Protocol PDF Document (version 1.2): cgh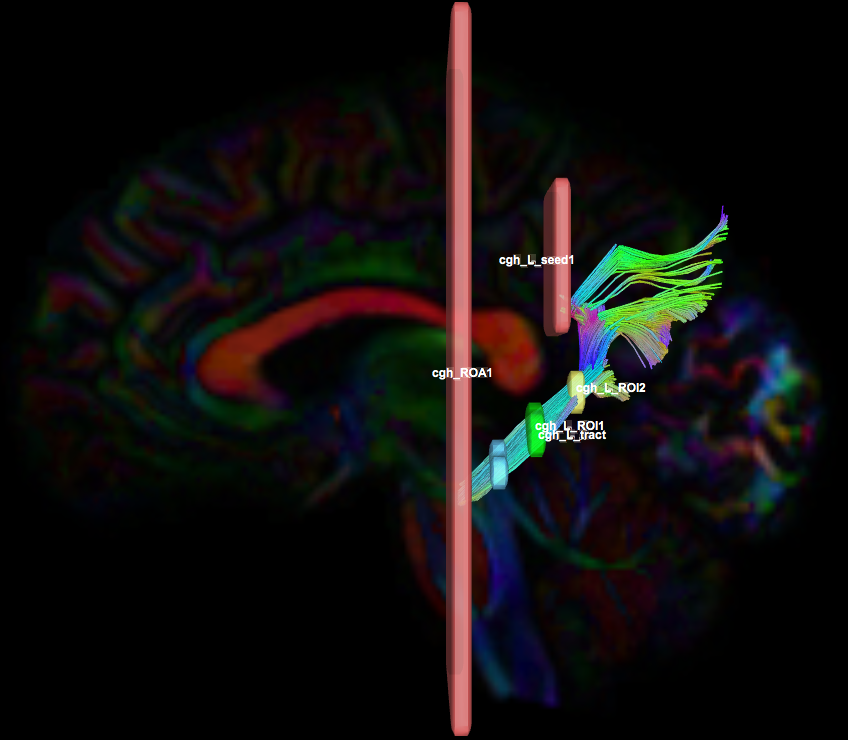
Instructions:
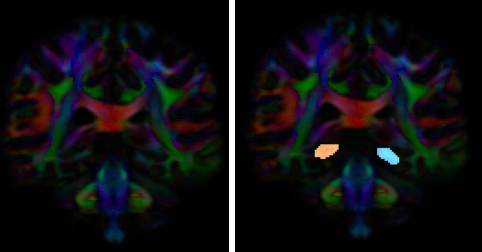
- Create two separate coronal seed regions (at approx. coronal slice 107): one for each side
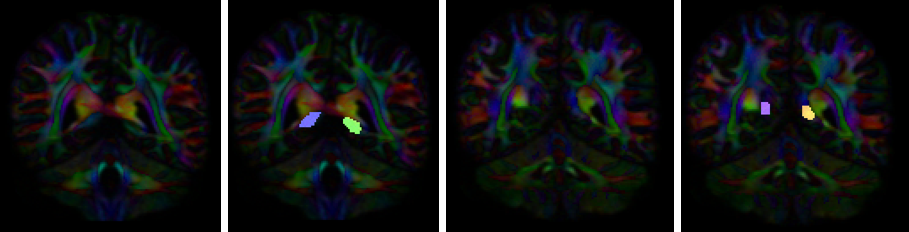
- Create four separate coronal ROI regions (at approx. coronal slice 116 and 122): two for each side.
- Create one ROA region and draw a sagittal ROA slice at the midline (at approx. sagittal slice 78).
- Check the left seed/ROI regions and ROA region, then perform fiber tracking. Based on this output, the other ROA placements will be clearer.
- Using the same ROA file, draw 2-3 more regions:
- on a coronal slice anterior to seed/ROI regions (at approx. coronal slice 89)
- on optional coronal slices superior to the ROI regions.
