%D0%B0%D0%B3%D0%B8%D1%81%D1%82%D1%80%D0%B0
https://keegandijig.blogstival.com/49288548/5-essential-elements-for-marketing
В нашем обществе, где диплом – это начало отличной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь диплом открывает двери перед каждым человеком, который собирается вступить в сообщество квалифицированных специалистов или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить любой необходимый документ. Вы можете приобрести диплом, что является отличным решением для всех, кто не смог завершить обучение, утратил документ или хочет исправить плохие оценки. диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим нюансам. На выходе вы сможете получить продукт, полностью соответствующий оригиналу.
Преимущество подобного решения состоит не только в том, что можно оперативно получить диплом. Процесс организован комфортно, с профессиональной поддержкой. Начав от выбора требуемого образца до грамотного заполнения личных данных и доставки по России — все под полным контролем опытных мастеров.
Всем, кто хочет найти быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать длительного процесса обучения и не теряя времени перейти к достижению собственных целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://forum.hi-def.ru/index.php?showtopic=24092
https://feeder.topbb.ru/viewtopic.php?id=1823#p67171
http://subarupleo.ixbb.ru/viewtopic.php?id=4722#p37754
http://building.ixbb.ru/viewtopic.php?id=3594#p5773
https://lms.nh.gov/liquor/portal/tag/index.php?tc=1&tag=%D0%91%D0%B0%D0%BA%D0%B0%D0%BB%D0%B0%D0%B2%D1%80%20%D0%BC%D0%B0%D0%B3%D0%B8%D1%81%D1%82%D1%80%20%D1%81%D0%B5%D0%BC%20%D0%BE%20%D1%87%D1%91%D0%BC%20%D1%80%D0%B5%D1%87%D1%8C%3F
В нашем мире, где диплом – это начало успешной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Наличие официального документа об образовании переоценить попросту невозможно. Ведь диплом открывает двери перед людьми, стремящимися начать трудовую деятельность или учиться в каком-либо ВУЗе.
В данном контексте наша компания предлагает оперативно получить этот важный документ. Вы сможете приобрести диплом, что является удачным решением для человека, который не смог завершить образование или потерял документ. Любой диплом изготавливается аккуратно, с особым вниманием к мельчайшим нюансам, чтобы в итоге получился 100% оригинальный документ.
Преимущество такого решения заключается не только в том, что можно оперативно получить свой диплом. Весь процесс организован удобно, с профессиональной поддержкой. От выбора подходящего образца диплома до грамотного заполнения личных данных и доставки по России — все будет находиться под полным контролем опытных специалистов.
Всем, кто ищет быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Заказать диплом – значит избежать длительного процесса обучения и не теряя времени переходить к достижению личных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://diplomsagroups.com/diplomy-po-specialnosti/diplom-medsestry.html
프라그마틱은 국내외에서 큰 사랑을 받고 있는데, 여기서 그 이유를 알 수 있어 좋아요!
PG 소프트
프라그마틱 관련 내용 감사합니다! 또한, 제 사이트에서도 프라그마틱에 대한 유용한 정보를 공유하고 있어요. 함께 서로 이야기하며 더 많은 지식을 쌓아가요!
https://hrbyszs.com/link/
https://www.iaz681.com
https://wqeeeeeee.weebly.com/
В нашем мире, где диплом – это начало успешной карьеры в любой области, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает дверь перед всеми, кто собирается вступить в профессиональное сообщество или учиться в каком-либо университете.
Мы предлагаем оперативно получить любой необходимый документ. Вы имеете возможность заказать диплом, и это является отличным решением для всех, кто не смог завершить образование или утратил документ. Все дипломы изготавливаются с особой тщательностью, вниманием ко всем нюансам. На выходе вы сможете получить документ, 100% соответствующий оригиналу.
Преимущество этого подхода состоит не только в том, что можно максимально быстро получить свой диплом. Процесс организовывается просто и легко, с профессиональной поддержкой. От выбора требуемого образца до консультации по заполнению персональной информации и доставки по России — все под абсолютным контролем опытных специалистов.
В итоге, для всех, кто пытается найти оперативный способ получить требуемый документ, наша услуга предлагает отличное решение. Купить диплом – значит избежать длительного обучения и сразу перейти к важным целям, будь то поступление в ВУЗ или старт карьеры.
Купить диплом фармацевта
geinoutime.com
여성들도 작업장에 들어가도록 격려를 받기 시작했습니다.
포츈 래빗
젊은 주인의 마음 속에는 심부름꾼이 있음을 보여줍니다.
geinoutime.com
Liu Jie는 14 명의 후배를 이끌고 아침 일찍 테스트 바구니를 들고 Fang의 집에 나타났습니다.
Hello, I think your site might be having browser compatibility issues. When I look at your blog in Opera, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, superb blog!
https://tandaseru.id/2017/09/06/usai-divonis-bebas-ustadz-alfian-tanjung-kembali-ditangkap/
http://www.matchfishing.ru/forumtest/1/member.php?u=23110&tab=activitystream&type=all&page=2
https://ravolutionfestival.com/
https://mixbookmark.com/story2474935/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%82%D0%B5%D1%85%D0%BD%D0%B8%D0%BA%D1%83%D0%BC%D0%B0
http://frugalmechanic.com/blog/user/fm
На сегодняшний день, когда диплом – это начало успешной карьеры в любой области, многие стараются найти максимально быстрый путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает двери перед каждым человеком, желающим вступить в сообщество квалифицированных специалистов или продолжить обучение в университете.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы сможете купить диплом, что становится удачным решением для человека, который не смог завершить обучение, потерял документ или хочет исправить свои оценки. диплом изготавливается с особой аккуратностью, вниманием ко всем элементам, чтобы в итоге получился документ, 100% соответствующий оригиналу.
Преимущества подобного решения заключаются не только в том, что можно максимально быстро получить диплом. Процесс организовывается комфортно и легко, с профессиональной поддержкой. Начиная от выбора подходящего образца до консультации по заполнению личной информации и доставки в любое место страны — все под абсолютным контролем квалифицированных мастеров.
В результате, для тех, кто хочет найти максимально быстрый способ получить требуемый документ, наша компания готова предложить отличное решение. Заказать диплом – это значит избежать длительного процесса обучения и сразу переходить к достижению собственных целей, будь то поступление в университет или начало трудовой карьеры.
http://diplomsagroups.com/drugie-dokumenty/sertifikat-specialista.html
Almabet üyeler sitedeki iletişim seçeneklerini Almabet diledikleri zaman kullanabilirler. Almabet şikayetlerinin nereye bildirileceği sorusu aynı zamanda yerinde destek ve müşteri hizmetleriyle de ilgilidir.
WOW just what I was looking for. Came here by searching for %keyword%
https://chanceruvvu.onzeblog.com/26908961/the-best-side-of-marketing
https://www.leukshoppen.nl/product/black-pants/
http://missatom.nuclear.ru/site_py/blogs/1/
https://real-amazon-promo-code79146.blogrelation.com/32827715/top-latest-five-marketing-urban-news
https://socialtechnet.com/story2524900/%D0%B3%D0%B4%D0%B5-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D1%8B-%D0%B2%D1%83%D0%B7%D0%B0
Puntobahis üye olmanın bir maliyeti var mı? Puntobahis Puntobahis Casino siteleri bu tür işlemler için herhangi bir ücret talep etmez.
Casinomhub Üyelik alanlarında herhangi bir ücret alınmaz Casinomhub ve destek işlemlerinde Casinomhub çekimlerinde herhangi bir kesinti yapılmaz. Casinomhub bahis sitesinde çok çeşitli oyunlar mevcuttur.
EGGC
잠시 생각에 잠긴 팡지판은 크게 웃었다. “하하하하…”
В современном мире, где диплом становится началом отличной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает двери перед всеми, кто собирается начать трудовую деятельность или учиться в высшем учебном заведении.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы можете приобрести диплом, и это становится удачным решением для человека, который не смог закончить обучение, потерял документ или желает исправить плохие оценки. дипломы выпускаются аккуратно, с особым вниманием к мельчайшим нюансам, чтобы на выходе получился 100% оригинальный документ.
Преимущество данного подхода состоит не только в том, что вы сможете быстро получить свой диплом. Процесс организовывается удобно, с нашей поддержкой. От выбора требуемого образца диплома до точного заполнения персональных данных и доставки по России — все под абсолютным контролем качественных мастеров.
Всем, кто хочет найти оперативный способ получения необходимого документа, наша компания может предложить выгодное решение. Купить диплом – значит избежать долгого процесса обучения и не теряя времени перейти к важным целям, будь то поступление в университет или старт профессиональной карьеры.
https://cbexapp.noaa.gov/tag/index.php?tc=1&tag=%D0%9A%D0%B0%D0%BA%20%D0%BD%D0%B5%20%D0%BF%D0%BE%D1%82%D1%80%D0%B0%D1%82%D0%B8%D1%82%D1%8C%20%D0%B2%D1%80%D0%B5%D0%BC%D1%8F%20%D0%B2%20%D1%83%D0%BD%D0%B8%D0%B2%D0%B5%D1%80%D1%81%D0%B8%D1%82%D0%B5%D1%82%D0%B5%20%D0%B7%D1%80%D1%8F%3F
https://vspmscop.edu.in/LRM/profile/LonnieKelley1/
http://fabrikaglamura.webtalk.ru/viewtopic.php?id=771#p39764
http://mamakrasnogorska.0pk.me/viewtopic.php?id=1415#p5366
https://qooh.me/JoseArnold2
На сегодняшний день, когда диплом – это начало удачной карьеры в любой области, многие ищут максимально быстрый путь получения качественного образования. Факт наличия официального документа сложно переоценить. Ведь диплом открывает двери перед любым человеком, желающим начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
Мы предлагаем быстро получить любой необходимый документ. Вы можете заказать диплом, что является удачным решением для человека, который не смог закончить обучение или потерял документ. диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим нюансам, чтобы в результате получился 100% оригинальный документ.
Преимущества данного решения заключаются не только в том, что вы сможете оперативно получить диплом. Весь процесс организован удобно, с нашей поддержкой. Начав от выбора нужного образца диплома до консультаций по заполнению личных данных и доставки в любое место страны — все под абсолютным контролем опытных специалистов.
Таким образом, для тех, кто хочет найти быстрый способ получить необходимый документ, наша услуга предлагает выгодное решение. Купить диплом – это значит избежать продолжительного обучения и сразу переходить к достижению личных целей: к поступлению в университет или к началу трудовой карьеры.
http://http://diplomsagroups.com/diplomy-po-specialnosti/diplom-stroitelya.html
geinoutime.com
번개 같은 속도로 장검이 허공에 완벽한 호를 그렸다.
Hello very cool website!! Guy .. Excellent .. Superb .. I will bookmark your web site and take the feeds additionally? I am glad to find numerous helpful info right here within the put up, we’d like work out extra techniques in this regard, thank you for sharing. . . . . .
https://oldforum.citysakh.ru/?talkid=26125
https://baiontap.com/
http://lsdsng.com/user/542212
https://anyreader.com/
https://medik-look.ru/diplomyi-na-vyibor-sdelayte-pravilnyiy-vyibor
Marlabet spor bahisleri kayıt her zaman şirketten yardım Marlabet alabilirler. Yeni Marlabet Adres Platformu bu hizmet ve işlevsellik için en etkili sitedir.
에그벳 주소
지친 수험생들은 흐르는 물처럼 자공학원을 빠져나왔다.
Betorro çok sayıda önemli ve çeşitli çevrimiçi Casino platformu bulunmaktadır. Betorro Bu yerler açık ve net politikalar izlerken lisans numaraları ve altyapı sunuyor. Temel olarak, kullanıcılar için yeni bir güven düzeyi oluşturur.
Run4bets mobil işlem Uygulamaya öncelikle Run4bets mobil internet tarayıcınıza giriş yaparak ulaşabilirsiniz.
Ruisbet güncellendi güncel Ruisbet adresinizi almak için Yeni Ruisbet</a Ruisbet Adresi linkine tıklayın. Ruisbet resmi giriş ve destek blogudur.
Meritlimancasino hesabınızı aktif tutmak hesabınızı aktif tutmak Meritlimancasino için aldığınız onay SMSini yanıtlayarak veya verilen numarayı arayarak üyeliğinizi doğrulamanız gerekecektir.
В нашем обществе, где диплом становится началом отличной карьеры в любом направлении, многие стараются найти максимально простой путь получения качественного образования. Факт наличия документа об образовании трудно переоценить. Ведь диплом открывает дверь перед людьми, желающими начать трудовую деятельность или учиться в университете.
Наша компания предлагает быстро получить этот важный документ. Вы имеете возможность купить диплом, и это будет выгодным решением для человека, который не смог закончить обучение, утратил документ или желает исправить плохие оценки. диплом изготавливается аккуратно, с особым вниманием ко всем элементам, чтобы в результате получился полностью оригинальный документ.
Превосходство такого решения состоит не только в том, что можно максимально быстро получить свой диплом. Весь процесс организовывается просто и легко, с нашей поддержкой. Начав от выбора нужного образца до грамотного заполнения персональных данных и доставки в любое место страны — все будет находиться под абсолютным контролем качественных мастеров.
Всем, кто пытается найти оперативный способ получения требуемого документа, наша компания может предложить отличное решение. Купить диплом – значит избежать длительного процесса обучения и не теряя времени перейти к достижению личных целей, будь то поступление в университет или старт карьеры.
Купить сертификат специалиста
Bet10line bahis menüsü ziyaret edin. Detayları bölümde bulabilirsiniz. Bet10line Her yeni kayıtta Bet10line güvenilir mi? ortaya çıkar.
whoah this blog is magnificent i love studying your posts. Stay up the great work! You know, a lot of persons are searching round for this information, you can help them greatly.
diplom-zakaz.ru/
Attractive section of content. I just stumbled upon your web site and in accession capital to assert that I get actually enjoyed account your blog posts. Anyway I’ll be subscribing to your augment and even I achievement you access consistently fast.
peoplediplom.ru/
It’s awesome to pay a visit this site and reading the views of all mates about this post, while I am also keen of getting familiarity.
http://diplom-profi.ru/
Hi everyone, it’s my first visit at this web page, and article is truly fruitful designed for me, keep up posting these articles.
http://www.diplom-profi.ru/
Hello it’s me, I am also visiting this web site daily, this web page is really fastidious and the users are actually sharing fastidious thoughts.
https://bisness-diplom.ru/
Wow that was unusual. I just wrote an very long comment but after I clicked submit my comment didn’t appear. Grrrr… well I’m not writing all that over again. Anyways, just wanted to say superb blog!
https://www.kupite-diplom0024.ru/
Simply want to say your article is as astonishing. The clearness in your put up is simply excellent and that i can suppose you’re knowledgeable in this subject. Fine along with your permission allow me to take hold of your feed to keep updated with imminent post. Thank you one million and please continue the enjoyable work.
http://www.sdiplom.ru/
Pretty! This was an extremely wonderful article. Thank you for providing this information.
http://diplom-city24.ru/
An outstanding share! I’ve just forwarded this onto a co-worker who was doing a little homework on this. And he actually bought me lunch because I stumbled upon it for him… lol. So let me reword this…. Thank YOU for the meal!! But yeah, thanx for spending the time to discuss this topic here on your web site.
https://zakaz-na-diplom.ru/
Hi exceptional website! Does running a blog like this require a great deal of work? I have absolutely no understanding of coding however I was hoping to start my own blog soon. Anyways, if you have any ideas or techniques for new blog owners please share. I know this is off topic but I simply needed to ask. Many thanks!
https://www.zakaz-na-diplom.ru/
Betanka bahis ajanslarında çalışan ve bahisçileri Betanka inceleyen bazı kişiler genellikle Betanka slot makinelerinin güvenilir olup olmadığını merak ediyor. Bu hedefe ulaşmak için kapsamlı bir inceleme yapıldı.
Kralpanda TVyi başlatmak için özel gereksinimler Kralpanda vardır. Bu mantığı kullanırsanız herkesin oyuna başlaması ve para kazanması kolay olacaktır.
Сегодня, когда диплом – это начало отличной карьеры в любой области, многие ищут максимально простой путь получения качественного образования. Факт наличия документа об образовании сложно переоценить. Ведь диплом открывает двери перед людьми, желающими вступить в сообщество профессионалов или учиться в ВУЗе.
В данном контексте мы предлагаем быстро получить этот необходимый документ. Вы сможете приобрести диплом нового или старого образца, что будет выгодным решением для человека, который не смог закончить обучение или потерял документ. Все дипломы выпускаются аккуратно, с особым вниманием ко всем нюансам, чтобы в результате получился полностью оригинальный документ.
Превосходство такого решения заключается не только в том, что можно максимально быстро получить свой диплом. Процесс организован просто и легко, с профессиональной поддержкой. От выбора необходимого образца до консультации по заполнению личной информации и доставки по стране — все находится под полным контролем наших мастеров.
Всем, кто пытается найти быстрый и простой способ получить необходимый документ, наша компания готова предложить отличное решение. Заказать диплом – это значит избежать долгого обучения и сразу перейти к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
https://connects.ctschicago.edu/forums/users/179606/
https://dzone.com/users/5143665/josearnold2.html
https://iot.ttu.edu.tw/members/elviraparks1/
http://poker.forum-top.ru/viewtopic.php?id=656#p2538
http://devushka.winbb.ru/viewtopic.php?id=684#p3322
Zoommobil güvenilir altyapıya sahip bir web sitesidir, Zoommobil dolayısıyla iyi altyapıya sahip bir web sitesi arıyorsanız Zoommobil dikkate almanız önerilir.
Sosabet bahis platformumuza girdiğiniz anda galibiyet Sosabet serinize başlayabilirsiniz. Sisteme giriş yapabilmek için yeni Sosabet giriş adresinizi girmeniz gerekmektedir Sosabet yeni giriş adresinizden almaya devam edeceksiniz.
Thank you for sharing your info. I truly appreciate your efforts and I will be waiting for your further post thanks once again.
https://institute-diplom.ru/
Larabahis para yatırma işlemi başvuru döneminde en çok ilgi çeken Larabahis konulardan biriydi. Süre kısa olabilir ancak sisteminize ve yaklaşımınıza göre dalgalanacaktır.
Сегодня, когда диплом является началом успешной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим вступить в сообщество профессиональных специалистов или учиться в каком-либо университете.
Наша компания предлагает оперативно получить этот важный документ. Вы сможете заказать диплом, что является выгодным решением для человека, который не смог закончить образование, потерял документ или хочет исправить свои оценки. диплом изготавливается аккуратно, с особым вниманием к мельчайшим элементам, чтобы в результате получился продукт, максимально соответствующий оригиналу.
Превосходство такого решения состоит не только в том, что можно быстро получить диплом. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора необходимого образца диплома до консультаций по заполнению личной информации и доставки по стране — все под полным контролем опытных специалистов.
Для тех, кто ищет максимально быстрый способ получить необходимый документ, наша услуга предлагает выгодное решение. Заказать диплом – значит избежать долгого обучения и сразу перейти к личным целям: к поступлению в ВУЗ или к началу удачной карьеры.
https://airguns.webtalk.ru/viewtopic.php?id=2747#p19081
https://sponforum.ixbb.ru/viewtopic.php?id=12316#p42418
http://automarket.topbb.ru/viewtopic.php?id=2007#p3647
https://ilm.iou.edu.gm/members/boyedoye4/
http://fishkaluga.0pk.me/viewtopic.php?id=1706#p11294
Betmabet birbirinin yerine geçebilir. Betmabet Tek pişmanlığım, herhangi bir sorun yaşamadan tekrar çevrimiçi olabilmiş olmamdır.
Hello, its nice post on the topic of media print, we all know media is a wonderful source of data.
https://www.diplom-kuplu.ru/
에그벳 계열
결국 … 사람이 너무 많습니다. 특히 승객이 가장 많은 경우에는 더욱 그렇습니다.
Do you have any video of that? I’d care to find out more details.
http://arvixe.ru/v/building/cotedge_1/P1070836.JPG.html
http://samara-fish.ru/konkursi?start=5
http://alter-energo.ru/topic2517.html
http://nad-dom31.ru/lenta-5.html
http://www.starlineworld.ru/index.php?links_exchange=yes&page=2&show_all=yes
Hello to all, how is the whole thing, I think every one is getting more from this web page, and your views are nice in favor of new people.
hellohome.ir/en/agent/mahyar-shahbazi/В
myteana.ru/forums/index.php?autocom=gallery&req=si&img=4490В
vadaszapro.eu/user/profile/1266221В
hellados.ru/texts/io.phpВ
http://www.blainesoccer.org/feesfutureВ
For hottest news you have to go to see internet and on web I found this website as a most excellent web site for latest updates.
http://risuemtut.ru/kak-narisovat-zhenskoe-telo-karandashom-poetapno/
http://pokatili.ru/f/viewtopic.php?f=10&t=70442
http://gravure-dore.ru/rMat2735_TheErectionOfTheCross.htm
http://auto-sale-msk.ru/policy
http://www.briefeducation.ru/bried-180.html
На сегодняшний день, когда диплом становится началом удачной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа переоценить невозможно. Ведь диплом открывает двери перед каждым человеком, который стремится вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы имеете возможность заказать диплом, что становится отличным решением для всех, кто не смог завершить образование или утратил документ. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам, чтобы на выходе получился документ, максимально соответствующий оригиналу.
Плюсы подобного подхода состоят не только в том, что можно максимально быстро получить диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начиная от выбора нужного образца диплома до консультаций по заполнению личной информации и доставки в любой регион страны — все находится под абсолютным контролем качественных специалистов.
В результате, для тех, кто ищет быстрый и простой способ получения требуемого документа, наша услуга предлагает выгодное решение. Заказать диплом – значит избежать длительного обучения и сразу переходить к достижению личных целей: к поступлению в университет или к началу трудовой карьеры.
http://novstan.ru
http://cbt-v.com.ua
http://kmsbryansk.ru
http://infolinia.org
http://thecrimea.org.ua
Bionbet kayıt olma kuralları bahis koyarken dikkate Bionbet alınması gereken çok sayıda faktör vardır. Bu web sitesi çeşitli video oyunlarına ilişkin kapsamlı eleştiriler sunmaktadır.
Betnblue bonus kampanyası işlemleri hızlı bir şekilde Betnblue gerçekleşiyor. Banka havaleleri yoluyla dünyanın her yerine dakikalar içinde para göndermek mümkün.
В нашем мире, где диплом – это начало удачной карьеры в любой сфере, многие ищут максимально простой путь получения качественного образования. Наличие официального документа об образовании переоценить попросту невозможно. Ведь диплом открывает двери перед людьми, желающими начать профессиональную деятельность или учиться в ВУЗе.
Наша компания предлагает оперативно получить любой необходимый документ. Вы можете купить диплом, что становится выгодным решением для всех, кто не смог завершить образование или потерял документ. дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим деталям, чтобы в результате получился продукт, максимально соответствующий оригиналу.
Преимущества данного решения заключаются не только в том, что можно быстро получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начиная от выбора необходимого образца диплома до консультаций по заполнению личных данных и доставки по стране — все находится под абсолютным контролем качественных специалистов.
Всем, кто ищет максимально быстрый способ получить требуемый документ, наша компания может предложить отличное решение. Заказать диплом – это значит избежать долгого обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в ВУЗ или старт карьеры.
http://sst-nnov.ru
http://girlzinweb.com
http://wathomejobs.com
http://xn—-8sbaagot1a4a4b8c.xn--p1ai
http://edmontonplumbers.net
Milyonluk web sitesi gelecekte Milyonluk kullanan Milyonluk web siteleri için daha basit ve daha kesin reklam engelleme önlemleri sunacağız.
Hodribet bonus türleri Bu, bonusların ve para yatırma Hodribet tekliflerinin bahis yükümlülüklerini hızlı bir şekilde tamamlamak isteyen oyuncular için faydalıdır.
Keep on writing, great job!
http://www.sixsigmaexams.com/mybb/member.php?action=profile&uid=263670В
freedomsolargenerators.com/index.php?do=/public/user/blogs/name_Alanpoe/page_7/В
http://www.find-topdeals.com/blogs/45974/D09AD0B0D187D0B5D181D182D0B2D0B5D0BDD0BDD18BD0B5-D0B4D0B8D0BFD0BBD0BED0BCD18B-D0BFD0BE-D0B4D0BED181D182D183D0BFD0BDD0BED0B9-D186D0B5D0BDD0B5-D092D0B0D188-D0BFD183D182D18C-D0BA-D183D181D0BFD0B5D185D183-D0BDD0B0D187?lang=tr_trВ
vague.social/blogs/31670/Why-is-the-popularity-of-universities-rapidly-declining-todayВ
http://www.ivedu.ru/forum/viewthread.php?forum_id=18&thread_id=46172В
Kingroyal kullanıcı işlemleri Bahis paranızı, işlem yaptıktan sonraki bir Kingroyal gün içerisinde Visa banka kartınızdan hiçbir ücret ödemeden çekebilirsiniz.
Hi, after reading this remarkable piece of writing i am also glad to share my experience here with friends.
http://www.lannach.eu/goa-arambol-beach/В
smartwatchcolombia.com/producto/78/smartwatch-serie-8-ultra-11-2023–manilla-de-regalo/7arusak-diploms.comВ
pipeclub.net/index.php?showuser=110605&k=880ea6a14ea49e853634fbdc5015a024&setlanguage=1&langurlbits=showuser=110605&cal_id=&langid=2В
http://www.datxanhhanoi.com/vinpearl-riverfront-condotel-vien-ngoc-quy-ben-bo-song-han/В
myworldoftank.ru/article/kak-vklyuchit-otobrazhenie-shansov-na-pobedu-xvm-v-world-of-tanks.htmlВ
Superb post but I was wanting to know if you could write a litte more on this subject? I’d be very grateful if you could elaborate a little bit further. Thank you!
http://delivery-containers.ru/photogallery/containers/containers_20futov/
http://nebesis.ru/6-fevralya-znak-zodiaka-vodolej/
http://crystal-pc.ru/chto-takoe-atlasnoe-plate
http://medsimptom.org/category/diety-pri-razlichnyh-zabolevanij/
http://preceptor.flybb.ru/topic4745.html
Hello, I think your website might be having browser compatibility issues. When I look at your blog site in Firefox, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, excellent blog!
kopd.ocdut.kr.ua/conference/topic/mentorska-diialnist-v-dytiachomu-samovriaduvanni/?part=4926В
aromatov.wooden-rock.ru/forum/topic.php?forum=1&topic=15351В
blouson-moto.net/Blousons/ixon.htmlВ
animalprotect.org/forum/index.php?action=profile;u=4779;area=showposts;sa=topics;start=330В
sportraketka.ru/izbavtes-ot-slozhnostey-s-ucheboy-kupite-diplomВ
Hello there! I know this is kinda off topic nevertheless I’d figured I’d ask. Would you be interested in trading links or maybe guest authoring a blog post or vice-versa? My website discusses a lot of the same topics as yours and I feel we could greatly benefit from each other. If you happen to be interested feel free to shoot me an email. I look forward to hearing from you! Excellent blog by the way!
http://www.downtowncookieco.com/shipping-policy/В
politikforum.ru/member.php?u=13387008В
bike.by/forum/viewtopic.php?f=84&t=30293В
levit.in.ua/about-us.htmlВ
dopul.ru/obzor-interneta/70-chto-takoe-diplom-ob-obrazovanii-zachem-on-nuzhen.htmlВ
В нашем мире, где диплом является началом удачной карьеры в любой отрасли, многие ищут максимально быстрый путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает дверь перед людьми, желающими начать трудовую деятельность или учиться в любом ВУЗе.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы можете приобрести диплом нового или старого образца, что будет выгодным решением для человека, который не смог завершить обучение, потерял документ или желает исправить плохие оценки. диплом изготавливается аккуратно, с особым вниманием к мельчайшим элементам, чтобы в итоге получился документ, 100% соответствующий оригиналу.
Плюсы данного решения состоят не только в том, что вы максимально быстро получите свой диплом. Процесс организовывается удобно, с нашей поддержкой. Начиная от выбора нужного образца до консультации по заполнению личной информации и доставки в любой регион России — все под абсолютным контролем наших специалистов.
Всем, кто ищет быстрый способ получить необходимый документ, наша компания готова предложить отличное решение. Купить диплом – значит избежать долгого обучения и сразу перейти к достижению личных целей: к поступлению в университет или к началу трудовой карьеры.
http://trudbud.ru
http://goprostore.in.ua
http://eon51.com
http://bgv-blasenschwaeche.de
http://contact-polis.ru
В современном мире, где диплом – это начало удачной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Наличие официального документа об образовании переоценить просто невозможно. Ведь именно диплом открывает двери перед всеми, кто стремится вступить в профессиональное сообщество или продолжить обучение в высшем учебном заведении.
Наша компания предлагает оперативно получить любой необходимый документ. Вы имеете возможность приобрести диплом, и это будет выгодным решением для всех, кто не смог завершить образование или потерял документ. дипломы изготавливаются аккуратно, с особым вниманием ко всем элементам, чтобы в результате получился продукт, 100% соответствующий оригиналу.
Преимущества данного подхода состоят не только в том, что вы сможете оперативно получить свой диплом. Процесс организован комфортно, с нашей поддержкой. Начиная от выбора нужного образца до правильного заполнения личной информации и доставки в любое место страны — все под полным контролем опытных специалистов.
Всем, кто пытается найти оперативный способ получения необходимого документа, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать долгого обучения и не теряя времени переходить к своим целям, будь то поступление в ВУЗ или начало профессиональной карьеры.
http://officesetupc.com
http://bc-pharmacy.com.ua
http://allluxuryhomes.com
http://daynghethanhnien.com
http://pharmacie-tarascon.fr
Thank you for sharing your thoughts. I really appreciate your efforts and I will be waiting for your further write ups thank you once again.
http://unforma.ru/rybalka/rybopromyshlennikam-razyasnyat-tonkosti-raboty-s-erzh.html
http://rusitemonitoring.ru/list/sockart.ru.html
http://mens-look.ru/modamuzhchiny.html
http://whirlpoolguide.de/whirlpool-hygiene-wasserpflege-reinigung/
http://100buketov.com/index.php?links_exchange=yes&page=2&show_all=yes
I’m gone to tell my little brother, that he should also pay a visit this blog on regular basis to obtain updated from most up-to-date reports.
danceway74.ru/users/36?page=28В
http://www.4-mobile.ru/index.php?links_exchange=yes&page=13&show_all=yesВ
http://www.livesportonline.org/football-news/news-details/Arsenal-handle-Dundalk-in-Europa-League-group-stage-win/35346/2/В
click2connectclubs.com/index.php?do=/public/user/blogs/view/name_Alanpoe/id_119636/title_/В
karabash.chelbusiness.ru/index.php?name=account&op=info&uname=unypavazeВ
http://vinhphatmobile.com
Hi there this is kind of of off topic but I was wondering if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding skills so I wanted to get advice from someone with experience. Any help would be greatly appreciated!
http://angelteam.uv.ro/memberlist.php?mode=joined&order=ASC&start=208150
http://mafiaclans.ru/topic3885.html
http://mypechen.com/alfa-normiks/
http://otradopt.ru/category/mollyuski/
http://stobolezni.ru/kishki/Appenditcit/appendicit_zhenshchin.html
Can you tell us more about this? I’d like to find out some additional information.
89.108.65.91/content/articles/65330/В
forumjizni.ru/member.php?u=120588В
http://www.9020blog.com/board/blogs/2763/Why-is-the-popularity-of-universities-decreasing-all-the-timeВ
alter-energo.ru/topic2861.html?view=previousВ
http://www.sonyericsson-championships.com/es/news/story/370.aspВ
Hi just wanted to give you a brief heads up and let you know a few of the images aren’t loading properly. I’m not sure why but I think its a linking issue. I’ve tried it in two different internet browsers and both show the same outcome.
flora-online.ru/glavnaya/tsvetyi-na-1-sentyabrya-2/В
here4deals.com/garderie-barny-horaires.phpВ
circuit-diagrams.com/pr4-PIC16F84A-discolight-effect-with-bass-beat-control.phpВ
wap-club.ru/guestbook.php?mode=quote&idmsg=210748В
http://www.bfauk.com/bfa_cms/userВ
I got this website from my friend who informed me about this web site and at the moment this time I am browsing this website and reading very informative articles or reviews here.
http://hvoinie.ru/xvojnye-derevya/listvennica.html
http://wild-animals.ru/systems/alp/
http://oopt-kurkino.ru/p1/history-10.php
http://forum.stockholdergame.com/user-83021.html
http://nmt200.ru/nfaq/faq036
Müşteri Hizmetleri Numarasının arkasındaki şirket olan Hatbet, Hatbet canlı çağrı hizmetinin yanı sıra en uygun slot ve bahis seçeneklerini sunmaktadır.
Hello! I realize this is somewhat off-topic but I needed to ask. Does running a well-established website such as yours take a massive amount work? I’m brand new to writing a blog however I do write in my diary every day. I’d like to start a blog so I can share my personal experience and views online. Please let me know if you have any ideas or tips for brand new aspiring bloggers. Appreciate it!
ant-france.eu/index.php/2016/08/06/communique-de-presse-du-6-aout-2016/В
vv.flybb.ru/topic1874.html?view=previousВ
eku.ru/comments/list/В
gamesfortop.ru/diplomyi-na-zakaz-vash-shans-na-novuyu-kareruВ
http://www.4-mobile.ru/index.php?links_exchange=yes&page=13&show_all=yesВ
Superb blog you have here but I was curious about if you knew of any discussion boards that cover the same topics discussed in this article? I’d really like to be a part of community where I can get feedback from other knowledgeable individuals that share the same interest. If you have any recommendations, please let me know. Appreciate it!
http://dom-culturi.ru/afisha/page/5/
http://russiahistory.ru/polyane/
http://vocerkovlenie.ru/stroy/16/127/
http://e-migdal.ru/motornoe-maslo/proizvodstvo-motornyh-masel.html
http://pro-torrent.net/knigi/15057-osobennosti-lovli-ryb-semeystva-lososevyh-elena-muradova-2013.html
Okyanusbahis spor bahisleri Diğer spor bahis Okyanusbahis seçeneklerini araştırın ve Avrupa Ligi ile başlayın. Eğer siz de temsilcilerimizle birlikte bu fırsattan yararlanmak istiyorsanız hemen harekete geçin.
İstekbet oyunlarda ve gerçek zamanlı etkinliklerde İstekbet yüksek kazanma şansıyla harika vakit geçirin. Sanal bahis en yeni ve en bilinen oyunları içerir.
Cazino güvenilir işlemler sağlamak için Cazino sitelerinin uluslararası bahis ve casino oyunlarına katılma lisansına sahip olması gerekir.
Betofwar müşteri destek ekibi web sitenizin Betofwar tüm güvenlik sorgularına yanıt verebilir. Betofwar, müşteri hizmetleri de dahil olmak üzere tüm üyelere ulaşmak için üç yöntem sunar.
В нашем обществе, где диплом является началом отличной карьеры в любой области, многие ищут максимально простой путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь именно он открывает дверь перед людьми, стремящимися начать профессиональную деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить любой необходимый документ. Вы можете заказать диплом старого или нового образца, и это будет отличным решением для всех, кто не смог завершить образование, потерял документ или желает исправить плохие оценки. диплом изготавливается аккуратно, с особым вниманием к мельчайшим деталям. В итоге вы получите продукт, максимально соответствующий оригиналу.
Плюсы этого решения заключаются не только в том, что можно быстро получить свой диплом. Весь процесс организован удобно, с нашей поддержкой. Начиная от выбора подходящего образца до консультации по заполнению персональной информации и доставки в любое место страны — все под полным контролем качественных мастеров.
Всем, кто ищет оперативный способ получения требуемого документа, наша компания предлагает отличное решение. Заказать диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к своим целям: к поступлению в университет или к началу успешной карьеры.
http://masseriacucuruzza.it
http://dmp-art.ru
http://pmk-polymer.ru
http://xn--logtervezs-j7a7i.hu
http://funhost.org.ua
더 도그 하우스 메가웨이즈
Hongzhi 황제의 눈은 즉시 Tian Jing에게 떨어졌습니다.Hongzhi 황제의 눈이 번쩍이고 그의 얼굴은 믿을 수 없다는 표정을지었습니다.
http://infovaccins.ch
В современном мире, где диплом является началом отличной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Наличие официального документа трудно переоценить. Ведь именно диплом открывает двери перед каждым человеком, желающим начать трудовую деятельность или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы имеете возможность купить диплом старого или нового образца, что будет отличным решением для человека, который не смог завершить образование или потерял документ. Все дипломы выпускаются с особой аккуратностью, вниманием ко всем нюансам, чтобы в итоге получился документ, 100% соответствующий оригиналу.
Преимущества подобного решения заключаются не только в том, что вы максимально быстро получите диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. От выбора необходимого образца диплома до правильного заполнения персональных данных и доставки в любой регион страны — все под абсолютным контролем качественных специалистов.
В результате, для тех, кто пытается найти максимально быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать долгого процесса обучения и не теряя времени перейти к достижению своих целей, будь то поступление в ВУЗ или старт трудовой карьеры.
http://philautarchie.net
http://lespilomaterial.ru
http://hdvideoboxs.ru
http://huulienasia.com.vn
http://arvine-restaurant.fr
В нашем обществе, где аттестат – это начало отличной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа об образовании сложно переоценить. Ведь диплом открывает дверь перед людьми, стремящимися начать трудовую деятельность или продолжить обучение в ВУЗе.
Предлагаем быстро получить этот важный документ. Вы можете купить аттестат, и это является отличным решением для всех, кто не смог завершить образование или утратил документ. Любой аттестат изготавливается аккуратно, с особым вниманием ко всем нюансам. В результате вы получите полностью оригинальный документ.
Преимущества подобного подхода состоят не только в том, что можно быстро получить аттестат. Весь процесс организовывается просто и легко, с нашей поддержкой. От выбора необходимого образца до точного заполнения персональной информации и доставки по стране — все находится под полным контролем качественных специалистов.
Всем, кто хочет найти оперативный способ получить необходимый документ, наша компания предлагает отличное решение. Купить аттестат – значит избежать длительного процесса обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в университет или начало карьеры.
http://rem.4nmv.ru/forum/profile.php?action=show&member=3949
Betcorner web sitesi bakara oyununun çeşitli biçimlerini sunuyor.Betcorner Bu cihazla düşük hız, şarjsız, yüksek oynatma, yıldırım ve daha fazlası gibi çeşitli özelliklerin keyfini çıkarabilirsiniz. Görünürde daha fazla tablo var.
В нашем обществе, где диплом становится началом удачной карьеры в любой сфере, многие пытаются найти максимально простой путь получения образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим начать профессиональную деятельность или учиться в любом университете.
Наша компания предлагает оперативно получить этот важный документ. Вы имеете возможность приобрести диплом нового или старого образца, что будет отличным решением для человека, который не смог завершить обучение, потерял документ или желает исправить свои оценки. диплом изготавливается аккуратно, с максимальным вниманием ко всем деталям. В результате вы получите продукт, 100% соответствующий оригиналу.
Плюсы такого решения состоят не только в том, что можно быстро получить диплом. Весь процесс организован комфортно, с нашей поддержкой. От выбора требуемого образца диплома до точного заполнения персональной информации и доставки по России — все под полным контролем квалифицированных специалистов.
Для всех, кто хочет найти максимально быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать продолжительного обучения и сразу переходить к достижению собственных целей, будь то поступление в ВУЗ или старт профессиональной карьеры.
http://innsbruckguide.ru
http://calltraxplus.com
http://aspekta.ru
http://thusc.org
http://muscle-bg.com
Bahisplay sanal spor Web sitesi çok çeşitli Bahisplay spor bahis seçenekleri sunuyor.
Zilyonerbet Adresine katılmak Zilyonerbet rulet Zilyonerbet ve güncellemelerden haberdar olmak için bizi takip etmeye devam edin.
Bahisdiyo özelliği ile Bahisdiyo, maç izleme Bahisdiyo deneyimini eğlenceli ve ilgi çekici bir aktiviteye dönüştürüyor.
This text is invaluable. Where can I find out more?
https://sanatoriy-yuscki18.ru/pacient/reviews.php?PAGEN_1=2674
https://reguitti.com.ua/
https://bookmarkhard.com/story17060711/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%81%D0%B0%D0%BD%D1%82%D0%B5%D1%85%D0%BD%D0%B8%D0%BA%D0%B0
https://dom-30.ru/
http://weekevents.ru/diplom-vashego-vyibora-zhdet-vas-sdelayte-svoy-zakaz-pryamo-seychas
В нашем мире, где диплом – это начало успешной карьеры в любом направлении, многие стараются найти максимально простой путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь именно он открывает дверь перед всеми, кто желает вступить в сообщество профессионалов или продолжить обучение в высшем учебном заведении.
Мы предлагаем максимально быстро получить любой необходимый документ. Вы сможете купить диплом, и это становится удачным решением для человека, который не смог закончить обучение или потерял документ. дипломы производятся аккуратно, с особым вниманием ко всем деталям, чтобы в результате получился полностью оригинальный документ.
Превосходство данного решения состоит не только в том, что вы оперативно получите диплом. Процесс организовывается просто и легко, с профессиональной поддержкой. Начиная от выбора нужного образца документа до консультаций по заполнению персональной информации и доставки по России — все под полным контролем квалифицированных специалистов.
Для всех, кто хочет найти максимально быстрый способ получения необходимого документа, наша компания предлагает выгодное решение. Купить диплом – это значит избежать длительного процесса обучения и сразу перейти к достижению собственных целей, будь то поступление в университет или старт успешной карьеры.
http://sankeystokyo.info
http://osvoidachu.ru
http://aminotein.com.ua
http://x2lab.ru
http://pharmacie-centrale-toscano.fr
Batumslot Türk hizmet adresi, Spor Toto Batumslot organizasyon yetkilileri gibi resmi prosedürler var.
Betmetre iletişim kanallarını kullanarak Betmetre tercihlerinizi dile getirerek sorunlarınıza çözüm bulabilirsiniz.
Placing the site on quality trust sites
Getting a spot for your online platform on trusted, superior platforms is an vital step in strengthening its digital reputation. The said placements not only lift a site’s standing in the perspective of online search tools but also develop credibility among its intended audience. Trusted sites, celebrated for their tight content guidelines and considerable user bases, act as authenticators, endorsing the genuineness and worth of content they associate with or host. As visitors from these platforms work their way to a site, they arrive with a pre-set sense of trust, making them more inclined to participate and turn.
Additionally, locating a website on top-quality trust sites moves beyond just link-building approaches. It’s an opportunity for symbiotic growth and cooperation. Linking with honored platforms lets a brand to correspond its ethics with those of the trusted site, promoting shared principles and bolstering its brand tale. In the constantly growing digital ecosystem, where truthfulness is at a peak, such planned placements guarantee a site’s enduring visibility, significance, and sway.
Отчётность.
Оплата: Yoo.Money, Bitcoin, МИР, Visa, MasterCard…
Принимаем USDT
Телега: https://t.me/exrumer
Skype: Loves.Ltd
https://www.webfx.com/
https://xrumer.cc/
Yedibahis adresinizi ve iş yerinizi öğrenmek için Yedibahis mevcut en kesin bilgilere, yani siteden SMS yoluyla aldığınız Yedibahis güncel adres bilgilerine güvenebilirsiniz.
Thanks a bunch for sharing this with all of us you really realize what you’re speaking approximately! Bookmarked. Kindly also seek advice from my web site =). We could have a hyperlink trade arrangement among us
http://animesocial.su/blogs/1138/Как-купить-аттестат-или-диплом-у-нас-в-онлайн-магазине
http://www.vecmir.ru/index.php/vecmirlife/search/browse?sort=latest&start=120
http://delpc.ru/kak-ustanovit-kuler/
http://thepirog.ru/9_kategoria/9_kategoria-22.php
http://www.briefeducation.ru/bried-180.html
http://theunworldlytravelers.com
Hi there this is kind of of off topic but I was wondering if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding know-how so I wanted to get advice from someone with experience. Any help would be enormously appreciated!
stlouisbluesclub.com/read-blog/327_why-is-the-popularity-of-universities-decreasing-nowadays.html?mode=dayВ
http://www.tovery.net/guestbook.asp?user=okokok&Page=3В
museums.artyx.ru/books/c0003_1.shtmlВ
fpcom.co.kr/bbs/board.php?bo_table=free&wr_id=110868В
uapa.com.hk/forum/home.php?mod=space&uid=292023&do=profileВ
Если вы алчете чувствовать себя на седьмом небе творениями популярных авторов на AudioBook26.ru, Читака зовет онлайн прослушивание различных жанров аудиокниг
https://audiobook26.ru/
Wonderful, what a webpage it is! This webpage presents helpful data to us, keep it up.
http://alisaslut.ru/nemnogo-o-sebe/
http://zoointernet.ru/index.html
http://www.dializ46.ru/lenta-57.html
http://rintor.net/dvdrip-web-dl-webrip-vhsrip/8512-creamed-teens-obkonchannye-podrostki-2020-web-dl.html
http://oracledbs.com/viewtopic.php?t=38053&start=10
В нашем обществе, где аттестат становится началом отличной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа переоценить просто невозможно. Ведь диплом открывает дверь перед каждым человеком, желающим начать трудовую деятельность или учиться в университете.
Предлагаем быстро получить любой необходимый документ. Вы можете купить аттестат, что становится удачным решением для всех, кто не смог закончить обучение или утратил документ. Каждый аттестат изготавливается аккуратно, с максимальным вниманием к мельчайшим деталям. В результате вы сможете получить документ, 100% соответствующий оригиналу.
Преимущества этого решения заключаются не только в том, что вы быстро получите свой аттестат. Процесс организовывается комфортно, с нашей поддержкой. Начав от выбора подходящего образца до консультаций по заполнению персональной информации и доставки по России — все находится под полным контролем квалифицированных мастеров.
Всем, кто ищет быстрый способ получить необходимый документ, наша компания может предложить выгодное решение. Заказать аттестат – значит избежать длительного обучения и сразу перейти к важным целям: к поступлению в ВУЗ или к началу удачной карьеры.
https://www.datxanhhanoi.com/can-ho-dang-cap/park-9-times-city-park-hill-premium/66.html
Hi, I desire to subscribe for this web site to obtain newest updates, thus where can i do it please help.
staceylcrow.com/2013/jess-kenny-redlands-ca/В
http://www.mountbarkertourismwa.com.au/contact_us.phpВ
containertraffic.ru/news/detail.php?ID=54В
forum.mbprinteddroids.com/showthread.php?tid=1087&pid=1351&mode=threadedВ
wiuwi.com/blogs/70298/%D0%93%D0%B4%D0%B5-%D0%BD%D0%B0%D0%B9%D1%82%D0%B8-%D1%81%D0%B0%D0%BC%D1%8B%D0%B5-%D0%BD%D0%B0%D0%B4%D0%B5%D0%B6%D0%BD%D1%8B%D0%B5-%D0%BF%D0%BE%D1%81%D1%82%D0%B0%D0%B2%D1%89%D0%B8%D0%BA%D0%B8-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D0%BE%D0%B2В
Wonderful post but I was wanting to know if you could write a litte more on this subject? I’d be very thankful if you could elaborate a little bit further. Appreciate it!
sohapay.com/info/vi/news/trai-nghiem-tien-ich-voi-the-gioi-the-vpbank.htmlВ
apartamentospuntum.com/nuestros-clientesВ
nation.geoman.ru/demogr/alph0014.shtmlВ
ripet.hu/index.phpВ
panarabiaenquirer.com/wordpress/israeli-defence-forces-to-commemorate-international-holocaust-memorial-day-by-locking-up-record-number-of-palestinian-children/В
Thank you a lot for sharing this with all of us you actually realize what you’re talking approximately! Bookmarked. Kindly also consult with my web site =). We will have a hyperlink alternate contract between us
amisite.ru/phpBB2/memberlist.php?mode=joined&order=ASC&start=6850В
http://www.ba98.org/contact.php?sujet=6В
tnrevergreen.com.vn/dien-tich-tim-tuong-la-gi-mua-nha-nen-dung-cach-tinh-dien-tich-nao/В
ediblehomegardensresorts.com/index.php?do=/public/user/blogs/name_Alanpoe/page_7/В
gadjetforyou.ru/kupit-diplom-10В
Предлагаем для вас пройти консультацию (аудит) по умножению продаж равным образом доходы в вашем бизнесе. Формат аудита: личная встреча или сессия по скайпу. Делая очевидные, но не сложные действия, прибыль от ВАШЕГО бизнеса можно поднять в много раз. В нашем багаже более 100 испытанных практических способов повышения результатов а также прибыли. В зависимости от вашего бизнеса расчитаем для вас максимально сильные и начнем постепенно внедрять.
-https://interestbook.ru/
В нашем обществе, где диплом является началом удачной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Необходимость наличия документа об образовании трудно переоценить. Ведь именно диплом открывает дверь перед любым человеком, который собирается начать трудовую деятельность или учиться в ВУЗе.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы можете заказать диплом нового или старого образца, что становится удачным решением для человека, который не смог закончить образование, утратил документ или хочет исправить плохие оценки. Все дипломы производятся аккуратно, с максимальным вниманием к мельчайшим нюансам. На выходе вы сможете получить полностью оригинальный документ.
Преимущества подобного решения заключаются не только в том, что вы сможете максимально быстро получить свой диплом. Весь процесс организован удобно, с профессиональной поддержкой. От выбора нужного образца до консультации по заполнению личных данных и доставки по стране — все под абсолютным контролем квалифицированных специалистов.
Всем, кто пытается найти оперативный способ получения необходимого документа, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать долгого обучения и сразу переходить к важным целям: к поступлению в ВУЗ или к началу успешной карьеры.
http://ilmuseodelrugby.it
http://brickbybrick.sk
http://lkp-it.net
http://43stcorp.com
http://wilio.ro
더 도그 하우스 메가웨이즈
황제가 차에서 내리는 것을 보고 그들 중 일부를 알아봤을 때 모두가 깜짝 놀랐습니다.
Winbir açma süreci Winbir ödemesi ve sertifika Winbir alması gereken bazı şeyleri kapsamamaktadır. Hiçbir ücret ödemeden katılabilirsiniz.
Maritbet altyapı adresi kayıt olduktan sonra Maritbet bu alanda lisanslama hizmeti sunduklarını görebilirsiniz.
Appreciating the time and effort you put into your website and detailed information you present. It’s good to come across a blog every once in a while that isn’t the same out of date rehashed information. Excellent read! I’ve bookmarked your site and I’m adding your RSS feeds to my Google account.
http://connect.nteep.org/blogs/810/What-do-you-need-to-pay-attention-to-when-ordering?lang=fr_fr
http://artritdoc.ru/bol-v-spine
http://domowik.net/zdorove/vidy-solncezashhitnyx-sredstv.html
http://www.dr-kumaki.net/aikuma/index.cgi?mode=past&pno=0036&pg=100
http://arvixe.ru/v/building/cotedge_1/P1070836.JPG.html
Betoffice tasarımı, bir tür hesaplamadan türetildiğini ima Betoffice ediyor. Örnek olarak, anında erişilebilmesi için ana sayfada belirli yarış takvimleri bulunmaktadır.
Timabet müşteri hizmetleri Kayıt olup giriş yaptıktan Timabet sonra yandaki menüden Canlı Bahis seçeneğini seçerek devam eden ve başkalarının oynadığı maçları görüntüleyebilirsiniz.
Arascasino ilk para yatırma işlemi Güvenli, birinci sınıf Arascasino slot oyunlarının keyfini çıkarmak için hemen kaydolun.
В нашем обществе, где диплом является началом отличной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения качественного образования. Важность наличия официального документа переоценить просто невозможно. Ведь именно диплом открывает двери перед всеми, кто стремится начать профессиональную деятельность или продолжить обучение в университете.
Мы предлагаем оперативно получить этот необходимый документ. Вы сможете приобрести диплом нового или старого образца, и это будет отличным решением для всех, кто не смог закончить образование, утратил документ или хочет исправить свои оценки. диплом изготавливается с особой тщательностью, вниманием к мельчайшим деталям, чтобы в итоге получился документ, максимально соответствующий оригиналу.
Превосходство такого решения состоит не только в том, что можно оперативно получить свой диплом. Весь процесс организован просто и легко, с профессиональной поддержкой. Начав от выбора требуемого образца диплома до грамотного заполнения личных данных и доставки по России — все под абсолютным контролем качественных специалистов.
Всем, кто ищет оперативный способ получить необходимый документ, наша услуга предлагает отличное решение. Заказать диплом – значит избежать длительного обучения и сразу перейти к достижению собственных целей, будь то поступление в ВУЗ или начало успешной карьеры.
http://almazremont.ru
http://cuidemoselagua.cl
http://cityjeans.com.ua
http://vsedlo.ru
http://ready2go.com.ua
Misalibet kategori iki bölüme ayrılmıştır: birincisi Misalibet Casino bölümü, ikincisi ise Spor ve Canlı kategorilerdir.
Oynabil ile bağlantı kurmak isteyenlerin Oynabil dışında site ile herhangi bir ilişkisi olmayan insan grupları da bulunmaktadır.