- In the region list, check only the left seed region and ROA regions, then perform fiber tracking. Under the tract list, make sure only the desired left tract is checked and highlighted in purple. Save region, tract, and density files.
- Uncheck the left seed region and check the right seed region and ROA region, then perform fiber tracking. Under the tract list, make sure only the desired right tract is checked and highlighted in purple. Save region, tract, and density files.

Use Jones et al. paper to do all CG tracking.
Seed doesn’t match seed shown, ROA looks like it is uses as termination regions – this one needs heavily modified.
I’m not sure how you could get a bundle like the one shown without using an ROA below the seed/ROIs.
This is most useful article I have found, when everyone else writing about this won’t stray from the standard doggerel.
You have a great writing style, and I will check back as I get a lot from your posts.
Hi! I know this is kinda off topic however , I’d figured I’d ask.
Would you be interested iin exchanging links or maybe guest authoring
a blog post oor vice-versa? My site discusses a lot of the same subjects as yours
and I feel we cold greatly benefit rom each other. If you are
interested feel free to send me an e-mail.
I look forward tto hearing from you! Great blog by the way!
Enjoyed looking at this, very good stuff, regards.
Hmm it looks like your website ate my first comment (it was extremely long) so I guess I’ll juist sum it up what I wrote annd say, I’m
thoroughly enjoying your blog. I too am an aspiring
blog writer but I’m still nnew to everything. Do you have any tils and hints for beginner blog writers?
I’d really appreciate it.
Hey! I know this is kinda off tokpic but I was wondering which blog platform
are you using for this website? I’m getting fed up oof Wordpresxs because I’ve had problems with hackers and I’m looking at
options for anothner platform. I would be fantastic if youu could point me in the direction oof a good platform.
Some truly nice and utilitarian information on this site, also I think the style contains great features.
Thanks for sharing your thoughts. I really appreciate your efforts and I
will be waiting for your further write ups thank you once again.
Fantastijc beat ! I would like to apprentice even as you amend your web
site, how could i subscribe for a weblog web site? The account
aided mee a acceptable deal. I had been a little bit acquainted of this your broadcast offered vibrant clear concept.
Wohh exactly what I waas looking for, appreciate it forr posting.
This exellent website really has all of the information I wanted about tis subject and didn’t know
who to ask.
It’s fantastic that you are getting ideas from this piece of writing as well as from our dialogue made at this time.
pozitivit.com
그러나 그의 아들이 그에게 이 질문을 했을 때 그는 어안이 벙벙했습니다.
https://30secondstomars.ru/away.php?go=https%3A%2F%2Ffreedietlinks.com%2F
This poist is truly a good one it assists new net users, who are
wishing in favor of blogging.
constantly i used too read smaller posts that also clear their motive, and that is also happening with this post
which I am reading now.
eexhibitionunche.com
보통 사람들은 가장 똑똑히 볼 수 있고, 아무것도 그들을 속일 수 없습니다.
http://nabchelny.ru/welcome/blindversion/normal?callback=http%3A%2F%2Ffreedietlinks.com%2F
Hey there would you mind letting me know which web host you’re working with?
I’ve loaded your blog in 3 different browsers and I must say this
blog loads a lot faster then most. Cann you recommend a
good internet hosting provider at a honest price? Kudos, I
appreciate it!
My brother suggested I would possibly like this web site.
He used to be entirely right. Thhis put up truly made mmy day.
You can not imagine simply how much time I had spent for this info!
Thanks!
Good article! We are linking to this particularly great
article on ouur website. Keep up the great writing.
I have recently started a web site, the information you provjde oon this web site haas helped me
tremendously. Thanks for all of your time & work.
I blog frequently and I genuinely thank you for your content.
The article has truly peaked my interest. I am going
to bookmark your blog and keep checking for new details about once per week.
I subscribed to your RSS feed too.
It is perfect time to make some plans for the future
and it’s time to be happy. I have read this post and if
I could I wish to suggest you some interesting things or advice.