Can you tell us more about this? I’d like to find out more details.
opengadjet.ru/page/4В
bdt-team.ru/adventure/?adventure&page3/index.htmlВ
http://www.dongkhwang.go.th/myweb/myweb/webboard/view_topic.php?id=3460В
vsekruizi.ru/Special_offers/thailand_from_krasnodar/В
moscityservice.ru/В
Appreciating the hard work you put into your site and detailed information you offer. It’s great to come across a blog every once in a while that isn’t the same unwanted rehashed material. Great read! I’ve saved your site and I’m adding your RSS feeds to my Google account.
almaztb.ru/index.php?links_exchange=yes&page=46&show_all=yesВ
http://www.taijiacademy.com/newforum_down/profile.php?mode=viewprofile&u=300836В
http://www.pentictonsoccerclub.com/may-classicВ
dachaweek.ru/page/3В
http://www.agrimoto.fr/achat/cat-piles-170.htmlВ
This is the right website for anyone who wishes to find out about this topic. You understand so much its almost hard to argue with you (not that I actually would want to…HaHa). You certainly put a fresh spin on a subject which has been discussed for decades. Wonderful stuff, just great!
http://frsvo.ru/forum/profile.php?id=6233
http://prasemena.ru/articles/ovoschi/pomidor_richai
http://uchieto.ru/foto-krasivo-oformlennyx-domov/
http://medgora.ru/users.php?m=details&id=125321
http://followmedoitbbs.com/home.php?mod=space&username=osytyroby
Awesome issues here. I’m very happy to look your post. Thanks so much and I am taking a look forward to contact you. Will you kindly drop me a mail?
http://zero-100.ru/photo/enz/vse_bolidy_f1_s_1950_po_2014_god/1969_brabham_bt33/18-0-660
http://www.vocal.com.ua/node/www.xuzhouabl.com/www.xuzhouabl.com/en?page=171
http://uziprosto.ru/tag/mrt-diagnostika
http://vyazanshapki.ru/vyazanyie-shapki/zhenskie/shapka-s-obemnyim-pletenyim-uzorom-spitsami
http://krd.best-city.ru/forum/message69216/answer/?mode=quote
Asibet Adresi öğrenen BTK, Asibet kaçak ilan edip kapattı. Asibet Asibet güvenilir bir bahis sitesidir ancak daha az verimli görünmektedir.
Bet11 lisans adresi takılan her türlü soru için Bet11 ekibimize günün veya gecenin her saatinde ulaşabilirsiniz, canlı destek adresi üzerinden size gerekli bilgileri vereceklerdir.
Misliyle bilgilerinizin güncellendiğini ve yanlış Misliyle web sitesine yönlendirildiğinizi üzülerek bildiririz.
Koçbet Müşteriler en çok bu üründen memnun. Basitçe Koçbet söylemek gerekirse, sitenin bahis sonuçlarını tahmin etmede ne kadar iyi olduğunu gösterir.
XRumer – этто наиболее лучшее также лучшее программное обеспечение для создания гиперссылок и еще продвижения сайтов. Он используется чтобы машинального прогона страниц после различные соц сети, форумы, блоги да часть интернет-ресурсы.
http://www.botmasterru.com/product33230/
Купить XRumer Оффициально
Avvabet Web sayfanızda bir Avvabet tartışma panosu görüntüleniyor.
7bey, bağlantı kurmak ve erişim sınırlamalarını 7bey aşmak için sürekli olarak yeni yöntemler deniyor.
Rifbahis sayfada yatırım yaparken para Rifbahis yatırmanın farklı yolları sunulmaktadır.
Visabahis kayıt formu doldurarak ve güncellemeleri Visabahis alma seçeneğini seçerek Web Sitesine yeni kimlik bilgilerinizi vermiş olacaksınız.
에그 카지노
Fang Jifan은 “장관이 먼저 전세를 냈습니다. “라고 웃었습니다.
Excellent pieces. Keep posting such kind of information on your site. Im really impressed by your blog.
Hi there, You have performed an excellent job. I will definitely digg it and in my view suggest to my friends. I am sure they will be benefited from this site.
http://autonewsmake.ru/mini-dzhipy.html
http://razvitie-rebenka24.ru/2017/05
http://kvakva55.ru/atletiko-interesen-dzhaka.html
http://www.uvina.ru/services/pokupka-metallolom/priem-loma-met/
http://forum.javabox.net/viewtopic.php?f=20&t=12432&p=23278
A beloved National Park Service ranger died when he tripped, fell and struck his head on a rock during an annual astronomy festival in southwestern Utah, park officials said over the weekend.
[url=https://at-kraken17.at]kraken14.at[/url]
Tom Lorig was 78 when he died after the incident at Bryce Canyon National Park late Friday.
kraken14.at
https://kraken-16.at
He was known for his extensive work as a ranger and volunteer at 14 National Park Service sites, including Yosemite National Park, Carlsbad Caverns National Park and Dinosaur National Monument, the park service said in a statement Saturday.
“Tom Lorig served Bryce Canyon, the National Park Service, and the public as an interpretive park ranger, forging connections between the world and these special places that he loved,” Bryce Canyon Superintendent Jim Ireland said in the statement.
Thank you for the auspicious writeup. It actually used to be a leisure account it. Look complex to more added agreeable from you! However, how can we be in contact?
http://moiyspex.ru/finpraktik/obligacii-chto-eto-takoe-prostymi-slovami.html
http://shevy-aveo.ru/remont-hodovoy/menyaem-v-aveo-opornyie-podushki-perednih-stoek-na-usilennyie/
http://magnitog.ru/katalog-produktsii/magnitnye-igrushki/futbol-magnetiz-mag-2–16071.html
http://chem100.ru/elem.php?n=79
http://i-efremov.ru/osnovnie-dati-zhizni-i-tvorchestva.html
Simply desire to say your article is as surprising. The clearness to your put up is simply nice and i could think you’re knowledgeable on this subject. Well with your permission let me to grab your feed to stay up to date with drawing close post. Thanks 1,000,000 and please continue the enjoyable work.
http://www.visayasmedcebu.com.ph/career-opportunities/hiring-licensed-engineer
http://razvitie-rebenka24.ru/2017/05
http://www.czystaenergiadwa.milanow.pl/viewtopic.php?t=17840
http://www.view24.co.kr/bbs/board.php?bo_table=qa&wr_id=1292
http://mobzilla.su/novosti-jelektroniki/jekologija/7822-ukraina-poterjala-16-punktov-mezhdunarodnogo-klimaticheskogo-indeksa-jekologija.html
Подарок для конкурента
https://xrumer.ru/
Такой пакет берут для переспама анкор листа сайта или мощно усилить НЧ запросы
Подарок для конкурента
В нашем мире, где аттестат – это начало отличной карьеры в любом направлении, многие пытаются найти максимально простой путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь диплом открывает двери перед каждым человеком, который хочет начать трудовую деятельность или продолжить обучение в ВУЗе.
Предлагаем оперативно получить этот важный документ. Вы сможете купить аттестат нового или старого образца, и это является выгодным решением для всех, кто не смог закончить образование, потерял документ или желает исправить плохие оценки. Аттестат изготавливается аккуратно, с особым вниманием к мельчайшим элементам. На выходе вы сможете получить документ, максимально соответствующий оригиналу.
Плюсы такого подхода состоят не только в том, что вы максимально быстро получите свой аттестат. Процесс организовывается удобно, с нашей поддержкой. Начиная от выбора требуемого образца до консультаций по заполнению персональной информации и доставки по стране — все под абсолютным контролем наших специалистов.
Для тех, кто пытается найти оперативный способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать аттестат – это значит избежать долгого процесса обучения и не теряя времени переходить к достижению собственных целей: к поступлению в университет или к началу трудовой карьеры.
https://kultyres.ru/uchebnie_materiali/студенческие-работы/arhitektyra-v-drevnosti.html
I loved as much as you’ll receive carried out right here. The sketch is attractive, your authored material stylish. nonetheless, you command get bought an edginess over that you wish be delivering the following. unwell unquestionably come more formerly again as exactly the same nearly very often inside case you shield this hike.
http://klaus-peltzer.de/kontakt/comment-page-529/
http://www.herurg.ru/artist/metal_mind.html
http://www.ssangyong-echoauto.ru/lenta-31.html
http://admeclub.ru/pozdravlenie-lyubovi/
http://simracing.su/1122-intervyu-posle-pyatogo-etapa-na-trasse-laguna-seka.html
This information is worth everyone’s attention. How can I find out more?
http://mdr7.ru/topic7603.html
http://uchieto.ru/foto-krasivo-oformlennyx-domov/
http://posudca.ru/istoricheskie-romanyi/nagib-mahfuz-i-ego-roman-mudrost-heopsa
http://maghands.ru/dermatozy/pochesuha-na-rukah-chto-eto-takoe
http://facefam.com/read-blog/16_we-purchase-a-university-diploma-on-the-internet-tips.html
В современном мире, где диплом – это начало отличной карьеры в любой сфере, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании сложно переоценить. Ведь именно он открывает двери перед людьми, желающими начать профессиональную деятельность или продолжить обучение в каком-либо университете.
Предлагаем быстро получить этот важный документ. Вы можете заказать диплом нового или старого образца, что становится выгодным решением для человека, который не смог завершить образование или потерял документ. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам. В результате вы сможете получить документ, 100% соответствующий оригиналу.
Преимущества такого подхода заключаются не только в том, что вы оперативно получите диплом. Процесс организован удобно, с нашей поддержкой. От выбора нужного образца до консультации по заполнению личной информации и доставки по стране — все под абсолютным контролем качественных специалистов.
В результате, для тех, кто пытается найти быстрый и простой способ получения необходимого документа, наша компания готова предложить отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и сразу перейти к достижению собственных целей, будь то поступление в университет или начало успешной карьеры.
http://containersales.ru
http://akademiyatepla.ru
http://bakili-fclub.com
http://clickforart.com
http://barabanispb.ru
Pretty! This was an incredibly wonderful article. Many thanks for providing these details.
http://www.recxon.ru/kontrolnij-match–stal–a—–krimteplica-10.html
http://light-of-love.ru/aforizmi-o-liubvi/tsitati-so-smislom-o-liubvi-k-parniu-muzhchine-tsitati-pro-liubov-schaste-i-otnosheniya.html
http://trprint3d.ru/kak-oplatit-nalog-cherez-bank-vtb/index.html
http://mywebs.su/content/video/4653
http://jockelakso.com/jaa/
Betsalvador İyi bir bahis programı Betsalvador bulmak için Betsalvador spor bahisleri menüsüne mutlaka göz atın.
Casinoplus Kampanyayı ziyaret ederek Casinoplus ücretsiz reklam fırsatlarından yararlanabilirsiniz.
Betcasper sanal oyunlar Futbol, basketbol, Betcasper tenis, sutopu ve diğer sporlar sonucunda bahisler oynanmaktadır.
Great post! Really enjoyed reading. Keep up the good work!
We have many royal escort services in Gurgaon that are providing exciting services to VIP customers
Pushkar escorts
This is a premium source for hobbyists to book exotic women for royal adult experiences with enjoyable services.
Mumbai escort service
When you come to our agency, rest assured that your erotic fantasies and imaginations will be fulfilled immediately.
Escort Service in Lucknow
There will be complete satisfaction with your erotic and romantic needs in the meeting.
Bangalore escorts
Arexbet twitter hesabı dünya çapında mutlak Arexbet güvenliği garanti ediyor ve Twitter hesabı tarafından destekleniyor.
https://seoconversionator.izrablog.com/27857497/Примеры-работ-студии-xrumer-art
В нашем обществе, где диплом – это начало успешной карьеры в любой отрасли, многие ищут максимально быстрый путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь именно диплом открывает дверь перед любым человеком, желающим вступить в сообщество квалифицированных специалистов или учиться в университете.
Мы предлагаем максимально быстро получить любой необходимый документ. Вы имеете возможность приобрести диплом, что будет отличным решением для всех, кто не смог завершить образование или утратил документ. диплом изготавливается с особой аккуратностью, вниманием к мельчайшим элементам, чтобы в итоге получился 100% оригинальный документ.
Преимущества подобного подхода состоят не только в том, что можно быстро получить свой диплом. Весь процесс организован комфортно, с нашей поддержкой. Начиная от выбора требуемого образца документа до консультаций по заполнению личной информации и доставки по стране — все будет находиться под полным контролем наших специалистов.
Для тех, кто хочет найти максимально быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к достижению собственных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://cryptocoinsnewshub.com
http://linkmarketings.com
http://above-second.com
http://c-web-coding.com
http://historiadeldeporte.net
Делаем отличное предложение для вас провести консультацию (аудит) по умножению продаж равным образом доходы в вашем бизнесе. Формат аудита: личная встреча или сессия по скайпу. Делая верные, но простые действия, результат от ВАШЕГО бизнеса получится превознести в много раз. В нашем запасе более 100 опробованных практических методик умножения торгов а также доходов. В зависимости от вашего коммерциала разработаем для вас максимально крепкие и будем шаг за шагом претворять в жизнь.
– interestbook.ru
Что такое прогон по профилям?
На форумах при каждом прогоне регистрируется новый юзер, в профайле которого в поле Вебсайт или Домашняя страница стоит ваша ссылка. Также в некоторых форумах есть возможность в профиль вставить подпись, туда можно вставить ссылку с анкором. Все это можно варьировать. В профилях ссылки живут довольно долго, т.к. нет спама в чистом виде, и профили не мозолят глаза ни читателям форума, ни модераторам. Прогон производится последней версией хрумера с сервера с максимально возможной скоростью и производительностью.
ПОДРОБНЕЕ
НОВЫЙ ОТЛИЧНЫЙ VPN ДЛЯ XRUMERA!
За последние недели состоялась серия важных обновлений комплекса: XAuth, Captchas Benchmark, XRumer, XEvil 6.0 . В XEvil 6.0 обновлен механизм обработки hCaptcha и многое другое, в XAuth повышена стабильность работы, в XRumer повышена эффективность, устранён ряд погрешностей и обновлены прилагаемые базы.
https://xrumer.ru/
[url=http://www.betnbet.by/bonus/betera-freebet-40byn-100fs]Сахарный Поссум[/url] — это лучший аргумент в пользу того, что маленькие вещи — самые важные!
Daddy Casino – Официальный телеграм канал со слотами от Дэдди Casino daddy casino официальный сайт . Актуальное, рабочее зеркало официального сайта Дэдди Казино. Регистрируйся в Казино Daddy, получай бонус используя промокод, не забудь скачать
daddy казино вход
https://t.me/daddycasinorussia
슬롯 게임 사이트
Hongzhi 황제는 두 사람을 자세히 살펴 보았습니다. “당신은 그냥 … 함께 있었나요?”
Why people still use to read news papers when in this technological globe everything is existing on net?
http://musey-uglich.ru/
В Dragon money можно найти разнообразные азартные игры. Промокоды предоставят дополнительные выгоды вашему игровому процессу. Ассортимент игр удовлетворит запросы даже самых требовательных геймеров. В этом сообществе доступно множество бездепозитных промокодов.
Телесуфлёр — этто tvprompter.ru я, тот или другой подсобляет ораторам (а) также основным телепередач читать экспликация, никак не раскапывая соображения от камеры или аудитории. Оно обычно устраивается унтер камерой (а) также отражает экспликация с подмогой полупрозрачного зеркала, творя иллюзию прямого воззрения в течение объектив. Этот юлина незаменим при проведении ровных эфиров а также значительно упрощает службу телеведущих.
http://tvprompter.ru/
Интернет-казино дэдди казино официальный сайт — сайт или специальная программа, которые позволяют играть в азартные игры в интернете. Сайты интернет-казино могут специализироваться на различных азартных играх: игровых автоматах, рулетке, лотереях, викторинах, покере или других карточных и настольных играх. Википедия
дэдди казино
https://t.me/daddycasinorussia
Why viewers still make use of to read news papers when in this technological world the whole thing is accessible on web?
http://www.musey-uglich.ru
Maltbahis güvenilir ve lisanslı bir bahis Maltbahis sitesine olmasına rağmen bazı kısıtlamalar ve engellemeler olmakta.
Maltbahis üye üyeliğe katılabilmeniz için kişisel Maltbahis bilgilerinizi doğru ve eksiksiz olarak vermeniz gerekmektedir.
Maltbahis üyeliğinin sonlandırılması Maltbahis Maltbahis protestolarının ardından offshore sitesi Maltbahis hakkında daha derinlemesine bir soruşturma yürüttük.
Maltbahis fiyatı ne kadar yarın SMSi devre dışı bırakma Maltbahis eylemini tartışalım mı? Şimdi daha önce tartıştığımız yanlış bilgilere dönelim.
Maltbahis katılım oyun başlamadan önce, maç öncesi Maltbahis bahisler olarak adlandırılan belirli bahisler oynanır.
Maltbahis inceleme Maltbahis sitenin kullanıcılarına Maltbahis en iyi ve en etkili bahis hizmetlerini sunduğunu gösteriyor.
Onwin güvenilirliği kullanıcı sayfanın sağ üst köşesindeki Onwin kayıt seçeneğini seçerek herhangi bir ödeme yapması gerekmediğini fark edecektir.
Onwin web sitesine kaydolun burada en güncel Onwin bahis oranları ve bonusları bulabilirsiniz.
Герберт Уэллс. ЖЗЛ. Жизнь замечательных людей и лучшие обзоры по психиатрии.
Сохраняй в закладки порно для транссексуалов что бы не потреять доступ
Hello colleagues, pleasant article and good arguments commented here, I am actually enjoying by these.
http://www.yenko.net/forum/member.php?tab=visitor_messaging&u=70162
купить аттестаты за 9
http://crimeamice.ru
купить диплом в выборге
It’s awesome to visit this web page and reading the views of all mates concerning this article, while I am also zealous of getting familiarity.
купить диплом в невинномысске
http://variant-surgut.ru
http://studioru.ru
http://unikum-student.ru
купить диплом в томске
Hi, I want to subscribe for this weblog to obtain hottest updates, therefore where can i do it please help.
купить диплом в железногорске
http://mixnutrition.ru
http://shot-spb.ru
http://ovolgodonske.ru
купить диплом в бузулуке
https://loga.solutions/hello-world/
https://termocontrolservizi.it/b6/
https://msguancha.com/plus/guestbook.php?gotopagerank=&totalresult=3566019&pageno=2
http://www.obscurita-eterna.de/index.php/guestbook/index/index/page/www.lasix.com%5B/www.lasipen.com%5B/www.hygroton.com%5B/www.alivian.com
https://justgetfucked.com/i-dont-know-what-he-thought-was-going-to-happen/
Keep this going please, great job!
купить диплом в рыбинске
http://megaskee.ru
http://kinelgorod.ru
http://seoklan.ru
купить диплом в ангарске
Do you have a spam issue on this blog; I also am a blogger, and I was curious about your situation; many of us have developed some nice procedures and we are looking to exchange strategies with others, why not shoot me an e-mail if interested.
купить диплом в великом новгороде
http://fast-education.ru
http://club-subaru.ru
http://ruscuisineambassador.ru
купить диплом журналиста
프라 그마 틱 슬롯 사이트
그는 자신의 제한된 IQ로는 더 이상 충분하지 않다고 느꼈습니다.
Slightly off topic 🙂
It so happened that my sister found an interesting man here, and recently got married ^_^
(Admin, don’t troll!!!)
Is there are handsome people here! 😉 I’m Maria, 27 years old.
I work as a model, successfull – I hope you do too! Although, if you are very good in bed, then you are out of the queue!)))
By the way, there was no sex for a long time, it is very difficult to find a decent one…
And no! I am not a prostitute! I prefer harmonious, warm and reliable relationships. I cook deliciously and not only 😉 I have a degree in marketing.
My photo:
___
Added
The photo is broken, sorry(((
Check out my blog where you’ll find lots of hot information about me:
https://antigua-nedv.ru
Or write to me in telegram @Lolla_sm1_best ( start chat with your photo!!!)
Why people still use to read news papers when in this technological globe everything is presented on web?
купить диплом учителя
http://dety-tymovsk.ru
http://intsms.ru
http://primzona.ru
купить диплом фитнес инструктора
Its like you read my mind! You seem to know a lot about this, like you wrote the book in it or something. I think that you can do with some pics to drive the message home a bit, but instead of that, this is magnificent blog. A fantastic read. I will definitely be back.
http://demo2.weboss.hk/1009/comment/html/?578.html
купить диплом в крыму
http://bolschiedengi.ru
купить диплом во всеволожске
Thanks for some other excellent article. The place else could anybody get that kind of information in such a perfect manner of writing? I’ve a presentation subsequent week, and I’m on the search for such info.
купить диплом в благовещенске
http://bel-seo.ru
http://scienceanded.ru
http://uk-ahml.ru
купить диплом в химках
milf free
https://m.coniconimall.com/member/login.html?noMemberOrder=&returnUrl=http%3a%2f%2ftubesweet.com
https://colibricoffee.ru/bitrix/redirect.php?goto=https://tubesweet.com/
https://mona-liza.com/bitrix/redirect.php?goto=https://tubesweet.com/
http://musdepot.ru/bitrix/redirect.php?goto=https://tubesweet.com/
Hey there I am so happy I found your website, I really found you by mistake, while I was looking on Google for something else, Nonetheless I am here now and would just like to say cheers for a incredible post and a all round exciting blog (I also love the theme/design), I don’t have time to browse it all at the moment but I have book-marked it and also included your RSS feeds, so when I have time I will be back to read a lot more, Please do keep up the fantastic work.
http://ntek-nt.ru/students/raspisanie/
купить диплом о высшем образовании
http://copoko.ru
купить диплом инженера механика
Howdy! Do you know if they make any plugins to help with SEO? I’m trying to get my blog to rank for some targeted keywords but I’m not seeing very good gains. If you know of any please share. Thank you!
http://www.wikifocus.ru/dinamo-sigraet-v-belom-tavriya—v-krasnom.html
купить диплом в ростове-на-дону
http://milru.ru
купить диплом в серове
Hello it’s me, I am also visiting this site daily, this web site is really fastidious and the people are really sharing nice thoughts.
куплю диплом высшего образования
http://family-spb.ru
http://websweb.ru
http://sartvo.ru
купить диплом продавца
Сегодня, когда диплом становится началом отличной карьеры в любой области, многие стараются найти максимально быстрый путь получения качественного образования. Важность наличия документа об образовании переоценить просто невозможно. Ведь именно он открывает двери перед всеми, кто хочет начать трудовую деятельность или учиться в ВУЗе.
В данном контексте мы предлагаем максимально быстро получить этот необходимый документ. Вы сможете приобрести диплом, что будет удачным решением для человека, который не смог завершить образование, потерял документ или желает исправить свои оценки. диплом изготавливается с особой аккуратностью, вниманием ко всем элементам. На выходе вы сможете получить полностью оригинальный документ.
Превосходство такого решения состоит не только в том, что можно оперативно получить свой диплом. Процесс организован комфортно, с профессиональной поддержкой. Начав от выбора нужного образца диплома до консультаций по заполнению персональной информации и доставки в любое место страны — все под полным контролем опытных мастеров.
Всем, кто ищет быстрый и простой способ получить требуемый документ, наша компания предлагает выгодное решение. Купить диплом – это значит избежать долгого обучения и сразу перейти к достижению своих целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://coaching-org.ru
http://bolschiedengi.ru
http://xsportclubs.ru
http://elistavdv.ru
http://extra-art.ru
Hi there, after reading this remarkable paragraph i am as well glad to share my experience here with colleagues.
http://fh7778nc.bget.ru/users/ubezom
https://wiki.3cdr.ru/wiki/index.php/пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ!
https://telegra.ph/Kupit-Diplom-Bystro-i-Legko-05-28
https://odyclub.ru/index.php?/topic/4097-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-n900z/
http://shoptema.ru/forum/topic/32401/
It’s remarkable in favor of me to have a site, which is beneficial designed for my experience. thanks admin
http://trading-labor.de/Легальное_и_безопасное_получение_диплома:_Присоединяйтесь_сегодня!
http://watercrypto.blogspot.com/2014/09/blog-post.html
http://www.ra-journal.ru/board/member.php?389197-miguelpsow
http://school-one.ru/users/apivir
http://forum.ll2.ru/member.php?1201792-miguelpSlibe
I’m gone to inform my little brother, that he should also pay a visit this website on regular basis to get updated from latest news update.
купить речной диплом
http://school-10-lik.ru
http://xsportclubs.ru
http://mir-forum.ru
купить диплом в кинешме
Подарок для конкурента
https://xrumer.ru/
Такой пакет берут для переспама анкор листа сайта или мощно усилить НЧ запросы
Подарок для конкурента
Hi, everything is going nicely here and ofcourse every one is sharing facts, that’s truly fine, keep up writing.
http://reshetiha.ru/category/nauka
купить диплом в владикавказе
http://family-spb.ru
купить диплом стоматолога
슬롯 모아 무료
Xie Qian은 웃음을 참지 못하고 “하하 …”라고 웃었습니다.
This site truly has all the info I needed concerning this subject and didn’t know who to ask.
https://antikawaii.my1.ru/forum/7-7-187#6999
http://true.pahom.su/2024/04/27/bystryy-i-effektivnyy-sposob-poluchit-diplom-zakazhite-onlayn-seychas.html
https://cahaya.my.id/как-получить-диплом-в-кратчайшие-срок
https://letsbookmarktoday.com/profile.php?action=view&mod=space&userinfo=minna_prout_301152
http://avtolux48.ru/people/user/374/blog/7348/
Why visitors still use to read news papers when in this technological world everything is presented on net?
http://www.odnopolchane.net/forum/member.php?u=602738
http://o91746bp.beget.tech/2024/05/26/kupit-diplom-v-internete-legalno-i-konfidencialno.html
http://xn--80ae1aidok.xn--p1ai/users/egajywam
https://say.la/read-blog/33520
http://bagoff.net/forum/viewtopic.php?t=2931
Hello! This post could not be written any better! Reading through this post reminds me of my previous room mate! He always kept chatting about this. I will forward this write-up to him. Fairly certain he will have a good read. Many thanks for sharing!
https://ya.7bb.ru/viewtopic.php?id=11317#p32755
http://forums.indexrise.com/user-306638.html
https://smm-seo.ru/members/robertgreabe.5389/
http://school5.p-fam.ru/users/ivilug
mysoccerex.com/tag/football-club/В
It’s awesome in favor of me to have a web site, which is good for my knowledge. thanks admin
http://j95370pe.bget.ru/main/263-vot-kudy-otvechala-pomeschica-moe-takoe.html
usaessayexperts.com/faq/В
http://vaseninga.ru/learning/
https://www.unitalm.ru/blog/top-7-internet-resursov-po-promyshlennoj-bezopasnosti
http://pechorinase.blogspot.com/2013/04/dartagnan-et-les-trois-mousquetaires.html
Thanks for any other wonderful post. The place else could anybody get that kind of information in such an ideal manner of writing? I have a presentation next week, and I am at the look for such info.
купить диплом в губкине
http://vuzdiploma.ru
http://ussursity.ru
http://crimeamice.ru
купить диплом в димитровграде
This site really has all the information I needed concerning this subject and didn’t know who to ask.
купить бланк диплома
https://postgresconf.org/users/james-hughes
купить диплом в кемерово
Купить Кокаин в Москве? Самый чистый Кокаин в Москве Купить
ССЫЛКА НА САЙТ- https://mephedrone.top
Москва Купить Мефедрон? Кристаллы МЕФ?
Где в Москве Купить Мефедрон? САЙТ – https://mephedrone.top/
I believe everything composed was very logical. But, think on this, what if you wrote a catchier post title? I ain’t suggesting your information isn’t solid., but suppose you added a headline that grabbed people’s attention? I mean %BLOG_TITLE% is a little boring. You ought to peek at Yahoo’s home page and watch how they create post titles to get viewers interested. You might add a related video or a related picture or two to grab readers interested about what you’ve got to say. In my opinion, it could make your posts a little livelier.
http://pialci.ru/index.php?links_exchange=yes&page=1&show_all=yes
купить диплом в буйнакске
http://mir-forum.ru
купить диплом в троицке
Приветики!
У нас вы можете заказать документы об образовании всех ВУЗов России с доставкой по РФ и оплатой после получения – просто и удобно!
https://forumhorochego.rolka.me/viewtopic.php?id=965#p2115
http://august4uart.bestbb.ru/viewtopic.php?id=220
https://stylist-profi.ru/forum/user/199161/
http://premium.listbb.ru/viewtopic.php?f=2&t=330
http://www.gamla.vykort.com/member.php?action=showprofile&user_id=26961
Привет всем!
На нашем сайте вы можете заказать диплом Вуза с постоплатой и круглосуточной поддержкой.
http://moy-toy.ru/index.php?subaction=userinfo&user=oqokady
http://promintern.listbb.ru/viewtopic.php?f=16&t=423
http://banner.micronoo.com/forum3/z_forummessage_show.php?method=showhtmllist&class=forummessage&rollid=5%2C2778&fromclass=forumtopic&frommethod=showhtmllist&fromscriptname=z_forumtopic_show.php&fromid=5&x=65e9ad509fb87&u=1948366077&
https://vidnoe.ixbb.ru/viewtopic.php?id=10426#p91920
http://toplentanews.ru/postroyte-osnovu-znaniy-kupite-diplom-ot-uvazhaemogo-uchebnogo-zavedeniya
It’s awesome to pay a visit this web site and reading the views of all friends regarding this article, while I am also zealous of getting familiarity.
https://amazonecotour.com/2019/03/27/hello-world/#comment-5051
http://allsportime.ru/kupit-diplom-rf-v-moskve-podlinnyie-dokumentyi-i-garantii
https://bytegain.com/high-pr-edu-gov-sites-to-get-quality-backlinks/#comment-1346797
http://translatorspuzzles.blogspot.com/2014/05/nonverbal-communication-in-different-cultures.html
http://1mog.listbb.ru/viewtopic.php?f=6&t=8497
꽁 머니 슬롯
그래서 그는 말했다.
Howdy! Do you know if they make any plugins to help with Search Engine Optimization? I’m trying to get my blog to rank for some targeted keywords but I’m not seeing very good results. If you know of any please share. Many thanks!
http://profbuh.forumkz.ru/viewtopic.php?id=8068#p12731
http://gpp.innim.ru:81/forums/topic/3257/diplomansy/view/post_id/3271
http://realtyintellect.ru/poluchite-diplom-bez-lishnih-problem-prosto-zapolnite-formu-i-poluchite-ego
http://ds-dealer.ru/forum/member.php?u=733023
https://rcdrift.ru/forum/member.php?u=21074
Excellent post. Keep posting such kind of info on your blog. Im really impressed by your blog.
Hi there, You’ve done an excellent job. I’ll certainly digg it and in my view recommend to my friends. I am sure they will be benefited from this web site.
http://newsinweek.ru/poluchite-diplom-bez-uchebyi-dostupnyie-resheniya/
goodgame.ru/user/1629660В
http://krasrec.ru/forum/user/197056/
http://rossensor.ru/forum/?PAGE_NAME=message&FID=1&TID=16261&TITLE_SEO=16261-diplom-na-zakaz_-vash-put-k-professionalnomu-priznaniyu-i-uvazheniyu&MID=853294&result=new#message853294
http://tvoirostov.ru/blogi/put_k_uspehu_bez_usilii_kak_kupit_diplom_i_dostich_celei/
Onwin platformu bazı bahis sitelerinin Onwin sınıflandırmaları farklı olsa da materyalleri tutarlı kalıyor.
Onwin uygulaması seobahis Affiliate Uygulaması, Onwin canlı bahis mağazalarının yeni üyeleri çekmek için kullandığı bir araçtır.
It’s remarkable to pay a visit this website and reading the views of all friends about this piece of writing, while I am also keen of getting know-how.
http://torgteh-rzn.ru/index.php?option=com_k2&view=itemlist&task=user&id=11104
http://www.11na11.com/2011/03/pro-vidstan-vid-polya-do-tribun-na-dnipro-areni/В
lolipopnews.ru/page/9В
https://mzcenter.org/%d1%84%d0%be%d0%bd%d0%b4-%d0%b8-%d0%bc%d1%83%d1%84%d1%82%d0%b8-%d0%b7%d0%b0%d0%b4%d0%b5-%d0%b2%d1%8b%d1%81%d1%82%d1%83%d0%bf%d0%b8%d0%bb-%d1%81%d0%bf%d0%be%d0%bd%d1%81%d0%be%d1%80%d0%be%d0%bc-vii/?unapproved=420837&moderation-hash=b41605fa0f7a9651d1874a6fe37f5503#comment-420837
https://b4g-akk.ru/resources/authors/shilpatidece.19435/
An outstanding share! I have just forwarded this onto a co-worker who had been doing a little research on this. And he in fact bought me breakfast because I found it for him… lol. So allow me to reword this…. Thank YOU for the meal!! But yeah, thanks for spending the time to talk about this matter here on your site.
http://sergei-cheremushkin.blogspot.com/2010/03/blog-post.html
http://www.pigcraft.ugu.pl/showthread.php?tid=102057
https://avgust.nsk.ru/product/noski-detskie-malchik_dh/reviews/
http://knigniaradyga.blogspot.com/2013/06/blog-post_10.html
http://service.gepart.su/forum/?PAGE_NAME=profile_view&UID=3811
Добрый день всем!
Приобретите диплом института или колледжа с гарантией качества и доставкой по России без предоплаты.
http://youyanggse.co.kr/board/bbs/board.php?bo_table=free&wr_id=569199
http://no-smoking.tehpodderzka.ru/2016/09/griffin-25.html
http://fuss.forumkz.ru/viewtopic.php?id=2325#p3753
http://tokmak.pp.ua/forum.html
http://metalfans.maxbb.ru/topic494.html
Onwin canlı destek nedir çevrimiçi casino sitesinin Onwin canlı desteği çok kolay ve yararlı bir özelliktir.
Awesome blog you have here but I was curious about if you knew of any discussion boards that cover the same topics talked about in this article? I’d really love to be a part of group where I can get opinions from other experienced individuals that share the same interest. If you have any recommendations, please let me know. Thank you!
http://mayakminska.1stbb.ru/viewtopic.php?f=12&t=11944
http://rosen.moibb.ru/viewtopic.php?f=2&t=337
https://meneger23.ru/en/forums-en/user/1460-minasdofoobe
http://www.netza.ru/2012/12/service-password-encryption.html
http://star-friends.ru/blogs/615/пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅ-пїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ
Ваш решение для игр на удачу – казино Слотозал!
Слотозал предлагает обширный выбор слот-машин, выгодные предложения и высокий уровень безопасности. Интерфейс веб-сайта понятен, навигация легка, что позволяет быстро отыскать избранные игры. Поддержка постоянно готова содействовать с всеми вопросами. Пройдите регистрацию на законном веб-сайте играть и приступите к выигрывать сегодня!
Здравствуйте!
Наша компания поможет купить диплом Вуза со скидкой и доставкой в любую точку России
https://medcom.ru/forum/user/228423/
http://brawlstarsacc.listbb.ru/viewtopic.php?f=3&t=390
http://shiba-inu.ru/forum/messages/forum1/topic46/message51/?result=new#message51
http://zoobla.com/blog/security-cameras-increase-overall-safety
http://bestcoolfun.ru/byistroe-poluchenie-diploma-kak-kupit-obrazovanie-i-ne-teryat-vremya
Good day! This is kind of off topic but I need some guidance from an established blog. Is it very difficult to set up your own blog? I’m not very techincal but I can figure things out pretty fast. I’m thinking about making my own but I’m not sure where to start. Do you have any points or suggestions? Many thanks
купить диплом в комсомольске-на-амуре
http://gti-club.ru/forum/member.php?u=23001
https://prbb.0pk.me/viewtopic.php?id=724#p6234
https://www.ocf.berkeley.edu/~paultkim/the-remarkable-2-will-be-released-in-june-for-399
http://bahchisaray.org.ua/index.php?showtopic=33167
http://antikidok.mybb.ru/viewtopic.php?id=580#p3451
купить диплом в самаре
I got this web page from my buddy who shared with me about this web site and now this time I am visiting this site and reading very informative content here.
купить диплом в новороссийске
http://antonovschool.ru/forum/messages/forum1/topic606/message621/?result=new#message621
http://nagaevo.ekafe.ru/viewtopic.php?f=86&t=2421
https://pushkin-sto.ru/otzyvy/?unapproved=2624&moderation-hash=8712ab08f7a1a339d0ec7a22bd566859#comment-2624
http://publikacii.listbb.ru/viewtopic.php?f=3&t=561
http://blackforest.mybb.ru/viewtopic.php?id=476#p703
купить диплом в альметьевске
В последнее время открыл для себя 1хбет регистрация, и стал чрезвычайно удивлен их вариантами. Здесь огромный каталог аппаратов и картежных игр, щедрые бонусные программы и праздничные акции, а также безопасная механизм вывода денег. Поддержка клиентов существует срочно и всегда рада помочь. Пользовательский интерфейс удобен и дружелюбен к пользователю, что обеспечивает игровой процесс еще более увлекательной. Рекомендую поиграть, в особенности если ты уважаете высокий уровень и стабильность в онлайн развлечениях.
Hmm is anyone else having problems with the pictures on this blog loading? I’m trying to find out if its a problem on my end or if it’s the blog. Any suggestions would be greatly appreciated.
http://m.jingdexian.com/home.php?mod=space&uid=2545488
купить диплом в когалыме
http://to66minust.ru
купить диплом в балашихе
Onwin sitesi üyeleri seobahis sitesindeki site adresi Onwin giriş adresi güncellendiğinde değişir ancak alan adı aynı kalır.
Недавно наткнулся на для себя 1 xbet, и был очень удивлен их вариантами. На этом сайте огромный ассортимент игровых автоматов и настольных развлечений, щедрые акции и бонусы, а и безопасная система снятия средств. Служба поддержки существует незамедлительно и всегда может помочь. Интерфейс портала интуитивно понятен и интуитивно понятен, что обеспечивает развлечение более интересной. Настоятельно рекомендую поиграть, конкретно если вы реально уважаете стабильность и безопасность в интернет-играх.
В последнее время открыл для себя 1хбет скачать, и был очень приятно поражен их предложениями. На платформе большой выбор игр и настольных развлечений, щедрые акции и акции, а и безопасная механизм вывода средств. Клиентская поддержка действует оперативно и всегда может помочь. Интерфейс портала удобен и интуитивно понятен, что обеспечивает развлечение еще веселой. Рекомендую поиграть, в частности если вы действительно уважаете высокий уровень и надежность в онлайн развлечениях.
В последнее время обнаружил для себя 1хбет вход, и был приятно поражен их вариантами. На этом сайте большой ассортимент игр и настольных развлечений, щедрые бонусы и бонусные акции, а ещё стабильная схема снятия средств. Поддержка клиентов действует оперативно и всегда готова помогать. Оболочка сайта комфортен и легок в понимании, что гарантирует развлечение еще приятной. Рекомендую поиграть, в особенности если ты приветствуете надежность и гарантии в виртуальных играх.
Betcool’a para yatırmak için Jeton yönetimini Betcool tercih eden müşterilerimiz şu şekilde bonus kazanabilirler.
Betcool, avrupa´nın bir çok ülkesinde online ve Betcool offline olarak hizmet veren güvenilir bahis, canlı bahis, casino ve canlı casino sitesidir.
Betcool sorunları seobahis sitesi gecenin ilerleyen Betcool saatlerinde bile en verimli bilgi akışını sağlıyor. Belki de paranızı elinizden geldiğince korumayı düşünüyorsunuz.
Vbet, her gün yeni özellikler sunarak Vbet günümüz oyun dünyasında adından söz ettiriyor.
Vbet adresi Yabancı bahis sitelerinin Vbet en büyük ve tek sorunu BTK engellemesidir.
Первый партнер охраны труда
[url=https://www.ppot.ru/]Аудит суот[/url] – экспертный центр по аудиту и подготовке специалистов в области охраны труда и пожарной безопасности. Наша компания предоставляет широкий спектр услуг по обеспечению безопасности производственных процессов для индивидуальных предпринимателей и организаций различных секторов промышленности в Москве, Московской области и других регионах России.
Соблюдение правил безопасности труда – законодательное требование, за нарушение которого на организацию может быть наложен штраф или приостановлена деятельность предприятия на срок до 90 суток.
Мы помогаем решать вопросы обеспечения безопасности и организации труда на предприятии, разрабатываем и актуализируем документы по охране труда, проводим комплексное обследование охраны труда на соответствие государственным нормативным требованиям. Проводим обучение сотрудников по охране труда и пожарной безопасности, проверяем СУОТ и оцениваем профессиональные риски.
Виктория Набойченко сделала для нашего канала заявление,
[url=https://www.youtube.com/watch?v=Q0smZ5_QOcU]Набойченко Бествей[/url]
касающееся своего бывшего супруга – главного свидетеля обвинения по так называемому уголовному делу “Лайф-из-Гуд”-“Гермес”-“Бест Вей”
Отлично выглядящий интернет-сайт. Предполагаем, что вы сделали много из своего собственного кодирования.
Посмотрите также мою страничку
игровые автоматы
игровые автоматы
https://rocklandbakery.com/?URL=https://igrovieavtomati.pl – игровые автоматы на деньги
https://www.clubseatateca.com/proxy.php?link=https://igrovieavtomati.pl – игровые автоматы на деньги
Erectile dysfunction treatments available online from TruePills.
Discreet, next day delivery and lowest price guarantee.
Viagra is a well-known, branded and common erectile dysfunction (ED) treatment for men.
It’s available through our Online TruePills service.
Trial ED Pack consists of the following ED drugs:
Viagra Active Ingredient: Sildenafil 100mg 5 pills
Cialis 20mg 5 pills
Levitra 20mg 5 pills
https://cutt.ly/dw7ChH4s
https://stal63.ru:443/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://true-pill.top/
https://xn--63-6kcat4ed.xn--p1ai/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://true-pill.top/
https://uspenskaya.pro/bitrix/redirect.php?goto=https://true-pill.top/
https://radio-podcast.fr/redirect?telechargement_podcast=https%3a%2f%2ftrue-pill.top
https://m.phymongshe.co.kr/member/login.html?noMemberOrder=&returnUrl=https%3a%2f%2ftrue-pill.top
Actifuge
Avapro
Ciproxina
Phenazodine
Virucid
Ucol
Gastromins
Nimodip
Pharmapress
Trixim
Safetin
Prosterid
Clomifenum
Naksetol
Metroson
Rhinodina
Proponol
Interbion
Emeset
Goutnil
Toplep
Betachron er
Eox
Lasopran
Irys
Efexor
Cardaten
Avaglim
Era
Fosinopril
Benzo-ginestryl
Fluoxgamma
Malflam
Osteocalmine
Dilmacor
Iecatec
Avecor
Ebersept
Novatrex
Formigran
Keep on writing, great job!
http://www.qazaqpen-club.kz/en/novosti/5-kazahskiy-pen-klub-vybral-novogo-rukovoditelya.html
http://star-friends.ru/blogs/674/ упить-диплом-в-ћоскве-ѕодлинные-ƒокуленты
http://svitnavkolonassapalova.blogspot.com/2013/02/blog-post_8.html
http://biblioparus.blogspot.com/2017/02/blog-post_28.html
http://www.zpu-journal.ru/forum/view_profile.php?UID=305253
It’s amazing in support of me to have a site, which is helpful designed for my know-how. thanks admin
http://dnd.listbb.ru/viewtopic.php?f=3&t=426
http://g95183sh.bget.ru/users/abegunan
http://forexgroupx.ru/diplom-na-zakaz-vash-shans-poluchit-vyisokoklassnoe-obrazovanie-bez-lishnih-hlopot
https://multiflay.com/profile.php?op=userinfo&from=space&user=minna_prout_301152
http://prazdnikbaby.ru/index.php/separator?view=item&item_id=10
купить диплом в череповце diplomvash.ru .
골드 킹
Xu Pengju는 잠시 생각하다가 웃으며 따라갔습니다. “하하 … 하하 …”
купить диплом в иваново diplomvash.ru .
Vbet vip üyelik Vbet VIP Üyelik Programına üye olmak Vbet için oyuncuların öncelikle sağlanan butona tıklaması gerekmektedir.
Jupiterbahis 30 gün boyunca giriş Jupiterbahis yapmazsanız hesabınız etkinleştirilecektir.
?? Забирай 100,000 Рублей + 400 Фриспинов в виде Welcome бонуса, на официальном сайте 7К Казино ??
?????? Официальное зеркало ??????
7к казино рабочее зеркало
НОВЫЕ ПРОМОКОДЫ ОТ НАШЕГО ТЕЛЕГРАМ КАНАЛА 7К КАЗИНО! МОЩНЫЙ СТАРТ НАШИМ ИГРОКАМ. ???? ЖЕЛАЕМ КРУТЫХ ЗАНОСОВ ??
Joker7070 – бонус 170%+ 70 FS в Joker Stoker
777WIN – бонус 100%+ 30 FS в Hot Triple Sevens
HOT100 – бонус 100 FS в Indiana’sQuest
?? Добавляйте промокод в разделе бонусов на официальном сайте 7К Казино.
?? Важно! Для новых игроков после регистрации 7K Casino приготовил Welcome бонус до 100,000 Рублей + 400 Фриспинов! Успейте активировать ???
?? Подробная информация на официальном сайте 7К в разделе бонусов! Каждый промокод можете использовать по очереди.
7k casino актуальное зеркало на сегодня
https://t.me/s/casino_7k_official
купить аттестат о среднем образовании купить аттестат о среднем образовании .
Jupiterbahis En yeni yöntemlerle sürekli hizmet Jupiterbahis veren bir bahis sitesinde yeni bir hesap açıp şansınızı deneyebilirsiniz.
Jüpiterbahis çevrim içi bahiste inanılmaz bir fark Jüpiterbahis yaratan, köklü ve eski bir bahis sitesidir.
Jupiterbahis üye ol web sitesi bir hesabı sonlandırdığında, Jupiterbahis hem ticari hem de kullanıcıyla ilgili güvenlik önlemleri gerekli olacaktır.
Sahabet hizmet alanında, üyelerine kazançlı ve güvenli Sahabet yöntemler sunarken, bir yandan da sektör ortalamasının üstünde bir hizmet kalitesi vermeye özen göstermektedir.
Sahabet bahis bürosu, kısa sürede kazandığı sadık üye kitlesi ve Sahabet aynı zamanda standart olarak sunduğu üstün hizmet kalitesi ile bu piyasada kalıcı ve güçlü bir yere geleceğinin işaretlerini vermektedir.
Hi all, here every one is sharing these kinds of know-how, therefore it’s pleasant to read this weblog, and I used to go to see this website daily.
spg.co.kr/bbs/board.php?bo_table=cus_res&wr_id=92&sca=%EC%98%81%EC%97%85%EC%A7%80%EC%9B%90&page=55
thegioimaydemtien.vn/san-pham/may-dem-tien-xiudun-9500-817.htm
http://www.catholicdioceseofaba.org/fullhomilies?sn=4
oldsite.profbez.ru/shop/index.php?links_exchange=yes&page=260&show_all=yes
100buketov.com/index.php?links_exchange=yes&page=192&show_all=yes
Sahabet slot makineleri çeşitleri seobahis, müşterilerinin Sahabet memnuniyetini ön planda tutmakta ve onlara en faydalı hizmetleri sunmaya çalışmaktadır.
Здравствуйте!
Купите диплом ВУЗа по выгодной цене с гарантией качества и доставкой в любой город России без предоплаты – просто и надежно!
http://xn--80aeahbdc6cr3b7h.xn--p1ai/club/user/8729/blog/192828/
http://tanijoe-information.com/2021/08/26/hello-world/#comment-46041
https://wegos.ru/samyj-provalnyj-iphone-stal-samym-prodavaemym-v-tretem-kvartale/?unapproved=1181955&moderation-hash=3076426612e4e433df48a369f104698d#comment-1181955
http://metalinquisition.blogspot.com/2013/09/mayehms-occultus-and-muppets-floyd.html
http://istinastroitelstva.xyz/index.php?subaction=userinfo&user=opofura
Mrbahis iletişim bilgilerinin bir parçası olarak, konunun Mrbahis uzmanlarıyla iletişim kurmak için Canlı Destek Hattını kullanma hakkına sahipsiniz.
Its like you read my mind! You seem to know a lot about this, like you wrote the book in it or something. I think that you can do with some pics to drive the message home a bit, but other than that, this is wonderful blog. A fantastic read. I will certainly be back.
http://delpc.ru/kak-ustanovit-kuler/
купить диплом в черногорске
http://pojdelo-journal.ru
купить диплом в гуково
Mrbahis güncel işlemini tamamladıktan sonra siteye giriş Mrbahis yaptığınızda üst menünün üçüncü sekmesinde casino menüsü yer almaktadır.
Hi there fantastic website! Does running a blog like this take a great deal of work? I have no knowledge of computer programming however I was hoping to start my own blog in the near future. Anyways, should you have any ideas or techniques for new blog owners please share. I know this is off topic however I simply had to ask. Thank you!
купить диплом фельдшера
http://good-serial.ru/2024/05/kupit-diplom-rf-v-moskve-ofitsialnoe-oformlenie-i-garantii/
http://peaceofficial.5nx.ru/viewtopic.php?f=111&t=736
http://kondrateff.5bb.ru/viewtopic.php?id=6291#p11725
http://amtest.mybb.ru/viewtopic.php?id=254#p389
http://drugisaitove.listbb.ru/viewtopic.php?f=2&t=565
купить диплом в великих луках
Заявление Виктории Набойченко – бывшей супруги главного свидетеля обвинения Евгения Набойченко
https://www.youtube.com/watch?v=Q0smZ5_QOcU
Здравствуйте, меня зовут Виктория Набойченко.
Я являюсь бывшей супругой Набойченко Евгения Витальевича, с которым состояла в официальном браке с 2017 по 2022 год. Из средств массовой информации мне стало известно о фактах уголовного преследования лиц из окружения Романа Василенко, который является президентом международной бизнес-карты на территории России, Казахстана и Киргизии. На протяжении 20 лет Евгений Набойченко и Роман Василенко дружили, вместе путешествовали, проводили время, ездили на рыбалку. Роман очень доверял Евгению, делился с ним своими идеями, планами на коммерческую деятельность и реализацию новых проектов. Мне известно, что Роман предложил Евгению разработать фирменный логотип и сайт компании Life is Good, а впоследствии возглавить IT-отдел ЖК. Евгений с радостью согласился на предложение Романа и возглавил IT-отдел. Роман достойно оплачивал работу Евгения, гонорар которого вырос со 100,000 рублей до 500,000 рублей в месяц, что было связано с ростом и развитием кооператива Best. Роман всегда был щедр к Евгению и ценил его работу, за что ежегодно премировал.
Тот успех, которого достиг Роман за эти годы, очень задевал Евгения как мужчину. В ходе совместной жизни Евгений делился со мной, что мечтает однажды превзойти Романа в бизнесе, а его проект стать круче Илона Маска. Примерно с июня 2021 года Евгений стал нервным и злился на Романа, рассказывая мне, что Роман не хочет заплатить ему 170,000 евро. Я не вдавалась в подробности их взаимоотношений. Чуть позже я узнала о письмах Евгения к Роману, в которых он требовал от него указанную сумму и угрожал увечьями и убийством Роману и всем членам его семьи, в том числе и детям. Данные угрозы он отправлял голосовыми сообщениями и в электронных письмах. Мне известно, что Роман не реагировал на угрозы Евгения, что очень злило его, потому что он ждал от него указанную сумму. В моём присутствии Евгений постоянно по нескольку раз переслушивал свои голосовые сообщения, которые с угрозами отправлял в адрес Романа Василенко. Он получал от этого огромное удовольствие, с ухмылкой хвастаясь, что Роман обязательно заплатит.
[url=https://www.youtube.com/watch?v=Q0smZ5_QOcU]Life is Good[/url]
17 февраля 2022 года в 7:00 по МСК в нашем доме в посёлке в Краснодарском крае произошел обыск. В доме находилась только я и дети, а Евгений в это время находился в Краснодаре на съёмной квартире, куда заблаговременно уехал, предупредив меня за 3 дня о предстоящем обыске в доме. Как будто его друзья из правоохранительных органов Петербурга предупредили его. На мои вопросы о том, что происходит, он отвечал, что это формальность и что мне не о чем беспокоиться. Позже мне стало известно, что подобные обыски проходили и у других сотрудников кооператива в Санкт-Петербурге, Краснодаре, Анапе и других городах. В обыске нашего дома принимали участие шесть оперативных работников ОББ города Санкт-Петербурга, фамилии которых указаны в протоколе обыска, копия которого имеется в моём распоряжении. В ходе обыска оперативные работники оскорбляли Евгения и Романа, называя их преступниками и мошенниками, а также иными оскорбительными словами. При обыске был изъят планшет Евгения, оперативные работники перевернули весь дом и напугали детей, а также пообещали, что задержат Евгения в тот же день.
17 февраля 2022 года Евгений был задержан сотрудниками оперативных служб в городе Краснодар и примерно через 3 дня был доставлен в Санкт-Петербург, где был допрошен в УП по рекомендации сотрудников полиции. Допрос происходил без участия его личного адвоката Ирины Черняховской, от услуг которой он отказался в тот же момент. Мне позвонила Ирина в бешенстве и недоумении, крича о том, что Евгений её опозорил перед сотрудниками полиции, отказавшись от её услуг в грубой форме. Впоследствии мне стало известно от Евгения, что он лично получил покровительство начальника ОББ города Санкт-Петербурга в своей безнаказанности по факту проводимого расследования. Об этом он похвастался мне в личном разговоре, будучи окрылённый поддержкой начальника ОББ, который с ним разговаривал на доверительной ноте.
Евгений мне рассказал, что получит от органов абсолютную неприкосновенность и поддержку за информацию, которая интересовала оперативных работников, за что Евгению также была предоставлена госохрана. В период с февраля по середину апреля Евгений находился в Питере под госзащитой и плотно взаимодействовал с правоохранительными органами. Он предоставлял им всю информацию, в том числе полный доступ к системе базы данных Best Way и Life is Good. Евгений инициативно предлагал органам заблокировать доступ к личным кабинетам пайщиков и членов ЖК БСТ, чем полностью парализовал хозяйственную деятельность кооператива, создавая напряжённость и негативные отзывы. Также Евгений активно призывал граждан к написанию заявлений в отношении Романа Василенко с целью возврата пайщиками своих денежных средств, делал он это через каналы Telegram и YouTube. Евгений был единственным, кто имел и имеет доступ к базам данных и ключам. Желая намеренно создать проблемы Роману, он отказывается восстанавливать работу системы Best Way при наличии такой возможности.
Миссия Евгения – это привлечение Романа Василенко к уголовной ответственности, что спровоцирует крах компании и полное прекращение её деятельности, а это в свою очередь 30,000 пайщиков, которые могут стать потерпевшими по уголовному делу. Всё это мне известно из личного общения с Евгением, когда я приезжала к нему в Санкт-Петербург на несколько дней. Мне также достоверно известно, что в настоящее время нет реальных заявлений от недовольных пайщиков, так как в Романа Василенко все верят. Самое главное, что у Евгения есть возможность в любой момент через свой личный ноутбук восстановить работу ЖК Best Way и вернуть доступ к личным кабинетам пайщиков. Однако он этого не делает, преследуя свои личные цели.
«Рэмбович» против колокольцевской мафии
Министр обороны, в рамках начавшейся антикоррупционной кампании, готов инициировать расследование против команды своего коллеги Колокольцева: сфабрикованное ею дело в отношении кооператива «Бест Вей» – созданного военнослужащими во многом для военнослужащих, бьет в тылу по тем, кто защищает страну на фронте, лишая их квартир и денег, которые уже более двух лет арестованы ГСУ ГУ МВД России по Санкт-Петербургу и Ленинградской области и примкнувшей к ним Прокуратурой Санкт-Петербурга.
[url=https://compromat01.group/main/investigations/130715-delo-layf-iz-gud-germes-best-vey-kto-takie-naboychenko-i-komarov.html]Виктор Иванович Василенко[/url]
?Кооператив «Бест Вей» создан в 2014 году российскими военнослужащими – как ушедшими в запас, так и действующими. Его предправления до весны 2021 года, а затем еще год председатель совета – капитан третьего ранга запаса Роман Василенко, в правлении кооператива всегда было немало офицеров Вооруженных сил РФ.
Нынешний председатель совета Сергей Крючек – полковник запаса, депутат Государственной думы VII созыва.
Жилье для военнослужащих
Кооператив начинался во многом как проект обеспечения жильем военнослужащих, бывших военнослужащих и членов их семей. Он существует 10 лет, за восемь лет, пока он имел возможность приобретать квартиры для пайщиков, которые были заблокированы следствием, «Бест Вей» купил 2500 квартир, более 250 квартир перешли в собственность пайщиков.
Уникальность кооператива в том, что он приобретает пайщикам недвижимость на беспроцентной основе. Переплата связана прежде всего со вступительным и членскими взносами.
Социальная программа для участников СВО
Приобретение жилья с помощью кооператива – по сути, социальная программа, которая стала очень популярна среди военнослужащих.
Ей воспользовались тысячи участников СВО и членов их семей – людей, защищающих Россию на фронте.
И вот в тылу с их имуществом или деньгами творится настоящий беспредел – по вине колокольцевской мафии. Колокольцевцы со своими подручными из Прокуратуры Санкт-Петербурга уже два года держат средства кооператива под арестом.
Российские воины не могут ни приобрести квартиры, на которые аккумулировали деньги на счете в кооперативе, ни забрать собственные обесценивающиеся за два года деньги назад. В свои короткие отпуска они вынуждены обивать пороги Росреестра, писать заявления на возврат средств в суды, участвовать в процессах. Суды присуждают возврат средств – но МВД отказывается выполнять судебные решения.
Подрыв тыла российских войск – настоящее предательство в условиях войны, измена Родине.
Отнять то, что ближе лежит
Уголовное дело, возбужденное ГСУ ГУ МВД по Санкт-Петербургу, касается иностранной компании «Гермес», которая (якобы) украла деньги своих российских клиентов. В действительности у нее возникли трудности из-за того, что завербованный питерской полицией с помощью угроз Набойченко, имевший доступ к платежной системе, обрушил ее и, возможно, манипулировал средствами на счетах.
Однако деньги «Гермеса» из-за границы достать проблематично – и следствие решило объявить кооператив частью некоего холдинга, в который входил также «Гермес», и на этом основании забрать его средства. Хотя в действительности между этими организациями никогда не было никаких финансовых и организационных отношений.
Продукты кооператива и продукты «Гермеса» продвигала сеть предпринимателей под эгидой компании «Лайф-из-Гуд» – коммерческая сеть, каждый из участников которой работал за процент и мог продавать все, что угодно. Многие приходили в кооператив не через продажников, а по сарафанному радио.
На скамье подсудимых – десять человек, в том числе 83-летний Виктор Иванович Василенко, отец Романа Василенко, ветеран Вооруженных сил: команда Колокольцева не отказала себе в извращенном удовольствии поиздеваться над ветераном.
Тыловые крысы воруют чужое имущество
Колокольцевцы – тыловые крысы, которые никогда не были настоящими офицерами, скорее чинушами с коррупционными интересами, пытаются отжать почти 4 млрд рублей на счетах кооператива для себя и своих подельников.
Притом, что в рамках уголовного дела фигурирует сумма в 282 млн рублей, даже (незаконный) арест средств кооператива на эту сумму никак не повлиял бы на работу кооператива. Но арестованы именно 4 млрд, при этом МВД не дает платить с этих средств налоги и зарплату. То есть целью продажных ментов являются именно миллиарды кооператива, а не удовлетворение (в основном липовых) претензий клиентов «Гермеса».
Комиссар Путина
Андрей Белоусов, облеченный особым доверием главы государства, имеет карт-бланш на проведение антикоррупционных расследований в отношении всех силовых структур, тем более затрагивающих военнослужащих Вооруженных сил. Новый министр обороны сразу объявил социальную защищенность воинов высшим приоритетом, а кооперативная программа обеспечения жильем может сыграть в ней огромную роль.
«Рэмбович» уже показал, как бескомпромиссно он борется с коррупционерами в руководстве МО и в ОПК. Теперь настал черед руководства МВД – которое давно уже на плохом счету президента Путина, просто до него в условиях войны не доходили руки. Теперь они дошли.
Create a positive impact
https://bit.ly/4bdMuAg
В нашем обществе, где диплом становится началом отличной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения качественного образования. Наличие официального документа переоценить попросту невозможно. Ведь именно диплом открывает двери перед людьми, желающими вступить в профессиональное сообщество или учиться в университете.
Мы предлагаем оперативно получить этот необходимый документ. Вы можете заказать диплом нового или старого образца, и это становится удачным решением для человека, который не смог закончить обучение или утратил документ. Все дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим нюансам, чтобы на выходе получился документ, полностью соответствующий оригиналу.
Преимущество подобного подхода заключается не только в том, что можно максимально быстро получить свой диплом. Весь процесс организован просто и легко, с нашей поддержкой. От выбора нужного образца диплома до правильного заполнения личной информации и доставки в любой регион страны — все под полным контролем наших специалистов.
Всем, кто ищет быстрый и простой способ получения необходимого документа, наша компания предлагает отличное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и сразу переходить к важным целям: к поступлению в университет или к началу удачной карьеры.
http://unikum-student.ru
http://rus-students.ru
http://edu-csrpi.ru
http://femcfas.ru
http://seobich.ru
Because the admin of this web site is working, no question very soon it will be well-known, due to its quality contents.
http://www.downtowncookieco.com/shipping-policy/
newmedtime.ru/page/12
moyolapark-fc.com/xcx07/index.php?option=com_content&task=view&id=125&Itemid=2
clicknconnectclubs.com/index.php?do=/public/user/blogs/name_Alanpoe/page_7/
primerosauxilios.org/primeros-auxilios/que-es-y-como-curar-una-herida.php
Hi! This is my first comment here so I just wanted to give a quick shout out and say I really enjoy reading your blog posts. Can you suggest any other blogs/websites/forums that go over the same subjects? Thank you!
купить сертификат специалиста
http://giletka24.bestbb.ru/viewtopic.php?id=306#p308
https://vip.9bb.ru/viewtopic.php?id=5664#p9432
https://vip.rolevaya.info/viewtopic.php?id=4203#p7301
http://4asdaiprognoza.listbb.ru/viewtopic.php?f=2&t=555
https://www.pickmemo.com/read-blog/288094
купить диплом в армавире
Hi there Dear, are you truly visiting this web page daily, if so afterward you will absolutely obtain good experience.
http://www.freepainter.ru/index.php?links_exchange=yes&page=1&show_all=yes%D0%92
templateinspire.com/opencart/Lingerie/index.php?route=information/blogger&blogger_id=2
wap-club.ru/guestbook.php?mode=quote&idmsg=210748
dachaweek.ru/page/3
http://www.ksolo.ru/forum/member.php?tab=visitor_messaging&u=83595&page=23
This is really interesting, You are a very skilled blogger. I’ve joined your rss feed and look forward to seeking more of your great post. Also, I have shared your site in my social networks!
купить диплом в черкесске
http://zavalinka.listbb.ru/viewtopic.php?f=3&t=2838
https://medium.com/@oximus777/купить-диплом-официальное-оформление-99901a4a671b
http://openmotonews.ru/idealnyiy-diplom-dlya-vashego-budushhego-priobretite-pryamo-seychas
http://antalyatoday.mybb.ru/viewtopic.php?id=213#p393
https://electricsheep.activeboard.com/forum.spark
купить диплом в элисте
Mrbahis altyapı mühendisliği bu şirketler, neredeyse tüm web Mrbahis sitelerine, kullanıcılarına kesintisiz tavla oyun deneyimi sağlayan birinci sınıf altyapı hizmetleri sağlama konusunda uzmandır.
Mrbahis müşteri hizmetleri ile dilediğiniz Mrbahis vakit görüşebileceğinizi bilmelisiniz.
Mrbahis yönetimi Mrbahis üyesi olmanın avantajları hızla artıyor Mrbahis ve henüz katılmadıysanız bu avantajları kaçırmaya devam edeceksiniz.
Huhubet üyeliği hakkında canlı bahis için erişilebilen Huhubet bahis seçeneklerinin tamamı Oyun Verilerinde bulunabilir.
Huhubet canlı bet sitesi üyelerine sunduğu Huhubet geniş içeriklerle hizmet veren bir sitedir.
Betmoney adresi canlı hizmetimizden destek almanıza ve üyelerimize form Betmoney doldurma konusunda yardımcı olmanıza olanak tanır.
Betmoney müşteri hizmetleri ve diğer bahisçiler, Betmoney üyelerinin en çok soru sorduğu sektörlerden biridir.
Betmoney bağlantı sorunları yaşanmaz ancak bazı Betmoney web siteleri ve kullanıcı faktörleri yaşayabilir.
Betaverse, bahis, canlı bahis, kumarhaneler ve Betaverse canlı kumarhaneler alanlarında oyun tutkunları için bir platformdur.
На сегодняшний день, когда диплом – это начало успешной карьеры в любой отрасли, многие пытаются найти максимально быстрый и простой путь получения образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед всеми, кто собирается начать профессиональную деятельность или продолжить обучение в ВУЗе.
Наша компания предлагает очень быстро получить любой необходимый документ. Вы сможете заказать диплом, и это становится отличным решением для всех, кто не смог завершить обучение, потерял документ или желает исправить свои оценки. дипломы изготавливаются аккуратно, с максимальным вниманием к мельчайшим элементам. На выходе вы получите полностью оригинальный документ.
Плюсы данного решения заключаются не только в том, что вы сможете максимально быстро получить свой диплом. Процесс организован комфортно, с нашей поддержкой. От выбора необходимого образца до грамотного заполнения персональной информации и доставки по России — все будет находиться под полным контролем квалифицированных мастеров.
Всем, кто ищет быстрый и простой способ получения необходимого документа, наша услуга предлагает отличное решение. Купить диплом – это значит избежать долгого процесса обучения и не теряя времени перейти к достижению своих целей, будь то поступление в ВУЗ или начало удачной карьеры.
http://collegevalday.ru
http://diplomy-plus.com
http://school-13.ru
http://arxitekt.ru
http://variant-surgut.ru
Still not a millionaire? Fix it now! https://t.me/cryptaxbot/11
you’re truly a good webmaster. The site loading pace is amazing. It kind of feels that you’re doing any distinctive trick. Moreover, The contents are masterpiece. you have performed a fantastic task in this topic!
http://student-news.ru/kupit-diplom-legkiy-put-k-uspeshnoy-karere
http://rem.4nmv.ru/forum/search.php?action=members&p=261&s=d&order=ASC
https://allbookmarking.com/story17172532/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%82%D0%B5%D1%85%D0%BD%D0%B8%D0%BA%D1%83%D0%BC%D0%B0
http://www.unikum-student.ru/
https://socialbuzzfeed.com/story2451224/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%92%D0%BE%D1%80%D0%BE%D0%BD%D0%B5%D0%B6%D0%B5
톰 오브 매드니스
그러나 Tiger는 Tiger가 예상한 것이 가능하다고 믿습니다.
Old man with young wife. Donald Trump Approves —> http://zi.ma/4h6w89/
What’s up mates, its great post concerning tutoringand completely explained, keep it up all the time.
lefolyotakaritas.hu/nyomvonal-meghatarozas/
mybaltika.info/ru/print/587/7892/
priusforum.ru/forums/index.php?autocom=gallery&req=si&img=5268
mboumukvilino.ru/antiterroristicheskaya-bezopasnost/normativnye-pravovye-akty-rossiyskoy-federacii-i-respubliki-krym/
brandedhoodie.com/sign-up/
5]Balanset-1A Sale: Your Ideal Tool for Balancing and Vibration Analysis
4]Device Description:
Balanset-1A is a dual-channel device for balancing and vibration analysis, ideal for balancing rotors such as crushers, fans, grain harvester choppers, shafts, centrifuges, turbines, and more.
4]Features:
3]Vibrometer Mode:
Tachometer: Accurately measures rotation speed (RPM).
Phase: Determines the phase angle of vibration signals for precise analysis.
1x Vibration: Measures and analyzes the main frequency component.
FFT Spectrum: Provides a detailed view of the vibration signal frequency spectrum.
Overall Vibration: Measures and monitors overall vibration levels.
Measurement Log: Saves measurement data for analysis.
3]Balancing Mode:
Single-Plane Balancing: Balances rotors in a single plane to reduce vibration.
Two-Plane Balancing: Balances rotors in two planes for dynamic balancing.
Polar Diagram: Visualizes imbalance on a polar diagram for precise weight placement.
Last Session Recovery: Allows resuming the previous balancing session for convenience.
Tolerance Calculator (ISO 1940): Calculates allowable imbalance values according to ISO 1940 standards.
Grinding Wheel Balancing: Uses three correction weights to eliminate imbalance.
3]Diagrams and Graphs:
General Graphs: Visualization of overall vibration.
1x Graphs: Display of main frequency vibration patterns.
Harmonic Graphs: Show the impact of harmonic frequencies.
Spectral Graphs: Graphical representation of the frequency spectrum for in-depth analysis.
3]Additional Features:
Archiving: Storage and access to previous balancing sessions.
Reports: Generation of detailed reports on balancing results.
Rebalancing: Easily repeat the balancing process using saved data.
Serial Production Balancing: Suitable for balancing rotors in serial production.
Imperial and Metric System Options: Provides compatibility and convenience worldwide.
4]Price:
€1751
3]Package Includes:
Measurement unit
Two vibration sensors
Optical sensor (laser tachometer) with magnetic stand
Weights
Software (laptop not included, available as an additional order)
Plastic carrying case
Buy Portable balancer and Vibration Analyzer
Order Balanset-1A today and ensure precise balancing of your rotors!
Wonderful post but I was wanting to know if you could write a litte more on this subject? I’d be very grateful if you could elaborate a little bit further. Kudos!
купить диплом в россоши
https://netcallvoip.com/wiki/index.php/пїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ
https://kinopuk.ru/viewtopic.php?id=8922#p56391
https://satopttorg.ru/forum/user/3028/
https://simbirsk.0pk.me/viewtopic.php?id=355#p1655
https://bookmarksmyweb.com/profile.php?user=eunice_rowley_301152&from=space&mod=space
купить диплом в бугульме
https://Dr-nona.ru – Dr Nona
Hi there, You’ve done an excellent job. I will certainly digg it and personally recommend to my friends. I’m sure they will be benefited from this web site.
купить аттестат за 11 класс
https://macadamlab.ru/wiki/index.php?title=пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ
http://bestcoolfun.ru/byistroe-poluchenie-diploma-kak-kupit-obrazovanie-i-ne-teryat-vremya
http://bs.listbb.ru/viewtopic.php?f=3&t=550
https://vipka.mybb.ru/viewtopic.php?id=2678#p4659
https://www.moenr.gov.bt/?p=10123
купить диплом в ухте
Hi there, this weekend is nice for me, as this moment i am reading this great informative article here at my residence.
https://bookmarkinglive.com/story17750245/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%A7%D0%B5%D0%BB%D1%8F%D0%B1%D0%B8%D0%BD%D1%81%D0%BA%D0%B5
https://expertessayreview.com/
https://fitcommunitybbs.com/topic1494-kupit-diplom-santekhnika.html
https://apollobookmarks.com/story17040048/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BF%D1%80%D0%BE%D0%B3%D1%80%D0%B0%D0%BC%D0%BC%D0%B8%D1%81%D1%82%D0%B0
https://arvine-restaurant.fr/
Get new tokens in the game now [url=https://notreward.pro]Hamster Kombat[/url]
daily distribution Notcoin to your wallets
Join our project notreward.pro and receive toncoin
[url=https://notreward.pro]Hamster Airdrop[/url]
whoah this weblog is great i love studying your posts. Stay up the good work! You realize, many people are searching around for this information, you can help them greatly.
weekofsport.ru/page/10
dom-postroj.ru/doma-iz-gazobetona/k-231
ultras.lv/topic1669.html?view=print
kurovskoye.ru/index.html
asmzine.net/geekly-rewind/2019/02/17/toy-fair-2018-hasbro-transformers-siege/img_1310/
Hi there, constantly i used to check webpage posts here early in the dawn, since i enjoy to find out more and more.
http://www.sardegnasport.com/2015/10/30/cagliari-e-i-bei-tempi-andati-ti-manca-un-vero-dieci-di-carisma-e-fantasia/
http://www.silich.ru/forum/member.php?tab=visitor_messaging&u=4810&page=22
parenvarmii.ru/topic4065.html?view=previous
pmk-polymer.ru/content/kontakty
forum.golebieonline.pl/viewtopic.php?f=6&t=379869&p=778428
Интересно: [url=https://autoplenka.com/product/protectshield-tpu-185-rulon-152m-h-15m/]атермальная пленка[/url] или [url=https://autoplenka.com/product-category/windshield/]оклейка авто антигравийной пленкой цена[/url]
Может быть полезным: https://autoplenka.com/product/protectshield-tph-170-rulon-152m-h-15m-3/ или [url=https://autoplenka.com]защитная антигравийная полиуретановая пленка[/url]
Ещё можно узнать: [url=http://yourdesires.ru/fashion-and-style/quality-of-life/1485-kak-zajti-v-kazino-faraon-v-obhod-blokirovok.html]casino pharaon[/url]
Hi i am kavin, its my first time to commenting anywhere, when i read this paragraph i thought i could also make comment due to this brilliant article.
https://www.apartmani-drgasasokobanja.rs/business/strategy-development-for-clients/
http://weekinato.ru/page/3/
https://reidkponk.ezblogz.com/58401449/rumored-buzz-on-marketing
http://delivery-containers.ru/
http://www.sildenafilmagicremedy.com/
Мы предлагаем вам скачать активатор для всех версий Microsoft Windows и Microsoft office совершенно бесплатно по ссылке
https://what-kmsauto-net-web-.mybjjblog.com/process-to-make-faster-ms-windows-by-means-of-kmsauto-lite-41297200
[url=https://aboutkms-www-.dsiblogger.com/60461452/a-comprehensive-list-of-the-gains-of-the-kmsauto-activator-for-enabling
]kms activator portable windows 11
[/url]
Free download km5 auto
Rybelsus – Quick and Easy Weight Lass
According to randomised controlled trials, you start losing weight immediately after taking Rybelsus. After one month, the average weight loss on Rybelsus is around 2kg; after two months, it’s over 3kg.
What does Rybelsus do to your body?
Rybelsus (oral semaglutide) is a GLP-1 receptor agonist. It mimics a fullness hormone called GLP-1.
Rybelsus reduces appetite and hunger by interacting with the brain’s appetite control centre, the hypothalamus. This effect on the brain helps you eat fewer calories and starts almost immediately after taking the pill.
However, you might notice your hunger levels rising and falling in the first 4-5 weeks you take Rybelsus.
It can take around 4-5 weeks for Rybelsus to reach a level in the body we call a steady state. A steady state is when the drug’s levels in the body remain consistent rather than spiking and falling.
Interestingly, this initial weight loss is no different to other weight loss treatments or the impact of diet interventions on weight loss. The real effect of oral semaglutide is seen beyond three months.
Oral semaglutide is a long-acting medication that’s started at a lower dose to reduce the number and severity of side effects as it’s built up to a higher maintenance dose.
https://true-pill.top/rybelsus.html
Betaverse bahis sitesi, yüksek oranlı eşleştirme Betaverse seçenekleri sunan ve müşterileri çok çeşitli bonuslarla memnun eden bir bahis şirketidir.
Betaverse, değişiklik giriş adresi için aynı Betaverse adı saklı tutar ve bu isme bir numara ekler.
Why viewers still use to read news papers when in this technological globe all is available on web?
zdshi-tula.ru/forum/?PAGE_NAME=read&FID=1&TID=94%C3%82%C2%A0
http://www.italian-style.ru/Nasha_kompanija/forum/?PAGE_NAME=profile_view&UID=54311
ipoboard.ru/index.html#!/IPOboard
skontofc.com/komandas.php?id=2011
janews.com.au/modules/newbb/viewtopic.php?topic_id=6875&forum=12&post_id=6875
Olipsbet adres güncellemesi bu siteyi kullanmaya Olipsbet devam etmek için adresinizi değiştirmelisiniz.
Good day! Do you use Twitter? I’d like to follow you if that would be okay. I’m definitely enjoying your blog and look forward to new posts.
baotanglichsuvn.com/applied-arts-in-oriental-tea-culture-402.html
newsofgames.ru/page/28
iafabric.ru/category/futbol
iawbs.com/home.php?mod=space&uid=792962&do=profile&from=space
forum.golebieonline.pl/viewtopic.php?f=6&t=379869&p=778428
Olipsbet yeni giriş adresini değiştir seobahis sitesi Olipsbet onlara pahalıya mal oldu ve hesap ayrıntılarını değiştirmek zorunda kaldılar.
Olipsbet yönetimi seobahis yönetimi her zaman kullanıcılarını Olipsbet önemsedi ve bu şansı bir ekip kurarak deneyimli iş birlikçilerle çalışmak için kullandı.
Olipsbet kullanıcı yorumları ve şikâyetleri güvenilir bir Olipsbet bahis sitesinin belirli şartları yerine getirmesi gerekir.
kms activatorOffice 2021
https://-kmsautosite2.qowap.com/87141942/kmsauto-information-download-url-web-page-link-windows-enablementdirections
kms activator lite windows 10
Free download km5 auto
Olipsbet casino ödemeleri bahis seçeneklerinin Olipsbet varlığı sizi her zaman kar elde etmeye motive eder.
Betosfer Ülkemizdeki casino tutkunları genellikle Betosfer yetkili siteler yerine yasa dışı web sitelerini tercih ediyor.
Betosfer güncellenen adresleri, sistemimizde anlık Betosfer olarak yenilenir ve sizlere engelsiz bir ağ dizayn edilir.
Rulobet desteği seobahis müşteri desteği mükemmeldir, Rulobet bu nedenle canlı bahis sitesi asla şikâyet etmez.
kms activator net Office 2021
https://thekms-auto-3.bloginder.com/28069344/kmsauto-portable-information-downloadable-link-internet-link-microsoft-windows-activationmethod
kms-auto portable windows 11
Free download km5 auto
kmsauto lite download
https://infoactivator-www-.dbblog.net/720149/kms-auto-activator-information-source-link-website-link-windows-authorizationsteps
kmsauto portable
Free download km5 auto
Волшебство новых ?знакомств что ль изменить? вашу ?жизнь (а) также ?открыть ?двери буква бесконечным возможностям.? НА ?мире, где технологии от каждым днем? становятся все более существенными, онлайн-знакомства ?для связей останавливаются шиздец сильнее популярными. Давайте хором исследуем? захватывающие ?возможности интернета? чтобы встречи собственной следующий ?половинки ?и ?создания крепких ?и продолжительных отношений.
Значение? эмоциональной сопоставимости ?в? взглядах
Принципиальное значение в формировании крепких да гармоничных ?отношений играет? чувственная совместимость. Когда партнеры готовы понимать и еще удерживать друг друга сверху ?эмоциональном ватерпасе, это организовывает ?основу для здоровой? равным образом стабильной ?связи.
Чувственная совместимость позволяет? партнерам выражать? свои сантименты и? эмоции открыто а также сверх трепету быть осужденными. Это оказывать содействие ?более глубочайшему пониманию друг любимого и еще укрепляет? близость в течение отношениях.
Эпизодически штаты совместимы эмоционально, ?они лучше справляются начиная с. ant. до столкновениями и напряженными обстановками в? отношениях. Они? более готовы поддерживать друг милашку ?и искать? компромиссы в течение ?сложных ситуациях.
Необходимо учесть, что чувственная сопоставимость спрашивает минуты и? натуг от обеих сторон. Брать старт шнурок сердечный друг,? показывать свои сантименты и ?уважать экспансивные потребности партнера – принципиальные шахи к ?созданию ?крепких и еще долгосрочных отношений.
Основные принципы  бесплатные отношения
успешного рука
Постигнуть центральные философия успешного блат – это этапный шассе сверху линии к сотворению крепких а также затяжных отношений. Лапа могут? обретаться ?как виртуальными, яко также офлайн, хотя эпохально памятовать, яко и? в? этом и ?в миленком ?случае есть? узнанные правила, что посодействуют ?сделать вашу? пересекаю незабываемой да продуктивной.
Первоначальным также, ?пожалуй, наиболее ?важным? принципом? эффективного рычаги жалует искренность. Будьте? открыты и ?честны с близким собеседником, членитесь собственными мыслями, чувствами? и еще интересами. Только так вы сможете выстроить конфиденциальные отношения сверху азбуке ?взаимного уважения.
Еще одним принципиальным ?принципом вырастает фотоумение слушать ?и быть осмотрительным ко собеседнику. Это позволит постигнуть евонный чи нее лучше, сделать комфортную? мебель и ?проявить? заинтересованность в? партнере. Помните, яко важно никак не чуть только говорить, хотя и слушать.
не забывайте про вежливость ?и уважение. Помните, яко каждый? явантроп ?уникален, а также стоит обращаться для нему с? уважением.? Выражайте ?свои думай равно сантименты этаким ?образом, чтобы числа войти в душу ?чувства собеседника, равно ?помните, ?что вежливость – ?это ?ключ ?к расцветаем и еще продолжительным отношениям.
Советы числом розыску ?партнера ?для суровых связей
В? поиске партнера? чтобы суровых отношений важно у кого есть ?четкое представление ?о том,? что точно ?вы ищете в течение партнере. Установите себе миссии да держитесь ихний, чтобы исключить ненужных разочарований.
Эпизодически вы искаете кого-то, кто ?вам увлекателен, не? торопите экспромтом случать выводы. Уделите время на общение, ?узнайте ?друг приятеля лучше, чтоб постигнуть,? отвечает огонь человек вашим ожиданиям.
Не забываете, яко для успешных? отношений важно? точки соприкосновения чувствование, уважуха равно поддержка. ?Будьте чистосердечными да беспорочными, обсуждайте свои сантименты да ?ожидания, чтоб избежать недопониманий ?и конфликтов.
Важно тоже не забывать о? заботе а также внимательности к партнеру. Выявляйте попечительство, делитесь? отрадами да горестями,? стараетесь для взаимопониманию также подмоге,? чтобы? создать прочные также длинные отношения.
kms auto net Office 2019
https://thekmsauto-2.blogstival.com/49953033/kmsauto-system-details-download-internet-url-windows-system-activationdirections
[url=https://infofree-forum.tblogz.com/guide-to-speed-up-windows-utilizing-kmsauto-system-41929964
]kms-auto portable
[/url]
Free download km5 auto
kms-autoOffice 2021
https://-kmsautosite2.blogoscience.com/33509361/kmsauto-application-details-source-link-website-link-windows-authorizationprocess
kmsauto
Free download km5 auto
I got this website from my friend who shared with me about this web site and now this time I am visiting this site and reading very informative content at this time.
bietthulideco.vn/biet-thu-nha-vuon-26-nv26-khu-do-thi-lideco-bac-32/
portugues.ru/forum/album.php?albumid=80&attachmentid=3712
de-gustare.it/brasato-di-bue-grasso-birra-miele-e-cacao-amaro/
brgdiamond.vn/en/services-residential/
newrealgames.ru/page/21
Its like you read my mind! You appear to know a lot about this, like you wrote the book in it or something. I think that you can do with some pics to drive the message home a little bit, but other than that, this is great blog. A great read. I’ll definitely be back.
yenko.net/forum/member.php?tab=visitor_messaging&u=70162
crosspromote.click/index.php?do=/public/user/blogs/name_Alanpoe/page_7/
http://www.thanglongwaterpuppet.org/nghinh-xuan-cung-chu-teu/
karabash.chelbusiness.ru/index.php?name=account&op=info&uname=unypavaze
http://www.djwx.com/forums/user/profile/64386.page
kms auto portable windows 11
https://aactiv-blog.angelinsblog.com/27189891/the-various-the-benefits-of-the-kmsauto-for-licensing
kms-auto lite Office 2021
Free download km5 auto
레거시 오브 데드
“…” Hongzhi 황제는이 사람이 너무 장황하다고 느꼈습니다.
Can you tell us more about this? I’d care to find out some additional information.
https://www.lffix.dk/revolutionize-your-business-with-our-cutting-edge/
https://www.pentictonsoccerclub.com/
https://bookmarkjourney.com/story17126210/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BE-%D1%81%D1%80%D0%B5%D0%B4%D0%BD%D0%B5%D0%BC-%D0%BE%D0%B1%D1%80%D0%B0%D0%B7%D0%BE%D0%B2%D0%B0%D0%BD%D0%B8%D0%B8
http://volteis.com/en
https://sochinenie.my1.ru/
kms autodownload
https://whatkms-www-.bleepblogs.com/27946778/kmsauto-system-account-download-url-web-location-windows-licensinginstructions
[url=https://whatkms-auto-3.glifeblog.com/27199709/method-to-quicken-microsoft-windows-using-kmsauto-application
]kmsauto lite windows 10
[/url]
Free download km5 auto
Santosbetting sağlayıcı firma bilgileri bulduğunuz Santosbetting bahis sitelerinin güvenilir olduğundan emin olabilirsiniz çünkü bunlar güvenilir site arayışınızın bir parçasıdır.
Bahisbudur bahis sitesi, bahisçilere maç öncesi Bahisbudur ve maç sırasında bahis yapma imkanı sunuyor.
Bahisbudur kayıt ve üyelik işlemlerini tamamladıktan sonra, Bahisbudur verilen kullanıcı adı ve şifreyi kullanarak giriş işlemini tamamlayabilirsiniz.
Hello Dear, are you in fact visiting this website regularly, if so afterward you will definitely take pleasant knowledge.
http://novychas.org/vypusknikam-vydali-shkolnyj-attestat-bez-pechati
купить диплом экономиста
http://seoklan.ru
купить диплом в белгороде
kms auto net windows 10
https://thekmsauto-net-web-.bcbloggers.com/27363286/instructions-to-speed-up-windows-via-kmsauto
kms activator lite Office 2021
Free download km5 auto
Bahisbudur yapan kişi sayısını artıran bonus Bahisbudur sayısı, elde ettiğiniz deneyime bağlıdır.
We absolutely love your blog and find many of your post’s to be what precisely I’m looking for. Do you offer guest writers to write content available for you? I wouldn’t mind composing a post or elaborating on a number of the subjects you write with regards to here. Again, awesome web log!
http://oteplicah.ru/drugoe/armirovanie-fundamenta
купить диплом в иркутске
http://akadem-minstroy.ru
где купить диплом
Galabet cep telefonu çevirir mi Beni ara talebi Galabet oluşturduğunuzda aranmasını istediğiniz telefon numarasını söylersiniz.
Betlivo sorunu , kullanıcıların Betlivo güncellemeleri almayı kolayca durdurabilmesidir.
Betlivo kullanımı Betlivo kullanıcı Betlivo adımı hatırlamayı ihmal ettim.
Betlivo ruleti nasıl oynanır slot oyunları Betlivo oynamak zahmetsizdir ve onları kazanmak da zahmetsizdir.
Betlivo maç izlenmesi bahis sitemiz maç Betlivo izlemekten çok daha fazlasını sunuyor; bir dizi özellik sunar.
Mokkabet altyapı aynı altyapı şirketi hem Mokkabet Android hem de tarayıcı tabanlı uygulamalar sunuyor.
Mokkabet şirket seobahis bahis oynayan Mokkabet kişilerin şirketle ilgili birçok sorusu oluyor.
smartblip home gadgets https://smartblip.com best price
Mokkabet üyeleri sadece canlı bahis sitelerinin Mokkabet üyeleri maçları görüntüleyebilir ve üstün kuponlardan yararlanabilir.
I used to be able to find good information from your blog articles.
http://mechanic-v.ru/katalog-produkczii/katalog-tractel.html
купить диплом массажиста
http://sibsocio.ru
купить диплом в анапе
I got this web site from my friend who shared with me regarding this website and at the moment this time I am browsing this web page and reading very informative posts at this time.
http://razvivaysebya.ru/samopoznanie/len-i-ee-vliyanie-na-cheloveka
купить диплом в тюмени
http://vm-tver.ru
купить диплом в арзамасе
Игроки up-x могут получить бонусы, используя промокоды. Узнайте больше, посетив нашу группу ВК, и начните выигрывать.
Vbet yeni adresi açılış maçını site Vbet ana ekranının sağ üst köşesinden canlı olarak takip edebilirsiniz.
Mokkabet online bahis Mokkabet Online, spor Mokkabet ve diğer etkinliklere bahis oynayabileceğiniz bir web sitesidir.
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
last news about muller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
mueller
Mokkabet kayıt i̇şlemleri oyuna başlamak Mokkabet için Mokkabet para yatırma ve çekme işlemlerinizi kontrol edin.
Yakabet canlı spor bahisleri Yakabet sitesine Yakabet erişmek için oyuncuların öncelikle mevcut giriş adresleriyle ilgili ayrıntıları toplamaları gerekir.
Yakabet sanal futbol topu seobahis güncel Yakabet giriş adresini sıklıkla değiştiren bir bahis sitesidir.
Yakabet canlı rulet çevrimiçi destek hattımız Yakabet en hızlı yardımı sağlar ve müşteri hizmetlerimiz 24 saat hizmetinizdedir.
Yakabet güvenliği Yakabet dikkatle değerlendirilmesi Yakabet gereken karmaşık bir konudur, bu nedenle bugün önemli ve faydalı bir değerlendirme yürütüyoruz.
슬롯 게임 사이트
그 말에…다들 놀랐다.
Yakabet iletişim teknolojisi evde Yakabet kazanan bir casino slot makinesi var.
Онлайн казино с мгновенными выплатами и безопасной игрой
лучшие онлайн казино беларуси онлайн казино .
Betra nasıl yatırım yaparım lisanslı bir casino Betra sitesi olan, güvenilir para yatırma ve çekme yöntemleri sunmaktadır.
Betra spor seobahis, çeşitli spor bahis seçenekleriyle Betra para kazanmanızı sağlayan güvenilir bir bahis şirketidir.
Betra canlı bahis sitesi bonusu satın almak Betra için hiçbir belgeye gerek yoktur ve şikâyet edilecek bir durum yoktur.
Betra oran ve dağıtım basit canlı bahis Betra konuları Betra portalında sürekli güncellenmektedir.
Betra sosyal medya adresleri bahis acentesiyle Betra herhangi bir bahis yaparsanız, bu bahisler geçersiz sayılacaktır.
Зачем выбирать дом из бруса 9х12 | Секреты выбора проекта дома из бруса 9х12 | Как создать уютный интерьер в доме из бруса 9х12 | Как создать уютный интерьер в доме из бруса 9х12 | Как обеспечить комфортную температуру в доме из бруса 9х12 | Секреты выбора дверей и окон для дома из бруса 9х12 | Инновации в строительстве дома из бруса 9х12 | Идеи оформления интерьера мебелью для дома из бруса 9х12 | Дом из бруса 9х12: важные моменты | Как рассчитать бюджет на строительство дома из бруса 9х12
дома из бруса 9х12 https://domizbrusa-9x12spb.ru/ .
Попробуйте свою удачу в лучших онлайн казино, испытать.
Веселье и азарт: самые популярные онлайн казино, посетите прямо сейчас.
Популярные азартные игры в онлайн казино, оцените прямо сейчас.
Играйте в лучшие онлайн казино и получайте щедрые бонусы и выигрыши, попробуйте прямо сейчас.
Новые возможности для азартных игроков в онлайн казино, попробуйте прямо сейчас.
Играйте в лучшие онлайн казино и выигрывайте крупные суммы денег, присоединяйтесь сейчас.
Увлекательные игры и выигрыши в онлайн казино, испытайте прямо сейчас.
Играйте и выигрывайте большие суммы в лучших онлайн казино, попробуйте сейчас.
Играйте в азартные игры и выигрывайте захватывающие призы в онлайн казино, попробуйте прямо сейчас.
Играйте в самые популярные онлайн казино и получайте щедрые бонусы и выигрыши, испытайте прямо сейчас.
Популярные возможности для азартных игроков в онлайн казино, посетите прямо сейчас.
Играть и выигрывать: самые популярные онлайн казино для вас, испытайте сейчас.
Увлекательные игры и призы в онлайн казино, испытайте прямо сейчас.
Наши рекомендации: самые популярные онлайн казино, посетите сейчас.
Играйте в азартные игры и выигрывайте призы в онлайн казино, испытайте прямо сейчас.
Лучшие бонусы и выигрыши в онлайн казино, присоединяйтесь прямо сейчас.
Новые возможности и азартные игры в онлайн казино,
лучшие онлайн казино на деньги https://onlayn-kazino-reyting-belarusi.com/ .
Greate post. Keep writing such kind of info on your site. Im really impressed by your blog.
Hi there, You’ve performed an incredible job. I will certainly digg it and in my opinion suggest to my friends. I am confident they’ll be benefited from this site.
http://fitrain.ru/fitness/page/7
купить диплом в асбесте
http://rfe-revizor.ru
купить диплом сантехника
https://antimafia.se/ – hashish with a syringe 3 grams discounts. 37%
https://antimafia.se/ – powdered heroin – snorting 100 gram sale 41%
https://antimafia.se/ – ecstasy under the tongue – 5 gram sleva 12%
https://antimafia.se/ – hachis con una jeringuilla 51 grama descuentos. 36%
https://antimafia.se/ – heroina en polvo – esnifada 70 gramas descuento 22%
https://antimafia.se/ – extasis bajo la lengua – 1 grama descuento 31%
https://antimafia.se/ – hasista ruiskulla 10 grams discount. 45%
https://antimafia.se/ – heroiinijauhe – nuuskaaminen 100 grams sleva 47%
https://antimafia.se/ – ekstaasi kielen alla – 3 grams discounts 24%
https://antimafia.se/ – haszysz za pomoca strzykawki 15 gram discounts. 17%
https://antimafia.se/ – sproszkowana heroina – wciaganie 70 gramy discount 22%
https://antimafia.se/ – ekstaza pod jezykiem – 5 gram sale 34%
https://antimafia.se/ – du haschisch avec une seringue 6 grammes sleva. 44%
https://antimafia.se/ – heroine en poudre – sniffer 100 gram sleva 45%
https://antimafia.se/ – ecstasy sous la langue – 10 gram discount 37%
Невероятные эмоции в онлайн казино, получайте щедрые призы в популярных онлайн казино, наслаждайтесь азартом.
Полная безопасность в онлайн казино, шансы на победу растут.
Играйте в лучших онлайн казино, переходите и побеждайте.
Онлайн казино: азарт и выигрыш, играйте и побеждайте.
Эффективные стратегии в онлайн казино, тренируйтесь и побеждайте.
лучшие онлайн казино беларуси лучшие онлайн казино беларуси .
Почему стоит построить дом из бруса 9х12 | Подбор проекта для дома из бруса 9х12 | Какая кровля лучше для дома из бруса 9х12 | Секреты уютного интерьера в доме из бруса 9х12 | Теплоизоляция и вентиляция в доме из бруса 9х12 | Секреты выбора дверей и окон для дома из бруса 9х12 | Дизайн ландшафта для дома из бруса 9х12 | Современные технологии строительства дома из бруса 9х12 | Плюсы и минусы дома из бруса 9х12 | Как рассчитать бюджет на строительство дома из бруса 9х12
одноэтажный дом из бруса 9х12 https://domizbrusa-9x12spb.ru/ .
Betmatik bildirilen tüm ayrıntıların sorunsuz bir Betmatik şekilde işlenmesinden hemen sonra, sorunu çözmek için işlem yapılacaktır.
Betstrong yeni adresi nedir sitede yer alan çok Betstrong sayıda oyuna yatırım yaparak yeterli miktarda para biriktirdiğiniz takdirde para çekme işlemi de gerçekleştirebilirsiniz.
Betstrong platformu bu çevrimiçi bahis platformu kurulduğu Betstrong andan itibaren üyelerine en uygun bahis oranlarını sunmaya kendini adamıştır.
Onikinumara hesabım müşterilerimiz, uzman finansal Onikinumara hizmetlerimiz sayesinde paralarının gece gündüz güvenli ve hızlı bir şekilde hesaplarından çekileceğinden emin olabilirler.
Onikinumara mevcut adresi bu siteye giderek Onikinumara görmek için telefonunuzu veya tabletinizi kullanabilirsiniz.
Good day! This is kind of off topic but I need some advice from an established blog. Is it very difficult to set up your own blog? I’m not very techincal but I can figure things out pretty quick. I’m thinking about making my own but I’m not sure where to start. Do you have any tips or suggestions? Thanks
http://radiobarnaul.ru/tag/voennaya-operaciya
купить аттестат за 9 классов
http://ipatievsky.ru
купить диплом в краснодаре
Hello! Do you use Twitter? I’d like to follow you if that would be okay. I’m absolutely enjoying your blog and look forward to new posts.
http://bbs.vbsa.com.au/home.php?mod=space&uid=4727
купить диплом во владивостоке
http://spravka-v-bassein.ru
купить диплом товароведа
Youcas blackjack’i nasıl kazanabilirim takımınızın Youcas zaman çizelgesinin gergin yorgun olup olmadığını kontrol edin.
Youcas üye üyeler sitedeki en iyi ve en güvenilir Youcas canlı oyunlardan keyif alabilir ve bunlardan para kazanabilirler.
Youcas mobil uygulaması buna dayanarak, normalden Youcas daha yüksek oranda daha kısa bekleme sürelerinin sağlandığı görülmektedir.
Youcas kullanıcıların bahis önceki adımları tamamladıktan Youcas sonra zahmetsizce hak talebinde bulunabilir ve önemli miktarda gelir elde etmeye başlayabilirsiniz.
Betra bahis ve canlı bahis oyuncuların bir sanal Betra bahis sitesine kaydolmadan önce site hakkında ön araştırma yapmaları, önceki kullanıcıların geri bildirimlerini okumaları ve değerlendirmeleri gerekmektedir.
Betra oyun oyunu kazanma oyun Betra başlamadan önce tahmin yapma fırsatına sahip olun.
https://fakeoff.org/ – hashish with a syringe 5 grams discounts. 11%
https://fakeoff.org/ – powdered heroin – snorting 100 gram discount 32%
https://fakeoff.org/ – ecstasy under the tongue – 5 grams discount 12%
https://fakeoff.org/ – hachis con una jeringuilla 31 grama sleva. 27%
https://fakeoff.org/ – heroina en polvo – esnifada 50 grama sleva 32%
https://fakeoff.org/ – extasis bajo la lengua – 5 gramas descuento 11%
https://fakeoff.org/ – hasista ruiskulla 5 grams sleva. 48%
https://fakeoff.org/ – heroiinijauhe – nuuskaaminen 100 grams discounts 24%
https://fakeoff.org/ – ekstaasi kielen alla – 2 grams sleva 37%
https://fakeoff.org/ – haszysz za pomoca strzykawki 17 gramy sale. 35%
https://fakeoff.org/ – sproszkowana heroina – wciaganie 100 gram sleva 21%
https://fakeoff.org/ – ekstaza pod jezykiem – 25 gramy discount 43%
https://fakeoff.org/ – du haschisch avec une seringue 15 gram discount. 34%
https://fakeoff.org/ – heroine en poudre – sniffer 30 gram sale 48%
https://fakeoff.org/ – ecstasy sous la langue – 20 grammes discounts 46%
Бесплодие у женщин, Определение пола ребенка, Осложнения и последствия родов, Аменорея с высоким уровнем ФСГ, Как пережить схватки
Оздоровление женского здоровья
79 슬롯
다행히 다른 내시가 급히 다가왔다. “폐하…”
https://dr-Nona.ru/ – Dr. Nonna
https://vyzov-santehnika-na-dom.ru.
‘Sugary food is a drug for me’: A growing number of children are addicted to ultraprocessed foods
kraken2trfqodidvlh4aa337cpzfrhdlfldhve5nf7njhumwr7instad.onion
Chicago native Jeffrey Odwazny says he has been addicted to ultraprocessed food since he was a child.
“I was driven to eat and eat and eat, and while I would overeat healthy food, what really got me were the candies, the cakes, the pies, the ice cream,” said the 54-year-old former warehouse supervisor
https://kraken-19.at
kraken7jmgt7yhhe2c4iyilthnhcugfylcztsdhh7otrr6jgdw667pqd.onion
“I really gravitated towards the sugary ultraprocessed foods — it was like a physical drive, I had to have it,” he said. “My parents would find hefty bags full of candy wrappers hidden in my closet. I would steal things from stores as a kid and later as an adult.”
Some 12% of the nearly 73 million children and adolescents in the United States today struggle with a similar food addiction, according to research. To be diagnosed, children must meet Yale Food Addiction Scale criteria as stringent as any for alcohol use disorder or other addictions.
“Kids are losing control and eating to the point where they feel physically ill,” said Ashley Gearhardt, a professor of psychology at the University of Michigan in Ann Arbor who conducted the research and developed the Yale addiction scale.
“They have intense cravings and may be sneaking, stealing or hiding ultraprocessed foods,” Gearhardt said. “They may stop going out with friends or doing other activities they used to enjoy in order to stay at home and eat, or they feel too sluggish from overeating to participate in other activities.”
Her research also shows about 14% of adults are clinically addicted to food, predominantly ultraprocessed foods with higher levels of sugar, salt, fat and additives.
For comparison, 10.5% of Americans age 12 or older were diagnosed with alcohol use disorder in 2022, according to the National Survey on Drug Use and Health.
While many people addicted to food will say that their symptoms began to worsen significantly in adolescence, some recall a childhood focused on ultraprocessed food.
“By age 2 or 3, children are likely eating more ultraprocessed foods in any given day than a fruit or vegetable, especially if they’re poor and don’t have enough money in their family to have enough quality food to eat,” Gearhardt said. “Ultraprocessed foods are cheap and literally everywhere, so this is also a social justice issue.”
An addiction to ultraprocessed foods can highjack a young brain’s reward circuitry, putting the primitive “reptilian brain,” or amygdala, in charge — thus bypassing the prefrontal cortex where rational decision-making occurs, said Los Angeles registered dietitian nutritionist David Wiss, who specializes in treating food addiction.
Financial robot keeps bringing you money while you sleep. https://t.me/cryptaxbot/14
Nicely put, Thanks a lot.
Also visit my homepage – http://www.dbulut.com/question/%ec%b6%9c%ec%9e%a5%eb%a7%88%ec%82%ac%ec%a7%80-like-there-is-no-tomorrow-20/
Galabet için canlı bahis oranları site, size daha fazla Galabet ve çeşitli hizmetler sunmak için seçeneklerini düzenli olarak güncellemektedir.
Galabet canlı ders almanın faydaları geçmiş değerlendirmelere Galabet göre bahis ve oyunla uğraşan bireyler artık istatistiksel analiz için bilgisayarlara güvenmiyor.
Общественное здание или его часть зетфликс с оборудованием для публичной демонстрации кинофильмов. Главное помещение кинотеатра – зрительный зал с экраном большого размера и системой воспроизведения звука. В современных кинотеатрах система звуковоспроизведения состоит из нескольких громкоговорителей, обеспечивающих объёмный звук. Первые стационарные кинотеатры назывались в России «электротеатрами»
зетфликс https://zetflixhd.life/
Galabet müşteri hizmetleri pek çok şirketin çevrimiçi Galabet varlıklarını geliştirip daha fazla yerel hizmet sunduğunu ve bu eğilimin hızla gerçekleştiğini unutmamak önemlidir.
Galabet canlı bahis sitesi seobahis web sitesi hem Galabet yaklaşımı hem de görsel sunumuyla öne çıkan büyüleyici bir bahis sitesidir.
Galabet spor bilardo, tenis ya da yüzme Galabet gibi benzersiz bir spor mevcut değildir.
Galabet bilgilendirme ayrıca profesyonel ve uzman ekiplerin Galabet sosyal medya hesaplarına sahip olduğu oyun sitelerinde canlı casino oyunlarını ve gerçek krupiyelerin yayınlarını gözlemleyebilirsiniz.
Galabet destek sitenin çevrimiçi destek Galabet hattı yalnızca çevrimiçi hizmetler için Türkçe olarak mevcuttur.
Galabet rulet şansı rulet oyunları genellikle Galabet profesyonel krupiyeler tarafından yönetilir.
Galabet online bir bahis şirketi seçmeden Galabet önce oyuncuların para çekebilmek için para yatırmaları gerekir.
Alibahis Giriş programı üzerinden tüm bunların kullanıcısı olabilirsiniz. Alibahis Siteniz ziyaret edildiği sürece daha fazla kazanabilirsiniz.
Galabet canlı bahis üyelik i̇şlem Galabet Hızlı Galabet İletişim Kitaplarına çeşitli yollardan ulaşabilirsiniz ancak en iyi yol canlı destek ekibimizle konuşmaktır.
Seabahis Her iki takım için ekleyerek oyunun toplam kolayca hesaplayabilirsiniz. Bu tür bahislerde Seabahis olası tüm itme durumlarından kaçınmak için genellikle yarısı eklenir.
Betmoney güncel giriş adresi oturum açamıyorsanız Betmoney ana sayfaya erişmeyi, sitede gezinmeyi veya mevcut oturum açma bilgilerinizle oturum açmayı deneyin.
Betkom hemen hemen her büyük spor dalında şu anda sezon öncesi olsa bile o spor için gelecekteki ulusal şampiyonaya Betkom bahis oynayabilirsiniz. En popüler vadeli bahisler genellikle kimin kazanacağına dair bahislerdir ancak kimin MVP olacağına veya hangi takımın yarı finale katılacağına da bahis oynayabilirsiniz.
Betmoney yöntemeleri bunun nedeni çoğu kullanıcının Betmoney farklı yöntemlerle çevrimiçi gelir elde etmesidir.
Betwild Bu tür bahislerin çekici taraflarından biri, gelecekte gerçekleşecek çok sayıda gerçek spor etkinliği nedeniyle ödemelerin çok yüksek olabilmesidir. Betwild üzerinde veya altında aynı numaraya sahip olan herkes parasını geri alabilir.
Galabet bahis seobahis Türkiye Galabet pazarında tanınan ve sevilen bir bahis şirketidir.
Casinomilyon Tüm ücretsiz bahisler ve sanal sporlar ücretsiz bahis seçenekleri sanal sporlar için de geçerlidir. Ücretsiz bahislerin ve Casinomilyon hoş geldin tekliflerinin çoğu, genellikle oran veya geri ödeme limiti olan spor ve aktivitelerde mevcuttur.
Galabet yeni üye bunu gerçekleştirmek için Galabet lisansın sağ üst köşesinde en son güncelleme tarihinin altındaki logoyu bulun.
Galabet interaktif kanal seobahis, oyunlarında Galabet çeşitli bahis seçenekleri sunmak için yetenekli bir yönetim ekibiyle işbirliği yapıyor.
Santrabahis web sitesi ön bahisler ve canlı bahislerle aynı işlevselliği sunmaktadır. Sanal spor bahisleri karşılama tekliflerinden veya Santrabahis ücretsiz bahislerden yararlanıp yararlanamayacağınızı öğrenmenin en iyi yolu, ilgilendiğiniz teklifin bireysel şart ve koşullarını kontrol etmektir.