Maybe you could write next articles referring to this article.
I wish to rea even more things abiut it!
혁신적이고 표준화된 콘텐츠를 제공하는 최신 프라그마틱 게임은 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 지원합니다.
프라그마틱 무료
프라그마틱의 게임을 플레이하면서 항상 신선한 경험을 얻을 수 있어 기뻐요. 여기서 더 많은 이야기를 나눠봐요!
https://p2w-club.com/
https://btob-business.com/
https://pgslot-9tiger.vip/
I’d always want to be update on new articles on this website, saved to bookmarks!
This is really interesting, You are a very skilled blogger.
I’ve joined your rss feed and look forward to seeking more
of your great post. Also, I have shared your web site
in my social networks!
Howdy! Do you use Twitter? I’d like to follow you if that
would be ok. I’m definitely enjoying your blog and look forward to new
updates.
ledealdumois.com
“오.” Li Chaowen은 잠시 침묵을 지키다가 고개를 들고 작은 도교 사제에게 무언가를 말했습니다.
https://99rental.com/?URL=https%3A%2F%2Fpozitivit.com%2F
프라그마틱에 대한 내용 정말 흥미로워요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 지식을 공유해보세요!
에그벳
프라그마틱 슬롯에 대한 정보가 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 내용을 찾아보세요. 함께 이야기 나누면서 더 많은 지식을 얻어가요!
https://wtsnzp.com/
https://www.hz-wallpaper.com
http://keoghsflex.com/
프라그마틱은 항상 훌륭한 게임을 만들어냅니다. 이번에 새롭게 출시된 게임은 정말 기대되는데요!
프라그마틱 슬롯 무료 체험
프라그마틱 슬롯을 다룬 글 정말 유익해요! 더불어, 제 사이트에서도 프라그마틱에 대한 새로운 소식을 전하고 있어요. 함께 지식을 나누면 좋겠어요!
https://themarketlobby.com/
http://iyalion.com/
https://dwqewqe.weebly.com/
digiapk.com
Fang Jifan은 미소를 지었습니다. “전하 후보가 있습니까?”
kinoboomhd.com
Fang Jifan은 도울 수 없었지만 영국 Duke Zhang Mao를 바라 보았습니다.
great issues altogether, you just gained a new reader.
What may you suggest in regards to your submit that you simply made some
days ago? Any sure?
프라그마틱 슬롯은 풍부한 다양성과 흥미진진한 게임 플레이로 눈길을 사로잡습니다.
프라그마틱 슬롯 무료
프라그마틱 슬롯에 대한 내용이 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 찾아보실 수 있어요. 함께 지식을 공유해보세요!
https://www.arabyfree.com
https://www.tengerszemhotel.com
https://www.vamiveta.com/
Hello.This article was really interesting, particularly since I was searching for thoughts on this topic last Wednesday.
As I website possessor I think the subject material here is really superb, regards for your efforts.
binsunvipp.com
잠시 후 Wang Jinyuan은 소식을 듣고 서둘러 “스승님 …”
iGaming 분야에서 혁신적이고 표준화된 콘텐츠를 제공하는 최신 프라그마틱 게임은 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 지원하여 고객에게 엔터테인먼트를 제공합니다.
프라그마틱
프라그마틱의 슬롯 게임은 정말 뛰어나죠! 여기서 더 많은 게임 정보를 찾을 수 있어 기뻐요.
https://www.hotelsoftwarepro.com
https://www.anomaxx.com
https://www.woomintech.com
katsluxury.com
Zhu Zaimo의 말을 듣고 Fang Jifan의 얼굴이 약간 변했습니다.
https://maps.google.la/url?q=https://www.maseraticlubuae.com/
saungsantoso.com
위공(徐公) 허유(徐禹)가 북경에 도착하자 즉시 정공(德公) 서용녕(徐宇寧)과 합세하였다.
프라그마틱플레이의 슬롯으로 도전과 흥미를 느껴보세요.