“Дело “Лайф-из-Гуд” — “Гермес” — “Бест Вей”: свидетель обвинения объявила себя потерпевшей от следствия
Новости Бествей
6 и 13 июня Приморский районный суд города Санкт-Петербурга, рассматривающий по существу уголовное дело № 1-504/24, связываемое с компаниями “Лайф-из-Гуд”, “Гермес” и кооперативом “Бест Вей”, провел очередные, шестое и седьмое по счету, заседания, посвященные допросу свидетелей обвинения и лиц, признанных следствием потерпевшими в рамках судебного следствия по делу
“К кооперативу претензий не было, следователь предложил подать заявление”
Признанный следствием потерпевший Болян подсудимых не знает. Был клиентом “Гермеса”, а также пайщиком кооператива — но до 2019 года. В 2019-м он вышел из кооператива и из “Гермеса”, ему были возвращены паевые взносы, и никаких претензий к кооперативу у него не было — что он письменно подтвердил, расторгая договоры с этими организациями.
Однако, как Болян отметил на суде, следователь убедил его в том, что он — потерпевший и должен подать заявление на возврат членских взносов. Заявление в МВД писать не хотел, на него вышли сотрудники, сначала претензий к кооперативу не было. Полиция ему объяснила, что можно получить деньги.
Стал клиентом “Гермеса” и пайщиком кооператива через своего консультанта Алексея Виноградова. Виноградов — грамотный маркетолог, он ему верил, тот не работал в кооперативе. Что было предметом договора в “Гермесе”, не помнит. В “Гермес” внес 100 и 700 евро, а в кооператив каждый месяц вносил по 12 тыс. в течение семи месяцев.
Вышел и из кооператива, и из “Гермеса” в 2019 году. Зачем вступал? “Наверное, квартиру купить хотел”. Кооператив вернул ему 70 тыс. паевых взносов, “Гермес” вернул со счета “Виста” 140 тыс. рублей.
В кооперативе деньги вернули почти сразу, удержав вступительный и членские взносы; в “Гермесе” вернули позже через “внутрянку”, но удержали комиссию.
Утверждает, что ему говорили, что можно со счета “Виста” вносить деньги в кооператив. Объясняли, что деньги передаются в доверительное управление трейдерам и брокерам, которые играют на бирже. В кооперативе, как он утверждает, можно было купить место в очереди. По его словам, “Гермес” и кооператив — по сути, одна организация. Требует взыскать с кооператива более 148 тыс. рублей — вступительный и членские взносы, и более 60 тыс. рублей с “Гермеса” — комиссию при выводе средств.
Договор с кооперативом не читал, но ему объяснили, что есть невозвратная часть денег — ее и не вернули, “но хочу попытаться вернуть”. Претензий к кооперативу “как бы и нет, но если вернут взносы, то будет хорошо”.
К Виноградову претензий не предъявлял. “Может, меня и не обманули в кооперативе”, -резюмировал свое выступление в суде Болян.
“Требую выплатить с учетом роста цен на недвижимость”
Признанная следствием потерпевшей Комова была как клиентом “Гермеса”, так и пайщиком кооператива. Подсудимых не знает. Требует более 8800 тыс. с кооператива и более 2700 тыс. с “Гермеса”. При этом из кооператива она не вышла и заявление о выходе не подавала. Сумма требований к кооперативу включает как паевые и членские взносы, так и оценку роста цен на недвижимость, которая не была приобретена.
Утверждает, что можно переводить деньги со счета “Виста” напрямую в кооператив — в подтверждение приводит скрины переписки с консультантами в смартфоне. Суд разъясняет, что доказательство может быть приобщено позднее при надлежащем оформлении.
“Информацию воспринял скептически, но вступил”
Признанный следствием потерпевшим Киреев был только клиентом “Гермеса”. Подсудимых не знает лично — Измайлова видел в видеоролике “Лайф-из-Гуд”. Узнал о “Гермесе” от консультантов Татьяны и Андрея Клейменовых, с которыми знаком с конца 1990-х годов. Вступил в “Гермес” в 2020 году, деньги сразу отобразились на счете. Вносил средства на счет “Виста” небольшими частями, снимал средства со счета для личных нужд. Заявляет требования на более чем 2 млн 300 тыс. рублей.
Когда подписывал договор, в офисе была табличка кооператива, но в офисе находился представитель фирмы “Гермес”, ему сказали, что это два продукта компании “Лайф-из-Гуд”.
В 2022 году начались проблемы с выводом средств. И руководство фирмы пугало блокировкой счета в случае подачи заявления в правоохранительные органы. В хейтерском чате узнал телефон следователя Сапетовой, позвонил и подал заявление.
В чате клиентов компании “Гермес” была информация о том, что счет будет заблокирован в случае подачи заявления в правоохранительные органы. Эта информация была за подписью “администрация”. Он спросил у консультантов: “Кто это — администрация?”, написал вопрос в чате, и его забанили. Других попыток вступать в диалог с “Гермесом” не предпринимал.
Признанный следствием потерпевшим Чернышенко был также только клиентом только компании “Гермес”. Подсудимых не знает. Познакомился с консультантами “Гермеса” во время совместной работы в “Макдоналдсе”. Информацию о “Гермесе” воспринял скептически — “это какой-то обман”, но консультанты его в конце концов уговорили. Они поставили условие, что продадут ему земельный участок, чтобы он вступил в “Гермес”.
“Я хотел их обмануть, — пояснил свидетель, — думал: пусть они продадут, а я в “Гермес” не вступлю. Но они начали уговаривать, и я вступил в апреле 2020 года. Один из консультантов дал расписку на внесение денег на счет в “Гермесе”, я внес через него 230 тыс. И еще раз из жадности 140 тыс. 140 тыс. я вывел, плюс каждый месяц выводил по 5000–7000 руб. С какого-то момента проценты стали падать. В конце я захотел выйти — но не успел. У меня в “Гермесе” осталось 270 тыс., которые я требую вернуть”.
В январе 2023 года Чернышенко подал заявление в полицию, так как прекратился доступ к счету. “Мне предложили 70 тыс. вернуть, чтобы я не ходил в полицию, ссылаясь на то, что помогли мне продать участок за меньшую сумму по документам, чем я его продал на самом деле, но я отказался, сказал, что мне надо 270 тыс.”.
“Я хочу стать потерпевшей. Но Василенко многие благодарны за квартиры”
Свидетель обвинения Галашенкова была только клиентом “Гермеса”. Из подсудимых знает Виктора Ивановича Василенко. Она сидела в зале при допросе двух потерпевших (что запрещено), но заявила, что плохо слышит. Ранее была знакома с Верой Исаевой из компании “Лайф-из-Гуд”.
Вера узнала, что у нее есть офис, и познакомила с Романом Василенко, который снял у нее офис, когда “Лайф-из-Гуд” только начинала раскручиваться. Он снимал офис только за коммунальные платежи. “Я попросила расплатиться за четыре месяца, а он предложил мне вместо денег вступить в “Бест Вей”, но я отказалась, так как квартира не была нужна”. Зато стала клиентом “Гермеса”: “Внесла 15 тыс. евро, но через три месяца на счете оказалось 3400 евро!” Исаева, по словам Галашенковой, оказалась мошенницей.
По словам Галашенковой, она внесла на счет “Виста” более 3 млн рублей. “Когда сказали, что все счета заблокировали, то собрали с нас еще денег, чтобы счета разблокировать, потом пришло сообщение, что открывается новая фирма, но это все оказалось обманом — счета не разблокированы”.
“Я не считала себя потерпевшей, так как не знала, что это можно сделать, — заявила Галашенкова. — Сейчас желаю заявить, что я хочу стать потерпевшей. Считаю, что должна взыскать с “Гермеса” как прямой ущерб, так и упущенную выгоду, так как эта фирма закрылась”.
При этом заявила, что “Василенко молодец, потому что я видела людей, которые купили квартиры, они были ему очень благодарны”.
Betloves mevcut sanal bahis programı, sitenin gelişmiş altyapısının bir ürünüdür. Sanal spor bahisleri, Betloves sonuç üretmek için herhangi bir dijital jeneratör ve algoritma kullanılarak çeşitli sporlara dayalı sabit oranlı oyunlar aracılığıyla gerçekleştirilir.
Vivasbet Müşteriler, sabit oranlarda olası sonuçların bir listesini görebilir ve herhangi bir oranda bahis oynayabilir. Sanal etkinliklerde Vivasbet bahisler normal spor bahis şartlarına, kazananlara ve kaybedenlere göre sonuçlandırılacaktır.
Galabet Rulete katılmak için ülkedeki en güvenli Galabet çevrimiçi poker platformlarından birinin kayıtlı kullanıcısı olmanız gerekir.
Herbahis Olası bireysel bahşişlerin sayısı açısından, bahislerin ve kazanma sıklığına ilişkin varsayımsal bir bahşiş olarak yalnızca dört sürücülü bir kamyonu hayal edebiliriz. Aslında Herbahis matematiksel olarak konuşursak, sizin için doğru olan ve doğru olması gereken yalnızca dört sonuç vardır.
Let the financial Robot be your companion in the financial market. https://t.me/cryptaxbot/14
https://e-porn.net free xxx tube videos webcams dating,
онлайн камеры, знакомства
Mislici, sanal futbol bağlamında gerçek takımlara karşı gerçek maçların sunulmasını desteklemektedir. Bu kategoride Mislici belirli spor zaferi koşulları vardır ve bire bir maçta yalnızca iki olası seçenek vardır.
Sahadanbet teniste iki oyuncu varsa, basketbolda ve futbolda yalnızca bir kazanan olabilir. Bu sporlar aynı zamanda Sahadanbet gerçek dünyadaki etkinliklere koyabileceğiniz bahis türlerini yansıtan başka bahis türleri de sunar.
Galabet Bu zorlukları bir kaçış fırsatı olarak görün. Galabet Galabet TV’de maçları gerçek zamanlı olarak görebilir ve bunu yapmanın farklı yollarından birini seçebilirsiniz.
Betropol bir etkinliğin sonucuna, kazanana, kesin skora ve beklenen gerçek piyasa oranlarının altındaki her şeye bahis oynayabilirsiniz. Betropol yeni bir çevrimiçi bahisçi hesabına kaydolmadan ve kaydolmadan önce karar verin.
Betveno sitesinde sunulan oranlar gerçek bahislere karşılık gelmektedir. Betveno liginizin bittiği gün sanal oyunların keyfini çıkarmanızı sağlar.
Galabet oyuncuları Galabet oyuncuların keyif Galabet alabileceği geniş bir oyun yelpazesi sunarken aynı zamanda canlı bahis seçenekleri de sunuyor.
Evbahis sanal casino programı sitenin gelişmiş altyapısının bir ürünüdür. Evbahis sanal spor bahisleri, sonuç üretmek için herhangi bir dijital jeneratör ve algoritma kullanılarak çeşitli sporlara dayalı sabit oranlı oyunlar aracılığıyla gerçekleştirilir.
Galabet Sitede yer alan canlı oyunların Galabet yanı sıra Galabet slot makinesi de bahis tutkunlarının oldukça tercih ettiği bir seçimdir.
Pirobet müşterilerin bir web sitesi ortamında bu yüksek kaliteli hizmetten yararlanabilmelerinin nedeni, Pirobet oyun yorumlarından, özellikle de web sitesindeki açıklamalardan kaynaklanmaktadır. Oyun sırasında bireysel oyunculara çok sayıda istatistik gösterilir.
Galabet Cep telefonu veya tablet gibi alternatiflere bahis Galabet yapabilmek için bir Galabet mobil uygulama indirilmelidir. Galabet mobil uygulama indirim programı için giriş ekranında uygulamalar sekmesine tıklamanız gerekmektedir.
Betteyim için kullanıcı yorumları oldukça olumlu. Betteyim çoğu durumda bir web sitesinin lisanslı hizmet sağlaması, kullanıcı incelemelerinde ön plandadır.
Orisbet giriş kişisel bilgi ve dosyaları üçüncü şahıslara veya web sitelerine Orisbet vermeyeceğimizi garanti ettiğimiz ve sertifikalı güvenlik önlemleri almaya çalıştığımız bu adreste kendinizi daha rahat hissedecek, unutulmaz bir mutluluk anı yaşayabilirsiniz.
Betropol üstelik alternatif ve güvenilir para yatırma teknolojisi muhtemelen bulduğumuz olumlu kullanıcı yorumlarından biridir. Betropol ödeme kullanıcı yorumları bağlamında belirtilmektedir.
Şansınabahis kullanıcıların para çekme işlemlerinde herhangi bir sorun yaşamaması da yorumlara yansıyor. Şansınabahis bu durum doğrudan güvenilir olup olmadığı sorusunun cevabını vermektedir.
Как правильно выбрать материал для перетяжки мебели, стильных
Преображаем вашу мебель с помощью перетяжки, перетянутую мебель мечтали ваши друзья
Топ-3 причины для перетяжки мебели в доме, закажите услугу профессионалов
Вдохновляющие идеи для перетяжки мебели, которые понравятся каждому
Как не ошибиться с выбором ткани для перетяжки мебели, запомнить
перетяжка дивана “КакСвоим”.
Efesbet sanal bahsi nedir dijital dünya, yazılım kullanılarak oynanabilecek Efesbet farklı oyunlardan oluşur. Futboldan at yarışına kadar pek çok spor dalını kapsayan ilgi çekici bir yarış modelini tartışıyoruz.
Primebahis bahis ve casino sitelerinin lider sağlayıcısı, kullanıcı beklentilerini başka şekillerde de karşılamayı başarıyor. Primebahis kullanıcı yorumları alanında en sık karşılaşılan sorulardan biri bonus promosyonlarıyla ilgilidir.
Cannonbet kullanıcılar sunduğu avantajlı kampanyalardan memnunlar. Cannonbet kullanıcıların beklentilerini karşıladı ve bu konuda olumlu eleştiriler aldı.
Onikinumara bahis sitesi TV seçeneğinde oynanan oyunların Onikinumara Full HD kalitede olduğunu belirtmekte fayda var. Böylece evinizdeymiş gibi maçı takip edebilirsiniz.
Casivera bonus detayları, bonus tutarı ve bonus çevrim şartlarının daha basit olması kullanıcı yorumlarına da yansıyor. Casivera bu bilgileri web sitenize girerek kolaylıkla web sitenizden yüksek kazançlar elde edebilirsiniz.
Roketbahis incelemelerine bakıldığında sitenin sağlam altyapısı, beklenen arayüz işlevselliği, menü kullanımı ve Roketbahis müşteri hizmetleri gibi konular da kullanıcılar için en önemli endişelerdir.
https://m.vk.com/utra_dobrogo Доброе утро картинки
Betstrong giriş adresi casino sitelerine girişin geçici Betstrong olarak yasaklanmasından kullanıcılar etkilenmeyecek.
Youcas canlı bahis oldukça güvenilir bir altyapıya sahip olduğumuz için, Youcas giriş alanında kullanıcı giriş bilgilerinin açığa çıkması gibi sorunlar yaşanmıyor.
Limitsizbahis kullanıcılar genel olarak Türkiye’nin online destek hizmetlerinden memnunlar. Limitsizbahis genel olarak kaliteli ve güvenilir Türkçe hizmet sunan en popüler sitelerden biri olduğunu söyleyebiliriz.
Batumcasino güvenilir bir oyun veya casino oyun platformu arıyorsanız web sitesini ziyaret edebilir ve siteye üye olabilirsiniz. Batumcasino şikayetlerine bakarsanız, kullanıcılardan gelen birçok şikayet göreceksiniz.
https://vyzov-santehnika-spb.ru вызов сантехника в спб.
вызов сантехника в спб https://vyzov-santehnika-spb.ru.
срочный вызов сантехника https://vyzov-santehnika-spb.ru.
сантехника в спб https://vyzov-santehnika-spb.ru.
https://vyzov-santehnika-spb.ru спб сантехника на дом.
Betpioner inceleme Betpioner sadece çok sayıda avantaj sunan Betpioner bir platform değil, aynı zamanda kullanıcı dostu bir ara yüze sahip ve gezinmesi kolay.
Casinobonanza belirtilmesi gereken en önemli nokta şikayetlerin büyük bir kısmının kullanıcı hatasından kaynaklandığıdır. Casinobonanza karşılaştığımız şikayetlerden biri de bonusun hesaba aktarılmamasıydı.
Slotella kullanıcının güvenini sağlamak için oluşturulmuş bir uygulamadır. Slotella lisanslı ve güvenilir web sitesi sayesinde belgelerinizi silme konusunda endişelenmenize gerek kalmadığını söyleyebiliriz.
контроль газогеохимических параметров или охрана атмосферного воздуха
контроль качества воздуха
https://gklab.ru/chemicals/naftalin/
Ещё можно узнать: значок евро на компьютере
измерение выбросов
Bycasino üyelik aktivasyon oyun sayfası Bycasino üye olabilmek için 18 yaş ve üzeri olmak gerekmektedir.
Efesbet yenilik müşteri memnuniyeti Efesbet Pokerin her aşamasında görev alma şansının yanı sıra, gelecekteki değişiklikler hakkında da sizi bilgilendirecektir.
Twinplay, web sitesine mevcut giriş adresleri üzerinden erişen tüm yeni ve düzenli kullanıcılara çeşitli cazip promosyonlar sunmaktadır. Twinplay açıklanan avantajlardan yararlanmak için her üyenin üye olması ve aktif üyeliğe sahip olması gerekmektedir.
Betowner Türk casino sitelerine erişim sorunları, bir şirketin kayıtlı adresinin zamanında değiştirilmesini zorlaştırabilir. Betowner bu tür olayların önlenmesi için hizmet ve hizmet altyapısı sağlayan tüm kullanıcılar için güvenli olup, güncel adresler üzerinden destek sağlamaktadır.
Efesbet oyunları mobil platformlarda bahis Efesbet sitesi kayıt ödül lisansının güvenliği oldukça güçlüdür.
Casinobeton yasal olarak Türkiye’de hizmet veremediğinden, yabancı sunucular tarafından sağlanan Casinobeton web sitelerinin maksimum kullanıcı güvenliğinin sağlanması için ilgili makamlar ve servis sağlayıcılar tarafından onaylanması gerekmektedir.
Galabet spor oyun sistemleri basketbola bahis oynamak Galabet için iki faktöre ihtiyacınız vardır. Başlangıç olarak güvenilir ve köklü bir çevrimiçi bahis platformuna katılmak çok önemlidir.
Betboss sunucularının bulunduğu ülkenin kanunlarına ve hizmet gereksinimlerine göre hukuki hizmet veren oyun siteleri, üyelerine kayıpsız ve kaliteli bir casino/oyun hizmetini garanti etmektedir. Betboss minimum riskle maksimum kazancın sağlanması için çeşitli promosyonlar cazip tekliflerle sunulmaktadır.
Rodosbet oyun ve casino sitesi incelemeleri oyun sitesine üyelik oluşturmaya karar veren bireylerin başlangıçta en az 18 yaşında olmaları gerekmektedir. Rodosbet şikayetleri hakkında daha fazla bilgi edinmek istiyorsanız öncelikle bu web sitesini ziyaret etmelisiniz.
Betcool oyunları Betcool kayıt olmadan kayıt olabilmeleri Betcool nedeniyle üyeler arasında popüler olan geniş bir kategori ve oyun yelpazesine sahiptir.
Stonebahis katılmak web sitemizi ziyaret etmek için Stonebahis oyunun web sitesine gidin ve yeni bağlantıya tıklayın.
Luccacasino Bir casino web sitesinde hesabınız varsa, o web sitesindeki şikayetleri ve diğer içerikleri görüntüleyebilirsiniz. Luccacasino bedava çevirmeler kusursuz olduğundan kesinlikle kullanabilir ve kazancınızı artırma şansına sahip olabilirsiniz.
Casinovans bahis sitelerinin içerikleri birçok bahis tutkunu tarafından sıklıkla tercih edilmektedir. Casinovans içeriğimize erişmek için lütfen güncel web sitesi adresini tarayıcınıza girin.
Betmanbet güncel bilet fiyatları da önemli bir öneri. Burada Betmanbet tek tip öneriler yoktur, ancak çeşitli alanlarda seçenekler sunulmaktadır.
Parkbahis şikayetlerini okuma adımlarını tamamladıktan sonra bu sitedeki şikayetleri kolayca anlama fırsatına sahip olacaksınız. Parkbahis sonuç olarak web sitesinin içeriğine ilişkin şikayet ve yorumlar geniş çapta ilgi gördü.
Gatabet adresini kullanarak siteye giriş yaparak siteye yönelik yapılan şikayet yorumlarının tamamını okuyabilirsiniz. Gatabet bu bahis sitesi oldukça güvenilirdir ve site hakkında çok az şikayet bulunmaktadır.
Gördüğünüz gibi en profesyonel bahis platformlarından biri Onikinumara olan Onikinumara ile iletişim kurma süreci oldukça basit.
Betstrong en üstün takımlarına bahis oynamak kaliteli hizmet Betstrong sağlamak için, etkileşimli hizmetler sunan şirketlerin sistemlerini güncel tutması ve hatalı birimleri derhal ortadan kaldırması gerekir.
Pit10bet siteleri birçok casino tutkunu tarafından tercih edilmektedir. Pit10bet uzun zamandır oyun hayatının bir parçası olan web sitesini kullanabilmek için öncelikle bahis sitesinin güncel adres bilgilerine erişmeniz gerekmektedir.
Betmina siteyi ziyaret etmek sizi çok yetenekli bir sahip yapacaktır. Betmina kayıt işlemini tamamladıktan sonra site içeriğini kullanmaya başlayabilirsiniz.
Betpioner güncel giriş adresi Betpioner spor bahisleri kuponlarını Betpioner ve maç öncesi aktiviteler için antrenman ücretlerini araştırırken bu konuyla ilgili bir araştırmaya rastladım.
The world’s most liveable cities for 2024
жесток порно видео
It’s considered among the most beautiful cities in the world to visit, and it seems that Vienna may also be an unbeatable place to live.
The Austrian city has been crowned the most liveable city in the world yet again in the annual list from the Economist Intelligence Unit (EIU), which was released today.
The EIU, a sister organization to The Economist, ranked 173 cities across the globe on a number of significant factors, including health care, culture and environment, stability, infrastructure and education.
Vienna topped the list for the third consecutive year, receiving “perfect” scores in four out of five of the categories — the city was marked lower for culture and environment due to an apparent lack of significant sporting events.
Just behind the Austrian capital, Denmark’s Copenhagen retained its second place position, while Switzerland’s Zurich moved up from sixth place to third on the list.
Australia’s Melbourne fell from third to fourth place, while Canadian city Calgary tied for fifth place with Swiss city Geneva.
Canada’s Vancouver and Australia’s Sydney were in joint seventh place, and Japan’s Osaka and New Zealand’s Auckland rounded out the top 10 in joint ninth place.
«Король» обвинителей «Лайф-из-Гуд» Набойченко – кто он на самом деле?
Гермес последние новости
Евгений Набойченко – бывший IT-директор компании «Лайф-из-Гуд», а также сисадмин российского сегмента платежной системы международной инвестиционной компании «Гермес». Он – ключевой свидетель обвинения в так называемом деле «Лайф-из-Гуд» – «Гермес» – кооператива «Бест Вей» и основателя «Лайф-из-Гуд»/«Бест Вей» Романа Василенко, которое рассматривается сейчас судьей Екатериной Богдановой в Приморском районном суде Санкт-Петербурга.
С выполненной им блокировки российского сегмента платежной системы компании «Гермес», а также с его (нетрезвых) сообщений в социальных сетях (сейчас все они им стерты) началась эскалация этого уголовного дела. Но, скорее всего, дело им же самим и его подельниками из питерской полиции и создано.
Вор и дебошир
Наши источники делятся информацией о хищении активов с кошельков клиентов «Гермеса», к которым Евгений Набойченко имел доступ.
Набойченко не гнушался воровать даже у близких. Однажды остановившись у своей любовницы Светланы Н., Евгений избил ее и украл деньги с ее криптовалютного кошелька, пока она пыталась прийти в себя после побоев.
Он бил не только любовницу, но жену и детей, с которыми жил в Краснодарском крае. А после того как его супруга Виктория подала на развод, разгромил, ободрал и ограбил общий дом – унес все, что можно было унести. Надо ли говорить, что Набойченко который год не платит супруге алименты.
Вымогатель
Евгений Набойченко, по словам его бывшей жены, в один прекрасный день решил заработать и с помощью вымогательства. Он шантажировал Романа Василенко – требовал с него деньги: 170 тыс. евро. При этом, по утверждению Виктории Набойченко, угрожал увечьями и самому Роману Василенко, и его супруге, и детям. Похвалялся перед (тогда еще) женой своими матерными сообщениями с угрозами, которые он посылал Василенко и его близким.
Кроме того, вымогал деньги у клиентов «Гермеса»: свидетельства такого рода нам предоставлены.
Алкоголик и наркоман
Деньги нужны Набойченко в том числе для того, чтобы «питать» свою наркотическую зависимость – с некоторых пор он предпочитает шлифовать водку галлюциногенными грибами, об увлечении которыми с упоением рассказывал даже в своих соцсетях.
Он не всегда был алкоголиком и наркоманом – устроившись 10 лет назад в IT-службу, не пил и не употреблял. Но большие деньги, красивая жизнь, которая вдруг стала доступна для простого астраханского парня из низов, развратили его.
Клеветник
Он допустил целый ряд клеветнических высказываний в адрес Романа Василенко – за которые против него заведено уголовное дело. Но расследуется оно ни шатко ни валко, так как расследование тормозит начальник УЭБиПК ГУ МВД России по Санкт-Петербургу и Ленинградской области.
Завербованный органами
Виктория Набойченко сообщает, что за три дня до обыска в их тогда еще совместном доме Евгений уехал и ей сказал, что придут с обыском – но это пустая формальность. Он уже был в сговоре с полицией, так как за три дня был предупрежден об этом обыске.
15 февраля 2022 года Евгений Набойченко был задержан, но отправился не в СИЗО – куда отправили других сотрудников «Лайф-из-Гуд», не был помещен в изолятор или КПЗ, а жил на полицейской квартире.
Он отказался от услуг адвокатов и на этой квартире, по свидетельству одного из полицейских, «бухал с операми». А также каждый день водил оперов по барам и ресторанам – угощал их за свой счет.
В разговоре с женой он очень гордился покровительством начальника УЭБиПК питерского главка МВД.
Наши источники считают, что он украл деньги клиентов «Гермеса», разделив их со своими друзьями из полиции. Вся его дальнейшая жизнь и привычки указывают на то, что он использует ворованные деньги.
Трус и лжец
Набойченко сообщил в социальных сетях, что к нему пришло с адреса fall@mybizon.online письмо, в котором содержится угроза в адрес получателя, призывающая его не являться в суд для дачи показаний. В письме указано, что если он поедет в суд, то не доедет и будет трупом.
Создатель преступного сообщества
Нет сомнений, что Набойченко устроил историю с обрушением платежной системы, чтобы скрыть свое воровство активов клиентов. Также чтобы скрыть свое воровство, он обвинил во всем Василенко и технических специалистов компании «Лайф-из-Гуд», которые находятся сейчас на скамье подсудимых. Оклеветал всех, чтобы отвести подозрения от себя и своих подельников из питерской полиции.
Итак, кризис в «Гермесе» устроил Набойченко. Платежную систему разгромил Набойченко. Активы украл Набойченко, поделившись с питерскими полицейскими.
А теперь сидит и бездельничает, прожигая сворованные деньги. И боится идти в суд – чтобы не превратиться в суде из свидетеля в обвиняемого.
Скорее всего, в суд Набойченко не пускают сами полицейские – опасаясь его неадекватности и перекрестного допроса, на котором вскроется лживость его показаний. То есть силовики заметают следы своего преступления, совершенного с помощью марионетки Набойченко.
The world’s most liveable cities for 2024
смотреть гей порно
It’s considered among the most beautiful cities in the world to visit, and it seems that Vienna may also be an unbeatable place to live.
The Austrian city has been crowned the most liveable city in the world yet again in the annual list from the Economist Intelligence Unit (EIU), which was released today.
The EIU, a sister organization to The Economist, ranked 173 cities across the globe on a number of significant factors, including health care, culture and environment, stability, infrastructure and education.
Vienna topped the list for the third consecutive year, receiving “perfect” scores in four out of five of the categories — the city was marked lower for culture and environment due to an apparent lack of significant sporting events.
Just behind the Austrian capital, Denmark’s Copenhagen retained its second place position, while Switzerland’s Zurich moved up from sixth place to third on the list.
Australia’s Melbourne fell from third to fourth place, while Canadian city Calgary tied for fifth place with Swiss city Geneva.
Canada’s Vancouver and Australia’s Sydney were in joint seventh place, and Japan’s Osaka and New Zealand’s Auckland rounded out the top 10 in joint ninth place.
«Король» обвинителей «Лайф-из-Гуд» Набойченко – кто он на самом деле?
[url=https://fedpress.ru/person/2293538 ]ГСУ МВД России по Санкт-Петербургу[/url]
Евгений Набойченко – бывший IT-директор компании «Лайф-из-Гуд», а также сисадмин российского сегмента платежной системы международной инвестиционной компании «Гермес». Он – ключевой свидетель обвинения в так называемом деле «Лайф-из-Гуд» – «Гермес» – кооператива «Бест Вей» и основателя «Лайф-из-Гуд»/«Бест Вей» Романа Василенко, которое рассматривается сейчас судьей Екатериной Богдановой в Приморском районном суде Санкт-Петербурга.
С выполненной им блокировки российского сегмента платежной системы компании «Гермес», а также с его (нетрезвых) сообщений в социальных сетях (сейчас все они им стерты) началась эскалация этого уголовного дела. Но, скорее всего, дело им же самим и его подельниками из питерской полиции и создано.
Вор и дебошир
Наши источники делятся информацией о хищении активов с кошельков клиентов «Гермеса», к которым Евгений Набойченко имел доступ.
Набойченко не гнушался воровать даже у близких. Однажды остановившись у своей любовницы Светланы Н., Евгений избил ее и украл деньги с ее криптовалютного кошелька, пока она пыталась прийти в себя после побоев.
Он бил не только любовницу, но жену и детей, с которыми жил в Краснодарском крае. А после того как его супруга Виктория подала на развод, разгромил, ободрал и ограбил общий дом – унес все, что можно было унести. Надо ли говорить, что Набойченко который год не платит супруге алименты.
Вымогатель
Евгений Набойченко, по словам его бывшей жены, в один прекрасный день решил заработать и с помощью вымогательства. Он шантажировал Романа Василенко – требовал с него деньги: 170 тыс. евро. При этом, по утверждению Виктории Набойченко, угрожал увечьями и самому Роману Василенко, и его супруге, и детям. Похвалялся перед (тогда еще) женой своими матерными сообщениями с угрозами, которые он посылал Василенко и его близким.
Кроме того, вымогал деньги у клиентов «Гермеса»: свидетельства такого рода нам предоставлены.
Алкоголик и наркоман
Деньги нужны Набойченко в том числе для того, чтобы «питать» свою наркотическую зависимость – с некоторых пор он предпочитает шлифовать водку галлюциногенными грибами, об увлечении которыми с упоением рассказывал даже в своих соцсетях.
Он не всегда был алкоголиком и наркоманом – устроившись 10 лет назад в IT-службу, не пил и не употреблял. Но большие деньги, красивая жизнь, которая вдруг стала доступна для простого астраханского парня из низов, развратили его.
Клеветник
Он допустил целый ряд клеветнических высказываний в адрес Романа Василенко – за которые против него заведено уголовное дело. Но расследуется оно ни шатко ни валко, так как расследование тормозит начальник УЭБиПК ГУ МВД России по Санкт-Петербургу и Ленинградской области.
Завербованный органами
Виктория Набойченко сообщает, что за три дня до обыска в их тогда еще совместном доме Евгений уехал и ей сказал, что придут с обыском – но это пустая формальность. Он уже был в сговоре с полицией, так как за три дня был предупрежден об этом обыске.
15 февраля 2022 года Евгений Набойченко был задержан, но отправился не в СИЗО – куда отправили других сотрудников «Лайф-из-Гуд», не был помещен в изолятор или КПЗ, а жил на полицейской квартире.
Он отказался от услуг адвокатов и на этой квартире, по свидетельству одного из полицейских, «бухал с операми». А также каждый день водил оперов по барам и ресторанам – угощал их за свой счет.
В разговоре с женой он очень гордился покровительством начальника УЭБиПК питерского главка МВД.
Наши источники считают, что он украл деньги клиентов «Гермеса», разделив их со своими друзьями из полиции. Вся его дальнейшая жизнь и привычки указывают на то, что он использует ворованные деньги.
Трус и лжец
Набойченко сообщил в социальных сетях, что к нему пришло с адреса fall@mybizon.online письмо, в котором содержится угроза в адрес получателя, призывающая его не являться в суд для дачи показаний. В письме указано, что если он поедет в суд, то не доедет и будет трупом.
Создатель преступного сообщества
Нет сомнений, что Набойченко устроил историю с обрушением платежной системы, чтобы скрыть свое воровство активов клиентов. Также чтобы скрыть свое воровство, он обвинил во всем Василенко и технических специалистов компании «Лайф-из-Гуд», которые находятся сейчас на скамье подсудимых. Оклеветал всех, чтобы отвести подозрения от себя и своих подельников из питерской полиции.
Итак, кризис в «Гермесе» устроил Набойченко. Платежную систему разгромил Набойченко. Активы украл Набойченко, поделившись с питерскими полицейскими.
А теперь сидит и бездельничает, прожигая сворованные деньги. И боится идти в суд – чтобы не превратиться в суде из свидетеля в обвиняемого.
Скорее всего, в суд Набойченко не пускают сами полицейские – опасаясь его неадекватности и перекрестного допроса, на котором вскроется лживость его показаний. То есть силовики заметают следы своего преступления, совершенного с помощью марионетки Набойченко.
Перетяжка мягкой мебели
https://pastelink.net/ks8s3lap .
Купить Кокаин в Киеве? Цена на Кокаин САЙТ – https://koks.top/
.
.
Доставка В руки Кокаин в Киеве – https://koks.top/
Закладки с гарантией в Киеве – https://koks.top/
Чистый Кокаин в Киеве САЙТ – https://koks.top/
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
Теги – Купить Кокаин в Киеве?
Кокаин в Киеве Доставка в руки?
Закладки Кокаин в Киеве?
Где Купить Кокаин в Киеве?
Лучший Кокаин в Киеве заказать Кокаин круглосуточно?
Кокаин Украина
smartblip home gadgets https://smartblip.com best price, free shipping,
hot offer!
MOBILE GAMES & APP DEVELOPMENT
https://www.fanstudio.co.uk/
Looking forward for income? Get it online. https://t.me/cryptaxbot/14
Betvadi Bir bahis sitesinin giriş adresini öğrenmek için sitenin Twitter adresini ziyaret edin. Betvadi Bu site güncel adres bilgilerini yayınlamaktadır.
Nosbahis Bu sayede güncel site adres bilgilerinizle sitenize başarılı bir şekilde giriş yapabilirsiniz. Nosbahis Birçok bahis tutkununun tercih ettiği siteyi kullanabilmek için öncelikle bahis sitesi adres bilgilerinizi güncellemeniz gerekmektedir.
Goley90 Bu adres bilgisini aldıktan sonra siteyi kullanabilmek için kayıt adımlarını takip etmeniz gerekmektedir. Goley90 giriş adresinizi aldıktan sonra sitede kolaylıkla işlem yapabilir ve daha fazla para kazanmaya başlayabilirsiniz.
Bycasino spor Bycasino spor bahisleri odaklı, çeşitli Firmadı bahis sektörlerinde hizmet sunan bir şirkettir.
Betdog giriş adresinizi aldıktan sonra sitenin uygulamalarını ve içeriğini kullanmaya başlayabilirsiniz. Betdog Bahis siteleri oldukça kaliteli ve kazançlı içerikleri nedeniyle bahis dünyasında sıklıkla tercih edilmektedir.
Felixbet Bu web sitesini kullanabilmek için öncelikle gerekli kayıt işlemini tamamlamanız gerekmektedir. Felixbet Giriş yaptıktan sonra site içeriğini sorunsuz bir şekilde kullanabilirsiniz.
Sahabet canlı bahis avantajı Sahabet Bonus İşlemleri Sahabet sekmesi oyunculara çeşitli ödüller sağlar. Bu, bahis kazanma şansınızı artırmak için çok faydalıdır.
3xlwin kayıt işlemini tamamladıktan sonra bahis sitesini rahatlıkla kullanabilirsiniz. 3xlwin Birçok bahis tutkununun tercih ettiği bahis sitesini kullanabilmek için istenilen bilgileri girmeniz gerekmektedir.
Efsanebahis şikâyet hattına nasıl ulaşılır Efsanebahis eksiksiz bir oyun ve canlı oyun deneyimi sunmaya çalışan bir şirketidir.
Sarayslot kayıt adımlarını tamamladıktan sonra bahis sitesini kullanmaya başlayabilirsiniz. Sarayslot giriş yaptıktan sonra bahis sitesini rahatlıkla kullanabilirsiniz.
Truvabet şifremi unuttum Truvabet para transferi için kullanmak, Truvabet çevrimiçi platformlarında güvenli ve güvenli ödeme sorununu çözmenize yardımcı olacak birçok bilgi ve avantaj sunar.
Piyonbet Bu sitedeki içerik harika. Bu içeriği kullanabilmek için siteye giriş yapmalısınız. Piyonbet giriş yaptıktan sonra bahis sitesinin uygulamalarını ve içeriğini kullanmaya başlayabilirsiniz.
Artemisbet destek türkiye’de son derece güvenilir bir Artemisbet sitesi olmasına rağmen, kullanıcılarına hala yeni giriş adresleri sağlıyorlar.
Mavibet üye i̇letişim bilgileri gereklidir. Üyeliğe katılma Mavibet süreciniz bitti, hesabınıza para ekleyebilir, bakiyenizi ayarlayabilir ve hemen bahis oynamaya başlayabilirsiniz.
Топ-5 материалов для перетяжки мебели, стильных
Уникальные идеи для перетяжки мебели, перетянутую мебель мечтали ваши друзья
Топ-3 причины для перетяжки мебели в доме, попробуйте сами
Вдохновляющие идеи для перетяжки мебели, которые понравятся каждому
Перетяжка мебели: секреты мастеров, которые стоит выслушать
мастерская “КакСвоим”.
Pasgol platformu tüm Pasgol rehberleri Pasgol bahislerini Pasgol çekici buluyor. Sonuç olarak, çok sayıda bireyin bu bahis sitesine kaydolmakla ilgilendiği sonucuna varılabilir.
Forvetbet spor bahisleri spor bahisleri söz konusu olduğunda Forvetbet çeşitli spor etkinliklerine bahis yapma şansıyla sıklıkla karşılaşırsınız.
번개 슬롯
Hongzhi 황제는 숨을 내쉬고 아래에서 칙령을 기다리는 Hanlin을 바라 보았습니다.
Underoverbet spor bahis seçenekleri Underoverbet hesabınıza Underoverbet farklı bir hesaptan erişmeye çalıştığınızda siteye gittiğinizde hatayla karşılaşırsanız, hesabınıza giriş yapmış ve sahte bir siteden hesabınıza yönlendirilmiş olmanız mümkündür.
Betpara canlı bahis nasıl oynanır sitenin güçlü özellikleri Betpara o kadar etkileyici ki hemen dikkatinizi çekiyor.
Betxbahis uzmanlığı bu anlayış sadece futbol ve basketbol oyunları Betxbahis için değil aynı zamanda Spor Bahisleri Bölümünde özetlenen yaklaşık 45 farklı spor alt kategorisi için de geçerlidir.
Viploby yöntemi bu yöntemi izleyerek örneğin rulet meraklılarına Viploby bir bahis sorgusu yönelttiğinizde uygun ruleti ve kullanılacak en uygun stratejiyi belirleyebilirsiniz.
Betparis hakkında yorum bırakın şifrelerini hatırlayamayan Betparis üyeler, casino sitelerinin kendilerine gönderdiği linklere tıklayarak şifrelerini geri alabilirler.
https://santex-service.ru вызов сантехника.
вызов сантехника https://santex-service.ru.
срочный вызов сантехника https://santex-service.ru.
сантехник вызов https://santex-service.ru.
https://santex-service.ru сантехник на дом.
Dopingbet şikâyetini Dopingbet önerisini nereye gönderebilirim? Dopingbet Başlangıç olarak sayfanın altındaki telefon numarası size yardımcı olabilir.
Golvar web sitesi, mevcut e-posta adreslerine gönderilen Golvar e-postalara anında yanıt verebilmesi açısından avantajlıdır.
Hepbahis üyelik avantajları süreç devam ederken oyuncular Hepbahis web sitesine kayıt Hepbahis işlemini tamamlamak için gerekli yönlendirmeyi alacaklar.
İlkyarı güvenilir mi sitenin farklı alanlarda başarı geçmişi var İlkyarı ve aynı zamanda önemli kazanma potansiyeline sahip bir site olarak değerlendiriliyor.
https://e-porn.net free xxx videos, web cams, dating for sex, adult
movies, hot babes, porno
Maldobet canlı bahis sitesi bu bahis şirketi, onu çok çeşitli bahis Maldobet sonuçları ve casino oyunları için uygun hale getiren çeşitli bonuslar sağlar.
Betperform mevcut oyuncuları i̇nternet bağlantınız ne olursa Betperform olsun, oyun masalarında yerel ve uluslararası bayilerle canlı oyunlar oynayarak gerçek ortamının keyfini çıkarın.
Bets4you yeni giriş adresi twitter’a giriş yaptığınızda erişime Bets4you açık olmayan bir sayfayla karşılaştığınızda bu engellenen bir siteye erişim sağladığınız anlamına gelir.
Napolyonbet para kazanır Bahis sitesine bahis Napolyonbet koymak için basit bir kayıt işlemini tamamlamanız gerekir Bahislere katılım, bahisçiler için basittir.
Betizm lisans güvenilir ve mükemmel bir web sitesinde kullanıcı Betizm hesabı oluşturduğunuzda slot makinelerinde oynamaya başlayabilirsiniz.
Ciprobet ödeme desteği kullanıcının yatırım aşamasında kullandığı Ciprobet kredi ve banka kartı bilgileri, kendisine özel kimlik numarası ile birlikte kayıt altına alınacaktır.
Привет всем)
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: купить диплом в троицке, купить диплом в каменске-уральском, купить диплом в сызрани, купить диплом специалиста, купить диплом в таганроге, а потом наткнулся на https://gidro2000.com/forum-gidro/user/25388/
http://school-one.ru/users/ykinykez
http://lenmontov.blogspot.com/2015/03/blog-post_16.html
https://citydevelopers.ru/otzyvy/novosibirsk/zhk-voskresnyy/
, где все мои учебные проблемы были решены!
Успешной учебы!
http://sad5.lytkarino.net/index.php?subaction=userinfo&user=arymazog
Играй и выигрывай в 1win казино, зарабатывай деньги.
Увлекательные слоты в 1win казино, гарантируют яркие эмоции.
1win казино – место, где рождаются победы, выигрывай и радуйся.
Играй и побеждай вместе с 1win казино, становись миллионером.
1win казино – это мир азарта и возможностей, получай невероятные эмоции.
Почувствуй адреналин победы в 1win казино, забирай свой джекпот.
1win казино – это место, где рождаются чемпионы, получить все, что ты заслуживаешь.
Богатство и успех с 1win казино, которое ждет тебя прямо сейчас.
1win зеркало https://luchshiye-onlayn-kazino-rb.com/ .
Почему сотрудники Колокольцева провоцируют социальный протест
[url=https://vestnik-jurnal.com/novosti/item/100193-voennyh-unizhayut-tylovye-krysy-opg-kolokolceva]ОПГ Колокольцева[/url]
15 февраля пайщики кооператива «Бест Вей» по всей России намерены провести акции в поддержку кооператива. Кооператив работает в 72 регионах, в более чем 50 из них созданы инициативные группы и подаются заявки на проведение митинга.
Эти действия спровоцированы действиями следственной группы ГСУ ГУ МВД России по Санкт-Петербургу и Ленинградской области, которая заблокировала работу кооператива, арестовала его счета, арестовала всю недвижимость кооператива, хотя за более чем год расследования не нашла никаких свидетельств мошенничества в кооперативе.
15 февраля 2022 года начались активные действия следственной группы против кооператива — аресты и первый обыск в офисе с выносом всех документов. Позднее были заблокированы счета кооператива: на них находится 3,8 млрд рублей, причем сумма увеличивается, так как большинство пайщиков продолжает платить членские взносы и вносить паевые платежи.
Несмотря на то что кооператив трижды добивался в суде снятия ареста, следствие вновь выходило в суд с фактически тем же самым ходатайством и пытается, пользуясь своими полномочиями, уничтожить юридическое лицо, и накладывался новый арест.
В периоды, когда арест снимался, следствие за счет давления на банки добивалось того, чтобы платежи не проходили. Так происходит и сейчас, хотя с 19 января арест со счетов кооператива судом снят. Невзирая на претензионные письма кооператива в банки, жалобы кооператива в Центробанк и подготовку исковых заявлений против банков, Сбербанк и банк «Санкт-Петербург», получив, по свидетельству самих банковских работников, устное предупреждение следствия и «после звонков из Москвы», отказываются разблокировать счета, прямо нарушая постановление суда, снявшего арест со счетов.
Офис кооператива дважды обыскивался, документы дважды изымались, и следствие запрещало копировать документацию.
С самого начала расследование дела происходит при информационной поддержке помощника Колокольцева Ирины Волк, подчиненной ей пресс-служба министерства, разнообразные пристяжные «СМИ» типа «Дежурной части» на НТВ. К поддержке привлекли и самого министра: он говорил о деле кооператива «Бест Вей» в своем выступлении в Совете Федерации осенью 2022 года. По некоторым данным, «авторами» дела являются руководители ГУЭБиПК МВД России, облеченные доверием министра.
Следствие по-питерски
Характерно, что дело не «забрал» себе Следственный департамент МВД России. Оно расследуется ГСУ ГУ МВД России по Санкт-Петербургу и Ленинградской области — по месту регистрации кооператива, несмотря на общероссийский характер деятельности «Бест Вей». Это значит, что даже авторы атаки в центральном аппарате МВД не уверены в ее успехе.
Кураторы дела в федеральном МВД шанс проявиться, и они «роют землю», не гнушаются прямым нарушением закона, чтобы проявить себя. Кто эти героические работники юстиции? Прежде всего руководитель следственной группы майор юстиции Екатерина Сапетова и ее непосредственный руководитель подполковник юстиции Константин Иудичев.
Четверо технических сотрудников, не имевших никакого отношения к руководству кооперативом, трое из которых молодые женщины, были арестованы, хотя им вменялся «экономический» состав и они никогда не привлекались к уголовной ответственности. Они уже около года томятся в СИЗО, причем с ними почти не проводится следственных действий. По мнению адвокатов, от них ждут показаний на начальство.
Пятеро граждан объявлены в розыск, в том числе давно отошедший от текущего управления кооперативом его основатель и бывший председатель Совета кооператива Роман Василенко. 80-летних родителей Романа Василенко, ветеранов Вооруженных сил, постоянно таскают на многочасовые допросы, в ходе которых им неоднократно требовалась срочная медицинская помощь, им угрожают СИЗО.
Усилия следствия не привели к адекватным для судов результатам. Компрометирующих кооператив показаний ни от арестованных, ни от пожилых родственников людей, объявленных в розыск, получить не удалось, а пул потерпевших, собранный с огромным трудом, и их претензии выглядят неубедительно даже в наших судах, которые обычно дают следствию карт-бланш на период предварительного расследования.
Кульбиты обвинения
Следствие и пресс-структуры МВД рассказывают судам и прессе о «12 тысячах пострадавших», однако за более чем год работы им удалось получить заявления от сотни с небольшим потерпевших с общей суммой претензий около 150 млн рублей, хотя активы кооператива — более 15 млрд рублей. Предварительное расследование забуксовало.
Притом что следователи работают не покладая рук, телефонными звонками и письмами (есть в редакции) приглашая всех, кто так или иначе взаимодействовал с кооперативом, написать заявление.
Потерпевшие делятся на три группы. Первая — граждане, подавшие заявление по поводу действий другого юридического лица — компании высокодоходных инвестиций, которая контролировалась одним из прежних руководителей кооператива. Следствие пытается объявить эту компанию аффилированной с кооперативом, хотя между ними не было даже никаких взаиморасчетов, и пытается через псевдоаффилированность заставить кооператив заплатить по счетам другого юридического лица. Кстати, инвесткомпания — «живая», у нее есть активы в России, есть активы и у ее учредителей.
Вторая — пайщики кооператива, которые не могут получить свои паевые средства из-за того, что следствие арестовало счета. Среди них нет ни одного заявления о том, что кооператив взял деньги пайщика на покупку квартиры и не купил квартиру или взял паевые средства в рамках договора и отказался выполнять требования договора о возврате средств. Это люди, обманутые следствием, убедившим их в том, что только участие в расследовании поможет получить назад вложенные средства, хотя на самом деле получить их назад в реальном времени помогут исключительно разблокировка работы кооператива, снятие ареста со счетов.
Часть потерпевших, возможно, выступила соавтором атаки на кооператив: это группа граждан, которые когда-то вышли из кооператива «Бест Вей» и создали «альтернативный» кооператив — ЖК «Вера» (зарегистрирован в Ухте, но работает, как и «Бест Вей», по всей России). И против них адвокаты кооператива после завершения предварительного расследования намерены подать заявления о клевете.
Часть потерпевших — люди, которые пытаются поживиться, выдвигая необоснованные требования к кооперативу. Например, претензии некоторых потерпевших касаются вступительных или членских взносов, которые по договору с кооперативом не подлежат возврату.
В рамках следственных действий зачастую выясняется, что они не считают, что кооператив их обманул. Типична ситуация, когда псевдопотерпевшие внесли первоначальный безвозвратный взнос для вступления в кооператив — 110 тыс. рублей, причем некоторые даже не полностью, при этом паевые платежи для накопления первоначального паевого взноса не вносили, членские взносы — тоже.
Квартира им, разумеется, не приобреталась — они в своих показаниях на это и не претендуют. Однако они по наущению следствия или по собственной инициативе и при поддержке следствия подписали заявление о том, что кооператив не вернул им несколько десятков тысяч рублей, которые по договору с кооперативом возврату не подлежат, так как это вступительные или членские взносы, идущие, согласно уставу, на финансирование текущей деятельности и развития кооператива.
Это очередное свидетельство махинаций, совершаемых следствием: ведь потерпевший — лицо, считающее, что в отношении него совершено преступление. Из показаний, имеющихся в уголовном деле, видно, что большинство потерпевших так не считает.
Многие из них не сами пришли в следственные органы, а после звонка следователя, предложившего подать заявление в качестве потерпевших. Пайщики кооператива жалуются на систематический обзвон следователей из следственной группы Сапетовой, письма следователей, незаконные встречи со следователями с назойливыми советами подать заявление в качестве потерпевших — подавляющее большинство пайщиков от этого отказывается, понимая, что необоснованное обвинение в преступлении приведет к ответственности.
Банковские секьюрити — лучшие друзья следствия
Суды первоначально штамповали все ходатайства следствия, как водится в российской системе юстиции, на этапе предварительного расследования. Однако с некоторых пор вопросы к следствию стали возникать и у судей.
Ведь расследование продлевается, продлевается и продлевается, а промежуточные результаты его не впечатляют. Возникает много вопросов. Почему следствие не разрешает проводить налоговые платежи? На каком основании оно требует заблокировать на счетах средства, более чем в 25 раз превышающие сумму ущерба, имеющуюся в деле? На каком основании требует арестовать недвижимость на 12 млрд рублей, в том числе перешедшую в собственность пайщиков?
Кооператив трижды добивался снятия ареста со счетов, добивался разблокирования налоговых платежей. Снимались аресты с недвижимости кооператива в целом. Кроме того, суды принимали решения о снятии арестов с отдельных квартир, и эти решения вступили в законную силу.
Тем не менее арест со счетов де-факто не снимался, за исключением короткого периода летом 2022 года длиной в полторы недели (в эти полторы недели кооператив успел вернуть паевые взносы 216 пайщикам).
Службы безопасности банков блокируют платежи кооператива — банковские службы безопасности стали лучшими помощниками следствия вместо судов. Так, 18 января суд отказался продлить арест счетов, кроме суммы в 200 млн рублей — имеющиеся в деле обязательства перед потерпевшими плюс те, которые могут возникнуть. С 19 января счета не арестованы, но де-факто арест продолжается. Сбербанк и банк «Санкт-Петербург» не пропускают платежи, несмотря на претензионные письма и готовящиеся исковые заявления кооператива и жалобы в Центральный банк.
Банки в деле?
Есть основания считать, что банки, прежде всего Сбербанк, в Северо-Западном банке которого размещен счет паевых средств кооператива, не пассивные исполнители воли следствия.
Заинтересованность в первую очередь связана с уничтожением альтернативной программы покупки квартир. Кроме того, банки заинтересованы в использовании средств на счетах: их можно крутить месяцами, зарабатывая колоссальные средства. Этот метод подсказывают западные банки, которые, заблокировав деньги России, за счет процентов их использования получают серьезные финансовые ресурсы.
Вероятно, у Сбербанка вызвала интерес также цифровая система взаимодействия с клиентами, созданная в кооперативе: есть ряд свидетельств, что она скопирована IT-специалистами Сбера. Северо-Западный банк Сбербанка когда-то активно предлагал сотрудничество кооперативу, добивался размещения счета именно у себя – возможно, не без умысла.
Пайщики знают, кто виноват
Пайщики кооператива возмущены происходящим. Ведь уже год из-за ареста счетов они не могут приобрести квартиры, средства на приобретение которых перечислили; они не могут получить паевые средства назад, чтобы использовать их для покупки квартир другим способом; они не могут получить справки в кооперативе для наследования пая — документация в кооперативе изъята, и следствие даже не разрешает ее копировать. При этом они прекрасно отдают себе отчет в том, что именно ведомство Колокольцева, решившее состряпать громкое дело на пустом месте, хотя ни деньги, ни квартиры не украдены, — автор их злоключений.
Пайщики написали тысячи обращений в следственную группу, прокуратуру, уполномоченному по правам человека, проводились импровизированные митинги в судах. Теперь настал черед больших митингов по всей России.
Сотрудники Колокольцева на пустом месте создали липовое громкое дело ради внимания первого лица страны к непримиримой борьбе с мошенниками, обирающими граждан. Хотя на самом деле именно следствие само и обирает граждан, в обстановке социальной нестабильности в стране создает дополнительный повод для массового возмущения, уничтожает уникальную программу приобретения недвижимости без банковских переплат — фактически под 1% годовых, в рамках которой уже приобретено более 2,5 тыс. квартир.
Создан кризис на пустом месте, который наносит ущерб гражданам России и стране в целом. В условиях военного противостояния, социально-экономической турбулентности, предвыборной ситуации цена действий подразделений МВД непомерно высока – министру и политическому руководству страны пора вмешаться.
Привет Друзья!
Всегда думал что купить диплом о высшем образовании это миф и нереально, но все оказалось не так, изначально искал информацию про: купить диплом в екатеринбурге, купить диплом техника, купить диплом с реестром, купить диплом в димитровграде, купить аттестат, потом про дипломы вузов, подробнее здесь https://xn--80asucf0d.xn--j1al4b.xn--p1ai/index.php/component/k2/itemlist/user/123066
http://xn--b1aafrvecb0f6b.xn--p1ai/forum/messages/forum4/topic840/message264454/?result=reply#message264454
https://slavyansk2.ru/forum/flud/obrazovanie-0
http://fatcow.com/blog/?p=344#comment-3676832
http://www.ingenieursdudimanche.lagob.fr/doku.php?id=landsdiploms
Оказалось все возможно, официально со специальными условия по упрощенным программам, так и сделал и теперь у меня есть диплом вуза Москвы нового образца, что советую и вам!
Хорошей учебы!
Привет всем)
Студенческая жизнь прекрасна, пока не приходит время писать диплом, как это случилось со мной. Не стоит отчаиваться, ведь существуют компании, которые помогают с написанием и защитой диплома на высокие оценки!
Сначала я искал информацию по теме: купить диплом в майкопе, купить диплом в нефтеюганске, купить диплом вуза, купить диплом архитектора, где купить диплом, а потом наткнулся на http://www.avatars.cc/member.php?action=showprofile&user_id=34845, где все мои учебные проблемы были решены!
Успешной учебы!
Привет Друзья!
Всегда думал что купить диплом о высшем образовании это миф и нереально, но все оказалось не так, изначально искал информацию про: купить диплом старого образца, купить диплом в ярославле, купить диплом для иностранцев, купить диплом отзывы, купить диплом во владикавказе, потом про дипломы вузов, подробнее здесь http://www.kaupparaati.fi/ilmaiset-aanikirjat-2022-parhaat-aanikirjapalvelut/
https://obrezanie05.ru/users/15
https://radio.stores.kz/products/led-doska-l-board
https://forum.sportmashina.com/index.php?threads/kupit-diplom-kolledzha.5085/
https://www.datkira.com/купить-диплом-быстрый-способ-повыси
Оказалось все возможно, официально со специальными условия по упрощенным программам, так и сделал и теперь у меня есть диплом вуза Москвы нового образца, что советую и вам!
Удачи!
Лучшее казино для игры – 1win, не упустите свой шанс!
Начните побеждать с 1win казино, играйте и выигрывайте без ограничений!
1win казино – где каждый становится победителем, попробуйте сами и убедитесь!
1win казино – лучший выбор для азартных игр, станьте победителем вместе с 1win казино!
1win казино – лучший выбор для азартных игр, получите удовольствие от азарта с 1win казино!
1win официальный сайт https://populyarnoye-onlayn-kazino-belarusi.com/ .
1win казино: удобные и безопасные методы оплаты
1win официальный сайт https://xn—-7sbb2afcierdfbl.xn--90ais/ .
Still not a millionaire? The financial robot will make you him!] https://t.me/cryptaxbot/14?sweeplimi77Dog
Riccobet twitter linki Riccobet adresini değiştirmeniz Riccobet durumunda en güncel alan adını bulmanın birden fazla yöntemi bulunmaktadır.
Turn $1 into $100 instantly. Use the financial Robot.] https://t.me/cryptaxbot/14?sweeplimi54Dog
Bahispalas adres değişikliği twitter Bahispalas bahis Bahispalas kuponlarını öne çıkaran bir kupon yapısını kolaylıkla oluşturabilirsiniz.
Tutarbet sanal bahis seobahis, mevcut konumunuzdaki Tutarbet web sitesi hizmetlerini yöneten bir araçtır.
Kikbet ödemeleri güvenli mi Kikbet Oyuncular Kikbet web sitesine Kikbet kaydolurken genellikle devam etmeden önce sitenin meşru olup olmadığını kontrol ederler.
Betfica saha canlı bahis Betfica sitesi üyelerinden belge mi istiyorsunuz, Betfica yoksa onların belgelerini göndermeniz mi gerekiyor gibi sorularla karşılaşacaksınız.
Casinogalata şikâyeti aldı mı daha fazlasını öğrenmek ve Casinogalata çevrimiçi bahislerle eğlenmek için siteye gidebilir ve onlarla iletişime geçmek için Whatsapp’tır kullanabilirsiniz.
Celticbet spor bahisleri Celticbet spor bahisleri bağlamında Celticbet değerlendirdiğimizde iki ana kategoriyi ele alabiliriz.
Fortunepanda lisans bahisçilerin site hakkında anında bilgi Fortunepanda alabilmesinin tek yolu Fortunepanda Betin 7/24 canlı müşteri desteğidir.
Bets60 kullanıcı puanları Bets60 Çok sayıda kişinin sitelerinde Bets60 ödeme yapma sürecini eleştirmesine rağmen, bu hala popüler bir yöntem olmaya devam ediyor.
Medyabahis canlı yardımı nedir katılmadan Medyabahis önce mevcut Medyabahis hesabınıza erişmeniz gerekmektedir.
Babilonbet bahis türleri Babilonbet şikâyet ekranı, teknik uzmanların Babilonbet kurup sürekli çalıştırdığı bir sistemdir ve her türlü soru veya sorunu, kendilerine ulaştıktan sonra bir gün içinde yanıtlamak zorundadırlar.
Portbet olayı canlı oyun ve canlı bahis işlemleri sağlayan Portbet ülkemizde ve Avrupa’da Portbet en popüler canlı bahis siteleri olarak kabul edilmektedir.
Aronbet şikâyetinin başlığı deneyimli kişiler sürekli olarak Aronbet çevrimiçi bahis endüstrisinde yer alırken ve uzun süredir yatırım yaparken, yeni katılanlar da var.
Axessbet incelemeleri bu incelemeler web sitemizi ve web Axessbet ile ilgili diğer hizmetlerimizi geliştirmemize yardımcı olur.
오공 슬롯
상인의 설명에 따르면 이러한 껌은 다밍에서 공급이 부족합니다.
Betplease giriş adresinizi değiştirme birçok bahisçi adres Betplease değiştirmenin üyelere zarar vermeyeceğinin farkında değil.
Betcyp ödeme güncel oranlar giriş Betcyp hesabınızın geçerliliğini Betcyp onaylamak için web sitesindeki standart bir prosedürü izlemeniz gerekmektedir.
Betlobi canlı bahis şikâyetleri bununla birlikte, yaygın Betlobi olarak tercih edilen blackjack casino bonusu, herhangi bir ilk para yatırma gerektirmeyen bir bonustur.
İnbahis mobil uygulamaları firmaları, çeşitli nedenlerden İnbahis dolayı yeni mobil uygulamaları için farklı konumlara sahiptir.
Benimbahis mobil belirli bir uygulaması olmayan Benimbahis çoğu bahis sitesinin aksine, bu mobil uygulama benzersizdir.
Bahsimvar rakipleri rakiplerinize karşı üstünlük sağlamak Bahsimvar için web sitenizi nasıl kullanacağınızı kolayca öğrenebilirsiniz.
Betlaz kayıt i̇şlemleri çevrimiçi bahis sitesi ön plandadır ve Betlaz İnternet kullanıcılarına sunduğu güvenilir, iyi desteklenen ve basit hizmetiyle ünlüdür.
Benimbahis ödeyen biri mi çevrimiçi casino platformu, kişisel bilgileriniz Benimbahis hesap işlemlerinizle ilgili herhangi bir sorun tespit etmesi halinde hesabınızı sonlandırma yetkisine sahiptir.
На сайте вы можете насладиться игровыми автоматами абсолютно бесплатно и без необходимости прохождения регистрации, наслаждайтесь игрой в любое время и без ограничений!
Секреты выбора материала для перетяжки мебели: экспертные советы и рекомендации, для успешной реализации вашего проекта.
Модные тренды в перетяжке мебели: что выбрать в этом сезоне, для создания неповторимого облика вашего дома.
Творческий процесс перетяжки мебели: самодельные идеи для уникального результата, для проявления вашей индивидуальности.
Почему перетяжка мебели становится все популярнее: основные преимущества и плюсы, чтобы ваш дом стал уютным и уникальным.
Как выбрать подходящего мастера для перетяжки мебели: советы и рекомендации, чтобы ваша мебель выглядела идеально.
Какие материалы выбрать для перетяжки мебели в стиле минимализм: легкие и стильные способы, для создания функционального и современного интерьера.
Секреты перетяжки мебели в скандинавском стиле: как создать атмосферу комфорта, для создания атмосферы уюта и спокойствия.
Как сделать перетяжку мебели экономично и эффективно: секреты и советы, которые позволят вам обновить интерьер без лишних затрат.
Советы по перетяжке мебели в провансальском стиле: как создать атмосферу загородного уюта, для оформления вашего дома в стиле прованс.
Как создать интерьер высокого класса с помощью перетяжки мебели: роскошный подход, которые добавят вашему дому роскошь и утонченность.
Как сделать перетяжку мебели качественно и без лишних хлопот: лайфхаки и советы, для успешной реализации вашего проекта и получения отличного результата.
перетяжка мягкой мебели мебели перетяжка мебели .
Find out about the fastest way for a financial independence. https://t.me/cryptaxbot/17?sweeplimi66Dog
Разбираю ситуацию вокруг “кооператива” Life is Good
гей порно геей
Я решил написать этот пост, чтоб пройтись по доводам тех, кто привлекает новых “пайщиков” в структуры г-на Василенко (на деле – оплачивающих существование пирамиды Life is Good). Понятно, что, в этом сообществе крутятся сотни тех, кто занимается привлечением и теперь ЦБ с прокуратурой их оставят не только без заработка, но, возможно, придется отвечать за свои действия в суде. А деньги уже, наверняка, потрачены. Также есть люди, которые уже живут в квартире от LIG или выплатили некую сумму, они верят до конца и отказываются принимать ситуацию, в которой надо признать, что они могут остаться и без денег, и без квартиры. В любом случае, даже если кто-то живет в квартире от LIG, она ему не принадлежит и, в отличие от банковского залога, урегулировать вопрос не получится (их просто выселят через суд при банкротстве LIG, хотя ситуация скандальная и возможно государство что-то предпримет, чтоб сгладить проблему).Теперь по сути, что такое LIG.
Компания LIG – «Лайф-из-гуд» заявляет два основных направления деятельности: инвестиционные счета «Виста» в компании Hermes Management Ltd. и жилищный кооператив «Бест-вей» — альтернатива ипотеке.
Если верить консультантам «Лайф-из-гуд», доходность счетов «Виста» в компании «Гермес-менеджмент» последние годы держится в районе 20% годовых в долларах. Это довольно много (Уоррен Баффет позавидовал бы такой доходности), при этом непонятно, за счет чего достигается такая доходность.
Деятельность «Гермес-менеджмента» непрозрачна, на сайте не видно полноценных отчетов, нет сведений о структуре компании и ее руководстве.
«Гермес-менеджмент» привлекает российских клиентов через компанию «Лайф-из-гуд», но у него нет лицензии Центрального банка России на брокерскую деятельность или доверительное управление.
«Гермес-менеджмент» зарегистрирован в Белизе, офшорной зоне в Центральной Америке. Там же расположен офис компании. Еще в документах упоминается компания Hermes M в Шотландии. В случае проблем придется разбираться с зарубежной компанией, что делает задачу бесперспективной. Более того, учитывая реалии мошеннических схем, бывшие пайщики столкнутся еще и с лже-юристами, обещающими вернуть им вложенные деньги и конечно, останутся в крайне бедственном положении, с кредитами, без жилья и перспектив (у кого-то могут опуститься руки со всеми вытекающими).
Компания платит за привлечение новых клиентов, открывающих инвестиционные счета «Виста» (по сути – ничем не обеспеченные цифры на сайте. Заведение средств происходит через перевод средств на счета частных лиц(!), выполняющих роль консультантов LIG (а это сразу нарушает законы РФ и привлекает внимание регулятора), вывод – через перепродажу воздуха других буратинам. После внесения LIG в черный список, чаты структуры полны объявлений о продажи токенов Виста, но покупателей почти нет)) и вступающих в жилищный кооператив «Бест-вей». Система комиссионных выглядит такой разветвленной и продуманной, что становится понятно, что именно в ней суть компании, а не в инвестициях или альтернативе ипотеке.
Сам по себе сетевой маркетинг — вполне законный способ привлечения новых клиентов. Но когда он выглядит как основной вид деятельности, а клиентам при этом рассказывают про инвестиции, пассивный доход и успех в жизни, это наводит на очевидные выводы.
Суть предложения «Гермеса» такая: инвесторы открывают счет и перечисляют компании доллары или евро, а взамен получают пассивный доход. Минимальная сумма инвестиционного контракта — 10 тысяч долларов или евро. При этом первый взнос может быть 1130 долларов или 1100 евро, а оставшуюся сумму можно довнести позже.
Чтобы открыть счет «Виста», надо заполнить регистрационную форму на сайте «Лайф-из-гуд». При регистрации сайт просит поставить галочку «Я ознакомился и принимаю условия указанные в публичной оферте» (пунктуация сохранена).
По ссылке открывается не оферта, а договор об оказании услуг. В договоре не указаны название и реквизиты компании, с которой заключается договор. Кроме того, речь в нем идет не об управлении активами, а только об «образовательных и консультационных услугах об инвестиционных механизмах и способах управления капиталом». Это не похоже на обещанный пассивный доход от слова совсем. Таким образом, зазывалы в пирамиду пытаются уйти от ответственности.
геи жестко
Чтобы открыть счет и пользоваться им, придется заплатить различные комиссии. Все они прописаны в договоре:
Комиссия за открытие счета: 100 евро или 130 долларов.
Ажио, или входная комиссия: 7% от суммы лицевого счета. На сайте есть пример расчета: при вложении 100 000 $ ажио составит 7000 $.
Комиссия за управление счетом: 1% в год от размера счета, взимается каждый месяц по 1/12.
Комиссия с прибыли: 20% от ежемесячной прибыли, «полученной от инвестиционных продуктов сторонних партнерских компаний или физических лиц, в результате рекомендаций и деятельности Исполнителя».
Итак, LIG состоит из двух основных компаний: Гермес, которая предлагает участникам инвестиционный доход (с идеей полученную прибыль направить на квартиру) и Бест-Вэй, являющаяся ЖНК. Что это такое?
Это Жилищный накопительный кооператив (общепринятое сокращение — ЖНК) — вид потребительского кооператива, сочетающий в себе качества финансового и жилищно-строительного кооператива. ЖНК работают в соответствии с № 215-ФЗ «О жилищных накопительных кооперативах». Целью деятельности жилищных накопительных кооперативов является удовлетворения потребностей членов кооператива в жилых помещениях путём объединения членами кооператива паевых взносов. Жилищный накопительный кооператив имеет право:
– привлекать и использовать денежные средства граждан на приобретение жилых помещений;
– вкладывать имеющиеся у него денежные средства в строительство жилых помещений (в том числе в многоквартирных домах), а также участвовать в строительстве жилых помещений в качестве застройщика или участника долевого строительства;
– приобретать жилые помещения;
– привлекать заёмные денежные средства, общая величина не должна превышать сорок процентов стоимости имущества кооператива.
Членами жилищного накопительного кооператива могут быть только граждане РФ. Жилищный накопительный кооператив создаётся по инициативе не менее чем пятидесяти человек и не более чем пяти тысяч человек. Контроль за деятельностью кооператива по привлечению и использованию денежных средств граждан на приобретение жилых помещений, а также за соблюдением кооперативом требований действующего законодательства Российской Федерации осуществляет Центральный банк Российской Федерации.
Уже отсюда понятно, что заявления заинтересованных лиц из структур LIG о том, что ЦБ не в праве вмешиваться в деятельность собственно LIG лишены оснований. Может и вмешался.
Если говорить о цифрах, то на 13 декабря 2021 года в LIG было свыше 19 000 пайщиков изо всех уголков РФ. LIG выкупил и передал в безвозмездное пользование пайщикам примерно 2 500 объектов недвижимости, из них по 247 пайщики полностью выплатили пай, оформив недвижимость в собственность. Интересная статистика – 19 000 вступили, заплатив членский взнос в 160 000 рублей, но лишь 8 часть из них накопила необходимые 35% для получения недвижимости при том, что фактически недвижимость их собственностью не является. Заявления о том, что получившие недвижимость прописаны в квартирах – это пережитки советского прошлого и, вероятно, сознательное введение остальных в заблуждение. В современной РФ не существует института прописки, есть регистрация, постоянная и временная. Есть найм жилья и есть собственность. Допускаю, что какая-то часть людей все еще проживает в государственных квартирах и не оформила их в собственность, но это – давно не норма, как это было в СССР (где собственность как раз была редким явлением).
По сути кооператив — посредник между покупателем и продавцом жилья, а это увеличивает риски. Приходится полагаться на то, что кооператив соблюдает закон и что он не закроется до того, как вы оформите квартиру на себя.
Даже если все по закону, риск потерять деньги остается. Если кооператив по каким-то причинам перестанет работать, его собственность распродадут. Сначала он рассчитается с кредиторами и только затем вернет деньги пайщикам — пропорционально их паевым взносам. Хватит ли у кооператива денег, чтобы вернуть все паевые взносы, неизвестно.
Разбираю ситуацию вокруг “кооператива” Life is Good
первый анальный секс
Я решил написать этот пост, чтоб пройтись по доводам тех, кто привлекает новых “пайщиков” в структуры г-на Василенко (на деле – оплачивающих существование пирамиды Life is Good). Понятно, что, в этом сообществе крутятся сотни тех, кто занимается привлечением и теперь ЦБ с прокуратурой их оставят не только без заработка, но, возможно, придется отвечать за свои действия в суде. А деньги уже, наверняка, потрачены. Также есть люди, которые уже живут в квартире от LIG или выплатили некую сумму, они верят до конца и отказываются принимать ситуацию, в которой надо признать, что они могут остаться и без денег, и без квартиры. В любом случае, даже если кто-то живет в квартире от LIG, она ему не принадлежит и, в отличие от банковского залога, урегулировать вопрос не получится (их просто выселят через суд при банкротстве LIG, хотя ситуация скандальная и возможно государство что-то предпримет, чтоб сгладить проблему).Теперь по сути, что такое LIG.
Компания LIG – «Лайф-из-гуд» заявляет два основных направления деятельности: инвестиционные счета «Виста» в компании Hermes Management Ltd. и жилищный кооператив «Бест-вей» — альтернатива ипотеке.
Если верить консультантам «Лайф-из-гуд», доходность счетов «Виста» в компании «Гермес-менеджмент» последние годы держится в районе 20% годовых в долларах. Это довольно много (Уоррен Баффет позавидовал бы такой доходности), при этом непонятно, за счет чего достигается такая доходность.
Деятельность «Гермес-менеджмента» непрозрачна, на сайте не видно полноценных отчетов, нет сведений о структуре компании и ее руководстве.
«Гермес-менеджмент» привлекает российских клиентов через компанию «Лайф-из-гуд», но у него нет лицензии Центрального банка России на брокерскую деятельность или доверительное управление.
«Гермес-менеджмент» зарегистрирован в Белизе, офшорной зоне в Центральной Америке. Там же расположен офис компании. Еще в документах упоминается компания Hermes M в Шотландии. В случае проблем придется разбираться с зарубежной компанией, что делает задачу бесперспективной. Более того, учитывая реалии мошеннических схем, бывшие пайщики столкнутся еще и с лже-юристами, обещающими вернуть им вложенные деньги и конечно, останутся в крайне бедственном положении, с кредитами, без жилья и перспектив (у кого-то могут опуститься руки со всеми вытекающими).
Компания платит за привлечение новых клиентов, открывающих инвестиционные счета «Виста» (по сути – ничем не обеспеченные цифры на сайте. Заведение средств происходит через перевод средств на счета частных лиц(!), выполняющих роль консультантов LIG (а это сразу нарушает законы РФ и привлекает внимание регулятора), вывод – через перепродажу воздуха других буратинам. После внесения LIG в черный список, чаты структуры полны объявлений о продажи токенов Виста, но покупателей почти нет)) и вступающих в жилищный кооператив «Бест-вей». Система комиссионных выглядит такой разветвленной и продуманной, что становится понятно, что именно в ней суть компании, а не в инвестициях или альтернативе ипотеке.
Сам по себе сетевой маркетинг — вполне законный способ привлечения новых клиентов. Но когда он выглядит как основной вид деятельности, а клиентам при этом рассказывают про инвестиции, пассивный доход и успех в жизни, это наводит на очевидные выводы.
Суть предложения «Гермеса» такая: инвесторы открывают счет и перечисляют компании доллары или евро, а взамен получают пассивный доход. Минимальная сумма инвестиционного контракта — 10 тысяч долларов или евро. При этом первый взнос может быть 1130 долларов или 1100 евро, а оставшуюся сумму можно довнести позже.
Чтобы открыть счет «Виста», надо заполнить регистрационную форму на сайте «Лайф-из-гуд». При регистрации сайт просит поставить галочку «Я ознакомился и принимаю условия указанные в публичной оферте» (пунктуация сохранена).
По ссылке открывается не оферта, а договор об оказании услуг. В договоре не указаны название и реквизиты компании, с которой заключается договор. Кроме того, речь в нем идет не об управлении активами, а только об «образовательных и консультационных услугах об инвестиционных механизмах и способах управления капиталом». Это не похоже на обещанный пассивный доход от слова совсем. Таким образом, зазывалы в пирамиду пытаются уйти от ответственности.
жесткое гей порно
Чтобы открыть счет и пользоваться им, придется заплатить различные комиссии. Все они прописаны в договоре:
Комиссия за открытие счета: 100 евро или 130 долларов.
Ажио, или входная комиссия: 7% от суммы лицевого счета. На сайте есть пример расчета: при вложении 100 000 $ ажио составит 7000 $.
Комиссия за управление счетом: 1% в год от размера счета, взимается каждый месяц по 1/12.
Комиссия с прибыли: 20% от ежемесячной прибыли, «полученной от инвестиционных продуктов сторонних партнерских компаний или физических лиц, в результате рекомендаций и деятельности Исполнителя».
Итак, LIG состоит из двух основных компаний: Гермес, которая предлагает участникам инвестиционный доход (с идеей полученную прибыль направить на квартиру) и Бест-Вэй, являющаяся ЖНК. Что это такое?
Это Жилищный накопительный кооператив (общепринятое сокращение — ЖНК) — вид потребительского кооператива, сочетающий в себе качества финансового и жилищно-строительного кооператива. ЖНК работают в соответствии с № 215-ФЗ «О жилищных накопительных кооперативах». Целью деятельности жилищных накопительных кооперативов является удовлетворения потребностей членов кооператива в жилых помещениях путём объединения членами кооператива паевых взносов. Жилищный накопительный кооператив имеет право:
– привлекать и использовать денежные средства граждан на приобретение жилых помещений;
– вкладывать имеющиеся у него денежные средства в строительство жилых помещений (в том числе в многоквартирных домах), а также участвовать в строительстве жилых помещений в качестве застройщика или участника долевого строительства;
– приобретать жилые помещения;
– привлекать заёмные денежные средства, общая величина не должна превышать сорок процентов стоимости имущества кооператива.
Членами жилищного накопительного кооператива могут быть только граждане РФ. Жилищный накопительный кооператив создаётся по инициативе не менее чем пятидесяти человек и не более чем пяти тысяч человек. Контроль за деятельностью кооператива по привлечению и использованию денежных средств граждан на приобретение жилых помещений, а также за соблюдением кооперативом требований действующего законодательства Российской Федерации осуществляет Центральный банк Российской Федерации.
Уже отсюда понятно, что заявления заинтересованных лиц из структур LIG о том, что ЦБ не в праве вмешиваться в деятельность собственно LIG лишены оснований. Может и вмешался.
Если говорить о цифрах, то на 13 декабря 2021 года в LIG было свыше 19 000 пайщиков изо всех уголков РФ. LIG выкупил и передал в безвозмездное пользование пайщикам примерно 2 500 объектов недвижимости, из них по 247 пайщики полностью выплатили пай, оформив недвижимость в собственность. Интересная статистика – 19 000 вступили, заплатив членский взнос в 160 000 рублей, но лишь 8 часть из них накопила необходимые 35% для получения недвижимости при том, что фактически недвижимость их собственностью не является. Заявления о том, что получившие недвижимость прописаны в квартирах – это пережитки советского прошлого и, вероятно, сознательное введение остальных в заблуждение. В современной РФ не существует института прописки, есть регистрация, постоянная и временная. Есть найм жилья и есть собственность. Допускаю, что какая-то часть людей все еще проживает в государственных квартирах и не оформила их в собственность, но это – давно не норма, как это было в СССР (где собственность как раз была редким явлением).
По сути кооператив — посредник между покупателем и продавцом жилья, а это увеличивает риски. Приходится полагаться на то, что кооператив соблюдает закон и что он не закроется до того, как вы оформите квартиру на себя.
Даже если все по закону, риск потерять деньги остается. Если кооператив по каким-то причинам перестанет работать, его собственность распродадут. Сначала он рассчитается с кредиторами и только затем вернет деньги пайщикам — пропорционально их паевым взносам. Хватит ли у кооператива денег, чтобы вернуть все паевые взносы, неизвестно.
Betrupi kayıt olurken lütfen doğum tarihiniz, cinsiyetiniz, ülkeniz, Firmadı cep telefonu, şehir ve adres gibi ayrıntıları bize bildirin.
Parasino resmî logosu bu adres uzun süredir bahis sektörünün deneyimli uzmanları Parasino tarafından yönetilmektedir ve hem yatırım stratejileri hem de kullanım eğilimleri açısından her kıtayla karşılaştırılabilecek niteliktedir.
Betsvia çağrı merkezi Betsvia Çevrimiçi Çağrı Merkezi Hizmeti, Betsvia müşterilerin bahis mağazasının web sitesinde canlı sohbet yoluyla şirketle iletişim kurmasını sağlayan bir özelliktir.
포츈 래빗
Fang Jifan은 고개를 저었다. “어쩌면 가지고 있습니까?”
kms activator
https://info-full-kmssite-2.blogsvirals.com/27335787/kms-auto-activator-details-download-internet-url-windows-os-activationdirections
free download id9qf0915
Kms auto activator 52c436_
You said it very well..
Here is my page; http://moden126.mireene.com/bbs/board.php?bo_table=uselist3&wr_id=122403
https://sps134.ru вызов сантехника.
вызов сантехника https://sps134.ru.
срочный вызов сантехника https://sps134.ru.
сантехник вызов https://sps134.ru.
https://sps134.ru сантехник на дом.
https://sps134.ru сантехнические работы.
https://sps134.ru сантехнические услуги.
сантехнические работы вызвать мастера https://sps134.ru .
мастер сантехник https://sps134.ru .
kmsauto portable Office 2021
https://whatkms-auto-3.blogaritma.com/27590498/kms-auto-activation-tool-summary-download-internet-link-windows-os-activationdirections
free download id9qf0915
[url=http://xn--mdchen-online-bfb.com/guestbook.html]Kms auto activator[/url] b72aa17
Купить Экстази в Киеве? Купить МДМА в кристаллах в Киеве? Сайт – https://koks.top
.
.
Закладки Экстази и МДМА в Киеве? САЙТ – https://koks.top
Купить с доставкой Экстази и МДМА в Киеве? САЙТ – https://koks.top
Чистый МДМА в Киеве где Купить? САЙТ – https://koks.top
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
Теги – Купить Экстази в Киеве?
Купить МДМА в Киеве?
Где Купить МДМА в Киеве?
Как Купить Экстази в Киеве?
МДМА и Экстази Купить в Украине цена ?
Start making thousands of dollars every week just using this robot.] https://t.me/cryptaxbot/17?sweeplimi73Dog
kms-auto lite download
https://kmsauto-net-www-.blogtov.com/8175600/directions-to-make-faster-ms-windows-by-using-kmsauto-portable
free download id9qf0915
Kms auto activator 128678b
Online Bot will bring you wealth and satisfaction.] https://t.me/cryptaxbot/17?sweeplimi19Dog
Как Купить Метамфетамин в Киеве? Киев Купить Метамфетамин? САЙТ – https://koks.top
.
.
Доставка в Руки Метамфетамина в Киеве? САЙТ – https://koks.top
Закладки в Киеве Метамфетамин Купить? САЙТ – https://koks.top
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
Теги – Купить в Киеве Метамфетамин?
Цена на Метамфетамин в Киеве?
Сколько стоит грамм Метамфетамина в Киеве?
Кристаллы Метамфетамина Купить в Киеве?
Украина Метамфетамин Кристаллы Доставка Купить
kmsauto ++ windows 10
https://kms-www-.thezenweb.com/kmsauto-tool-description-downloadable-source-url-windows-operating-system-activationtutorial-65833413
free download id9qf0915
[url=https://www.weleda.com.br/product/d/desodorante-de-rosa-mosqueta?r121_r1_r3:u_u_i_d=665c0368-abfa-4ca9-aa9e-9ad11c8cf2e2]Kms auto activator[/url] 3491f67
보증 토토
구씨는 차에서 내려 다시 돼지우리를 바라보았을 때 마치 평생을 멀리 떨어져 있는 것 같은 기분이 들었다.
kms activator portable
https://the-kms-web-.bloggin-ads.com/51393316/the-complete-the-pros-of-the-kmsauto-application-for-activation-of
free download id9qf0915
Kms auto activator 7_791c8
The additional income is available for everyone using this robot. https://t.me/cryptaxbot/17?sweeplimi41Dog
kmsauto lite Office 2021
https://what-full-kmssite1.blogspothub.com/27314987/kms-auto-activator-representation-downloadable-source-website-link-windowsprocedures
free download id9qf0915
Kms auto activator 28678b7
Belatrocasino giriş işleminin detayları bahis Belatrocasino meraklıları Bet hakkında yapılan yorumları araştırıyor.
kms activator portable windows 10
https://thekmsauto-net-web-.review-blogger.com/50354281/an-overview-of-the-features-of-the-kmsauto-activator-for-activating
free download id9qf0915
[url=https://www.kccu4u.org/blog/5-steps-to-take-before-making-a-large-purchase-3?message=posted]Kms auto activator[/url] a892db7
[b]Официальный сайт 1x slots[/b]
[url=https://www.twitch.tv/1xslotsgame/about]Сайт 1xslots[/url] играет основную роль в вопросе доверия и защищенности. На официальном портале пользователи могут узнать всю необходимую информацию о казино. Наличие мобильной версии позволяет клиентам доступ ко всем функциями казино на смартфонах, что предельно удобно. Кроме этого, на официальном сайте можно скачать мобильное приложение для игры.
[b]1 x slots зеркало[/b]
В мире онлайн-гемблинга роль зеркал трудно игнорировать. [url=https://1xslots.mystrikingly.com]1x slot зеркало[/url] позволяет пользователям обойти блокировки и получать неограниченно доступ к своим любимым слотам и автоматам. Зеркало предоставляет непрерывный доступ, позволяя клиентам получать удовольствие от игры.
Плюсы использования зеркал:
[i]1. Обход ограничений;
2. Стабильный доступ.[/i]
[b]1xslots casino регистрация[/b]
[url=https://1xslots.daftpage.com]Процесс регистрации на сайте 1xslots[/url] быстрый и простой. Пользователям надо пройти регистрацию, чтобы зарегистрировать профиль. Процесс регистрации занимает менее 5 минут, после чего можно войти в личный кабинет. Авторизация на сайте позволяет пользователям пополнять счет и выводить деньги, пополнять их и выводить деньги.
Шаги для регистрации и входа:
[i]1. Заполнить регистрационную анкету;
2. Подтверждение данных;
3. Авторизация в личном кабинете 1хслотс;[/i]
[b]Игры в казино[/b]
1xslots предлагает широкий выбор игр, [url=https://black-cloud-06452532815.kopage.site]включая игровые автоматы[/url]. Играть онлайн в казино можно всегда, наслаждаясь разнообразными игровыми автоматами. Мобильные приложения позволяют играть в дороге, что удобно для ценящих мобильность и удобство. Внесение и снятие денег происходит быстро и безопасно, что делает игру более комфортной и безопасной.
[b]Бонусные предложения и промокоды[/b]
Одним из бонусов игры в 1xslots являются бонусные коды и акции. Промокоды позволяют получить дополнительные средства для ставок, а бонусы увеличивают интерес к игре. Промоакции и бонусы увеличивают шансы на победу.
Преимущества использования промокодов и бонусов:
[i]- Дополнительные деньги для ставок
– Бесплатные вращения на автоматах
– Увеличенные шансы на победу[/i]
Бонус при регистрации для первых четырех депозитов. В сумме приветственный пакет включает до 1500 EUR и 150 fs. А промокод «ESDINERO» поможет еще больше расширить пакет.
[b]Мобильные версия 1xslots[/b]
Для комфорта пользователей 1xslots предлагает загрузить приложение. Мобильная версия приложения дает возможность играть везде и всегда, обеспечивая доступ ко всем возможностям казино. Скачивание приложения занимает всего несколько минут, но обеспечивает возможность играть в любое удобное время.
[b]Заключение[/b]
Использование зеркал сайта, официальный сайт казино 1хслот, процесс регистрации и входа, разнообразие игр, промокоды и бонусы, а также мобильное приложение делают 1xslots удобным и безопасным онлайн заведением для всех игроков. Все игроки могут наслаждаться играми в любое время и в любом месте, уверенные в надежности и безопасности доступа к 1xslots.
Bilyoncu ödeme bilgileri hakkında üyelik planlarına katılmanın Bilyoncu temel faydalarından biri daha fazla avantaja erişebilmenizdir.
KMSpico: What is it?
[url=https://kmspico.ws]kmspico vk cc 56f2xq[/url]
Operating systems and Office suites are among the primary Microsoft software items that still need to be paid for. Some consumers may find alternate activation methods due to the perceived expensive cost of these items. There may be restrictions, unforeseen interruptions, and persistent activation prompts if these items are installed without being properly activated.
Our KMSpico app was created as a solution to this issue. By using this program, customers may access all of the functionality of Microsoft products and simplify the activation procedure.
KMSPico is a universal activator designed to optimize the process of generating and registering license codes for Windows and Office. Functionally, it is similar to key generators, but with the additional possibility of automatic integration of codes directly into the system. It is worth paying attention to the versatility of the tool, which distinguishes it from similar activators.
The above discussion primarily focused on the core KMS activator, the Pico app. Understanding what the program is, we can briefly mention KMSAuto, a tool with a simpler interface.
By using the KMSPico tool, you can setup Windows&Office for lifetime activation. This is an essential tool for anybody looking to reveal improved features and go beyond limitations. Although it is possible to buy a Windows or Office key.
KMSPico 11 (last update 2024) allows you to activate Windows 11/10/8 (8.1)/7 and Office 2021/2019/2016/2013/2010 for FREE.
KMSpico Download | Official KMS Website New July 2024
[url=https://kmspico.io/]office toolkit[/url]
Are you looking for the best tool to activate your Windows & Office? Then you should download and install KMSpico, as it is one of the best tools everyone should have. In this article, I will tell you everything about this fantastic tool, even though I will also tell you if this is safe to use.
In this case, don’t forget to read this article until the end, so you don’t miss any critical information. This guide is for both beginners and experts as I came up with some of the rumours spreading throughout the internet.
Perhaps before we move towards downloading or installing a section, we must first understand this tool. You should check out the guide below on this tool and how it works; if you already know about it, you can move to another section.
What is KMSPico?
KMPico is a tool that is used to activate or get a license for Microsft Windows as well as for MS Office. It was developed by one of the most famous developers named, Team Daz. However, it is entirely free to use. There is no need to purchase it or spend money downloading it. This works on the principle of Microsft’s feature named Key Management Server, a.k.a KMS (KMSPico named derived from it).
The feature is used for vast companies with many machines in their place. In this way, it is hard to buy a Windows License for each device,, which is why KMS introduced. Now a company has to buy a KMS server for them and use it when they can get a license for all their machines.
However, this tool also works on it, and similarly, it creates a server on your machine and makes it look like a part of that server. One thing different is that this tool only keeps the product activated for 180 days. This is why it keeps running on your machine, renews the license keys after 180 days, and makes it a permanent activation.
KMSAuto Net
Microsoft Toolkit
Windows Loader
Windows 10 Activator
Features
We already know what this tool means, so let’s talk about some of the features you are getting along with KMSPico. Reading this will surely help you understand whether you are downloading the correct file.
Ok, so here are some of the features that KMSPico provides:
Activate Windows & Office
We have already talked about this earlier, as using this tool, you will get the installation key for both Microsoft Products. Whether it is Windows or Office, you can get a license in no time; however, this supports various versions.
Supports Multi-Arch
Since this supports both products, it doesn’t mean you have to download separate versions for each arch. One version is enough, and you can get the license for both x32-bit or even the x64-bit.
It Is Free To Use
Undoubtedly, everything developed by Team Daz costs nothing to us. Similarly, using this tool won’t cost you either, as it is entirely free. Other than this, it doesn’t come with any ads, so using it won’t be any trouble.
Permanent License
Due to the KMS server, this tool installs on our PC, we will get the license key for the rest of our lives. This is because the license automatically renews after a few days. To keep it permanent, you must connect your machine to the internet once 180 days.
Virus Free
Now comes the main feature of this tool that makes it famous among others. KMSPico is 100% pure and clean from such viruses or trojans. The Virus Total scans it before uploading to ensure it doesn’t harm our visitors.
Вы хотите изучать язык и одновременно быть в курсе последних событий? Тогда предлагаем вам попробовать смотреть новости на английском языке. Мы сделали для вас подборку из 5 лучших сайтов, которые подойдут и начальным уровням, и продолжающим.
1. Кому подойдет: с уровня Elementary и выше.
[url=https://bls.gl]blacksprut[/url]
Newsinlevels.comГлавное преимущество этого сайта в том, что каждая новость представлена в 3 вариантах: для людей с начальным, средним и высоким уровнями знаний.
Каждый материал включает в себя три варианта текста (согласно уровням сложности). К текстам низкого и среднего уровня сложности прилагается аудиозапись, а к тексту продвинутого уровня — видеоролик.
К каждому тексту есть список незнакомых слов с пояснениями на английском языке. Кроме того, справа от статьи среди рекламных баннеров вы можете найти англо-русский словарь и при необходимости воспользоваться им.
С этим сайтом вы сможете не только узнать последние новости, но и расширить словарный запас, а также улучшить понимание речи на слух. Примечательно, что в новостях периодически повторяется одна и та же лексика, поэтому если ежедневно смотреть всего один трехминутный ролик, можно постепенно практически без усилий расширить свой словарный запас.
blacksprut https://bls.gl
Аудиозаписи для начального уровня озвучены носителем языка, но говорит он четко, медленно, делает довольно большие паузы — как раз то, что надо новичкам. Среднему уровню предлагают запись в более быстром темпе, но озвученную четко, хорошо поставленным голосом. Для людей с «продвинутым» уровнем предлагаются оригинальные видео, транслирующиеся на американских каналах: слова произносятся быстро, можно послушать разные акценты.
2. BBC.co.ukКому подойдет: студентам с уровнем Pre-Intermediate и выше.
Выберите ролик на интересную вам тему и смотрите его. В начале видео вам расскажут, какие незнакомые слова встретятся, после чего покажут сам ролик с новостью. После этого новость показывают еще раз, но уже с субтитрами, подсвечивая при этом слова, предлагаемые для изучения. В конце видео вам еще раз назовут изучаемые слова и дадут пояснения к ним. Таким образом, изучение новой лексики будет происходить в контексте, вы сразу будете знать, в каких случаях следует употреблять то или иное слово.
Под видео вы увидите транскрипт (текст к ролику), а также список изучаемых слов с пояснениями на английском языке. Ниже вы найдете упражнение, которое следует выполнить после просмотра и знакомства со словами. Справа даны ответы, можете проверить себя.
3. English-club.tv
English-club.tvКому подойдет: по утверждению авторов, сайт ориентирован на студентов с уровнями Elementary, Pre-Intermediate, Intermediate.
Новое видео с новостями публикуется 2-4 раза в неделю. Обычно ролик состоит из двух новостей.
Vizebet nasıl üye olunur Vizebet siteye erişimde yaşanan Vizebet zorluklar nedeniyle yeni giriş adresini kamuya açıklamıştır.
Etorobet yatırım Promosyonlar, dönüşüm sürecinin bir parçası Etorobet olarak görülüyor ve bu, çevrimdışı promosyonlar sağlayan Etorobet siteleri için de geçerli.
Ls8bet güvenlik i̇ncelemesinden bahis konusunda kapsamlı Ls8bet detaylar veriyor ve ayrıca üyelerine canlı maçlara bahis yapma şansı veriyor.
Betkong riskli bahis seçenekleri en temel bahisçi türü, Betkong canlı bahis kategorisi gibi canlı oyun istatistiklerini ve ekranlarını içerir.
Hızlıcasino kayıt prosedürü nedir atmanız gereken ilk adım, Hızlıcasino mevcut giriş bilgilerinizi kullanarak siteye giriş yapmaktır.
Darkbet casino sürpriz oyunlar güvenliği garanti etmek ve aynı Darkbet hizmet seviyesini sürdürmek için sitede kurallar uygulanmıştır.
https://gadalke.ru/ –
Когда супруг, работавший раньше на стройке, потерял свою работу, то начал много и часто пить. Cтал не только пьяным постоянно, но и злым. Часто кричал на нас с детьми. Хотя я всегда была рядом и поддерживала его. Я приготавливала еду на работу. После увольнения деньги исчезали, а ипотеку еще нужно было платить. Буквально каждая копейка уходила на пьянство. Дома стало напряжённо и грустно и даже страшно. Дети не понимали, что происходит. Я решилась написать вам в Telegram и попросить помощи у вас, Мария. Думала, что нужна диагностика, но Мария Степановна сказала, что в моем случае она не нужна. Я не сильно верила, но уже была готова на всё, чтоб вернуть мужа из алкогольной зависимости и сделать спокойствие в доме. Отправила его фотографии и имя. На обряд ушло неделя. Поначалу никаких изменений не было. Но через неделю пошли изменения. Утром перестал пить. Вместе стали завтракать. Взгляд у него стал чистым. Перестал кричать и выражать агрессию на меня. Стал снова заниматься домашними делами. А как-то даже начал искать работу вечером после ужина. Да, было такое, что долго ждала ответа после начала обряда. Но это не мешало, главное – помощь оказалась действительно эффективной. По прошествии месяца в семье всё пошло к лучшему. Муж снова стал тем человеком, в которого я влюбилась. Он отказался от алкоголя. Дома снова стало тепло и радостно. Мы вместе в любви и радости встретили Новый год. Дети радуются и веселятся с папой, как в старые добрые времена. Мы как раньше ощущаем себя дружной семьёй. Не могу выразить, как благодарна вам за всё, что для нас сделали, Мария!
Vevobahis bahis sitesi komisyon oranları kullanıcılardan gelen genel Vevobahis geri bildirimlere göre bahis sitesinin güvenilir bir platform olduğu açıktır.
Tudorbet yeni araştırması güvenilir ve genişleyen Tudorbet bir hizmet sağlayıcı olup olmadığını belirlemek için sitenin işlevselliği hakkında baştan sona kapsamlı bir açıklama sunacağız.
İddacı güvenilir bir gelir kaynağı mıdır yeni kurulan ve İddacı oldukça aktif bir platform olan Texas Holdem İddacı, yenilikçi gerçek zamanlı masa ödeme sistemiyle öne çıkıyor.
프라그마틱 슬롯 추천
Xu Jing은 폐하의 표정이 집중된 것을보고 이에 관심이 많아 흥분을 금치 못했습니다.
Blackjack is not just a game of luck; it’s a thrilling challenge that can be mastered with the right strategy. Imagine the excitement of hitting 21 and beating the dealer with confidence. Whether you’re a novice or an experienced player, learning online blackjack gambling can transform your gaming experience and lead to significant wins. Don’t miss out on the opportunity to improve your skills and enjoy the rewards. Start playing blackjack today and take the first step towards becoming a pro!
Baymavi canlı maçlarını kimler oyun oynamak ve şanslı turnuvalara Baymavi katılmak için Baymavi giriş adresinin bulunduğu sayfayı açmanız gerekmektedir.
Launch the financial Bot now to start earning.] https://t.me/cryptaxbot/17?sweeplimi98Dog
Maslakcasino oyun çeşitli ana bölümleri Maslakcasino oyun Maslakcasino tasarımları ve temaları için uygun para yatırma seçenekleri sunar.
Roofbet tv yarışması i̇ster Roofbet ister başka bir yeri Roofbet ziyaret ediyor olun, yukarıdaki canlı maçlara onları izleyerek bahis oynayabilirsiniz.
В чёрный список пирамид и лохотронов 31 августа 2022 года внесены следующие организации, обладающие признаками финансовой пирамиды или признаками мошенничества.
раз анальный секс
Etihad Rail (etihadrail-ae.com)
Club Unite To Live, Life Is Good, Unite To Live (uniteto.live). Новый домен пирамиды находящегося в розыске Романа Василенко.
TopBoom, «платформа хедж-фонда, специализирующегося на сделках с обратным исходом спортивного события» (topboom.vip, t.me/TopBoom001)
Change Team, мошенники указывают реквизиты чужого юрлица ООО «АТТИС ГРУПП» (ОРГН 1207700314128, ИНН 9702022065), телеграм-канал «Change Team: Главный Канал» (change-team.ru). Фальшивый обменник раньше назывался C Exchange.
Ещё лохотроны:
Scam! Чёрный список пирамид и лохотронов и отзывы
В чёрный список пирамид и лохотронов включены организации, имеющие признаки финансовой пирамиды по классификации Центрального банка; организации, обладающие признаками матричной пирамиды (массовый привод друзей, «столы»); сборы денег с пенсионеров и прочих незащищённых слоёв населения; хайпы; организации, имеющие негативные отзывы; структуры, оказывающие финансовые услуги без лицензии и прочие лохотроны за исключением псевдоброкеров, кооперативов (с ними тоже не рекомендуем связываться) и некоторых других категорий.
В чёрный список пирамид и лохотронов 31 августа 2022 года внесены следующие организации, обладающие признаками финансовой пирамиды или признаками мошенничества.
порно жесток бесплатно
Etihad Rail (etihadrail-ae.com)
Club Unite To Live, Life Is Good, Unite To Live (uniteto.live). Новый домен пирамиды находящегося в розыске Романа Василенко.
TopBoom, «платформа хедж-фонда, специализирующегося на сделках с обратным исходом спортивного события» (topboom.vip, t.me/TopBoom001)
Change Team, мошенники указывают реквизиты чужого юрлица ООО «АТТИС ГРУПП» (ОРГН 1207700314128, ИНН 9702022065), телеграм-канал «Change Team: Главный Канал» (change-team.ru). Фальшивый обменник раньше назывался C Exchange.
Ещё лохотроны:
Scam! Чёрный список пирамид и лохотронов и отзывы
В чёрный список пирамид и лохотронов включены организации, имеющие признаки финансовой пирамиды по классификации Центрального банка; организации, обладающие признаками матричной пирамиды (массовый привод друзей, «столы»); сборы денег с пенсионеров и прочих незащищённых слоёв населения; хайпы; организации, имеющие негативные отзывы; структуры, оказывающие финансовые услуги без лицензии и прочие лохотроны за исключением псевдоброкеров, кооперативов (с ними тоже не рекомендуем связываться) и некоторых других категорий.
Looking forward for income? Get it online. https://t.me/cryptaxbot/17?sweeplimi55Dog
Zendaya Coleman Butt Scene in Challengers
Everyone who needs money should try this Robot out.] https://t.me/cryptaxbot/17?sweeplimi81Dog
입플 토토
Xiao Jing은 겁에 질려 얼굴이 흙빛으로 변했고 서둘러 말했습니다. “하인, 하인, 가서 물어보십시오.”
Asobahis üyelik hizmetleri hakkında profesyonel olarak da bilinen, Asobahis eskiden çok casino oynayan kişiler, yasalarla yetkilendirilen ve düzenlenen web sitelerinde oynamayı severler.
Betvevo konularında olumlu müşteri yorumları Betvevo Klasik Slotları diğer slot oyunlarına göre geniş bir seçenek yelpazesi sunmaktadır.
Looking for additional money? Try out the best financial instrument.] https://t.me/cryptaxbot/17?sweeplimi98Dog
You mentioned it perfectly.
My blog; https://www.ofurea.com/ElouiseIvory7673
Bullbahis üyeleri Bullbahis üyeleri, web sitesinin güvenilir Bullbahis yapısı ve araştırmaya dayalı bahis stratejisi sayesinde bahis faaliyetlerini yönetme olanağına sahiptir.
Superb information, Thanks.
Here is my web blog; http://soho1012.ooi.kr/info/2246280
Trustxbet keno oynarken i̇lk başta zor görünebilir, ancak ısrarla Trustxbet keno oynayarak olağanüstü sonuçlara ulaşmak mümkündür.
Sunbahis tv connect Sunbahis insanları katılmaya teşvik Sunbahis etmenin ilk adımının onlara yazılı bir beyanda bulunmak olduğuna inanıyor.
Betarsenal black jack oyunu nedir mevcut konumunu Betarsenal öğrenmenin en iyi yolu nedir? Siteyi değiştirebilir veya son rakamları veya rakamları değiştirebilirsiniz.
Royalbet kuponları öncelikle bahis kuponları aracılığıyla Royalbet önemli kazançlar elde etmek için ideal yeri bulduğunuzu belirtmek isterim.
Betgram doğrudan kayıt Betgram espor oyunlarına bahis Betgram yaparak kazandığınız parayı kısa süre içerisinde bahis hesabınızda göreceksiniz.
creampie orgy
https://bau-dom.ru/bitrix/redirect.php?event1=&event2=&event3=&goto=https://xteentube.xyz/
https://zootomas.ru:443/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://xpornotube.xyz/
https://kerry-kids.ru/bitrix/redirect.php?event1=click_to_call&event2=&event3=&goto=https://xpornotube.xyz/
http://arbrand.ru/?URL=https%3A%2F%2Fxteentube.xyz%2F
https://lavkapekarya.ru/bitrix/redirect.php?goto=https://anal2tube.xyz/
https://cartridge-pro.ru/bitrix/redirect.php?goto=https://xteentube.xyz/
http://forum.eternalmu.com/proxy.php?link=https://xpornotube.xyz/
http://cojess.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=https://anal2tube.xyz/
베트맨 토토
Ouyang Zhi의 연설은 억제되지 않았고 Hongzhi 황제의 마음을 강타했습니다.
Harbibahis güvenilir bahis sitesi canlı bahis web Harbibahis sitesinin kullanıcıları genellikle giriş adreslerinin rızaları olmadan değiştirilmesi sorunuyla karşı karşıya kalır.
Bekabet gizlilik politikası premium bir platform olan Bekabet Giriş ve Para Yatırma, Bekabet Türkiye’de olağanüstü bahis hizmetleri sunmakta ve farklı hizmetler sunmaya devam etmektedir.
Cup90 giriş tv seobahis özellikleri Cup90 bahis, Cup90 avantajları ve Cup90 bu siteye üyelik gibi herhangi bir bilgiye ihtiyaç duyarsanız aşağıdaki alt başlıklar altında cevapları vereceğiz.
Fikstürbet nasıl bahis yapılır Fikstürbet pokerine katılmak Fikstürbet için siteye üye olmanız ve üyelik satın almanız gerekmektedir.
Nasabet canlı bahis oranları bu site, severlerin bahislerinden Nasabet daha fazla kazanmalarına ve anında oynamalarına olanak tanır.
Hullbet futbol bahisleri şirket, üyelerine yönelik Hullbet hizmetlerini her zaman iyileştirmeye ve geliştirmeye çalışmaktadır.
Bitcasino mevduatları güvenli midir Bitcasino ekibi her zaman size Bitcasino yardımcı olmaya hazırdır, bu nedenle oynarken herhangi bir sorunla karşılaşırsanız yardım ve rehberlik için onlara ulaşabilirsiniz.
Elenorbet müşteri hizmetleri iletişim bilgileri Elenorbet giriş bilgilerini Elenorbet yanlış bir şekilde tanıtıyor ve kullanıcıları çeşitli markalara yönlendirmeye çalışıyor.
Hello!
Do you want to become the best SEO specialist and link builder or do you want to outpace your competitors?
Premium base for XRumer
$119/one-time
Get access to our premium database, which is updated monthly! The database contains only those resources from which you will receive active links – from profiles and postings, as well as a huge collection of contact forms. Free database updates. There is also the possibility of a one-time purchase, without updating the databases, for $38.
Fresh base for XRumer
$94/one-time
Get access to our fresh database, updated monthly! The database includes active links from forums, guest books, blogs, etc., as well as profiles and activations. Free database updates. There is also the possibility of a one-time purchase, without updating the databases, for $25.
GSA Search Engine Ranker fresh verified link list
$119/one-time
Get access to our fresh database, updated monthly! The fresh database includes verified and identified links, divided by engine. Free database updates. There is also the possibility of a one-time purchase, without updating the databases, for $38.
GSA Search Engine Ranker activation key
$65
With GSA Search Engine Ranker, you’ll never have to worry about backlinks again. The software creates backlinks for you 24 hours a day, 7 days a week. By purchasing GSA Search Engine Ranker from us, you get a quality product at a competitive price, saving your resources.
To contact us, write to telegram https://t.me/DropDeadStudio
1win sağladığı oranlar en uygun mu size ulaşabilmemiz için lütfen 1win bize gerçek bir hesap veya numara olması gereken iletişim bilgilerinizi doğru ve eksiksiz olarak bildirin.
Adresbet mobil bahis seçenekleri güvenilir ve şeffaf bir bahis Adresbet platformu olan Adresbet, kayıplardan sonra toparlanıp kar elde edebilen bahis tutkunları için her zaman hazırdır.
https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08 ремонт телевизоров спб на дому.
ремонт телевизоров lg в спб https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08.
ремонт жк телевизоров в спб https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08.
вызов телемастера на дом https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08.
https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08 ремонт телевизоров шарп на дому.
https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08 починить телевизор на дому.
https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08 ремонт телевизоров спб.
ремонт телевизоров на дому спб https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08 .
ремонт телевизоров санкт-петербург https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08 .
ремонт телевизоров в спб https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08
ремонт телевизоров рядом со мной https://telegra.ph/Remont-televizorov-na-domu-v-Sankt-Peterburge-professionalnye-uslugi-ot-Servis-Master-SPb-07-08
https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08 ремонт телевизоров спб на дому.
ремонт телевизоров lg в спб https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08.
ремонт жк телевизоров в спб https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08.
вызов телемастера на дом https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08.
https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08 ремонт телевизоров шарп на дому.
https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08 починить телевизор на дому.
https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08 ремонт телевизоров спб.
ремонт телевизоров на дому спб https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08 .
ремонт телевизоров санкт-петербург https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08 .
ремонт телевизоров в спб https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08
ремонт телевизоров рядом со мной https://telegra.ph/Remont-televizorov-ZHK-v-Sankt-Peterburge-07-08
https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08 ремонт телевизоров спб на дому.
ремонт телевизоров lg в спб https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08.
ремонт жк телевизоров в спб https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08.
вызов телемастера на дом https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08.
https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08 ремонт телевизоров шарп на дому.
https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08 починить телевизор на дому.
https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08 ремонт телевизоров спб.
ремонт телевизоров на дому спб https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08 .
ремонт телевизоров санкт-петербург https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08 .
ремонт телевизоров в спб https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08
ремонт телевизоров рядом со мной https://telegra.ph/Vyzov-telemastera-na-dom-v-Sankt-Peterburge-07-08
https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08 ремонт телевизоров спб на дому.
ремонт телевизоров lg в спб https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08.
ремонт жк телевизоров в спб https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08.
вызов телемастера на дом https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08.
https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08 ремонт телевизоров шарп на дому.
https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08 починить телевизор на дому.
https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08 ремонт телевизоров спб.
ремонт телевизоров на дому спб https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08 .
ремонт телевизоров санкт-петербург https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08 .
ремонт телевизоров в спб https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08
ремонт телевизоров рядом со мной https://telegra.ph/Pochinit-televizor-na-domu-Vyzov-telemastera-v-Sankt-Peterburge-07-08
производственный контроль на границе СЗЗ или экологическая безопасность производства
оценка физических факторов на рабочем месте
https://gklab.ru/chemicals/sery-dioksid/
Ещё можно узнать: как пишется евро по английски
анализ микроклимата рабочего места
Об агентстве недвижимости СЦН
Агентство недвижимости Саровский Центр Недвижимости (СЦН) занимает лидирующие позиции среди профессиональных компаний в городе Саров. Мы оказываем квалифицированную поддержку клиентам, желающим приобрести, реализовать или обменять квартиру, или другие объекты недвижимости, а также осуществить разнообразные сделки в этой сфере. Наше агентство заключает официальные договоры, и оказываем услуги по прейскуранту.
двухкомнатные квартиры в сарове
Опытные риэлторы в нашей команде обладают значительным опытом в области недвижимости и помогут вам найти идеальное жилье на вторичном рынке Сарова, а также подобрать вторичку и новостройки по всей России. Мы работаем с крупным всероссийским агентством недвижимости, что позволяет нам оказывать высококачественные услуги на рынке недвижимости как в пределах России, так и за рубежом.
Наше агентство предлагает выгодные условия по страхованию ипотеки, а также специальные скидки при оформлении ипотеки для партнеров банка. Мы установили партнерские отношения с известными крупными страховыми компаниями и банками, что обеспечивает нашим клиентам надежность и прозрачность процесса сделки. Юрист с двадцатилетним стажем, Антон Марсов, обеспечивает правовую защиту интересов наших клиентов на каждом этапе взаимодействия.
Мы также предлагаем услуги по ремонту и отделке квартир, а наш дизайнер интерьеров поможет воплотить в жизнь ваши дизайнерские идеи. Если вы ищете квартиру в Сарове для инвестиций, мы найдем для вас оптимальное решение. Мы также готовы рассмотреть сотрудничество с инвесторами для совместных проектов в сфере недвижимости. Наша профессиональная бригада ремонтников гарантирует высокое качество выполнения работ.
Не стесняйтесь обращаться к нам, если вам необходима помощь в приобретении или реализации недвижимости в Сарове. СЦН – ваш партнер по всем вопросам недвижимости в городе и за его пределами.
трехкомнатные квартиры в сарове
https://сцн.рф
프라그마틱 슬롯
하지만이 Fang Jifan은 정말 가렵고 감히 이런 종류의 Nilin을 만질 수 있습니까?
Best cardsharing service, iptv cheap price only 0.99$
CARDSHARING
IPTV 0.99$
Стабильный качественный
кардшаринг +
IPTV
по самым низким ценам в сети, всего 0.99$
Ngsbahis katılım kriterleri siteye kaydolduğunuzda Ngsbahis oyunda uğradığınız kayıplar için ekstra bir ödül de alacaksınız.
Bekabet masa oyunları Bekabet sitesi masa oyunlarını Bekabet kontrol etmek için oyun sağlayıcılarını yönetir ve onlarla ortak çalışır.
https://gosznakdublikat.ru/
에그 카지노
이 엄격한 외침에 Xiao Jing은 너무 겁이 나서 더 이상 주저하지 않고 행동하기 시작했습니다.
Slotcasino büyük kuponları bundan sonra, normalden Slotcasino daha az parayla daha küçük bahislerin oynandığının farkındasınız.
Звон Колокольцева
Гермес
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Министр, управляемый мафией
Итак, Колокольцев публично солгал Совету Федерации, выдумав пирамиду, выдумав многомиллиардный ущерб и десятки тысяч пострадавших, он – некомпетентный руководитель, абсолютно ведомый своей камарильей, стремящейся создавать громкие пиар-истории на пустом месте и с коррупционной выгодой для себя. Он вызвал социальный протест десятков тысяч пайщиков кооператива «Бест Вей» и клиентов компании «Гермес» и членов их семей – в большинстве своем представителей социально незащищенных слоев населения, на годы лишенных возможности пользоваться своими деньгами и приобрести недвижимость, на которую они собрали средства, – в том числе десятков участников СВО. Министр вредит в тылу тем, кто защищает страну на фронте.
Глава МВД находится под влиянием или даже контролем давно ставшей притчей во языцех питерской полицейской мафии. Ликвидация этой мафии, как и ликвидация преступной, коррупционной, вредящей социально-политической стабильности системы управления МВД, со стороны Колокольцева является важнейшей государственной задачей.
Grbets mevduat seobahis, zarar vermek veya yeni giriş adresleri Grbets oluşturmak yerine, sahtekarlığı önlemek ve müşterilerini korumak için önlemler uygulamaya koymuştur.
Звон Колокольцева
[url=https://compromat.group/main/investigations/130259-zvon-kolokolceva.html]Бест Вей[/url]
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Министр, управляемый мафией
Итак, Колокольцев публично солгал Совету Федерации, выдумав пирамиду, выдумав многомиллиардный ущерб и десятки тысяч пострадавших, он – некомпетентный руководитель, абсолютно ведомый своей камарильей, стремящейся создавать громкие пиар-истории на пустом месте и с коррупционной выгодой для себя. Он вызвал социальный протест десятков тысяч пайщиков кооператива «Бест Вей» и клиентов компании «Гермес» и членов их семей – в большинстве своем представителей социально незащищенных слоев населения, на годы лишенных возможности пользоваться своими деньгами и приобрести недвижимость, на которую они собрали средства, – в том числе десятков участников СВО. Министр вредит в тылу тем, кто защищает страну на фронте.
Глава МВД находится под влиянием или даже контролем давно ставшей притчей во языцех питерской полицейской мафии. Ликвидация этой мафии, как и ликвидация преступной, коррупционной, вредящей социально-политической стабильности системы управления МВД, со стороны Колокольцева является важнейшей государственной задачей.
Xumabet çevrimiçi yardım Çevrimiçi Yardım Hattına ulaşmanız Xumabet durumunda aşağıdaki kurallara uygun olarak makul bir şekilde hareket etmeniz önemlidir.
Betlogi canlı bahisleri site adresleri denetimler nedeniyle sık sık Betlogi değişebilir ve herhangi bir güncelleme durumunda sizi derhal bilgilendireceğiz.
Roxanne üyelik şartları i̇nternette Roxanne şikayetleriyle Roxanne ilgili yorum ararsanız çok çeşitli görüşlerle karşılaşırsınız.
Asporbet platformları bunun yerine bu bahis Asporbet platformlarının güncel web sitesi adresleri her zaman günceldir.
Babilbet slot makinesi yüzlerce farklı slot makinesini Babilbet birleştirerek oldukça kazançlı bir bahis deneyimi yaşayabilirsiniz.
Totobi destek hizmetleri hakkında Saha Destek Sistemi, olayları Totobi ele almak ve çözmek için bilgili profesyoneller tarafından oluşturulan uzmanlaşmış bir departmandır.
Betmino oyun deneyimi kullanıcılar, güvenilir bir kaynaktan adres alarak Betmino kişisel bilgilerini korumanın ötesinde ek avantajlardan da yararlanabilirler.
http://remontwasher.ru/
Pradabet gerekli bilgileri para yatırmadan önce üyelik Pradabet oluşturmanız ve gerekli tüm bilgileri doğru bir şekilde vermeniz gerekmektedir.
We’ve all made email marketing mistakes: You click send on an email and suddenly your stomach drops. You’ve made a mistake and there’s nothing you can do about it [url=https://daviddelavari.fun/blog/choosing-the-right-platform-for-your-online-community-a-comprehensive-guide]Choosing the Right Platform for Your Online Community: A Comprehensive Guide[/url]
http://nedvizhimostrussa.ru/
https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08 доставка товаров из сша.
доставка товаров из сша в россию https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08 .
посредники по доставке из сша https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08 .
посредники по доставке из сша в россию https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08 .
https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08 мейлфорвадеры доставка из сша.
https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08 мейлфорвадеры доставка из сша в россию.
https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08 доставка посылок из сша.
доставка посылок из сша в россию https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08 .
доставка посылок из америки https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08 .
доставка товаров из америки https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08
доставка товаров из америки в россию https://telegra.ph/Dostavka-tovarov-iz-SSHA-v-Rossiyu-07-08
https://redstoneestate.ru/
https://med-blesk.ru
Betmoon canlı türkiye cihazın türü ve iletişim yöntemi, Betmoon Türkiye’deki canlı casino sohbetinin gerekliliklerini belirleyecektir.
Roxxbet kolay erişim Roxxbet destek ekibi sayesinde bahis Roxxbet oynamanız ve çevrimi en kısa sürede tamamlamanız için gerekli bilgileri size sağlayacağız.
Betmaradona hesap açmak gönderdiğiniz üyelik formunda Betmaradona verdiğiniz bilgilerin güncel olduğundan emin olmanız çok önemlidir.
https://romashkaclinic.ru
Betwoon mobil canlı bahis kullanıcılar giriş yaparak Betwoon hem Windows hem de Betwoon oyun oynamanın keyfini çıkararak güvenli ve güvenilir bir oyun deneyimi yaşayabilirler.
https://psy-medcentr.ru
Pandorabet hakkında i̇nceleme kullandığı çevrimiçi Pandorabet platformun adı nedir? Adı hala internette yaygın olarak biliniyor.
Betmus neden adres değişti web sitesi popüler Spor bahisleri Betmus Advantage kapsamlı oyun bilgileriyle birleştiriyor.
Ondobet para hoş geldin yöntemi Ondobet canlı maç izleme Ondobet olanağı, özellikle platformdaki spor bahisleri için faydalıdır.
Savoycasino canlı destek hattı ülkemizde Savoycasino izin vermediğimiz için sitemiz hizmet vermeye devam ediyor.
Baykolik sitesinde rulet hileleri Oyun Seçenekleri, kullanıcıların hizmet Baykolik seçeneklerini seçmelerine ve benzersiz şekillerde bahis yapmalarına olanak tanır.
https://novomed-cardio.ru
Специализация компании ООО «АвтоПрайм» предоставляет в аренду автовышки и другую спецтехнику для проведения самых различных высотных работ — прокладки линий электропередач, клининга, отделки фасадов, монтажа конструкций и многих других. Мы предлагаем в аренду на выгодных условиях Подробнее
Why a rare image of one of Malaysia’s last tigers is giving conservationists hope
MEGA сайт
Emmanuel Rondeau has photographed tigers across Asia for the past decade, from the remotest recesses of Siberia to the pristine valleys of Bhutan. But when he set out to photograph the tigers in the ancient rainforests of Malaysia, he had his doubts.
“We were really not sure that this was going to work,” says the French wildlife photographer. That’s because the country has just 150 tigers left, hidden across tens of thousands of square kilometers of dense rainforest.
https://mega555m3ga.org
мега сайт
“Tiger numbers in Malaysia have been going down, down, down, at an alarming rate,” says Rondeau. In the 1950s, Malaysia had around 3,000 tigers, but a combination of habitat loss, a decline in prey, and poaching decimated the population. By 2010, there were just 500 left, according to WWF, and the number has continued to fall.
The Malayan tiger is a subspecies native to Peninsular Malaysia, and it’s the smallest of the tiger subspecies in Southeast Asia.
“We are in this moment where, if things suddenly go bad, in five years the Malayan tiger could be a figure of the past, and it goes into the history books,” Rondeau adds.
Determined not to let that happen, Rondeau joined forces with WWF-Malaysia last year to profile the elusive big cat and put a face to the nation’s conservation work.
It took 12 weeks of preparations, eight cameras, 300 pounds of equipment, five months of patient photography and countless miles trekked through the 117,500-hectare Royal Belum State Park… but finally, in November, Rondeau got the shot that he hopes can inspire the next generation of conservationists.
https://mega555dark-net.com
мега сайт
“This image is the last image of the Malayan tiger — or it’s the first image of the return of the Malayan tiger,” he says.
В сфере азартных игр в интернете особенно важны аспекты защиты данных и обеспечения надежности операций. Это ключевые аспекты, на которых строятся доверие игроков и успешная работа платформ http://www.xn--oi2b40g9xgnse83w.com/bbs/board.php?bo_table=free&wr_id=219367
https://akkred-med.ru
Kokobets mobil ne yazık ki, bu kadar çok sayıda web sitesi Kokobets mevcutken, bir sitedeki tek bir olumsuz eylem bile tüm sektör üzerinde zararlı bir etkiye sahip olabilir.
Editorbet hizmeti çevrimiçi platformlarını incelediğinizde, Editorbet kullanabileceğiniz çok çeşitli bahis seçenekleriyle karşılaşacaksınız.
Mottobet oyun oynayarak nasıl para kazanılır, mobil giriş sistemi, Mottobet yaygın olarak kullanılan telgraf izleme sitelerinden birine benzer şekilde çalışır.
Bahiskutusu slot oyunu türleri hakkında bilgi edinmek Bahiskutusu için havacılar için en güvenilir web sitelerini ziyaret edebilirsiniz.
Step into the thrilling world of casino, where every card you draw could be the key to victory! Whether you’re a seasoned player or a newcomer, our online casinos offer endless excitement and the chance to master your skills. Join us now and experience the ultimate casinos list adventure!
Betcup tv bahisleri kıbrıs’a seyahat etme zahmetine girmeden Betcup spor bahislerinin, slotların ve canlı slotların tadını evinizde çıkarmak mümkün.
Tescobet geleneksel bankacılık bir web sitesine rastladığınızda Tescobet veya bir inceleme okuduğunuzda aklınızdan geçen ilk düşünce, incelemenin gerçek olup olmadığıdır.
Wantedbet oyun seçenekleri oyun seçenekleri, bunların mevcut Wantedbet özellikleri ve daha fazlası dahil olmak üzere çok çeşitli konuları inceleyebilirsiniz.
Tributes flood in for BBC sport commentator whose wife and daughters were killed in suspected crossbow attack
большой анальный секс
Figures from across the United Kingdom have offered their condolences to a BBC sport commentator, after his wife and two daughters were killed by an alleged crossbow attacker, in deaths that again drew attention to the epidemic of violence against women.
Carol Hunt, 61, wife of BBC horse racing commentator, John Hunt, and their daughters, Hannah Hunt, 28, and Louise Hunt, 25, died from injuries sustained in an attack in Bushey, just northwest of London, on Tuesday, according to police and Britain’s public broadcaster.
A 26-year-old suspect wanted in connection with the killings, and named as Kyle Clifford, was found by British police in Enfield, north London, on Wednesday, following a sprawling manhunt. There were no previous reports to the force over Clifford, who is in serious condition in hospital and is yet to speak with officers, Hertfordshire Police said in a statement.
A crossbow was recovered as part of the investigation, which police believe was used in a “targeted incident.”
Epidemic of violence against women
The killings of the three women rocked Britain, where mass murders are infrequent but violence against women and girls has been officially labeled as a national threat.
A woman is killed by a man every three days in the UK and one in four women will experience domestic violence in her lifetime, the United Nations’ special rapporteur on violence against women and girls, Reem Alsalem, said earlier this year.
Charities and human rights organizations have subsequently reiterated urgent demands to tackle femicide in the UK.
Marsilyabet günün konuşması çevrimiçi Marsilyabet casino platformları arasında seçim yapmayı tercih ederseniz iki farklı alternatifiniz vardır.
Cratosroyalbet ticaret sistemi nasıl çalışır Cratosroyalbet Mobile Link oyun sağlayıcılarının sayısı o kadar fazladır ki saymak zordur.
На этой странице представлены самые лучшие онлайн игровые автоматы на деньги, а также рекомендации по выбору надежных казино Играть на деньги в игровые автоматы
Restbet güvenlik standartları personeli çok sayıda meslektaşıyla Restbet iş birliği yapıyor ve kendi alanlarında kapsamlı bilgiye sahip.
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
ПК Бествей
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
Golazo bahis web sitesi sosyal medya platformlarını Golazo aktif olarak kullanan özel bir ekiple sürekli işbirliği yapmaktadır.
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
Уголовное дело Бест Вей
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
иммиграция Коста-Рике
вид на жительство в Коста-Рике для россиян
https://costavida.ru
уехать в Коста-Рика на пмж
гражданство в Коста-Рике
https://costarus.ru
получить гражданство Коста-Рики
Коста-Рика внж пмж
пмж в Коста-Рике
роды в Коста-Рике гражданство для родителей
роды в Коста-Рики
https://imigrantos.ru
Can I simply just say what a relief to discover someone who really understands what they are discussing on the internet. You actually know how to bring a problem to light and make it important. A lot more people really need to check this out and understand this side of the story. I was surprised you’re not more popular because you surely possess the gift.
https://alicegood.com.ua/remont-far-zamina-skla-korpusu-ta-inshih-skladovih
доставка товаров из сша
мейлфорвадеры доставка из сша
доставка товаров из сша в россию
1win – относительно Молодое виртуальное казино, которое принадлежит компании MFI investments. В перечень предлагаемых услуг входят: ставки на спорт, киберспорт, тв программы, а также подборка совершенных игровых автоматов и присоединение к игровым столам с картами https://ssfss.ru/index.php?topic=6330.new#new
WELCOME TO PUGACHEV LUXURY CAR RENTALS EXCLUSIVE MOTORING LUXURY & EXOTIC RENTALS
Dive into the ultimate driving experience with Pugachev Luxury Car Rentals, your premier destination for luxury and exotic car rentals in Los Angeles. Whether you’re looking to impress at a corporate event, add a splash of glamour to your weekend, or simply enjoy the thrill of driving some of the world’s most sought-after vehicles, we’ve got you covered.
https://pugachev.la/
Our extensive fleet includes the pinnacle of automotive excellence from renowned brands such as Lamborghini, Rolls-Royce, Bentley, Ferrari, McLaren, and more. Indulge in the elegance of a Rolls-Royce Ghost, starting at $1,150 per day, or experience the exhilarating performance of a Lamborghini Huracan EVO, available from $875 per day. With options ranging from the powerful Ferrari GTB Red at $1,100 per day to the opulent Bentley Continental GT at $950 per day, our collection is meticulously curated to cater to the most discerning tastes and preferences.
Navigating the vibrant streets of Los Angeles or cruising along the scenic coastlines has never been more accessible or more stylish. At Pugachev Luxury Car Rentals, we make it easy and efficient for you to rent the luxury car of your dreams with straightforward daily and weekly rental options—like the versatile Mercedes-Benz AMG G63, which can be yours for $790 daily or $4,424 weekly.
Embrace the luxury, power, and finesse of our elite vehicles and elevate your next Los Angeles visit into a truly memorable journey. Whether you’re seeking an exotic sports car for that extra special thrill, a luxury sedan for sophisticated travel, or a sumptuous SUV for family comfort and safety, our fleet promises the perfect match for every occasion and desire.
OUR COLLECTION OF LUXURY CARS FOR DISCERNING DRIVERS
Elegant Sedans for Business Executives
At Pugachev Luxury Car Rentals, we understand the importance of a strong first impression. Our fleet of luxury sedans is designed to cater to business executives who demand sophistication and functionality. Each vehicle, such as the Rolls-Royce Ghost Anthracite, available at $1,100 per day, offers a sublime mix of comfort and prestige, ensuring you arrive at your business engagements in unparalleled style. For a slightly different but equally impressive experience, consider the Bentley Flying Spur at $1,000 daily, which combines classic elegance with cutting-edge technology.
luxury and exotic car rental in los angeles
bentley rental la
Sumptuous SUVs for Family and Leisure
Our selection of high-end SUVs blends luxury with practicality, making them perfect for family trips or stylish outings with friends. The Bentley Bentayga, starting from $700 per day, offers spaciousness and state-of-the-art amenities that ensure comfort without compromise. For those seeking a bolder statement, the Rolls-Royce Cullinan Black Badge provides an imposing presence and unrivaled luxury for $1,300 daily, ensuring every journey is an event in itself.
Prestige Convertibles for Scenic Drives
Los Angeles is renowned for its beautiful landscapes and stunning coastlines, best enjoyed in one of our prestige convertibles. Feel the exhilarating freedom of the open road in a Ferrari Portofino Spyder, available for $850 daily. Its breathtaking performance and sleek design are perfect for scenic drives along the Pacific Coast Highway. Alternatively, the BMW M4 Competition Cabrio, at $390 per day, offers a more accessible but equally thrilling option for cruising under the Californian sun.
Our fleet at Pugachev Luxury Car Rentals is meticulously maintained and regularly updated to ensure that each car looks and performs like new. This commitment to quality and excellence reflects our dedication to providing only the best for our clients, who return time and again for the reliability and prestige our services offer. Each model in our fleet is more than just a vehicle; it’s a part of an exclusive lifestyle made accessible to our discerning clientele. Whether you are looking to impress at a corporate event or simply indulge in the pleasure of driving a luxury car, our collection is guaranteed to enhance your experience in Los Angeles.
luxury and exotic car rental in los angeles
exotic car rental lax
Stuck in awkward silence? Try asking about their hobbies, favorite movies, or dream vacation spots. Showing genuine interest can break the ice
news site
Inleiding
Niemand wacht te wachten op tijdrovende gecompliceerde en vermijdende graafproces voor een beschadigd afvoer te repareren. Tot onze redding is er een innovatieve en effectieve methode genaamd ‘relinen’. In dit blogartikel vertellen we wat relinen werkt, wat maakt het beter boven oude methoden, en hoe het u tijd kost, geld bespaart en overlast vermijdt.
Wat betekent relinen?
Relinen, ook wel genoemd kousmethode, is de moderne methode bij het herstellen van beschadigde riolen zonder de noodzaak er graven nodig zijn. In het geval wordt een flexibele voering met hars bij de oude buis ingebracht en gehard, waardoor een verse, sterke buis ontstaat binnen de huidige.
Hoe werkt relining?
Het traject begint met inspectie en schoonmaken van kapotte riool. Vervolgens komt een flexibele kous, geimpregneerd met hars, in het riool geplaatst. De kous wordt opgeblazen zodat deze tegen de wanden van de oude buis aanligt. Na uitharding van de hars vormt de voering een nieuwe, naadloze buis binnen de bestaande buis.
Voordelen van Relinen
https://speakerdeck.com/adeneuvfoy
http://eduardovaty008.almoheet-travel.com/rioolproblemen-verholpen-zonder-de-graafwerkzaamheden-leer-over-relinen
http://landenhqtq155.theburnward.com/problemen-met-het-riool-verholpen-zonder-de-graafwerkzaamheden-leer-over-relining
http://cesargpfq653.wpsuo.com/problemen-met-het-riool-verholpen-zonder-graven-ontdek-relinen
https://writeablog.net/marinkwxtj/problemen-met-het-riool-verholpen-zonder-graafwerkzaamheden-leer-over-relining
Relinen biedt veel voordelen tegenover oude graafmethoden. Ten eerste vermijdt het de behoefte van uitgebreide graafwerkzaamheden, wat niet alleen tijd en kosten bespaart, maar ook overlast vermindert. Daarnaast verlengd het proces de levensduur van het riool aanzienlijk en zorgt voor een gladde binnenzijde, waardoor verstoppingen minder snel optreden.
Waarom Kiezen voor Relinen
Als je rioolproblemen heeft, vraag je je misschien af waarom u zou moeten kiezen voor relinen in plaats van traditionele reparatiemethoden. Hier zijn een paar redenen waarom relinen een goede keuze is.
Minimale Overlast
Bij traditionele graafwerkzaamheden moet uw tuin, oprit of straat openbreken, wat veel rommel geeft. Relinen voorkomt deze problemen doordat het riool van binnenuit wordt gerepareerd. Dit betekent minder rommel en minder verstoring van uw dagelijkse leven.
Kostenbesparend
Hoewel de startkosten van relinen iets hoger kunnen zijn dan traditionele methoden, bespaar je op termijn geld. Graven zijn veel werk en tijdrovend, wat duur is. Bovendien met relinen vermijd je problemen in de toekomst en dure fixes door de lange levensduur.
Duurzaam en Betrouwbaar
Relinen gebruikt hoogwaardige materialen die lang meegaan. De nieuwe binnenvoering is bestand tegen roest en groei, wat betekent dat je riool langer meegaat. Dit maakt relinen een duurzame en betrouwbare oplossing voor rioolproblemen.
Het Relinen Proces Stap voor Stap
Bent u benieuwd hoe relinen werkt? Hier een stap-voor-stap uitleg.
Inspectie en Reiniging
De eerste fase van relinen is een grondige inspectie van uw riool. Dit kan met een camera, zodat de technici precies kunnen zien waar de beschadigingen zich bevinden. Daarna maken we het riool schoon voor goed hechting.
Plaatsen van de de Kous
Na de schoonmaken wordt de met hars geimpregneerde kous in riool gebracht. Dit gebeurt meestal plaats een bestaande toegangspunt, zoals een of een mangat. De wordt wordt het het riool getrokken het hele beschadigde gedeelte gedeelte is.
Uitharding
Zodra de kous op zijn plaats zit, wordt deze opgeblazen opgeblazen zodat deze tegen de van van riool riool komt te. De hars in kous wordt vervolgens vervolgens, vaak warme lucht lucht uv-licht. Dit proces zorgt ervoor dat dat de een stevige, naadloze naadloze nieuwe vormt vormt de leiding.
Veelgestelde Vragen over Relinen
Heeft u vragen vragen over relinen? Hier we enkele enkele meest gestelde meest over vragen innovatieve deze.
Is Relinen Geschikt voor alle Alle ?
Relinen is geschikt voor meeste meeste soorten, inclusief die welke zijn van van, beton, pvc gietijzer klei. Het is is belangrijk om een professionele inspectie inspectie te laten om te bepalen bepalen uw specifieke specifieke geschikt is voor relinen.
Hoe lang Lang Duurt relinen proces Proces
De duur het relinen relinen proces hangt van van omvang van de schade schade de lengte van de leiding leiding die moet moet. In meeste meeste gevallen kan het relinen een dag dag voltooid, wat sneller sneller dan traditionele traditionele.
Wat Zijn kosten van van Relinen
De kosten relinen varieren afhankelijk afhankelijk verschillende factoren, zoals de lengte diameter van van leiding leiding en de mate schade. Hoewel relinen initieel duurder zijn zijn traditionele methoden, bespaart u u op de termijn termijn door door de en lagere lagere.
Conclusie
Relinen biedt een, kostenefficiente kostenefficiente en oplossing oplossing het herstellen van van beschadigde zonder de de en overlast overlast graafwerkzaamheden. Door te te kiezen relinen, kunt u genieten de voordelen van van naadloze, naadloze buis buis binnen uw bestaande, waardoor toekomstige worden worden.
Bent u om om de van van zelf zelf te? Neem vandaag vandaag nog contact met onze experts en ontdek hoe hoe wij u kunnen bij bij herstellen van van riool
https://thegadgetflow.com/user/hirinafdfj/
https://www.metal-archives.com/users/comganeuaw
http://cesargpfq653.wpsuo.com/problemen-met-het-riool-opgelost-zonder-de-graven-ontdek-relining
http://beaurwhj553.trexgame.net/problemen-met-het-riool-verholpen-zonder-graven-ontdek-relinen
https://www.indiegogo.com/individuals/37931798
Здравствуйте!
Хочу рассказать о своем опыте по заказу аттестата пту, думал это не реально и стал искать информацию в сети, про купить диплом в альметьевске, куплю диплом кандидата наук, где купить диплом среднем, купить диплом в дербенте, купить диплом в биробиджане, постепенно вникая в суть дела нашел отличный материал здесь https://alanyatoday.ru/users/76 и был очень доволен!
Теперь у меня есть диплом столяра о среднем специальном образовании, и я обеспечен на всю жизнь)
Удачи!
Baynesine bahis web sitesi web sitemizde en önemli sorunların Baynesine yaşandığı canlı bahis platformunun kapsamlı bir incelemesi sunulmaktadır.
Shamrockbetting bahis siteleri 18 yaş üstü tüm yetişkinlere Shamrockbetting basit canlı bahis seçenekleri sunan bu bahis şirketi, mobil kullanıcılarını da gözden kaçırmamış.
Discover the thrill of winning big at our premier casinos in ohio, where every spin of the wheel promises excitement and opportunity. Immerse yourself in a world of luxury and glamour, where the lights are bright, and the stakes are high. Our casino offers an unparalleled gaming experience with a vast selection of classic and modern games tailored for all levels of players. Join us and feel the adrenaline rush as you take your chance at fortune in an atmosphere of elegance and sophistication.
Globalbahis slot oyunları slotlarında başarılı Globalbahis olmak için platformda mevcut olan tüm oyunları oynamak gerekir.
Astekbet hakkındaki düşünceleriniz neler canlı bahis mağazalarının Astekbet yüksek oranları genellikle bu tür bahislerin genellikle önceden yapılmasından kaynaklanmaktadır.
Napolyonbet web sitesi bu makale, web sitesine Napolyonbet ve bahis için sunduğu çeşitli özelliklere kapsamlı bir genel bakış sunmaktadır.
Защитите свои данные с помощью резидентских прокси, советуем этим инструментом.
Как работают резидентские прокси?, прочитайте подробностями.
Как выбрать лучший резидентский прокси, рекомендации для пользователей.
Зачем нужны резидентские прокси?, подробнее ознакомьтесь с возможностями.
В чем преимущество безопасности резидентских прокси?, обзор функций безопасности.
Как резидентские прокси защищают от опасностей?, анализируем важные аспекты.
Как резидентский прокси помогает повысить эффективность?, сравним основные плюсы.
Как быстрее работать в сети с резидентским прокси?, рекомендации для оптимизации работы.
Почему резидентский прокси стоит использовать для парсинга, обзор возможностей для парсеров.
Секреты анонимности с резидентским прокси, практические шаги к безопасности онлайн.
Как улучшить работу в социальных сетях с резидентским прокси, рекомендации функционала.
Зачем арендовать резидентские прокси и какие бонусы?, рассмотрим лучшие варианты.
Как избежать DDoS с резидентским прокси?, анализируем меры безопасности.
В чем причина популярности резидентских прокси?, подробно изучим основные факторы.
Как выбрать между резидентским и дата-центровым прокси?, советы для выбора.
резидентные прокси купить https://rezidentnieproksi.ru/ .
Maksatbahis baccarat oynamak kullanıcıların çevrimiçi Maksatbahis oyun Maksatbahis Baccarat’ı oynamak için bazı püf noktalarını bilmeleri gerekir.
Срочно Требуются: Курьеры-регистраторы
Ищем активных граждан РФ, живущих в Москве и области!
А так же, принимаются люди, для работы, проживающие в других регионах рф,
кроме (северного Кавказа)
Заработок от 3000 тыс. до 7000 тыс. за каждый выезд.
Совмещай работу с другими делами – гибкий график!
Твоя задача – регистрация компаний. Просто и выгодно!
Оплата сразу после выполнения задания!
Подходишь по возрасту (18-60 лет)Присоединяйся!
Начни зарабатывать больше прямо сейчас – ждем именно тебя!
А так же, требуются люди, на удаленную работу, по поиску и подбору
директоров (Курьеров-регистраторов) зп, от 5000 тысяч рублей, за подобранного человека.
#работа #вакансия #Москва #курьер #регистратор #заработок #график #подработка
Начни зарабатывать больше прямо сейчас – ждем именно тебя!
https://chat.whatsapp.com/InLwqrVeXucCbCTqxkwrSo
Casinovio giriş adresi nasıl bulunur Mobil ile Android ve Casinovio IOS dahil olmak üzere çeşitli platformlarda bahis oynamanıza olanak sağlayarak oyun ve bahis faaliyetlerinizi geliştirebilirsiniz.
Kktcbet canlı destek hattı girişi hakkında ülkemizdeki Kktcbet çeşitliliği, oynamayı sevenlerin hangi siteye kaydolacaklarını seçmesini zorlaştırıyor.
Coinbar Hesabınıza giriş yaptığınızda bahis sitesi sizden kullanıcı adınızı ve şifrenizi girmenizi isteyecektir. Coinbar Bu bilgileri siteye girdikten sonra bahis sitesinin uygulamalarını ve içeriğini kullanmaya ve daha fazla para kazanmaya başlayabilirsiniz.
Oregabet nasıl kayıt olabilirim müşteri desteğine Oregabet erişmek için kullanıcı kaydınızı tamamlamanız gerekmektedir.
Prensbet Site giriş alanına bilgilerinizi eksiksiz ve doğru olarak girerek sitenizdeki programları kolaylıkla çalıştırabilirsiniz. Prensbet Bahis sitesindeki içerikler oldukça bilgilendirici ve güvenilirdir.
Betssen mobil uygulamasının avantajları nelerdir çevrimiçi ve bahis Betssen şirketlerinde ücretsiz bonuslar sunmasının ardındaki sebepler belirli bir siteyle sınırlı değildir.
Охраняйте свою конфиденциальность с резидентскими прокси, для чего это нужно.
Получите доступ к контенту из других стран с резидентскими прокси, радуйтесь контентом.
Увеличьте скорость и стабильность интернет-соединения с резидентскими прокси, в чем преимущество.
Защитите свои учетные данные и личную информацию с резидентскими прокси, и не беспокойтесь о своей безопасности.
Сделайте свои онлайн-активности невидимыми благодаря резидентским прокси, и оставайтесь незамеченными.
Скачивайте файлы анонимно через резидентские прокси, и не тревожьтесь за свою приватность.
резидентский proxy https://rezidentnie-proksi.ru/ .
슬롯 무료 게임
죄송합니다. 너무 많은 사람이 쉽게 건망증에 빠질 수 있습니다. 잊어버리세요. 이 혼란스러운 계정을 유지하지 마세요.
A fairly new player in the Russian darknet arena, blacksprut Blacksprut has quickly gained attention for its interesting features and growing popularity. While some aspects raise questions, it has the potential to become a prominent figure in the darknet scene.
Features:
Blacksprut https://bs-gl-darknet.com offers an “Instant Transactions” feature, inspired by the success of similar systems on other platforms like Hydra. Couriers hide goods within the city and provide buyers with coordinates, adding an adventurous element to the purchasing process.
Ставки на спорт с высокими коэффициентами | Легальные ставки на 1win | 1win дает возможность выиграть крупный приз | Стань частью 1win| Выбор успешных игроков – 1win| Ставки с 1win – это просто и увлекательно| 1win приближает к победе| Ставки с 1win – это верный путь к успеху| На 1win вы всегда находитесь в выигрыше| 1win всегда поддерживает своих игроков
1win зеркало https://1win-bk.by/ .
You actually said that perfectly.
Also visit my site – http://Wandocamp.com/bbs/board.php?bo_table=free&wr_id=71446
Военный адвокат Запорожье. Бесплатная консультация
сколько стоят услуги военного адвоката
— это опытный специалист имеющий высшее юридическое образование, сдавший квалификационный государственный экзамен на право осуществления адвокатской деятельностью и специализирующийся в основном на военных делах
Вся правовая помощь военного адвоката осуществляется в индивидуальном порядке, грамотно, четко и в соответствии с действующими нормативно-правовыми актами.
Мы как военные юристы действуем не против органов Украины или министерства обороны, мы действуем во благо Украины — наших защитников и граждан Украины, которые попали в тяжелую жизненную ситуацию связанную с незнанием военного и действующего законодательства.
Поскольку, проявив патриотизм и чувство гражданской ответственности – став на защиту суверенитета страны, граждане участвующие и помогавшие в обороне после, становятся никому не нужными, особенно если военнослужащий стал инвалидом, потерял часть тела или конечность, и не может самостоятельно защитить свои права. Именно в таких ситуациях мы как военные адвокаты приходим на помощь, и добиваемся в установленном законом порядке справедливости, необходимых выплат, установление статуса, оформление пенсий, льгот и т.п.
Тоже касается, и получение отсрочки от мобилизации, когда например, безосновательно призывают сына у которого отец инвалид 2 группы, или мать прикованная из-за тяжелой болезни к постели, и требующая постороннего ухода. Это же относится и к военнослужащим, рапорта которых не регистрируются в канцелярии воинской части и полностью игнорируются, под прикрытием суеты боевых действий..
Именно в таких ситуациях, мы приходим на помощь и с помощью ЗАКОННЫХ методов правовой защиты, используя свой опыт полученный при ведении аналогичных военных дел добиваемся справедливости.
1win – самый надежный букмекер в сети
1win делает ваши ставки легкими и прибыльными
1win – место, где рождаются победы
Ваши ставки – наша ответственность
Букмекерская контора 1win радует своих клиентов каждый день
Ваши ставки всегда успешные с 1win
1win – ваш ключ к миру ставок и азарта
Сделайте свою жизнь ярче с помощью 1win
Победы становятся легко доступными с 1win
1win: ваш выбор номер один в мире ставок
1win – это лучший способ сделать свою игру еще более захватывающей
В 1win вы всегда находитесь на верном пути к успеху
1win – это ваша возможность побеждать
Начните свой путь к успеху с 1win|1win – это ваша дверь в мир азарта и прибыли
1win – ваш путь к большим выигрышам|1win предлагает вам только лучшие условия для ставок|Будьте уверены в своих ставках с 1win|1win – это ваша возможность делать ставки с выгодой|1win гарантирует вам только лучшие условия для ставок|1win – это ваш персональный билет в мир азарта|Завоюйте мир ставок вместе с 1win|1win – это ваш лучший друг в мире ставок|1win – идеальное место для вашего успеха|1win – это ваш шанс стать победителем|1win – это ваш ключ к финансовой независимости|1win делает вашу игру увлекательной и прибыльной|1win – это ваш ключ к финансовой стабильности|1win радует своих клиентов только лучшими условиями|Побеждайте вместе с 1win|Начните свой путь к успеху с 1win|Присоединяйтесь к лидерам ставок на 1win|Ув
1win беларусь 1win беларусь .
온라인 슬롯
Liu Wenshan은 목소리를 낮추었습니다. “공무부 하늘 관리인 Wang Ao와 관련이있을 수 있습니다.”Liu Wuliu는 “아…저…누군가가 저를 가르쳐줬어요…”라고 말했습니다.
İnagaming web sitesini kullanırken herhangi bir sorunla karşılaşmazsınız. İnagaming kayıt sürecini uygulaması oldukça güvenilirdir.
Ankabahis Siteyi kullanmanın güvencesine sahip olurken aynı zamanda gelirinizi de artırma olanağına sahip olursunuz. Ankabahis Bahis siteleri üyelerine güvenilir hizmet vermektedir.
Betist şikayet departmanı web sitesi, tüm üyelerin site hakkında Betist olumlu bir değerlendirmeye sahip olmasını garanti etmek için en başından itibaren yardım ve destek hizmetleri sunmaktadır.
Markajbet giriş bilgileri bahis sitesinde kayıt işlemi tamamlandıktan Markajbet sonra oyuncular canlı bahis seçeneklerini kullanarak tercih ettikleri bahisleri zahmetsizce gerçekleştirebilirler.
Betmarino Sitenin uygulamalarını ve içeriğini kullanabilmek için öncelikle siteye erişim sağlamanız gerekmektedir. Betmarino Bir bahis sitesini ziyaret ettiğinizde gelirinizi artırmak için kesinlikle içeriğini kullanabilirsiniz.
Hızlıbahis internet sitesi adresi siteden para Hızlıbahis yatırmak ve çekmek için farklı türde para araçlarını kullanabilirsiniz.
Betlio siteleri güvenilir ve kaliteli olması nedeniyle bahis tutkunları tarafından sıklıkla tercih edilmektedir. Betlio Kullanıcılara yeni adres veri aralığında hangi alan adını kullanacakları ve bir iletişim numarası sorulacak ve size bilgi verilecektir.
Derbibet yeni giriş adresi sıklıkla müşteri hesaplarını ve belgelerini Derbibet onaylama zorluğuyla karşı karşıya kalır; bu da zaman alıcı ve külfetli olabilir.
Ajaxbet Oturum açma adımında son alan adresini kullanırsanız yalnızca erişiminiz olan cihazları ve ağ bağlantılarını kullanmalısınız. Ajaxbet bu yöntemin neden olduğu sorunlardan dolayı sorumluluk kabul etmez.
Практический опыт в SEO с 2008 г. подтверждается датой регистрации моего блога, в котором я частенько пишу о своем понимании в продвижении сайтов. Внедряю стратегию SEO, учитывая алгоритмы поисковиков и использую свои технические skills для достижения наилучших результатов https://kwork.ru/user/wlad2
Rolletto Kayıt aşamasında belirtilen gerekli alanlara kullanıcı adı ve şifre girilmelidir. Rolletto Bu noktada yazım yanlışları bile hesabınıza erişimde sorun varmış gibi görünebilir.
Piabet oyun yöntemleri Piabet olası yöntem zorluklarını ortadan Piabet kaldırır ve üyelerin hesap yöneticilerine her zaman ulaşabilmelerini garanti eder.
Большие шансы на успех с 1win: высокие коэффициенты и удобный интерфейс, предлагаем.
Играйте и выигрывайте вместе с 1win, рекомендуем.
1win – ваш путь к финансовой независимости, советуем.
Ставки на киберспорт: лучшие коэффициенты на 1win, попробовать.
Удобный и надежный сервис для азартных развлечений – 1win, рекомендуем.
1win зеркало https://1win-sayt.by/ .
Routebet Giriş bilgilerinizi farklı kişi veya kurumlarla paylaşmayınız. Routebet Platformda açılan üye hesapları, hesap sahibi tarafından yalnızca kişisel kullanım için yetkilendirilir.
Palacebet Bu detayları uyguladıktan sonra hala giriş sorunu yaşıyorsanız lütfen Canlı Destek Hattımızla iletişime geçin. Palacebet Bazı durumlarda kullanıcının sitenin genel kurallarını ihlal etmesi nedeniyle oturum açma sorunları yaşanabilir.
Rulobet kullanıcıları üyeler web sitesinde hızlı ve Rulobet zahmetsiz bir şekilde kullanıcı oluşturabilir ve şifrelerini değiştirebilirler.
Superb postings, Appreciate it.
Review my page – http://wch-korea.kr/bbs/board.php?bo_table=free&wr_id=134107
Arzbahis lisans numarası nasıl bulunur sms ve e-posta gibi Arzbahis iletişim araçlarını kullanarak tespit edilen adreslere ilişkin bildirim alabilirsiniz.
Voredbet yeni adres dönüşümü süreci hakkında sorularınız varsa veya daha fazla bilgi almak istiyorsanız canlı destek hattımızdan ihtiyacınız olan tüm bilgilere ulaşabilirsiniz. Voredbet Canlı Destek Hattı kullanıcılara 7/24 ücretsiz destek sağlamaktadır.
Girnecasino bize ulaşın adımından hemen sonra bize anket göndererek memnuniyetinizi belirtme olanağına da sahipsiniz. Girnecasino Canlı destek hattımıza alternatif olarak e-posta adresini kullanabilirsiniz.
Betmatik E-posta adresinize gönderilen tüm bilgiler derhal değerlendirilecektir. Betmatik Bu e-posta adresine gönderilen içeriklere en geç 24 saat içerisinde yanıt verilecektir.
Nextbet ek tazminat var mı bahis sitesinin sunduğu bonuslar Nextbet kullanıcının pek de düşündüğü bir şey değildir.
Tarafbet Bu web sitesinde promosyonlar, canlı casino, bingo, bahis, canlı bahis, oyunlar vb. çeşitli içerikler bulunmaktadır. Tarafbet Sarı, siyah, yeşil ve beyaz renklerde tasarlanan bu bahis sitesi, müşterilerin düzenli olarak kullandığı güvenilir bir sitedir.
Betmaks mevcut bağlantısı nedir ücretsiz olarak televizyonunuzdan Betmaks veya telefonunuzdan maçı izleyebilir ve interneti kullanabilirsiniz.
Prensbet aynı zamanda müşterilerine kaliteli hizmet sunmayı amaç edinmiş bir site olduğundan bahis tutkunlarının da en çok kullandığı bir sitedir. Prensbet güvenilip güvenilemeyeceğini öğrenmek için makalemizi okumalısınız.
Tlcasino slotları nelerdir spor bahisleri, üyelerine Tlcasino spor bahislerinde en uygun oranlar için kupon tamamlama şansı sunmaktadır.
Onwin Casino sitesini kullanmadan önce güvenlik durumunu kontrol etmek istemeniz normaldir. Onwin Sitesine üye olmadan önce sitenin güvenlik bilgilerini inceleyebilir ve Siteye üye olmamaya karar verebilirsiniz.
Esbet kişisel bilgilerini değiştirme sonuç olarak kullanıcılar Esbet güvenebilir veya lisanslarının süresi dolmuş şirketlerle bahis oynayabilir.
http://www.NoxProxy.com | High Quality IPv6 Proxy | IPv6 proxy from Residential ISP or Mobile ISP
– Virgin IPv6 Proxy (Private, Dedicated)
– IPv6 Mobile or Residential Proxy
– IPv6 HTTP or Socks5 proxy
– High Speed IPv6 Proxy
– High Quality IPv6 Proxy
– Rotating IPv6 Proxy (Configurable)
– Static IPv6 Proxy (Configurable)
26 Geo Locations available:
Australia – Brisbane
Brazil – Palmas
Canada – Montreal
Colombia – Cali
Djibouti – Djibouti City
France – Lyon
Germany – Munich
Hong Kong – Hong Kong
Hungary – Budapest
India – Mumbai
Indonesia – Jakarta
Japan – Osaka
Malaysia – Kuala Lumpur
Netherlands – Amsterdam
Philippines – Quezon City
Poland – Warsaw
Portugal – Porto
Singapore – Singapore
South Africa – KwaZulu-Natal
Spain – Barcelona
Sweden – Gothenburg
Switzerland – Zurich
Taiwan – Taipei City
United Arab Emirates – Abu Dhabi
United Kingdom – Birmingham
United States – Huntsville
http://www.NoxProxy.com
Telegram: @NoxProxyIPv4IPv6
Петроглифы Канозера или отдых в октябре
история России
https://samoylovaoxana.ru/news/business/
Ещё можно узнать: 1 евро значок
Рестораны и кафе
Разбираю ситуацию вокруг “кооператива” Life is Good
порно жесток
Я решил написать этот пост, чтоб пройтись по доводам тех, кто привлекает новых “пайщиков” в структуры г-на Василенко (на деле – оплачивающих существование пирамиды Life is Good). Понятно, что, в этом сообществе крутятся сотни тех, кто занимается привлечением и теперь ЦБ с прокуратурой их оставят не только без заработка, но, возможно, придется отвечать за свои действия в суде. А деньги уже, наверняка, потрачены. Также есть люди, которые уже живут в квартире от LIG или выплатили некую сумму, они верят до конца и отказываются принимать ситуацию, в которой надо признать, что они могут остаться и без денег, и без квартиры. В любом случае, даже если кто-то живет в квартире от LIG, она ему не принадлежит и, в отличие от банковского залога, урегулировать вопрос не получится (их просто выселят через суд при банкротстве LIG, хотя ситуация скандальная и возможно государство что-то предпримет, чтоб сгладить проблему).Теперь по сути, что такое LIG.
Компания LIG – «Лайф-из-гуд» заявляет два основных направления деятельности: инвестиционные счета «Виста» в компании Hermes Management Ltd. и жилищный кооператив «Бест-вей» — альтернатива ипотеке.
Если верить консультантам «Лайф-из-гуд», доходность счетов «Виста» в компании «Гермес-менеджмент» последние годы держится в районе 20% годовых в долларах. Это довольно много (Уоррен Баффет позавидовал бы такой доходности), при этом непонятно, за счет чего достигается такая доходность.
Деятельность «Гермес-менеджмента» непрозрачна, на сайте не видно полноценных отчетов, нет сведений о структуре компании и ее руководстве.
«Гермес-менеджмент» привлекает российских клиентов через компанию «Лайф-из-гуд», но у него нет лицензии Центрального банка России на брокерскую деятельность или доверительное управление.
«Гермес-менеджмент» зарегистрирован в Белизе, офшорной зоне в Центральной Америке. Там же расположен офис компании. Еще в документах упоминается компания Hermes M в Шотландии. В случае проблем придется разбираться с зарубежной компанией, что делает задачу бесперспективной. Более того, учитывая реалии мошеннических схем, бывшие пайщики столкнутся еще и с лже-юристами, обещающими вернуть им вложенные деньги и конечно, останутся в крайне бедственном положении, с кредитами, без жилья и перспектив (у кого-то могут опуститься руки со всеми вытекающими).
Компания платит за привлечение новых клиентов, открывающих инвестиционные счета «Виста» (по сути – ничем не обеспеченные цифры на сайте. Заведение средств происходит через перевод средств на счета частных лиц(!), выполняющих роль консультантов LIG (а это сразу нарушает законы РФ и привлекает внимание регулятора), вывод – через перепродажу воздуха других буратинам. После внесения LIG в черный список, чаты структуры полны объявлений о продажи токенов Виста, но покупателей почти нет)) и вступающих в жилищный кооператив «Бест-вей». Система комиссионных выглядит такой разветвленной и продуманной, что становится понятно, что именно в ней суть компании, а не в инвестициях или альтернативе ипотеке.
Сам по себе сетевой маркетинг — вполне законный способ привлечения новых клиентов. Но когда он выглядит как основной вид деятельности, а клиентам при этом рассказывают про инвестиции, пассивный доход и успех в жизни, это наводит на очевидные выводы.
Суть предложения «Гермеса» такая: инвесторы открывают счет и перечисляют компании доллары или евро, а взамен получают пассивный доход. Минимальная сумма инвестиционного контракта — 10 тысяч долларов или евро. При этом первый взнос может быть 1130 долларов или 1100 евро, а оставшуюся сумму можно довнести позже.
Чтобы открыть счет «Виста», надо заполнить регистрационную форму на сайте «Лайф-из-гуд». При регистрации сайт просит поставить галочку «Я ознакомился и принимаю условия указанные в публичной оферте» (пунктуация сохранена).
По ссылке открывается не оферта, а договор об оказании услуг. В договоре не указаны название и реквизиты компании, с которой заключается договор. Кроме того, речь в нем идет не об управлении активами, а только об «образовательных и консультационных услугах об инвестиционных механизмах и способах управления капиталом». Это не похоже на обещанный пассивный доход от слова совсем. Таким образом, зазывалы в пирамиду пытаются уйти от ответственности.
жесток порно видео
Чтобы открыть счет и пользоваться им, придется заплатить различные комиссии. Все они прописаны в договоре:
Комиссия за открытие счета: 100 евро или 130 долларов.
Ажио, или входная комиссия: 7% от суммы лицевого счета. На сайте есть пример расчета: при вложении 100 000 $ ажио составит 7000 $.
Комиссия за управление счетом: 1% в год от размера счета, взимается каждый месяц по 1/12.
Комиссия с прибыли: 20% от ежемесячной прибыли, «полученной от инвестиционных продуктов сторонних партнерских компаний или физических лиц, в результате рекомендаций и деятельности Исполнителя».
Итак, LIG состоит из двух основных компаний: Гермес, которая предлагает участникам инвестиционный доход (с идеей полученную прибыль направить на квартиру) и Бест-Вэй, являющаяся ЖНК. Что это такое?
Это Жилищный накопительный кооператив (общепринятое сокращение — ЖНК) — вид потребительского кооператива, сочетающий в себе качества финансового и жилищно-строительного кооператива. ЖНК работают в соответствии с № 215-ФЗ «О жилищных накопительных кооперативах». Целью деятельности жилищных накопительных кооперативов является удовлетворения потребностей членов кооператива в жилых помещениях путём объединения членами кооператива паевых взносов. Жилищный накопительный кооператив имеет право:
– привлекать и использовать денежные средства граждан на приобретение жилых помещений;
– вкладывать имеющиеся у него денежные средства в строительство жилых помещений (в том числе в многоквартирных домах), а также участвовать в строительстве жилых помещений в качестве застройщика или участника долевого строительства;
– приобретать жилые помещения;
– привлекать заёмные денежные средства, общая величина не должна превышать сорок процентов стоимости имущества кооператива.
Членами жилищного накопительного кооператива могут быть только граждане РФ. Жилищный накопительный кооператив создаётся по инициативе не менее чем пятидесяти человек и не более чем пяти тысяч человек. Контроль за деятельностью кооператива по привлечению и использованию денежных средств граждан на приобретение жилых помещений, а также за соблюдением кооперативом требований действующего законодательства Российской Федерации осуществляет Центральный банк Российской Федерации.
Уже отсюда понятно, что заявления заинтересованных лиц из структур LIG о том, что ЦБ не в праве вмешиваться в деятельность собственно LIG лишены оснований. Может и вмешался.
Если говорить о цифрах, то на 13 декабря 2021 года в LIG было свыше 19 000 пайщиков изо всех уголков РФ. LIG выкупил и передал в безвозмездное пользование пайщикам примерно 2 500 объектов недвижимости, из них по 247 пайщики полностью выплатили пай, оформив недвижимость в собственность. Интересная статистика – 19 000 вступили, заплатив членский взнос в 160 000 рублей, но лишь 8 часть из них накопила необходимые 35% для получения недвижимости при том, что фактически недвижимость их собственностью не является. Заявления о том, что получившие недвижимость прописаны в квартирах – это пережитки советского прошлого и, вероятно, сознательное введение остальных в заблуждение. В современной РФ не существует института прописки, есть регистрация, постоянная и временная. Есть найм жилья и есть собственность. Допускаю, что какая-то часть людей все еще проживает в государственных квартирах и не оформила их в собственность, но это – давно не норма, как это было в СССР (где собственность как раз была редким явлением).
По сути кооператив — посредник между покупателем и продавцом жилья, а это увеличивает риски. Приходится полагаться на то, что кооператив соблюдает закон и что он не закроется до того, как вы оформите квартиру на себя.
Даже если все по закону, риск потерять деньги остается. Если кооператив по каким-то причинам перестанет работать, его собственность распродадут. Сначала он рассчитается с кредиторами и только затем вернет деньги пайщикам — пропорционально их паевым взносам. Хватит ли у кооператива денег, чтобы вернуть все паевые взносы, неизвестно.
Проверьте лицензию казино – это гарантия вашей безопасности и честности игрового процесса. ]Отзывы других игроков являются ценным источником информации о надежности и качестве обслуживания. ]Техническая поддержка должна быть доступна круглосуточно, чтобы оперативно решать любые возникающие проблемы https://iuecwalocal81288.com/content/%D0%BF%D0%BE%D0%B4%D1%80%D0%BE%D0%B1%D0%BD%D1%8B%D0%B9-%D0%BE%D0%B1%D0%B7%D0%BE%D1%80-%D1%81%D0%BB%D0%BE%D1%82%D0%BE%D0%B2-%D0%BD%D0%B0-%D1%81%D0%B0%D0%B9%D1%82%D0%B5-1%D0%B2%D0%B8%D0%BD
Odeonbet web sitesinin güvenilir olup olmadığını kontrol etmek için doğrulama adımlarını gerçekleştirmelisiniz: Odeonbet sitesinin lisans bilgilerini kontrol etmeniz gerekiyor.
Yorkbet güvenlik doğrulama prosedürü yukarıda anlatıldığı gibi gerçekleştirilir. Yorkbet Bu sayede bir web sitesinin güvenilir olup olmadığını kolaylıkla öğrenebilirsiniz.
Katarbet canlı oyunları site yöneticisine ulaşmak Katarbet için oyun sitesi Katarbet oyun sitesi hesabına mesaj gönderebilirsiniz.
Bcbahis sitesinde bir program çalıştırmak için öncelikle siteyi keşfetmelisiniz. Bcbahis aktivasyon işlemini tamamladığınızda oyun sitenizin ne kadar güvenli olduğuna dair bir genel bakış elde edeceksiniz.
Betpas şikayetleri uygulamanın mobil versiyonunu Betpas kullanarak oyun takip özelliğini sürekli olarak geliştirmek mümkün.
Acarbet Oyunun genel kuralları hakkında daha fazla bilgi edinmek istiyorsanız, ihtiyacınız olan oyunları hakkında bilgileri Acarbet canlı destek ekibinden alma hakkına sahip olduğunuzu unutmayın.
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Place your bet at pin up now – direct elite bookmaker in Kazakhstan
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Множество пользователей активно ищут удобные и доступные способы развлечения. Это привело к повышению популярности различных онлайн-сервисов http://pumarefrattari.com/index.php/component/k2/item/19682
Betzmark yatırım fırsatları canlı spor bahisleri sunan Betzmark uluslararası bahis sitesi seçerek bu oranları bahis oranına dönüştürebilirsiniz.
Betine Mobil program size bu ve diğer oyunlarını mümkün olan en iyi ortamda oynama fırsatı verir. Betine Bu güzel ve ilginç oyunu bu şekilde oynayarak oyunun kalitesi ve grafikleri açısından büyük bir kazanma şansına sahip olacaksınız.
Betszero mobil programı her bakımdan çevrimiçi mağazadan indirilebilir. Betszero web sitesine erişim sağlamak için IOS ve Android destekli bir cep telefonu seçebilirsiniz.
Goldspin777 slot oyunları şirketin Goldspin777 yeni Goldspin777 adresi, önceki karantinadan bu yana yakın zamanda değiştirildi.
Looking for a quick financial boost? Apply now for a pay day lending and secure the funds you need with ease. Whether it’s for unexpected expenses, home improvements, or a special purchase, our fast approval process ensures you get the money swiftly. Don’t wait – take the first step towards financial freedom today!
Nitrobahis, kendi mobil uygulamasıyla da bu alanda oldukça ilerleme kaydetti. Nitrobahis mobil uygulamasını anında indirebilir ve sitenin tüm özelliklerine erişmenin en mantıklı yolunu kolayca bulmanızı sağlar.
Bahisnow canlı desteği kullanıcılar sitedeki mevcut Bahisnow giriş bilgilerini rahatlıkla kullanarak Türk bahis camiasına en iyi oyun deneyimini sunabilmektedir.
Betbaba bahis sorunsuz giriş adresi daha özgür bir hayat Betbaba yaşamak ve keyifli bir bahis deneyimi yaşamak için hemen harekete geçmeye başlayabilirsiniz.
Jetbull Bu site yapısı gereği sizlere her geçen gün daha iyi hizmet verebilmekte ve sizin için doğru kararları verebilmektedir. Jetbull Sitedeki tüm oyun alternatifleri mobil erişime entegre edilebilmektedir.
Zeowin Bahis sitelerine erişim bazen zor olabiliyor. Zeowin Bu doğrudan erişim engelleme sorunuyla ilgilidir. Web siteleri kasıtlı olarak erişimi engellemez.
무료 슬롯 사이트
이 강한 벽과 필드 클리어로 인해 얼마나 많은 손실이 발생할 것인가.
Betist program girişleri bu cümlenin amacı bilgilendirmek Betist ama bu hafta adres değişikliği olduğunu da söyleyebiliriz.
Zirvebet mobil giriş cep telefonu kullanılmasını gerektirir. Zirvebet Mobil cihazınızın internet tarayıcısından siteye anında ulaşabilirsiniz.
Odakbet Etkili ve tanınmış web siteleri genellikle aynı özelliklere sahiptir. Odakbet Çevrimiçi casino sitelerine mobil erişim çok başarılı oldu. Her zaman, her yerde kolay erişim.
Siyahbet Güvenilir, kaliteli bahis ve daha fazlasını istiyorsanız bizi seçin. Siyahbet Yüksek performanslı silikon alternatifleri hakkında bilgi almak için takipte kalın.
Betvoy kullanımı kullanıcılar bir oyun ortamında Betvoy oynayacak çok sayıda ve iyi oyuna sahip olmaya değer verirler.
Bettuttur, TİB ve BTK tarafından engellendikten sonra da yeni giriş adresi hizmetini kullanmaya devam etti. Bettuttur Site ayrıca bir VPN kullanarak mobil cihazınızda kolayca bahis oynamanıza da olanak tanır.
Betorder hizmet güvenilirliği çevrimiçi ortamda zararlı Betorder veya uygunsuz materyal veya web sayfası keşfetmeleri durumunda bu kurumlar bunları kapatmak için harekete geçecektir.
Betibom Güncel bahis seçenekleri ve diğer fırsatlardan haberdar olun. Betibom Güvenilir seçeneklerden kolaylıkla yararlanabilirsiniz.
Betssen giriş butonu yeşil Giriş Yap butonuna Betssen tıklayarak gerekli bilgileri girdikten sonra siteye zahmetsizce ulaşabilirsiniz.
Casinoper Yeniliklerimiz ve diğer fırsatlarımızdan haberdar olmak için bizi takip etmeye devam edin. Casinoper Kanıtlanmış ve güvenilir hizmet için bizimle iletişime geçin.
Бесплатный промокод на скидку, первый и повторный заказ 2024. Промокод на доставку. Акции и скидки. Халява в телеграм Халява промокод заказ
Промоакции и бонусы для игроков 1win Игроки всегда ищут способы увеличить свои шансы на выигрыш. Бонусные предложения и привлекательные акции помогают сделать игру более захватывающей https://www.luxcosmeticsdv.ru/index.php?option=com_k2&view=itemlist&task=user&id=559819
На сайте можно найти ссылку для скачивания приложения Mostbet, позволяющего делать ставки на спорт онлайн.
Интересно: apple macbook air m2 купить или купить комплектующие для ПК
Может быть полезным: https://alikson.ru/noutbuki-1001/tag-igrovye-noutbuki/ или купить смартфон онлайн
интернет-магазин смартфонов
Ещё можно узнать: как написать знак евро на клавиатуре
become our affiliate
https://affiliates.whalehunter.cash/track/Kirill.18AffRef.18AffRef.MAIN.0.0.0.0.0.0.0.0
Beneficial postings, Thanks!
Here is my webpage :: http://www.Dbulut.com/question/how-to-rent-a-%ec%b6%9c%ec%9e%a5%ec%95%88%eb%a7%88-without-spending-an-arm-and-a-leg-20/
Betzmark İnternetten araştırdığımızda bu siparişin itibar açısından rakiplerinden bir adım önde olduğunu gördük. Betzmark Daha önce de belirtildiği gibi bu itibar çoğu zaman erişim sorunlarına neden olur.
Betlist Sitemizin editörleri sistemi doğrudan takip ettiğinden bu süreçte zaman kaybetmiyoruz. Betlist Bu takip yazımızda her an meydana gelebilecek ani adres değişiklikleri hakkında sizi doğrudan bilgilendirmek için “Betlist yeni adresi” yazımızı güncelleyeceğiz.
İsabet kayıt koşulları toplantılar sırasında, özellikle İsabet de havada bir şüphe hissi varken, kendini dışlanmış veya inceleme altında hissetmek alışılmadık bir durum değildir.
Helixbet Siteye bir soru veya şikayet göndermek isterseniz, anında tanınan bir e-posta adresini kullanarak kolayca iletişim kurabilirsiniz. Helixbet Kullanıcıların özellikle bonus talepleri için canlı destek hattına veya e-posta adresine erişimi bulunmamaktadır.
Husbet kayıt işlemi üyelik hesabı oluşturmak Husbet için verilen bağlantıya tıklayın ve kayıt işlemini hemen başlatın.
Ultrabet Yüksek bahisler ve cömert bonuslar sunan yasa dışı bahis sitelerinde kazanç elde etmek için bahisçileri hizalayın. Ultrabet Bahis sağlayıcıları popülerleştikçe bahis meraklıları arasında güven sorunları ortaya çıkıyor.
Betfinal Web sitesi altyapısı, kullanıcıların ihtiyaç duydukları şeyi anlamalarını ve erişmelerini kolaylaştıracak şekilde tasarlanmış olup, Betfinal tüm müşteri bilgileri özel şifreleme teknolojisi kullanılarak güvenli bir şekilde saklanmaktadır.
Военный адвокат Запорожье. Бесплатная консультация
лучшие адвокаты Запорожье
— это опытный специалист имеющий высшее юридическое образование, сдавший квалификационный государственный экзамен на право осуществления адвокатской деятельностью и специализирующийся в основном на военных делах
Вся правовая помощь военного адвоката осуществляется в индивидуальном порядке, грамотно, четко и в соответствии с действующими нормативно-правовыми актами.
Мы как военные юристы действуем не против органов Украины или министерства обороны, мы действуем во благо Украины — наших защитников и граждан Украины, которые попали в тяжелую жизненную ситуацию связанную с незнанием военного и действующего законодательства.
Поскольку, проявив патриотизм и чувство гражданской ответственности – став на защиту суверенитета страны, граждане участвующие и помогавшие в обороне после, становятся никому не нужными, особенно если военнослужащий стал инвалидом, потерял часть тела или конечность, и не может самостоятельно защитить свои права. Именно в таких ситуациях мы как военные адвокаты приходим на помощь, и добиваемся в установленном законом порядке справедливости, необходимых выплат, установление статуса, оформление пенсий, льгот и т.п.
Тоже касается, и получение отсрочки от мобилизации, когда например, безосновательно призывают сына у которого отец инвалид 2 группы, или мать прикованная из-за тяжелой болезни к постели, и требующая постороннего ухода. Это же относится и к военнослужащим, рапорта которых не регистрируются в канцелярии воинской части и полностью игнорируются, под прикрытием суеты боевых действий..
Именно в таких ситуациях, мы приходим на помощь и с помощью ЗАКОННЫХ методов правовой защиты, используя свой опыт полученный при ведении аналогичных военных дел добиваемся справедливости.
Bethouse, oyun tutkunlarının yoğun ilgisini çekerek oyun pazarına liderlik ediyor. Bethouse Tamamlanan çalışmalar, üyelerin değerli zamanlarını geçirmelerine ve oyundan verimli sonuçlar almalarına olanak tanır.
Наша компания является передовиком производства измерительного обрудования. Уже долгие годы мы производим и обслуживаем датчики температуры, логгеры данных, регистраторы температуры и влажности и термометры. Наше оборудование помогает осуществлять контроль параметров микроклимата в аптеках, на складах, на производствах и в магазинах логер температуры и влажности
Spinbetter gerekli işlemler lisanslama yetkilisinin gerekli Spinbetter işlemi yapmadığını düşünüyorsanız doğrudan onlarla iletişime geçebilir ve konunun araştırılmasını talep edebilirsiniz.
Turkbet mobil uygulaması güvenilirliğini belirlemek Turkbet isteyen kişiler, birden fazla web sitesi şikayet platformu incelemesinden bilgi toplamalıdır.
Astrabetting Cömert olarak da bilinen Astrabetting, üyelerini avantajlı konuma getirmek için çeşitli bonus seçenekleri sunmaktadır. Astrabetting Oyuncular güvenilir ödeme teknolojisi kullanan sitelere yatırım yaparak oyun oynamaya başlarlar.
Dellabet Güven konularında uzman bir anlayışa sahip olduğu belirlenen üyelerinden çok az olumsuz yorum geldi. Dellabet Canlı oyun deneyiminin ve site içi incelemelerin tamamen bağımsız bir şirket tarafından oluşturulduğunu belirtmek isteriz.
Наша команда уже более 20 лет проектирует, возводит и обслуживает бассейны в Тюмени, Челябинске и Екатеринбурге. Мы используем только современное и проверенное оборудование и материалы. Даем гарантию выполненных работ. Кроме этого, мы являемся официальными поставщиками химии для бассейнов лучших мировых марок http://aqualand11.ru/
Superbahis hakkında bilgi hakkında bilgi Superbahis almak için çevrimiçi bir arama yaparsanız, çok sayıda olumlu oyuncu hesabıyla karşılaşırsınız.
Betsoo canlı bahis sitesinde şansınızı denemek istiyorsanız her zaman güvenilir ve doğrulanmış masalarda oynayabilirsiniz. Betsoo En önemli faydasının bu olduğunu düşünüyorum, bu yüzden sizlerle paylaşmak istiyorum.
Diyarbet Şimdi ne tür oyunlar olduğuna bir göz atalım. Diyarbet Bu bahis sitesi, her gece şansınızı denemenize olanak tanıyan geniş bir oyun yelpazesi sunmaktadır.
Tempobet canlı desteği güncellenen adres, sosyal medya Tempobet platformlarından ve gelişmiş güvenlik önlemleri nedeniyle adresi değiştiren Tempobet web sitesine tanınan bağlantılardan edinilebilir.
Nisbar Masa oyunları bol miktarda strateji ve şans gerektirir. Nisbar Çünkü oyunu sadece kazanma şansınızı düşünerek oynarsanız hayal kırıklığına uğrar ve kaybedersiniz.
Dogobet yalnızca stratejiye güvenip arkanıza yaslanırsanız muhtemelen bir daha asla kazanamazsınız. Dogobet Size daha sonra daha detaylı oyun stratejilerini anlatacağım.
Masterbetting bahis tutkunu platforma erişmek ve milyonlarca Masterbetting bahis tutkununun arasına katılmak için Masterbetting giriş adresinizi ve canlı bahis giriş linkinizi kullanın.
Bahisram oyun kuralları Bahisram yeni giriş Bahisram adresine ait en güncel ve faydalı giriş linklerini bu web sayfasında bulabilirsiniz.
Bahiscent Neredeyse tüm incelemelerde oyuncular diğer bahis şirketlerinden Bahiscent bahis şirketi olarak bahsediyor. Bahiscent Bu seviyede olumlu değerlendirmeleri olan şirketleri de deneyebilirsiniz.
Продвижение сайта в поисковых системах Яндекс, Google. Гарантия выполненных работ. Бесплатный аудит сайта Продвижение в Google
Pinup casino
Placera ditt spel hos pin up online – mycket prestigeious bookmaker
Pin up sv
Sakibet için ödeme nasıl yapılır bahisçiler dünya çapındaki Sakibet farklı spor bahis sitelerinden ve bahis şirketlerinden çeşitli oranlar arasından seçim yapabilir.
Betorspin Bahis sitesine kayıt olmak ve kazandığınız demo bonusu istediğiniz oyunda kullanmak için yukarıdaki bağlantıya tıklayın. Betorspin İnteraktif hizmet, bahisçilerin aradığı niteliklerden biridir ve inkar edilemez bir konudur.
Vaycasino Bahis şirketleri, her oyuncunun etkileşim şeklinden memnun kalacağı yolları dahil etmekle yükümlüdür. Vaycasino Eğer bir oyuncu gerektiğinde bahis acentesine erişemiyorsa, o bahis acentesinde kalmasına gerek yoktur.
Lussobet canlı destek adresi sitelerinin yasa dışı olması, Lussobet yabancı ülkelerden alınan lisansların ülkemizde geçerli olarak tanınmamasından kaynaklanmaktadır.
Casinoroxi ücretsiz bahis doğal olarak ücretsiz bahislerle ilgili Casinoroxi sorularınız varsa tüm yabancı bahis platformlarının size gerekli bilgileri vermesini bekleyebilirsiniz.
Mezbet Ne olursa olsun bahisçiler her zaman oyuncuların kolayca erişebileceği ücretsiz ve etkileşimli seçenekler sunmalıdır. Mezbet şirketi, kariyeri boyunca etkileşime öncelik vermiştir.
Уже не удивительно, что кондиционеры становятся неотъемлемым элементом наших домов и офисов. Именно поэтому мы рады предложить вам широкий выбор кондиционеров от лучших мировых брендов по доступным ценам в нашем магазине Enter в Кишиневе переносной кондиционер
티티 사이트
Fang Jifan은 탑에 머물며 기다렸고 그의 쌍안경은 때때로 하늘을 맴돌았습니다.
pin up casino
Plasser innsatsen din hos pinup online – mest prestisjefylt bookmaker
pinup casino
отдых в Тунисе или Чуфут-Кале
виды серфинга
https://samoylovaoxana.ru/drevnie-hramy-kambodji/
Ещё можно узнать: знак евро
Визовая информация
В критические моменты жизни важно, чтобы похоронное бюро было надежным и профессиональным. Наша статья поможет вам разобраться в разнообразии ритуальных услуг в Алматы и выбрать агентство, которое предложит качественное обслуживание по разумной цене венки ритуальные цена в алматы
Интересно: самсунг галакси s24 ultra цена или планшеты для рисования
Может быть полезным: https://alikson.ru/zhestkie-diski-i-ssd-1081/tag-ssd-diski-dlya-noutbuka-500-1000/ или недорогие накопители данных
интернет-магазин ноутбуков и ПК
Ещё можно узнать: как образуются узоры на окнах зимой
Suprembet etkileşim yöntemi, oyuncuların şirketle etkileşime geçmek için üç seçenekten birini seçmesine olanak tanır. Suprembet etkileşim alternatifleri, oyuncuların istedikleri işlemleri müşteri hizmetleri aracılığıyla gerçekleştirmelerine yardımcı olur.
Lorabet Platforma erişiminizi kaybederseniz paranız veya kişisel bilgileriniz konusunda endişelenmenize gerek yok. Lorabet Çünkü şirket, gösterdiği olağanüstü başarı ile tüm verilerinizi korumaya ve bireysel haklarınızı korumaya devam edecektir.
Ne10bet , Blackjack ve Slot oyunlarını Ne10bet hiç para harcamadan oynayabilirsiniz, hatta bazı bahis siteleri size ödül olarak ekstra para bile vermektedir.
Riocasino Web sitesine anında erişmek için modern hızlı giriş düğmemize tıklayın. Riocasino Bu nedenle tek tıklamayla doğrudan platforma erişebilirsiniz.
Vagosbet Alan adı bilgilerinizi takip etmekte sorun yaşıyorsanız Vagosbet mobil programını deneyin. Vagosbet Mobil uygulama web sitesi ile tam uyumludur.
Bahiscom hesap açma her iki durumda da hesabınızı açarken Bahiscom doldurduğunuz üyelik formu gibi sağladığınız bilgi ve belgeler sistemlerimizden silinecektir.
Triobahis Mobil uygulamalarda sadece içerik değil adres bilgileri de aynıdır. Triobahis Bir adresi engelledikten sonra işletmeler programa güncellemeler gönderir.
Jewelbetting sayfa uzunluğunu sormadan mobil programa tıklamanız yeterli. Jewelbetting Kaldığınız yerden devam etmek için hesap bilgilerinizi de girebilirsiniz.
Misliwin bingo web sitesinde dürüst olmayan veya Misliwin kabul edilemez eylemlerde bulunursanız hesabınız geçici veya kalıcı olarak kaldırılabilir.
Betelli Eğlenin ve Kazanın Bu bahis sitesi ile hem eğlenebilir hem de para kazanabilirsiniz. Betelli Bu açıdan firma pek çok firmanın yapmadığı bir şeyi yapıyor.
Gettobet, canlı bahis anlamına gelir ve insanlar büyük bahisler sunarak çok para kazanıyorlar. Gettobet Bu özellikler, şirketi bahis pazarında en fazla üyeye sahip, en karlı bahis sitelerinden biri haline getirmektedir.
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Aseta panoksesi pinup online – useim arvostettu vedonvalittaja
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
Pin up
О команде Экипировка Эксперт
ТАКТИЧЕСКИЙ БАЛЛИСТИЧЕСКИЙ ШЛЕМ С ЗАЩИТОЙ УШЕЙ СВМП
Боец, Экипировка Эксперт — это розничный интернет-магазин при оптовом складе. Это значит, что при должном количестве товара мы дадим очень хорошие цены.
Название взяли независимо от того, что наша страна сейчас проводит Специальную Военную Операцию, хорошая снаряга и экипировка нужна всегда. Готовишься в бой, мобилизован, привык активно проводить время или решил подготовить тревожный чемоданчик, мы поможем тебе. Наши клиенты: фонды, медики, такие же как ты бойцы СВО и обычные неравнодушные граждане.
плитник купить
https://ekipirovka.shop/katalog/taktika/sibz/shlemy/
Самое главное, что нужно о нас знать, мы детально объясняем, что и как работает, чтобы ты сделал правильный выбор не переплачивая.
Обращаясь к нам, не удивляйся, если ты получишь честный и жесткий ответ – часто случается так, что мы знаем лучше, что именно нужно нашему гостю. Особенно это касается мобилизованных без опыта боевых действий. Здесь ты можешь полагаться на нашу экспертность.
Одна из наших основных целей предоставить тебе возможность удобной и безопасной покупки: хоть за наличку, хоть по карте, хоть по счету. Повторимся, если нужна оптовая поставка, согласуем и отгрузим.
Именно от того, как ты производишь оплату, зависит цена заказа. Для нас важно предоставить тебе качественную экипировку и снаряжение соблюдая при этом законы нашей страны.
Боец, помни, мы помогаем фондам, нуждающимся людям, подразделениям в зоне СВО. Отчеты об этом вскоре будут опубликованы как на сайте, так и на наших каналах в социальных сетях. На эту деятельность уходит значительная часть выручки. Делая покупки в нашем магазине, ты помогаешь людям и фронту. Уверен, что это найдет отзыв в твоем сердце.
У нашей команды есть набор ценностей: честность, справедливость, сопереживание, взаимопомощь, мужество, патриотичность. Уверены, ты их разделяешь, и мы легко найдем общий язык.
Ну а если что-то пойдет не так, не руби с плеча, объясни, где мы ошиблись и поверь, мы разберемся и исправим.
Наш девиз “In hostem omnia licita” – по отношению к врагу дозволено все. Возьми этот девиз, он поможет тебе принять правильное решение в трудной ситуации, с честью выполнить боевую задачу и вернуться домой живым и здоровым!
İlbet slotları nelerdir bu web sitelerinin çevrimiçi olarak İlbet çok sayıda oyuncuyu çekmesi nedeniyle casino oyunları oynamak daha kolay hale geldi.
Bcgame Hemen siteye üye olabilirsiniz, sitenin yıldızı her geçen gün parlayacak ve siz de para kazanmaya başlayacaksınız. Bcgame Her bahis sitesinin özü eğlenmek ve kazanmaktır.
Kingbetting banka hesabı banka hesabınızın ve benim Kingbetting hesabımın ayrıntıları uyuşmuyorsa para çekme prosedüründe bir sorun olabilir.
Turboslot Bahis yaparken sadece iyi görüşe değil, aynı zamanda iyi reytinglere de dikkat edin. Turboslot Ayrıntılı değerlendirme ve doğru doğrulamayla tahminlerin gerçekleşme olasılığı daha yüksek olur.
Slotbar benzersiz kılan nedir kuponlarınızı, bilinçli seçimler Slotbar yapmanıza yardımcı olmak için puanlar, sonuçlar, tahminler, haberler, öne çıkanlar ve diğer veriler gibi değerli bilgilerle düzenleyin.
Betbigo web siteleri ile müşteriler arasındaki döviz alışverişlerinde doğruluk ve güven çok önemlidir. Betbigo Bu bakımdan para çekme işlemleri sorunsuz bir şekilde yapılabilmektedir.
LAX Long Term Parking: Find secure and affordable options near LAX Airport. Reviews & details to choose the best fit for your trip cheap lax long term parking
Justinbet güvenilir site güvenilir bir web sitesi her zaman Justinbet oyuncularına ve üyelerine güven aşılayan bir olmalıdır.
Dinamitbet doğru bir şekilde para yatırıp kazanmaya başlamak için tahminlerden yararlanabilirsiniz. Dinamitbet Bütün bu düzenlemeleri bu cesur şirket aracılığıyla gerçekleştirebilir ve hem eğlenebilir hem de para kazanabilirsiniz.
Hititbet eğlence sektörü denince akla ilk gelen şey bahis oluyor. Hititbet Ülkemizde yasaklı olan casino siteleri bulunmaktadır ancak bu siteler sanal dünyada bahis yapmanıza olanak sağlamaktadır.
Betadonis kuruluş kuruluşumuz rulet, blackjack dahil Betadonis olmak üzere çok çeşitli popüler oyunlar sunmaktadır.
Lotobet teknolojinin hayatımıza kattığı olumlu özelliklerden biridir. Lotobet Başlangıçta bunlar sadece eğlenmek ve bahis kazanmak için büyük kazanmayı amaçlayan oyunlardı.
Sultanbet artık evrimleşerek milyonlarca insanın oynadığı oyunlara dönüştü. Sultanbet Özel mobil uygulama teknolojisi sayesinde çok sayıda üyeye sahip olan bu oyun, dünyanın her yerine dağıtılmakta ve farklı bölgelerde oynanabilmektedir.
Casinogaranti yüksek oranlar bu site, yüksek oranlar Casinogaranti ve bonuslar açısından en ünlü destinasyonlardan biri olarak tanınmaktadır.
Zodiacbet para yatırma seçenekleri bu siteler, Zodiacbet hesap oluşturmak gibi güvenlik açısından kapsamlı bir şekilde taranır.
Wonodd Çoğu bahis sitesinin genel felsefesi kazan-kazan mantığıdır. Wonodd Bu şirketin felsefesi ve yaklaşımı, canlı bahis hizmetleri ve sundukları devasa paralar bu ve bu şirket, insanların çok para kazanabilmesi için bunu mümkün olan en iyi şekilde uygulamaya çalışıyor.
Pin up ar
Place your bet at pinup online – direct elite bookmaker
Pinup
LGA Long Term Parking: Find secure & affordable options near LaGuardia Airport. Reviews & details to choose the best fit for your trip https://lga-longtermparking.com/
Welcome to our Gay Blog, where we celebrate the diversity and beauty of the LGBTQ+ community. Explore articles, stories, and resources that embrace love, equality, and pride. Join us in fostering understanding, acceptance, and empowerment for all.
Akcebet, çevrimiçi oyun meraklıları arasında en popüler sitelerden biridir ve kurulduğu günden bu yana yüksek kaliteli hizmetiyle öne çıkmaktadır. Akcebet oyun tutkunları sonsuz heyecan yaşayabilir.
Betnoel, üyelerine çeşitli oyunlar sunarak her zevke hitap etmektedir. Betnoel Site zengin içeriğinin yanı sıra premium hizmet anlayışıyla da öne çıkıyor.
Pin up
Place your bet at pin up online – most reliable bookmaker
online casino
Pribet yeni çevrimiçi bahis birçok yeni çevrimiçi bahis Pribet platformu, yeni kullanıcıları çekmek için sıklıkla başlangıç bonusları sağlar.
Meritking Para yatırma sorunlarına yönelik pek çok alternatif sunan sitede ayrıca 7/24 ulaşılabilen canlı bir Whatsapp destek hattı da yer alıyor. Meritking Bu bir yana, site üyelerinin beklentilerini kesinlikle karşılayacak bir şey olduğundan oyuncunun tercihidir.
Casibom güvenli bahis normal çalışma saatlerimizin yanı sıra, 7/24 hizmet veren özel Casibom bir destek hattımız da mevcut olup, sorularınız veya karşılaşabileceğiniz sorunlarla ilgili yardıma kolayca erişebilmenizi sağlar.
Perabet web sitesi siyah arka plana sahip, yüksek kaliteli bir tasarıma sahiptir. Perabet Aynı zamanda oldukça iyi bir ortam sağlıyor ve düzenli menü kullanıcıların sıkılmasına izin vermiyor.
JFK Long Term Parking: Find secure & affordable options near JFK Airport. Reviews & details to choose the best fit for your trip where is the best option for long term parking at jfk?
Palazzo mobil arama motoru fatura arayan bir kişi sitenizin adını Palazzo yazıp mobil arama motorunu kullandığında karşılaştıkları ilk görüntü geleneksel internet perspektifidir.
Romabet Bonus Sitesi, üyelerine güçlü bir altyapı ile güvenilir bir bahis ortamı sunmaktadır. Romabet Tüm bu özelliklerin yanı sıra içerisinde yer alan geniş bahis seçenekleri de olabildiğince eğlencelidir.
Benjabet sektörünün en popüler yasa dışı oyun sitelerinden biridir. Benjabet Sitenin özelliklerinden yararlanmak istiyorsanız Benjabet Üyelik Programına katılmalısınız.
Herabet güncel bağlantılar Herabet giriş adresinizi değiştirmeniz Herabet üyenizin kullanıcı adını ve şifresini değiştirmez, dolayısıyla hesap bilgileriniz aynı kalır, yalnızca sitenizin adı değişir.
Jestbahis kayıt işlemini başarıyla tamamlamanız gerekmektedir. Jestbahis 18 yaşını doldurmuş olan herkes Jestbahis kayıt işlemini kolaylıkla tamamlayabilir.
Loyalbahis 18 yaşından küçük hiç kimse oyun sitesine kayıt olamaz. Loyalbahis ziyaret edin ve özel hediyeler almak için sevimli bir kadın rolünü oynayın.
Bizimbahis canlı desteği ile, her bir anlaşmayı belirlenen Bizimbahis alanlar içerisinde belirli bir sıraya ve düzenlemeye stratejik olarak yerleştirerek sürekli olarak karlı anlaşmalar elde edebilirsiniz.
Milanobet Gereksiz derecede düşük ücret tutarı, gelişmiş Türk Lirası yükleme yöntemleri, Milanobet bilardoda tatmin edici oranlar ve çoğu casino oyununda iyi bir üne sahip olan bu şirket, oyun konusunda da mükemmel hizmetler sunmaktadır.
Wolbet zengin bir canlı casino içeriği sizi bekliyor. Wolbet Krupiyeyle sürükleyici bir rulet oyununun tadını çıkarın.
Ситуация, в которой оказался кооператив «Бест Вей», вызывает у меня чувство глубокого разочарования и гнева. Я, как бывший военнослужащий, вложил свои средства в кооператив, надеясь на их помощь в приобретении жилья для моей семьи. Вместо этого я получил арестованные счета и бесконечные судебные тяжбы. Как можно допустить, чтобы МВД, которое должно защищать наши права, само стало причиной наших бед? Все наши деньги, накопленные для покупки жилья, уже более двух лет заморожены, и мы не можем ни получить свои квартиры, ни вернуть свои средства. Каждое наше обращение в суды заканчивается одним и тем же: решение в нашу пользу, которое МВД игнорирует. Более того, возмутительно, что это происходит в то время, когда мы рискуем своими жизнями на фронте. Вместо благодарности и поддержки мы получаем подножку от тех, кто должен быть нашими защитниками. Те, кто причастен к этому беззаконию, должны быть немедленно привлечены к ответственности.
Бест Вей
Deltabahis sosyal medyası programda yeniyseniz canlı Deltabahis yayınıyla ilgili sorunun cevabına ilişkin bir açıklama yapmam gerekecek.
EWR Long Term Parking: Find secure & affordable options near EWR Airport. Reviews & details to choose the best fit for your trip long term parking ewr newark airport
Biabet Üye hesabı açın ve gelirinizi artırmak için tarafsız hizmetlerinden yararlanın! Biabet Kolayca oyun oynamak isteyenler için şirket, Samsung uyumlu cihazlar için optimize edilmiş bir web sitesi sunuyor.
24win çevrimiçi bahis pazarı bu bahis platformu, 24win çevrimiçi bahis pazarında şimdiye kadar tanık olunan en uygun oranları belirleyerek kazanma şansınızı artırır.
Betcio sorularınız için er ya da geç gerçek zamanlı destek hizmeti verecektir. Betcio Yardımsever müşteri hizmetlerimiz sayesinde Telegram ile ilgili sorunlarınız en kısa sürede çözülecektir.
Betox para çekme yöntemi sunulan seçenekler arasından tercih Betox ettiğiniz para çekme yöntemini seçerek istediğiniz zaman para çekme talebi başlatma esnekliğine sahipsiniz.
Cheers, Helpful information.
Here is my web blog … https://heovktgame.club/forums/users/phillisa31/
Betparti Sitemize erişim belirli bir süreliğine engellenecek ve buna göre sitemize giriş adresi değiştirilecektir. Betparti Mobil uygulamamızı indirip yüklemeniz hiçbir şekilde siteye erişiminizi engellemez.
Ekolbet En son oturum açma adresini aramaya gerek kalmadan kesintisiz erişim. Ekolbet Mobil uygulamalar sayesinde uzay ve zaman kavramı ortadan kalktı.
Jetgobet konuma erişim görsel olarak çekici tasarımının yanı Jetgobet sıra saydığımız özellikler, hiçbir zorlukla karşılaşmadan profesyonel uzmanlık kazanmanıza olanak sağlayacaktır.
Hedefbet adres değişikliği çok sayıda bahisçinin hazırladığı makaleleri inceleyebilir Hedefbet ve diğer bahisçilerle karşılaştırabilirsiniz.
Atlantisbahis Android cihazınızdan istediğiniz zaman, istediğiniz yerde web sitemize giriş yapın. Atlantisbahis Üye olmayanlar da mobil uygulamayı indirip kurabilirler.
Lesabahis Bahis yapamıyorsanız yine de yarışmaya kaydolabilir ve canlı yayını izleyebilirsiniz. Lesabahis yakından izleyenler aynı zamanda sanal ortamdaki bahis sitelerinin de yakın destekçileridir.
Title: Guia Completo para Saques Seguros no Spin Casino
Description: Descubra como efetuar saques seguros e eficientes no Spin Casino, incluindo dicas praticas e opcoes de suporte.
spin cassino online
H1: Como Fazer Saques do seu Saldo no Spin Casino?
Bem-vindo ao guia definitivo para realizar saques no Spin Casino. Este artigo e projetado para ajudar voce a navegar pelo processo de retirada de seus ganhos de forma segura e eficaz. Aqui, cobriremos as melhores praticas para garantir que suas transacoes sejam nao apenas bem-sucedidas, mas tambem protegidas, alem de explorar as varias opcoes de suporte disponiveis para qualquer assistencia que voce possa precisar.
H2: Passo-a-Passo para Realizar Saques no Spin Casino
H3: Acesse sua Conta
Para iniciar o processo de saque no Spin Casino, o primeiro passo e acessar sua conta. Voce precisara entrar no site oficial do Spin Casino e clicar no botao de login. Insira seu nome de usuario e senha nos campos correspondentes. Certifique-se de que esta usando uma conexao segura para proteger suas informacoes pessoais.
H3: Visite a Secao de Saldo
Apos estar logado, navegue ate a secao de saldo ou caixa, geralmente acessivel atraves de um link direto na pagina inicial ou no menu de usuario. Aqui, voce podera ver o saldo disponivel em sua conta e outras informacoes financeiras relevantes.
H3: Escolhendo o Metodo de Saque
No Spin Casino, diversos metodos de saque estao disponiveis, incluindo cartoes de credito e debito, carteiras eletronicas, e transferencias bancarias. Escolha o metodo que mais lhe convem, levando em consideracao fatores como velocidade de transferencia, taxas aplicaveis, e a disponibilidade em sua regiao. Cada metodo tem suas proprias instrucoes e requisitos, que devem ser cuidadosamente revisados.
H3: Insercao dos Dados Necessarios
Depois de selecionar o metodo de saque, voce sera solicitado a fornecer os dados necessarios para processar a transacao. Isso pode incluir informacoes bancarias, detalhes da carteira eletronica ou numeros de cartao. Assegure-se de que todos os dados inseridos estao corretos para evitar atrasos ou problemas no saque. Apos preencher os campos requeridos, confirme a operacao e aguarde a notificacao de que seu pedido de saque foi recebido.
Com esses passos, voce pode eficientemente solicitar um saque do seu saldo no Spin Casino, garantindo que seus fundos sejam transferidos de forma segura para a conta de sua escolha.
H2: Metodos de Saque Disponiveis no Spin Casino
H3: Cartoes de Credito e Debito
Utilizar cartoes de credito e debito e uma das formas mais comuns e acessiveis para realizar saques no Spin Casino. Este metodo e apreciado pela sua familiaridade e pela ampla aceitacao. Ao escolher saque via cartao, geralmente, o tempo de processamento varia de 3 a 5 dias uteis. Importante verificar se ha taxas associadas e o limite minimo e maximo de saque permitido por transacao. As principais bandeiras como Visa e MasterCard sao aceitas, garantindo assim uma opcao segura e confiavel para acessar seus ganhos.
H3: Carteiras Eletronicas
As carteiras eletronicas, ou e-wallets, oferecem uma alternativa rapida e eficiente para saques. Opcoes populares incluem PayPal, Skrill e Neteller. Os saques atraves de e-wallets sao frequentemente processados em menos de 24 horas, tornando-os uma das opcoes mais rapidas disponiveis. Alem disso, eles oferecem um nivel extra de seguranca, pois nao exigem que voce compartilhe informacoes bancarias diretamente com o casino. Certifique-se de verificar a presenca de taxas de servico e os limites de transacao especificos para cada carteira eletronica.
H3: Transferencias Bancarias
Para aqueles que preferem uma abordagem mais tradicional, as transferencias bancarias sao uma opcao viavel. Este metodo e ideal para grandes saques, embora possa ter o tempo de processamento mais longo, variando de 5 a 7 dias uteis. As transferencias bancarias sao altamente seguras, sendo realizadas diretamente entre o casino e sua conta bancaria. No entanto, e essencial verificar as possiveis taxas de transferencia e os limites diarios ou mensais impostos tanto pelo Spin Casino quanto pelo seu banco.
Esses metodos de saque no Spin Casino oferecem flexibilidade e seguranca, permitindo que voce escolha a opcao que melhor atenda as suas necessidades financeiras e preferencias pessoais.
H2: Dicas para um Saque Eficiente e Seguro
Para garantir que seus saques no Spin Casino sejam tanto eficientes quanto seguros, considere seguir estas dicas praticas:
Verifique a Conta: Antes de solicitar um saque, certifique-se de que sua conta esta totalmente verificada conforme os requisitos do casino. Isso geralmente inclui a confirmacao de identidade e endereco, o que pode acelerar significativamente o processo de saque.
Leia os Termos e Condicoes: Conhecer as politicas de saque do casino, incluindo limites de saque, taxas e tempos de processamento, pode ajuda-lo a evitar surpresas desagradaveis.
Mantenha suas Informacoes Atualizadas: Assegure-se de que todas as informacoes em sua conta estao atualizadas, especialmente seus dados de contato e metodos de pagamento. Isso minimiza o risco de atrasos devido a dados desatualizados ou incorretos.
Escolha o Metodo Adequado: Selecione o metodo de saque que melhor se adapta as suas necessidades, considerando fatores como velocidade, seguranca e facilidade de acesso aos fundos.
Utilize Conexoes Seguras: Sempre acesse sua conta e realize transacoes financeiras atraves de conexoes seguras para proteger suas informacoes pessoais e financeiras.
H2: Contato e Suporte
O Spin Casino oferece varias opcoes para contato e suporte, garantindo que voce possa resolver qualquer questao ou problema relacionado aos seus saques de maneira rapida e eficaz:
Chat ao Vivo: Disponivel 24/7, o chat ao vivo e a forma mais rapida de entrar em contato com o suporte. Voce pode acessar esta opcao diretamente pelo site do casino.
E-mail: Para questoes menos urgentes ou para enviar documentos, o e-mail e uma excelente opcao. O Spin Casino geralmente responde dentro de 24 horas.
FAQ: Antes de entrar em contato direto, vale a pena conferir a secao de Perguntas Frequentes (FAQ) no site. Esta secao cobre uma ampla gama de topicos, incluindo saques, e pode fornecer respostas imediatas as suas duvidas.
Telefone: Alguns jogadores preferem o contato telefonico para discutir suas questoes. Verifique se ha um numero disponivel para sua regiao.
Utilizando esses recursos de suporte, voce pode assegurar que qualquer desafio ou duvida seja resolvido de maneira eficiente, permitindo uma experiencia de jogo tranquila e agradavel no Spin Casino.
?? Fazer saques no Spin Casino pode ser um processo simples e seguro, desde que voce siga as diretrizes adequadas e utilize os recursos de suporte disponiveis. Lembre-se de verificar sua conta, ler atentamente os termos e condicoes, manter suas informacoes atualizadas, escolher o metodo de saque adequado e sempre usar conexoes seguras. Com essas praticas em mente, voce esta pronto para desfrutar de seus ganhos com tranquilidade e seguranca.
FAQ:
Quanto tempo leva para processar um saque no Spin Casino?
Geralmente, o processamento pode levar de 24 horas a 5 dias uteis, dependendo do metodo escolhido. Carteiras eletronicas sao mais rapidas, enquanto transferencias bancarias podem demorar mais.
Existem taxas para saques no Spin Casino?
O Spin Casino pode aplicar taxas dependendo do metodo de saque utilizado. E crucial verificar os termos e condicoes especificos para evitar surpresas.
Como posso garantir que meus saques sejam seguros?
Sempre use conexoes seguras, verifique sua conta e escolha metodos de pagamento confiaveis. Alem disso, siga as diretrizes de seguranca fornecidas pelo casino.
O que fazer se tiver problemas com meu saque?
Nao hesite em contatar o suporte ao cliente atraves do chat ao vivo, telefone ou e-mail. Eles estao disponiveis para ajuda-lo a resolver qualquer questao rapidamente.
Люки Шагма – идеальное сочетание функциональности и эстетики для вашего интерьера. Инновационные решения от SHAGMA обеспечивают надежный доступ к коммуникациям, сохраняя безупречный внешний вид помещения. Наши люки под плитку не только решают проблему провисания, но и гармонично вписываются в любой дизайн. Выбирая SHAGMA, вы выбираете качество, долговечность и индивидуальный подход https://www.pushkino.org/ipb/index.php?showtopic=76485
Betstra ödeme ve finans i̇os uyumlu apple cihazı kullanarak Betstra siteye giriş yapan kullanıcılarına yönelik bir uygulama maalesef mevcut değildir.
Concordeslot sosyal medyası her zaman tüm bahisçilerin Concordeslot ligde bahis oynamak için gerekli paraya erişmesini sağlayacaktır.
Hayalbahis Müşterileri çekmek için yüksek oranlar ve çeşitli bahis oyunları sunun. Hayalbahis Bir diğer sebep ise bahis sitelerinin sıklıkla verdiği bonuslardır.
Meritslot Bahisçiler çoğu kategoride hediyeler sunan siteleri inceleyerek çok fayda sağlayabilirler. Meritslot Koşulların karşılanması durumunda bonus otomatik olarak hesabınıza aktarılacaktır.
Nightwin poker bahis seçenekleri kaliteli mi yatırımınızı Nightwin yaptıktan hemen sonra başka hiçbir şey yapmanıza gerek kalmadan canlı destek hattımıza ulaşarak bonusunuzu talep edebilirsiniz.
https://gosznakdublikat.ru/
Альпы или обзорные экскурсии по Парижу
поездки на море в Дивноморск
https://samoylovaoxana.ru/tag/ostrov-saona/
Ещё можно узнать: как сбросить настройки internet explorer
Туристические советы
Cheers! Numerous info!
Here is my web blog https://Nilecenter.online/blog/index.php?entryid=905651
https://xrumer.ru/ – роблокс r34
https://xrumer.xyz/ – роблокс r34
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
i-guru – Ремонт Iphone и другой техники Apple в Минске
Перепрошивка apple watch ultra
Профессиональный ремонт любых устройств из линейки Apple, без посредников
80% ремонтов iPhone занимает около 20 минут
Несложные ремонты делаются быстро, модульный ремонт в большинстве случаев занимает не более 20 минут.
Почему выбирают i-guru
Ремонт в тот же день
Даже большинство сложных ремонтов выполняем день в день.
Гарантировано низкие цены
Мы беремся за сложные задачи, не передавая их сторонним исполнителям, и зарабатываем не как посредники.
Честная гарантия
Предлагаем честную гарантию на выполненные работы до 12 месцев в зависимости от вида работ.
https://sormol.ru/
https://sormol.ru/
https://sormol.ru/
Виртуальные карточные и настольные игры также ждут своих поклонников. От блэкджека до покера – испытайте удачу и стратегию https://total-rating.ru/2393-revizionnye-lyuki-skrytye-pod-plitkoy-sovremennye-resheniya-i-tendencii.html
Betitbet şikayetlerine dayanarak canlı bahis ajansına başvurursanız sitenin kalitesi hakkında daha iyi bir fikir edinirsiniz. Betitbet Şikayetler, liberal platformlar ve diğer tüm şikayet siteleri taranır.
Essbahis Alınan verilere göre bu bahis sitesine yönelik neredeyse hiç şikayet yok. Essbahis Bu şikayetlerin çoğu üyelerin kendi mali veya kişisel sorunlarından kaynaklanıyor gibi görünüyor.
Hazbet sanal oyun seobahis sanal oyun üyelik kayıt Hazbet işlemini tamamlayabilir ve üyelik kayıt butonuna tıklayarak üye olabilirsiniz.
Pablosbet Bu sitenin ödeme yapmadığına veya üyelerini dolandırmadığına veya dolandırdığına dair herhangi bir iddiaya rastlamadık. Pablosbet Müşteri memnuniyeti söz konusu olduğunda kullanıcı yorumlarına bakıldığında bu canlı bahis sitesinin üyelerini mutlu etme konusunda başarılı olan modern bir şirket olduğu görülmektedir.
Xslot Önemli noktaları kendiniz incelemek ve siteyle ilgili çeşitli sorunlardan şikayetçi olmak istiyorsanız şikayette bulunmanız gerekmektedir. Xslot Ücretsiz hesap açma konusunda şikayette bulunmak için atabileceğiniz adımlar aşağıda verilmiştir.
Suvbet adresi hakkında türkiye’de çok sayıda yasa dışı sitesi Suvbet bulunmaktadır ancak en güvenilir ve güvenilir seçeneklerden biri olarak öne çıkmaktadır.
Mislibet hakkında düşünceler premier Lig üyeleri, Mislibet üyelik hesaplarına giriş yaparak Mislibet kuponundan yararlanabilirler.
Galabet Hesap açarken TC kimlik numaranız ve telefon numaranızı girmeniz istenecektir. Galabet Belirttiğiniz cep telefonunuza SMS kodu gönderilecek ve hesabınız bu SMS kodu kullanılarak aktif hale getirilecektir.
Galabet Şikayet oluşturmada sorun yaşıyorsanız Galabet adresine yönelik şikayetleri, diğer canlı bahis siteleri veya Galabet canlı bahis ofisleri hakkındaki şikayetleri kontrol edin ve “Benzer şikayet oluştur” butonuna tıklayın.
Ngsbahis doğrudan destek ve güvenilirlik bahisçiler bu Ngsbahis seçeneklerin yanı sıra canlı bahis söz konusu olduğunda birçok zorlukla karşılaşırlar.
Galabet Bir şikayette bulunduğunuzda şikayetiniz genellikle 48 saat içinde yayınlanır. Galabet Artık şikayetler hakkında daha fazla bilgi edinmek ve siteyi daha iyi anlamak istiyorsanız soruların yanıtları büyük ilginizi çekecektir.
Noxwin oyun kuralları oyun kurallarının veya ev kurallarının Noxwin açıklığa kavuşturulması dahil, karşılaşabileceğiniz her türlü küçük sorun için anında yardım sunuyoruz.
Galabet çevrimiçi iletişimi, şikayetin müşteri hizmetleri aracılığıyla çözülememesi durumunda bahisçilerin ve Galabet oyuncuların çevrimiçi iletişim platformu “Şikayetler”e şikayetlerini göndermelerine olanak tanır.
Limosbet güvenilebilir mi bu temaları web sitesinde Limosbet tanımlayabilirseniz, bunlara erişimde herhangi bir sorun yaşamayacaksınız.
Galabet Telefonunuza gönderilen kimlik numarası ve SMS kodu yeterlidir. Galabet Sitenizin adresi değiştiğinde kaç tane canlı bahis sitesinin karşınıza çıkacağından emin olabilirsiniz.
Ladesbet tüm şartları tüm bu şartların yerine Ladesbet getirilmesi durumunda promosyon web sitenizde görüntülenecektir.
Galabet, canlı bahis şirketlerinin, adresinizi değiştirdiğinizde adresinizi belirlemek için adınıza bazı sayılar eklemesidir. Galabet Verilen adresi kullanarak siteye tekrar giriş yapabilirsiniz.
Bluebahis için tarayıcı bildirimleri Bluebahis güvence sağlar. Bluebahis Burada beklediğinizden daha fazlasını keşfedecek ve doyum duygusunu yaşayacaksınız.
Sonicbet Canlı Bahis Ofisini öne çıkaran birkaç önemli tema var. Sonicbet Altyapı kalitesi bunlardan biri ve çok önemli.
Logocasino Web sitesi internet üzerinden hizmet verdiği için fiziki bir ofisi bulunmaktadır. Logocasino güvenilebilir mi? sorusunda kısaca değinilen altyapının kalitesi gündeme geldi ve detaylı bir şekilde incelendi.
Formenbet incelemelerinde açıkça işaretlenmiştir kullanıcılara Formenbet kesintisiz bir oyun deneyimi vaat eden oyun web sitesine adı veriliyor.
토토 분석 사이트
“이해합니다.” Liu Jian은 Hongzhi 황제를 깊이 들여다 보았습니다.
Betbino Sitede hesap açmayı düşünüyorsanız ve altyapıyla ilgili sorularınız varsa vereceğiniz bilgiler çok önemlidir. Betbino altyapısının güvenilir olup olmadığını sorarak canlı bahis ofisleri için önemli olan seçenekleri de ele alacağız.
Luvibet güvenilir bahis sitesi kullanıcıların çoğu Luvibet sunduğu hizmetten oldukça Luvibet memnun kalıyor ve tadını çıkarırken gönül rahatlığı yaşıyor.
Betxbet altyapı şirketinin program desteği başarısız olursa doğrudan lansmanı eksik kalacaktır. Betxbet İnternet üzerinden yayın yapma zorunluluğu, bu sansürü ülkemizde de uygulamamızı gerektiriyor.
Grandbetting i̇nterneti giriş sorunlarını çözmek için yeni Grandbetting giriş adresinizi kullanarak işlem başlatabilirsiniz.
Takvimbet Detaylı bir analiz bahis sitesinin sağlam bir altyapıya sahip olduğunu ortaya koymaktadır. Takvimbet Bu tasarımın sonucu sağlam bir web sitesidir.
Faffbet üyelerimize en kaliteli kazanç fırsatlarını sunarken, sitenin altyapısı gereği para yatırma ve çekme gibi ödeme tekniklerinden de ödün vermiyoruz. Faffbet alanında sunulan bu sistemleri kaliteden ödün vermeden profesyonelce kullanabilirsiniz.
Betpipo mobil spor bahisleri bir web sitesinin yasal Betpipo sayılması için devlet tarafından tanınan bir otoriteden lisans alması gerekir.
Sesbets Özel bonuslardan yararlanın ve etkinlik gereksinimlerinize uygun profesyonel çalışmayı garanti edin. Sesbets Sadece siteye üye olarak basit temasından ve kullanım kolaylığından faydalanabilir ister bahis alanında ister oyun alanında daha büyük bir sistemin parçası olabilirsiniz.
Tulipbet şu anki konumu bahis sitesi uygulamasını ve Tulipbet içeriğini kullanabilmek için sitenin güncel adresini girmeniz gerekmektedir.
Baji Live Casino offers a dynamic and immersive gaming experience with a wide range of games, exciting bonuses, and a user-friendly interface that caters to both novice and seasoned players [url=https://baji-999.org/]https://www.baji-999.org[/url]
С каждым годом растет количество людей, выбирающих этот вид досуга. Онлайн-платформы предлагают не только развлечение, но и шанс испытать удачу https://iuecwalocal81288.com/content/%D0%BE%D1%81%D0%BD%D0%BE%D0%B2%D0%BD%D1%8B%D0%B5-%D0%BF%D1%80%D0%B5%D0%B8%D0%BC%D1%83%D1%89%D0%B5%D1%81%D1%82%D0%B2%D0%B0-%D0%B8%D1%81%D0%BF%D0%BE%D0%BB%D1%8C%D0%B7%D0%BE%D0%B2%D0%B0%D0%BD%D0%B8%D1%8F-%D0%B7%D0%B5%D1%80%D0%BA%D0%B0%D0%BB%D0%B0-%D1%80%D0%B8%D0%BE%D0%B1%D0%B5%D1%82
Компания Септик-Нара-купить септик
занимается продажей, установкой и обслуживанием септиков в Наро-Фоминске Наро-Фоминском районе. Основное направление деятельности нашей компании именно установка под ключ септиков любых видов и размеров. С момента основания нашей компании, мы произвели монтаж более 1000 септиков по Московской и Калужской области. Благодаря этому у нас огромный опыт работы с любыми станциями, представленными в нашем регионе!
Если Вы решили купить септик для дома или дачи, мы с радостью поможем Вам с выбором модели, доставим и установим септик на вашем участке в кратчайшие сроки.
Мы занимаемся продажей септиков таких марок: Топас Юнилос Астра Евролос Тверь Аквалос Дочиста Фекалов Волгарь Удача. Мы работаем напрямую с производителями септиков, поэтому Вы можете быть уверены, что не переплачиваете ни копейки. Вся продукция в нашей компании имеет соответствующие сертификаты и лицензии. Время выезда на осмотр, установки или привоза оборудования согласовывается с клиентами и выполняется в срок. Мы заботимся о своей репутации, поэтому выполняем работу надежно и быстро.
Если Вы не знаете, какой септик больше всего Вам подходит, мы предоставим консультацию и выезд специалиста на Ваш участок абсолютно бесплатно!
Благодаря приобретению качественных септиков в нашей компании, каждый клиент получает большое количество преимуществ:
» компактные размеры и небольшой вес устройства;
» доступная стоимость очистной установки;
» быстрый и простой монтаж;
» невысокая стоимость эксплуатации;
» отсутствие неприятных запахов;
» высокая производительность;
» длительный срок эксплуатации;
» высокая степень очистки;
» практически полностью автономный режим работы.
Sahnebet Hepimizin bildiği gibi oyun sektöründe altyapı sağlayıcılarının online oyun sitelerine katkısı büyüktür. Sahnebet Bir bahis sitesi iyi bir altyapı sağlayıcısıyla ortaklık kurduğunda sunulan hizmet oldukça olumlu yönde gelişir.
Nissbet Zayıf bir altyapı sağlayıcısıyla çalışmak, kötü web sitesi hizmeti ve performansıyla sonuçlanacaktır. Nissbet bahis siteleri kendi alanlarında köklüdür ve mükemmel bir canlı oyun deneyimi sunmak için tasarlanmış TV bahis altyapısı sunmaktadır.
Adaxbet bahis canlı oyunlar müşterilerine karşı sarsılmaz güvenilirliği Adaxbet ve maksimum müşteri memnuniyeti sağlama konusundaki kararlılığıyla tanınmaktadır.
Egeslot canlı oyun altyapı sağlayıcısıdır. Egeslot Bet, ülkemizdeki diğer büyük canlı bahis siteleri tarafından da tanınan, canlı oyun alanında kapsamlı bir marka ve kaliteli hizmete sahip bir altyapı sağlayıcısıdır.
https://tezfiles.cc/
Betingo bahis sitesi, diğer tüm Bet altyapı siteleri gibi, firmanın tecrübesi, Betingo güvenilirliği ve çok yüksek oyun ödülleri nedeniyle üyelerine oldukça karlı ve güvenilir bir hizmet sunmaktadır.
Metabahis giriş hakkında kullanıcılar bir bahis Metabahis sitesinin alan adına giderek Metabahis giriş yapmayı kafa karıştırıcı bulabilirler.
Piabellacasino blackjack şirketin diğer rakipleriyle ikramiye Piabellacasino konusunda yoğun rekabeti, şirketin genel yapısını etkileyen en önemli faktörlerden biri olarak değerlendiriliyor.
Adiosbet adresiniz ile yüksek kazançlı ve son derece eğlenceli canlı oyunlara güvenli bir şekilde erişmenizi ve oynamanızı sağlar. Adiosbet Login online bahis sitesi, canlı oyunlar bölümünde üyelerine ve bahis meraklılarına oldukça tatmin edici sayıda oyun sunmaktadır.
Rekorbet online bahis sitesinin güncel giriş adresinde daha birçok canlı oyun sizi bekliyor. Rekorbet Oyun oynamak ve para kazanmak için bu siteye üye olmanız yeterli.
Nevacasino son güncellemesi doğal olarak bu aktiviteye katılmak için Nevacasino çalışan bir internet bağlantınızın olması gerekir. Oyun uygulamasını son sürüme güncellemek gerekmektedir.
Betvole Poker, canlı oyunlarda en popüler oyunlardan biridir ve oyun tutkunları tarafından en çok oynanan oyundur. Betvole Bahis siteleri bu oyunu çok ciddiye alıyor çünkü diğer birçok oyuna göre daha eğlenceli ve pahalı.
https://www.themoviedb.org/movie/261903-real-gangsters
Lotobet canlı pokeri sitedeki en popüler canlı oyunlardan biridir ve sitenin odaklandığı şey de budur. Lotobet Klasik pokerden farkı olmayan bir oyunu oynamak ve kazanmak için Lotobet online bahis sitesine üye olmanız gerekmektedir.
Good manners go a long way. Being punctual, polite, and respectful during your date sets a positive tone and shows you value their time
news site
Betofbet çeşitli mevcut çeşitli oyunları arasında rulet, Betofbet blackjack, slot makineleri, canlı bahis ve bahis seçeneklerinin yanı sıra sitenin casino bölümü en çok öne çıkanlardır.
Sultanbet Üye olmadan önce güncel giriş adresinizi kullanarak web sitesini ziyaret edebilir, Sultanbet canlı oyunlar bölümünde oyun mantığını ve nasıl oynanacağını görebilir, görüşlerinizi belirtebilirsiniz.
Betwon güncel adres bahisleri neredeyse tüm spor, canlı bahis, canlı oyunlar ve sanal oyunları içermektedir. Betwon Bizi diğer internet tabanlı bahis sitelerinden ayıran özelliğimiz, canlı oyunları sunmamızdır.
Gamabet canlı bahis nedir mevcut adresinizi Gamabet girin ve bu harika bahis seçenekleriyle kaldığınız yerden devam edin.
Top News Sites for Article Post Permanent
aljazeeranewstoday.com
australiannewstoday.com
bbcworldnewstoday.com
bloombergnewstoday.com
bostonnewstoday.com
britishnewstoday.com
Dont hesitate to Contact me.
Gorabet kayıp sürekli ve gereksiz kayıplar yaratmayan Gorabet bir web sitesi olarak sektörün önde gelen firmaları arasındaki yerini sağlamlaştırmıştır.
Betvole Canlı destek hattımız yaşadığınız sorunları en hızlı, sorunsuz ve modern şekilde çözüme kavuşturuyor. Betvole Şirket, sistemi kurmak için çeşitli uygulama şirketleriyle sözleşmeler imzaladı ve sözleşmelerle ilgili geri bildirimleri geri çekti.
Trendbet kullanıcı incelemeleri sitesi sunduğu promosyonlar Trendbet sayesinde bahisçilerin ihtiyaçlarına en etkili şekilde cevap vermektedir.
Norabahis Güncel yeniliklerle birlikte çoğu müşteri ayın bahis sitesini seçiyor. Norabahis Şirket, diğer platformlara örnek olmaya devam ederek hak ettiği rakamlara ulaşıyor.
İstanbulbahis yatırımları akıllı telefonlar ve tabletler gibi İstanbulbahis çeşitli cihazlardan kullanılabilmekte ve geniş bir tasarım yelpazesine sahiptir.
Toobet mevcut giriş adresi ihtiyati tedbirdir. Toobet adres değişiklikleri BT K’daki erişim engellerinden kaynaklandı.
Redfoxbet Mevcut giriş adreslerini değiştiren Redfoxbet Canlı Poker Firmaları bu önlemlerle sorunlarla karşılaştı ancak Redfoxbet sorunlar artık ortadan kalkmaya başladı. İşletmeler alan adlarını korur.
Ultibahis betting farklı bahis türlerine sahip olan Ultibahis bahis sitelerinin spor bahisleri bölümü de kullanım alanlarından biridir.
İnterbahis bahis canlı oyunlar sonuç olarak, oyuna İnterbahis bahis koymakla ilgilenen kişiler, bahis ihtiyaçlarını karşılayan en saygın Türk online şirketlerini arıyor.
Tipbet Adreslerinizden birinin bloke olması halinde yedeklerinizden yeni keşfedilen bir adresi açıp o adres üzerinden hizmet vermeye devam edeceğiz. Tipbet Fark edeceğiniz tek değişiklik adres adında bir değişiklik olacaktır.
Ritimbet mevcut giriş modunuzu ve eski giriş adresinizle kullandığınız kullanıcı adı ve şifreyi kullanarak giriş yapabilirsiniz. Ritimbet Bu gibi durumlarda şirketin müşteri hizmetleri ekibi bu tür sorunları çözmek için eğitilir.
Dumanbet web oyunları türkiye’den katılımcı kabul Dumanbet eden tek site sitesi olup, en fazla katılımcıya sahip olan site ise Dumanbet.
Dinamobet şikayetleri oyun hakkında daha fazla bilgi Dinamobet edinmek isteyen kişiler bu bilgilere web sitesinden rahatlıkla ulaşabilirler.
A fairly new player in the Russian darknet arena, blacksprut Blacksprut has quickly gained attention for its interesting features and growing popularity. While some aspects raise questions, it has the potential to become a prominent figure in the darknet scene.
Features:
Blacksprut https://bs-gl-darknet.com offers an “Instant Transactions” feature, inspired by the success of similar systems on other platforms like Hydra. Couriers hide goods within the city and provide buyers with coordinates, adding an adventurous element to the purchasing process.
Регистрация на различных ресурсах, форумах или интернет-магазинах сегодня осуществляется только с применением номера телефона. Ранее для этой цели могла использоваться электронная почта, но сегодня правила изменены https://pribylwm.ru/raznoe/virtualnye-nomera-onlayn.html
News Sites for Article post
frenchnewstoday.com
germaynewstoday.com
guardiannewstoday.com
headlinesworldnews.com
huffingtonposttoday.com
irishnewstoday.com
Dont hasitate to contact us
Hello there! Do you know if they make any plugins to assist with Search Engine Optimization? I’m trying to get my blog to rank for some targeted keywords but I’m not seeing very good success. If you know of any please share. Cheers!
https://thetvdb.plex.tv/movies/real-gangsters
슬롯 나라 무료
최후의 수단이 아니었다면 오늘 밤 그렇게 많은 신하들이 소환되지 않았을 것입니다.
Vidobet Özel bahisler, topluluk bahisleri ve E-spor bahislerinin yanı sıra bahis teklifi de oldukça iyi yapılandırılmıştır. Vidobet Bu yazıda yeni bir adrese giriş, Vidobet giriş ve kayıt işlemlerine odaklanılacaktır.
Favexbet Futbol bahis pazarının zirvesinde dünyanın en ünlü sporuyla bağlantılı çok sayıda Favexbet promosyonu da bulunmaktadır. Forvet bahisleri sonuçlandırılırsa para iadesi.
Ирина Волк: расследование уголовного дела пирамиды Life is Good завершено
порно секс жесток
Официальный представитель МВД России Ирина Волк сообщила об окончании предварительного расследования уголовного дела Life is Good. Материалы с утвержденным обвинительным заключением в отношении десяти человек направлены в Приморский районный суд Санкт-Петербурга для рассмотрения по существу. Они обвиняются в организации деятельности по привлечению денежных средств и мошенничестве, совершенных в особо крупном размере в составе преступного сообщества. Всего, по версии следствия, в состав криминальной организации входили 35 соучастников. Потерпевшими по уголовному делу признаны 221 человек – люди заключили договоры с целью получения пассивного дохода. Общая сумма материального ущерба составила свыше 280 млн рублей.
«Кроме того, в деятельность финансовой пирамиды были вовлечены более 18 тысяч россиян. Они вложили в жилищный кооператив более 15 миллиардов рублей, два миллиарда из которых организаторы криминальной схемы присвоили. Десять предполагаемых участников преступного сообщества были задержаны в феврале 2022 года оперативниками ГУЭБиПК МВД России совместно с коллегами из Санкт-Петербурга и сотрудниками ФСБ России. Остальные обвиняемые в противоправной деятельности объявлены в розыск. Возбуждено уголовное дело по признакам преступления, предусмотренного частью 3 статьи 210, частью 4 статьи 159, частью 2 статьи 172.2 УК РФ. Расследование было взято на контроль руководством Следственного департамента МВД России», – сообщила Ирина Волк.
По информации МВД, в общей сложности полицейскими и сотрудниками ФСБ проведено более 100 обысков по местам проживания фигурантов и в офисах подконтрольных им организаций. Изъята компьютерная техника, средства связи, документы и другие предметы, имеющие доказательственное значение.
«В качестве обеспечительной меры по возмещению причиненного материального ущерба наложен арест на более чем 2300 находящихся в собственности кооператива квартир, а также на денежные средства на счетах компании в сумме свыше 3,7 млрд рублей. Также арестованы активы фигурантов на общую сумму порядка 1 млрд рублей», – рассказала генерал-майор полиции.
По версии следствия, противоправная деятельность велась с 2014 года на территории девяти регионов России. Злоумышленниками были созданы подконтрольные организации с сетью региональных филиалов, которые работали под единым брендом «Лайф из Гуд». Гражданам предлагали стать пайщиками кооператива, обещая либо новое жилье, либо получение дохода в размере до 30% годовых. При этом реальным инвестированием вкладов фигуранты не занимались. Приобретение недвижимости и обещанные выплаты осуществлялись за счет привлечения средств новых клиентов. Кроме того, граждан вводили в заблуждение об уровне доходности вложений, указывая в договорах параметры гораздо ниже обещанных. Полученные от вкладчиков деньги похищались.
Ранее стало известно, что Приморский районный суд Санкт-Петербурга заочно арестовал топ-лидера инвестиционного проекта Life is Good в Набережных Челнах Сергея Санникова и его жену Юлию. Также суд избрал меру пресечения в виде заключения под стражу и в отношении директора холдинга Игоря Маланчука. В июле этого года они были объявлены в федеральный розыск – соответствующая информация появилась в базе данных министерства.
Betzula, bahisçilere haftalık bedava dönüşler sunuyor. Betzula Spor bahisçileri, canlı sohbet aracılığıyla müşteri destek ekibimizle 7/24 etkileşim kurabilir.
https://www.publico.es/sociedad/publico.es-nancy-fraser-no-dejar-temor-ultraderecha-lleve-feminismo-liberal.html
Zurihbet Acil olmayan sorunlar için mesai saatleri içerisinde isteğe bağlı olarak telefon hattı veya e-posta iletişim formu da sunuyoruz. Zurihbet Markanın kökleri göz önüne alındığında Zurihbet çevrimiçi hizmetinin birinci sınıf olması şaşırtıcı değil.
Лучшие автомобили из Кореи | Секреты выбора авто из Кореи | Что нужно знать о корейских авто | Пятерка лучших корейских автомобилей | Как правильно ухаживать за авто из Кореи? | Популярные модели корейских авто | Топ-7 подержанных авто из Кореи | Ценовая политика корейских автомобилей | Выбор лучшего корейского автомобиля | Лучшие внедорожники из Кореи | Как правильно страховать авто из Кореи? | Корейские авто для долгих поездок: комфорт и безопасность | Электромобили из Кореи: перспективы и реальность | Проверка корейского автомобиля перед приобретением | Как сделать уникальным свой корейский автомобиль
автомобиль из кореи https://avtodom63.ru/ .
Kronosslot Etkileyici bir slot ve oyun yelpazesine ek olarak, masa oyunları, jackpotlar ve Kronosslot evrim oyunları gibi gösterişli isimlerin yer aldığı canlı bir casinoya de sahiptir.
Probet Bazı web siteleri canlı oyun seçenekleri için özel kategoriler oluşturur. Probet Tüm canlı maçların tek bir yerde listelendiği bu sayfaya erişmek için bu sayfaya giriş yapın ve ‘Maçları İzle’ seçeneğini kullanın.
Betsibet “Canlı Oyun” seçeneğine tıklamak sizi ilgili oyunun oynandığı sayfanın ekranına götürecektir. Betsibet Bu sayfa farklı bir formatta ve alan adı uzantısında açılacak ve eşleştirme tamamlanana kadar aktif kalacaktır.
Devrebet Maç bittikten sonra yayın iptal edilecek. Devrebet Ancak programlaması zayıf olan siteler oyunun yayınını aksatabilir.
Laligabetting canlı casino bölümü ile iş Laligabetting yaparken karşılaşabileceğiniz küçük sorunlar için 7/24 destek hattımızı kullanmayı unutmayın.
Heybet, oyun meraklıları için özgün lisanslara sahip en önemli oyun şirketlerinden biridir. Heybet Özellikle oyun çeşitliliğinin fazla olması ve fiyatların yüksek olması nedeniyle kullanıcı memnuniyeti yüksektir.
Vebahis bahis yelpazesinde hem spor bahisleri hem de oyunlarında canlı bahis mevcuttur. Vebahis iletişim kanallarını kullanarak üyeleriyle birden fazla kanal üzerinden iletişim kurar ve etkileşim yönetimi politikalarına uyar.
Casinoturka Bu politika çerçevesinde üyelerimizden gelen birçok soru, şikayet, öneri ve Casinoturka tavsiyeyi çok ciddi ve profesyonel bir şekilde takip ediyor ve olası çözümleri tespit ederek konuyla ilgili çok aktif araştırmalar yürütüyoruz.
Gececasino üyelerimizin görüşlerini dikkatle dinliyor ve ortaya çıkan sorunların çözümüne yönelik yenilikçi çözümler sunuyoruz. Gececasino Bu web sitesi aracılığıyla canlı destek ekibine 7/24 ulaşabilirsiniz.
Olaybahis turnuvası dolandırıcılık ve Olaybahis hileli web sitelerinin kurbanı olmamak için resmi adresi bilmeniz çok önemlidir.
Rodezbet, herhangi bir kaza anında olay yerine anında müdahale eden kalifiye profesyonellerden oluşuyor. Rodezbet Ayrıca ekibinize para çekme ve yatırım yatırımları konusunda yardımcı olabilir ve bu süreçte ortaya çıkabilecek sorunları giderebilirsiniz.
Tuabet şirket üyeleri şirket, üyelerine UEFA ligi, Türkiye Süper Ligi, Tuabet İspanya ligi, İngiltere ligi, Almanya ligi, İtalya ligi, Fransa ligi ve Belçika ligi gibi popüler ligler sunmaktadır.
Papelgo Oyun sitesi ülkenin pazarına girdi. Papelgo Bu site altyapı açısından oldukça tecrübeli bir sitedir.
Pekinbet giriş bahisleri cep telefonları hayatımızda Pekinbet önemli bir yere sahip olup birçok aktiviteyi gerçekleştirmemize olanak sağlamaktadır.
Bayabetcasino Özellikle müşteri hizmetleri söz konusu olduğunda oldukça yapılandırılmıştırlar. Bayabetcasino bahis sitesi, müşterilerine sunduğu mükemmel hizmet ve katma değer nedeniyle bahis pazarında kendine yer edinmiştir.
Dipbet avantaj kampanyası kesintisiz hizmet ödül kampanyası katılımcıları, Dipbet ödül kampanyasının faydalarını en üst düzeye çıkarma şansına sahiptir.
Zegabet bahis sitesi en popüler oyun sitelerinden biridir. Zegabet Site aynı zamanda sınırsız para çekme olanağı da sunuyor.
Pusulabet mobil programı adresinizi değiştirmek Pusulabet bir zorunluluktur ve bu soru rehberlerini ilgilendirmiyor.
Betfokus bu sitenin gerekli görmesi halinde ilk para çekme anında belge talep edebiliriz. Betfokus bahis sitesi kolay ve hızlı bir şekilde paranızı çekebileceğiniz bahis sitelerinden biridir.
Goodbahis canlı hizmeti spor bahisleri bölümünde futbol, Goodbahis basketbol, voleybol, hentbol ve daha birçok spor dalına bahis oynayabilirsiniz.
Casinomavi incelemeleri ve desteği her kartın asla Casinomavi gözden kaçırılmaması gereken kendine ait üst ve alt limitleri vardır.
Betcombet Siteden para çekmek için ana sayfadaki para çekme bölümüne giriş yapmanız gerekmektedir. Betcombet Daha sonra işlemi tamamlamak için uygun yöntemi, hesap bilgilerini ve tutarı seçmelisiniz.
Privebet şirketi şirket herkese yardımcı olmak için Privebet geniş bir oyun yelpazesi, yüksek faiz oranları ve bonuslar sunuyor.
Bonus bez depozytu lub innymi slowami darmowy bonus to nic innego jak gratis oferowany przez kasyna online dla swoich graczy vavada bonus bez depozytu
Looking for a quick financial boost? Apply now for a installment loans and secure the funds you need with ease. Whether it’s for unexpected expenses, home improvements, or a special purchase, our fast approval process ensures you get the money swiftly. Don’t wait – take the first step towards financial freedom today!
Защитите свою конфиденциальность с резидентским прокси, использовать этим инструментом.
Какие преимущества у резидентских прокси?, ознакомьтесь с подробностями.
Какой резидентский прокси выбрать?, советы для пользователей.
Зачем нужны резидентские прокси?, ознакомьтесь с возможностями.
Почему резидентские прокси безопасны?, анализ функций безопасности.
Защита от каких угроз обеспечивает резидентский прокси?, анализируем важные аспекты.
Зачем нужны резидентские прокси и какой их выигрыш?, сравним основные плюсы.
Как быстрее работать в сети с резидентским прокси?, практические советы для оптимизации работы.
Зачем использовать резидентский прокси для сбора информации?, разбор возможностей для парсеров.
Как обеспечить конфиденциальность в Интернете с резидентским прокси?, шаги к безопасности онлайн.
Как расширить свои возможности в соцсетях с резидентским прокси?, рекомендации функционала.
Зачем арендовать резидентские прокси и какие бонусы?, проанализируем лучшие варианты.
Способы защиты от DDoS с помощью резидентского прокси, подробно изучим меры безопасности.
Почему резидентские прокси пользуются популярностью, подробно изучим основные факторы.
Как выбрать между резидентским и дата-центровым прокси?, подсказки для выбора.
аренда резидентных прокси https://rezidentnieproksi.ru/ .
Sandalsbet bahis sitesi kolaylıkla para yatırabileceğiniz bahis sitelerinden biridir. Sandalsbet Ayrıca bahis sitelerinin farklı para yatırma yöntemleri ve esnek limitleri vardır.
Uncover the future of gaming with Musk Empire – a revolutionary Web3 adventure! Build your business empire free of in-app purchases. Upgrade your assets to amplify your revenue stream. Engage, accumulate, and eventually exchange in-game wealth for actual cash. Join the Musk Empire now and mold your digital fortune! invitation link https://tinyurl.com/muskempre
Hektorbet web sitesi, oyuncunun hesabını doğrulamak için belgelere ihtiyaç duyar. Hektorbet Bir bahis şirketinin müşteri hizmetleri, özellikle yatırım müşterileri için çok önemlidir.
Rinabet giriş sorunları sms mesajı göndermeyi bırakmanız Rinabet gerekirse bunu istediğiniz zaman kolayca yapabilirsiniz.
https://e-porn.net
https://analwife.net
free xxx tube videos webcams dating,
онлайн камеры, знакомства
Levabet Bahisçiler internet üzerinden oyunlara katıldıkları için müşteri temsilcileriyle sürekli iletişim halinde olmak isterler. Levabet Ayrıca müşteri hizmetleri faaliyetlerine yeterince dikkat etmeyen şirketler üyelerini yabancılaştırabilir.
Lensbahis çevrimiçi gizliliği ayrıca çevrimiçi gizliliğinizi nasıl Lensbahis koruyacağınız ve herhangi bir gizlilik sorunuyla karşılaştığınızda ne yapmanız gerektiği konusunda bazı ipuçları ve tavsiyeler de sağlıyor.
Neobet Bahis Sitesi müşteri hizmetleri faaliyetlerini çok ciddiye almaktadır. Neobet Bu amaçla proaktif müşteri danışmanları 7/24 sahada bulunmaktadır.
Betunlim işlemleri hızlı mıdır poker tutkunları, kazanma ve Betunlim kaybetme marjının diğer oyunlara göre daha küçük olması nedeniyle en kazançlı çevrimiçiyi seçmelidir.
Betsound katılımcılar dört farklı dil konuşabilmektedir. Betsound Bahisçiler müşteri temsilcileriyle üç farklı şekilde iletişime geçebilirler.
Alobet yöntemler arasında e-posta, şikayet hatları ve müşteri temsilcileri yer almaktadır. Alobet Son olarak bu yöntemlerle bildirilen sorunlar en kısa sürede değerlendirilip çözüme kavuşturulacaktır.
Anındabet canlı slot makinesi çok sayıda bahisçiyi cezbeden bir işletmeden Anındabet gelir elde etmek için, sitedeki slot makineleri gibi seçenek yelpazesini sınırlamadan farklı arazi seçeneklerini birleştirebilirsiniz.
Unibahis bahis sitesi uzun süredir piyasada olan bir bahis şirketidir. Unibahis Site tecrübesini pazara aktararak yenilikçi bir şekilde gelişmeye devam etmektedir.
Trwin güvenilir hizmeti özellikle kullanıcılara yönelik en yeni oyun platformunda, Trwin mevcut oyun ağının tamamı deneyimli bahisçilerden büyük övgü aldı.
Casinoburada sitesi altyapısıyla güven veren bir site. Casinoburada Ancak sitede sunulan aktiviteler müşteri memnuniyetini sağlamada başarılıdır.
톰 오브 매드니스
장 황후는 “지금은 황후를 위한 시간이 아닙니다. 폐하께서는…”이라고 말을 하지 않을 수 없었습니다.
Kazandra web sitesi yenilikçi bir yaklaşım benimsiyor ve en son teknolojiyi deneyimlerken müşterilerini şaşırtmayı başarıyor. Kazandra bahis sitesi farklı yaklaşımları ve çığır açan yenilikleriyle bahisçiler arasında ilgi çekmeyi başarmıştır.
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
Детская одежда оптом из Турции
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
детская одежда оптом
Axacasino oyun siteleri güvenlik önlemlerini ön planda tutarak müşterilerine sağladıkları değeri en iyi yansıtan siteler arasında yer alıyor. Axacasino Son yıllarda oyun sitesinin kullanıcı sayısı arttı.
Pokerbeta lisanslıdır takımınızın oyun içerisinde biriktirdiği Pokerbeta puanları dikkate alarak sıralamadaki yerini tahmin edebilirsiniz.
승무패 사이트
하지만 앞으로 다가올 큰 변화를 생각하면 수많은 사람들이 마음속으로 눈물을 흘리지 않을 수 없었습니다.
Кооператив для военных
Как пайщики — участники СВО относятся к событиям вокруг «Бест Вей»
Полковник юстиции А.Н. Винокуров
Потребительский кооператив «Бест Вей» оказался затронут уголовным делом, касающимся в основном иностранной инвесткомпании «Гермес», которое сейчас рассматривается Приморским районным судом Санкт-Петербурга. Более двух лет более 3,5 млрд рублей на счетах кооператива почти непрерывно арестованы по ходатайству сначала ГСУ ГУ МВД по Санкт-Петербургу, а затем Прокуратуры Санкт-Петербурга: пайщики не имеют возможности ни приобрести недвижимость, ни вернуть средства.
По данным совета потребительского кооператива «Бест Вей», в числе его пайщиков, страдающих от блокировки средств, тысячи военнослужащих, в том числе сотни участников СВО, часть из которых успела приобрести квартиру, часть собирает или собрала первоначальный взнос, а часть — планировала вступить в кооператив. «СП» пообщалась с некоторыми из них и их родственниками, чтобы узнать отношение к «Бест Вей» и событиям вокруг кооператива.
«К кооперативу отношение очень хорошее»
Александр Голдман на СВО с мая 2022 года как доброволец, три ранения. Пришел на СВО рядовым, сейчас — начальник штаба батальона. Был пайщиком кооператива, сейчас пайщик — его мама.
«С помощью кооператива в 2019 году приобретена двухкомнатная квартира во Владивостоке, в которой проживает мама, — рассказывает он. — Расплачиваемся за нее, в ближайшие месяцы намерены погасить задолженность перед кооперативом и оформить квартиру в собственность. К кооперативу отношение очень хорошее, полностью его поддерживаю — он дает возможность без больших переплат приобрести недвижимость. К действиям в отношении кооператива отношусь отрицательно, так как „Бест Вей“ — единственная возможность приобрести жилье в рассрочку».
«Происходящее вокруг кооператива вызывает шок»
Гвардии рядовой Глушков Иван Васильевич — пайщик кооператива из Челябинской области. Танкист, мобилизованный, проходил службу в 15-й отдельной гвардейской мотострелковой Александрийской бригаде 2-й гвардейской общевойсковой армии ЦВО. Погиб 11 октября 2023 года.
Рассказывает его вдова Татьяна Неручева:
«Мы внесли первоначальный паевый взнос и во второй половине 2021 года встали в очередь на приобретение квартиры в Челябинске, когда начались события вокруг кооператива — была заблокирована его возможность приобретать недвижимость и заблокированы его счета. Наша очередь должна была подойти примерно через год. Мы заявили для приобретения небольшую квартиру, но планировали при покупке увеличить ее стоимость до 3 млн и купить двухкомнатную квартиру — с увеличением первоначального паевого взноса: уставом кооператива это позволяется, а затем переехать из Коркино в Челябинск. Сейчас 3 млн хватит только на небольшую квартиру-студию метров 25, то есть мы понесли материальный ущерб из-за блокирования деятельности кооператива, так как лишены были возможности приобрести квартиру, на которую собрали первоначальный паевый взнос. Работа кооператива заблокирована, счета арестованы уже более двух лет. В наследование пая я только вступаю, так как муж очень долго считался пропавшим без вести — долго шла экспертиза, и свидетельство о смерти мы получили только 18 июня этого года».
Татьяна — юрист: «Как у юриста у меня происходящее вокруг кооператива вызывает шок, и любой непредвзятый юрист вам скажет то же самое». «Кооператив, — подчеркивает она, — абсолютно прозрачен, он полностью соответствует законодательству о кооперации — что и подтверждалось многократно государственными органами и судами. Если бы мы с мужем не были уверены, что все прозрачно и законно, мы бы не вкладывали в него деньги. Мы не подавали заявление о выходе из кооператива. Я, несмотря ни на что, жду счастливого завершения рукотворного кризиса вокруг кооператива. Для меня это еще и память о муже — он продал долю в квартире, которая ему принадлежала, чтобы вложиться в кооператив. Кроме того, мне хочется понять — до какого маразма может дойти ситуация у нас в государстве в плане незаконных действий в отношении организации, которая по тем или иным причинам не понравилась каким-то чиновникам?».
«Рассчитываю, что ситуация закончится благополучно»
Сергей Логинов, рядовой, мобилизованный, представлен к награде «Честь и доблесть».
«Пять лет назад я стал пайщиком кооператива, участвовал в накопительной программе, планировал прибрести однокомнатную квартиру в Самарской области. Уже нужно было подбирать объект недвижимости и вставать в очередь на покупку, как работа кооператива была заблокирована по инициативе правоохранительных органов, и это продолжается уже более двух лет. По-прежнему надеюсь получить квартиру и рассчитываю, что ситуация закончится благополучно. Поддерживаю кооператив».
«В кооперативе минимум переплат — несопоставимо с ипотекой»
Егор Ивков, офицер флота, дирижер военного оркестра, пайщик кооператива с 2019 года.
«Я нахожусь в накопительной программе — планировал покупку квартиры в Санкт-Петербурге. До постановки в очередь на покупку дело не дошло, но сумма внесена серьезная — и на два года все зависло. Есть друзья-военнослужащие, которые также являются пайщиками и тоже накапливали деньги на первоначальный паевый взнос — они, как и я, не могут ни продолжать накапливать, ни получить деньги обратно, потому что счета арестованы. Отношение наше к ситуации, создавшейся вокруг кооператива, крайне негативное».
«Кооператив поддерживаю, — говорит пайщик. — Моя сестра живет в квартире, приобретенной с помощью „Бест Вей“ — у нее многодетная семья, сейчас кооперативная квартира переходит в ее собственность. Минимум переплат — несопоставимо с ипотекой. Надеюсь, что ситуация с арестом счетов разрешится в ближайшее время».
Slotbahis twitter ödülleri için hangi alternatiflerim var Slotbahis oyunları bireysel tercihlere göre düzenleyerek daha fazla tasarruf edebilirsiniz.
Abrabet Kalite, güvenilirlik ve hızlı hizmet ile karakterizedir. Aynı zamanda bahis sitesindeki oyun seçenekleri de oldukça geniştir. Abrabet Siteyi kullananlar oldukça memnun. Bu oyun çeşitliliğiyle tanınır.
Admiralcasino Bu sitenin kurucu ekibi deneyimli kişilerden oluşmaktadır. Admiralcasino Aslında bu deneyimler kullanıcılara büyük ayrıcalıklar sağlıyor.
Top News Sites for article Post
chinaworldnewstoday.com
chroniclenewstoday.com
cnbcnewstoday.com
cnnworldtoday.com
crunchbasenewstoday.com
dailyexpressnewstoday.com
Don’t hesiate to conatct us.
Exxenbet sanal futbol sanal futbol bahisleri, son yıllarda Exxenbet severlerin ve oldukça ilgisini çeken sanal bahis veya sanal oyun türlerinden biri olarak tanımlanmaktadır.
Tambet Oyun çeşitliliği de kullanıcıların ilgisini çekti. Tambet Ülkemizde de oldukça popüler bir bahis sitesidir.
Palmibet Öncelikle bu bahis sitesi kişisel bilgilerinizi ve finansal işlemlerinizi korur. Palmibet Güvenilirliği ve yüksek kalitesiyle dikkat çekiyor.
Elitroyal günlük güncellemeleri i̇lgi alanlarınıza ve Elitroyal tercihlerinize uygun her türlü faaliyette bulunma özgürlüğüne sahipsiniz.
Merkezbahis Uzun yıllardır birçok lokasyonda kesintisiz hizmet sağlıyoruz. Merkezbahis Tüm avantajlardan yararlanmak için öncelikle bu bahis sitesine üye olmanız gerekmektedir.
Marsbahis ödeme işlemleri kapsamlı sektör tecrübesine Marsbahis ve sağlam bir başarı geçmişine sahip bir müşteridir.
Meybet Üye olarak çok para kazanabilirsiniz. Meybet Hatta web sitenizle ilgili herhangi bir sorun yaşarsanız, profesyonel yerinde destek ekibimiz her zaman parmaklarınızın ucundadır.
Erisbet Menünün tasarımı son derece kullanıcı dostudur, arayüzü basit ve anlaşılması kolaydır; Erisbet sol üstte arama butonu, sağ üstte sıralama seçenekleri, açık seçenekler ve oyun sağlayıcı bulunmaktadır.
Бесплатный сервис по подбору и ведению сделок с недвижимостью в новостройках Москвы и Московской области! Новые квартиры для жизни, дохода или инвестирования от топовых застройщиков с защитой государства купить квартиру в новостройке выгодно
Vangabet oyun bahisleri bu sitedeki oyun seçenekleri, Vangabet oyuncuların bu sitedeki oyunlardan daha fazla kazanmasını sağlayan yüksek faizli bonuslar içermektedir.
Moldebet oyunlar kategorisine baktığınızda artık bambaşka bir oyun türü göreceksiniz. Moldebet Ayrıca oyunlara alternatif olarak sitede yer alan çeşitli etkinlik ve promosyonlar da üyelerin ilgisini çekecektir.
Casinovale giriş ücretleri sanal hesap yatırım limitleri henüz Casinovale kesinleşme aşamasında olup, uygulama canlı bahis ofisleri arasında en ucuz olanlardan biri olarak kabul edilmektedir.
Kordonbet sitenin oyun kataloğu sürekli güncellendiğinden popüler ve yeni kategorilerin paylaşımı da sürekli güncellenmektedir. Kordonbet Böylece daha fazla oyun türü bulabilirsiniz.
Casinoslot ana temaları web sitesinin ana temaları arasında Casinoslot genel bakış, skor tablosu, canlı çoklu görünüm, canlı takvim ve istatistiksel sonuçlar yer alır.
Safirbet Sanal bahis söz konusu olduğunda oyun sektöründe tercih edilebilecek güvenli ve kaliteli firmalardan biridir. Safirbet Yayın faaliyetlerini AB konseptlerine göre sürdürmekte olan şirket, farklı dil seçenekleriyle dünya çapında çeşitli faaliyetler yürütmektedir.
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
Оформи заявку на кредитную карту от лучших банков России онлайн
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
оформить кредитную карту
Защитите свои данные с помощью резидентских прокси, преимущества.
Смотрите зарубежные сериалы с резидентскими прокси, радуйтесь контентом.
Оптимизируйте работу сети благодаря резидентским прокси, как это работает.
Скройте свой реальный IP-адрес от хакеров с резидентскими прокси, и чувствуйте себя спокойно.
Защитите свою личную жизнь и данные с резидентскими прокси, и чувствуйте себя невидимкой.
Скачивайте файлы анонимно через резидентские прокси, и не бойтесь за свою приватность.
резидентские прокси https://rezidentnie-proksi.ru/ .
Звон Колокольцева. Питерская полицейская мафия виляет Министерством внутренних дел
Руководитель следственной группы Винокуров
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
Уголовное дело компании Гермес
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Звон Колокольцева. Питерская полицейская мафия виляет Министерством внутренних дел
ГСУ Санкт-Петербурга беспредел
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
Суд Бествей
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Top News Sites for article post
dailymirrornewstoday.com
dailystarnewstoday.com
dailytelegraphnewstoday.com
dutchnewstoday.com
dwnewstoday.com
europeannewstoday.com
Don’t hasitate to conatct us.
МОСКВА, 7 мая — РИА Новости. Правительство России, возглавляемое Михаилом Мишустиным, уйдет в отставку во вторник.
Сразу после инаугурации президента Владимира Путина нынешний кабинет министров сложит полномочия перед вступившим в должность главой государства.
Прогон по комментария
cp loli adversity
==> biturl.top/qeAJJf rlys.nl/6epap3 <==
Flyinbet kaliteli canlı bahis sitelerinden biridir ve spor bahisleri ve canlı bahis söz konusu olduğunda mutlak otoritedir. Flyinbet Sitenin portföyünde yer alan sanal bahisler de üye ve kullanıcılar tarafından yüksek puan almaktadır.
В нашем салоне красоты мы заботимся о ваших волосах, создаем безупречный макияж и подчеркиваем вашу естественную красоту, чтобы вы чувствовали себя уверенно и великолепно каждый день https://ekostil-salon.ru/
Yanbahis sanal bahis altyapısını daha derinlemesine inceledikten sonra yazılımlarını bir kez daha keşfettik. Yanbahis Bu nedenle sanal bahis altyapısı oldukça güvenilir kabul edilmektedir.
Trinkbet spor bahis platformları seobahis spor bahis platformları Trinkbet üzerinden hem kaliteli hem de popüler bahis seçeneklerine ulaşabilirsiniz.
Интерьерная печать на самоклеющейся пленке – это процесс создания качественных полноцветных изображений на самоклеющейся пленке, которая может быть наклеена на различные поверхности печать интерьерная на ткани
Alibahis Altyapı kalitesinin yanı sıra oyunun 3D grafikleri de önemli ölçüde iyileştirildi. Alibahis Oyun içeriklerinin ses kalitesi de kullanıcılar tarafından oldukça beğenildi.
Seabahis kaliteli altyapıları kullanarak üyelerimize en kaliteli hizmeti sunmak çok kolaydır. Seabahis Spor bahisleri veya canlı bahislerde boşluklar veya farklılıklar varsa sanal bahis hizmetlerini de tercih edebilirsiniz.
Southbet bahis mobil giriş i̇ndirme i̇şlemi Türkiyeyi ziyaret Southbet eden tüm ziyaretçileri erişim adresinin en güncel versiyonu hakkında bilgilendirdik.
Betkom sanal bahis seçeneklerine daha yakından bakıldığında kullanıcılara hem klasik hem de yeni modern sanal oyunlar sunulmaktadır. Betkom Sanal futbol, sanal basketbol ve sanal tenisin bu sitede sunulan en klasik seçenekler olduğunu söyleyebiliriz.
Betexper ile iletişime geçin bunun için bahis şartlarını Betexper yerine getirmeniz ve ‘finansal işlemler’ menüsünden ‘para çekme’ butonuna tıklamanız gerekmektedir.
Betwild oyunlara pek çok sitede rastlamak mümkündür ancak canlı bahis sitelerinin diğer sitelerden temel farkı, akıllıca yeni seçenekler sunmasıdır. Betwild Sanal Penaltı Atışları ve Sanal Yarışlar bu kategoride oldukça yenidir ve bu özellikler bu sitede çok nadirdir.
Neyine video poker bahisleri Neyine katılmak için Neyine gerekli tüm detayları doğru ve eksiksiz olarak vermeniz gerekmektedir.
Casinomilyon sanal köpek yarışları, sanal at yarışları gibi yarış seçenekleri de üyelere üstün görsel kalite sunmaktadır. Casinomilyon Bu oyunları denediğinizde gerçek bir yarış veya yarışma oynuyormuşsunuz gibi görünebilir.
Santrabahis Oyunun başlangıcından oyunun başlangıcına kadar yalnızca bir dakikanız var, böylece günün 24 saati sanal bahisler oynayabilirsiniz. Santrabahis şikayet analizinin yanı sıra canlı bahis ofisinde de uzun bir tarama yapıldı.
Shotbet iş yaklaşımı etkin, hızlı ve sorunsuz bir iş Shotbet yaklaşımı sunan yüksek verimli özelliklerin aynı zamanda karlılığı da artırdığını göreceksiniz.
Golden90 güvenilir site bonusların sitenin kendi Golden90 bütçesinden sağlanması sitenin güvenilir olduğunun açık bir göstergesidir.
Betloves Şikayetleri incelerken öncelikli olarak “şikayetim var” gibi sayfalar inceleniyor. Betloves Bağımsız şikayet sayfamızda binlerce farklı yorum bulunmaktadır.
Vivasbet Canlı bahis siteleri o kadar popüler ki, bağımsız bahis sitelerini ve platformlarını inceliyoruz. Vivasbet Bu tür web sitelerinde kullanıcıların yorumlarına ve şikayetlerine rastlayacaksınız.
Setrabet şikayetleri kullanıcıların en çok vakit geçirdiği alanlar Setrabet da dahil olmak üzere sürekli güncellenen ve geliştirilen oyun sitemizde uzun süreli oyun oynamanın keyfini çıkarın.
Herbahis, bahis sitelerini araştıran oyuncular için bahisleri hakkında ayrıntılı veri sağlamaya çalışmaktadır. Herbahis Twitter bahis sitesi, Avrupa ülkelerinde gerçek dünya lokasyonlarında ve internet üzerinden bahis oyunları yayınlayan bir şirkettir.
Mislici Avrupa ülkelerinin oyun deneyimini Çinli oyuncuların oyun deneyimiyle birleştirdik. Mislici Bu popüler ve beğenilen bahis şirketi kısa sürede binlerce oyuncuya hizmet vermeye başladı.
Vippark mobil giriş denemesi bu çevrimiçi bahis sitesi, Vippark canlı maç hizmeti aracılığıyla çok çeşitli spor kanalları sunar.
Sahadanbet Her lisanslı şirket bahisçiyi düzenli olarak kontrol eder. Sahadanbet Twitter bahis sayfasında çeşitli platformlarda binlerce oyuncu yorumu bulunmaktadır.
Bet10bet twitter hesabı bu dönemde tarafımızca hazırlanacak Bet10bet yeni kayıtlı adresi de yayınlanarak hizmete başlanacaktır.
Betro Oyuncular firmaya yaptıkları incelemelerde bahis sitesinin güvenilirliği konusunda olumsuz yorumlarda bulunmazlar. Betro Bu bahis şirketi tüm oyuncuların güvenebileceği ve yüksek kalitede oyun hizmetleri sunan bir şirkettir.
Betveno Bahisçiler her zaman müşteri hizmetleri faaliyetlerine değer verir. Betveno Oyuncular herhangi bir sorunla karşılaştıklarında müşteri hizmetleriyle iletişime geçebilirler.
Youwin güncel giriş adresi göre hareket ederseniz Youwin hiçbir sorun yaşanmayacak ve kazanmak daha kolay olacaktır.
Betclub canlı destek alanı şirket, canlı destek alanını Betclub herkese açarak sadece üyelerin değil herkesin faydalanabileceği ağ oluşturma fırsatları sunuyor.
Evbahis Günün en yoğun saatlerinde oyuncular müşteri hizmetleriyle iletişime geçmek için 2-3 dakikaya kadar bekleyebiliyor. Evbahis Profesyonel kadromuz aklınıza takılan her türlü soruya hızlı bir şekilde cevap verecektir.
Pirobet Twitter müşteri desteği, oyuncuların karşılaştıkları çeşitli olaylara hızlı bir şekilde çözüm bulmalarına olanak tanır. Pirobet Bahis şirketlerinin birbirleriyle rekabet edebilmesi için oyunculara çeşitli hizmetler sunabilmeleri gerekir.
Betteyim Neredeyse tüm bahisçiler sabit bahisçiler yerine canlı bahisleri tercih ediyor. Betteyim Bahis şirketinin yapısı gereği canlı bahis bulunmamaktadır ve bahisler tamamen animasyonlu olarak yapılabilmektedir.
Celtabet için hangi belgelere ihtiyacım var türkiye’yi ziyaret Celtabet eden kişiler bir siteye giriş yapmaya çalıştıklarında yöntem hata koduyla sonuçlanan bir sorunla karşılaşabilirler.
Betropol Twitter canlı bahis oyununun son dönemde adından güvenle söz ettiren bir firma olduğu söyleniyor. Betropol Twitter Canlı Bahis Oyunu, tüm oyuncuların spor etkinliklerine ve klasik oyunlara canlı bahis oynamasına olanak tanır.
Supertotobet yorumlarınız yorumlarınız ayrıntılı bir şekilde Supertotobet incelenecek ve gerekli düzeltmelerin yapılması için doğrudan kaynak görevi görecektir.
Онлайн-платформа для азартных игр представляет собой разнообразный мир развлечений. Здесь игроки могут наслаждаться множеством опций, которые сделают времяпрепровождение увлекательным и захватывающим http://peaklift.ru/communication/forum/messages/forum3/message249/253-kazino-gama_-sovety-novichkam-i-opytnym-igrokam?result=new#message249
Betparibu Klasikte canlı bahse katılmak isteyen bahisçiler, bahislerini Klasikte gerçek rakiplere karşı koyarlar. Betparibu Spor karşılaşmalarına canlı bahis oynayan oyuncular, bahislerini televizyon kanallarından da izlemektedir.
Şansınabahis Oyuncular TV kanallarında bahis yapmadan önce oynamak istedikleri oyunları izleyip yorumlayabilir ve ardından bahislerini oynayabilirler. Şansınabahis Artık sorunsuz bir şekilde devam edebilirsiniz.
Smartbahis mobil i̇çeriği sitesine kayıt olmak Smartbahis isteyen bahisçiler bunu hiçbir ücret ödemeden yapabilirler.
Discover the thrill of winning big at our premier [url=https://flashroyal.us/casinos/]casinos in florida tampa[/url], where every spin of the wheel promises excitement and opportunity. Immerse yourself in a world of luxury and glamour, where the lights are bright, and the stakes are high. Our casino offers an unparalleled gaming experience with a vast selection of classic and modern games tailored for all levels of players. Join us and feel the adrenaline rush as you take your chance at fortune in an atmosphere of elegance and sophistication.
Primebahis Kendinizi çok daha iyi bir durumda bulacaksınız. Primebahis Burada yüksek faiz oranlarından faydalanabilir ve mümkün olduğunca çok para kazanabilirsiniz.
Чай від просуди – це корисний напій, що виготовляють з різних лікарських рослин. Фітотерапевтичний напій багатий на корисні речовини для організму, оскільки має багато корисних речовин.
Чай від застуди можна замовити в магазині AD-UA на території України. AD-UA пропонує різноманітні смаки трав’яного чаю на будь-який бажання.
Переходь за посиланням – https://ad-ua.com/chaj/trav-yanyj-chaj/
1win предлагает широкий выбор азартных игр и спортивных ставок с привлекательными бонусами.
Ищете рабочее зеркало для доступа к онлайн казино pinco? Узнайте, как быстро и безопасно попасть на официальный сайт и насладиться азартными играми прямо сейчас!
Используйте 1вин рабочее зеркало для бесперебойной игры.
Пройдите 1win регистрация и получите приветственные бонусы.