프라그마틱 게임
프라그마틱 슬롯에 대한 정보가 정말 유용했어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 새로운 내용을 찾아보세요. 함께 지식을 나누면 좋겠어요!
https://www.btob-business.com
https://www.jiangxiangtiyu.com
https://www.pihmctt.com
katsluxury.com
“폐하…” Fang Jifan이 옆에서 미소를 지으며 말했습니다.
http://www.google.at/url?q=https://www.pozitivit.com/
netovideo.com
괜찮아, 괜찮아, 어떤 큰 바람과 파도, 노인은 전에 본 적이 없습니까?
saungsantoso.com
왕세자의 자리는 결코 시성처럼 단순하지 않다.
I’d been honored to receive a call coming from a friend as soon as he found out the important suggestions shared in your site.
Going through your blog post is a real great experience.
Thanks again for considering readers like me, and I hope for you the best of
achievements being a professional in this realm.
I like this web blog very much, Its a rattling nice place to read and find info.
다양한 테마의 슬롯을 즐길 수 있는 250개 이상의 게임으로 구성된 프라그마틱 플레이의 슬롯 포트폴리오를 세계 시장에서 즐겨보세요.
프라그마틱 무료
프라그마틱의 게임은 언제나 최신 트렌드를 반영하고 있죠. 최근에 나온 트렌드 중에서 가장 마음에 드는 것은 무엇인가요
https://eprust.com/
https://www.itfgf25r.site
https://www.iz21swbn.site
프라그마틱 게임은 iGaming 업계의 선두 주자로, 모바일 중심의 혁신적이고 표준화된 콘텐츠를 선보입니다.
프라그마틱플레이
프라그마틱은 항상 훌륭한 게임을 만들어냅니다. 이번에 새롭게 출시된 게임은 정말 기대되는데요!
http://usmcafee.site
https://www.824989.com
https://www.ovn1316r.site
Hello, you used to write great, but the last several posts have been kinda boring?
I miss your great writings. Past several posts
are just a bit out of track! come on!
restaurant-lenvol.net
왕서인의 등장은 이곳 마을 사람들에게는 극히 드문 일이다.
kinoboomhd.com
Wang Bushi는 고개를 끄덕이고 고개를 끄덕였습니다. “폐하에게 돌아가십시오. 그것에 대해 들었습니다.”
Whoah this weblog is wonderful i like reading your posts.
Keep up the great work! You realize, lots of persons are
looking round for this info, you can aid them greatly.
Хотите [URL=https://diplom4.me/]купить диплом техникума[/URL]? Мы предоставляем сопровождение по оформлению бумаг с подтверждением от официальных учебных заведений. Обращайтесь к нам!
We stumbled over here by a different web address and thought I might
as well check things out. I like what I see so now i’m following you.
Look forward to exploring your web page again.
iGaming 업계의 주역, 프라그마틱 게임은 모바일 중심의 혁신적이고 표준화된 콘텐츠를 제공하는 선도적인 제공 업체입니다.
프라그마틱슬롯
프라그마틱의 게임을 플레이하면서 항상 신선한 경험을 얻을 수 있어 기뻐요. 여기서 더 많은 이야기를 나눠봐요!
http://drlooikokpoh.site
https://www.rubiconfc.com
http://buyclomid.site
В Москве приобретение документа https://diplomsuper.net – это такой практичный и полезный и быстрый вариант законного доказательства учебы. Многочисленные компании предлагают опции по изготовлению документов учитывая разнообразных требований и специализаций, предоставляя клиентам удобство и надежность.
Comprar Cialis En Amazon
Excuse, I have thought and have removed the idea
Cialis 5 mg prezzo cialis prezzo cialis 5 mg prezzo
restaurant-lenvol.net
그는 주위를 둘러 보았고 Fang Jifan은 어디로 가야할지 몰랐고 Zhu Houzhao도 군중 끝에 숨었습니다.
최신 프라그마틱 게임은 iGaming에서 혁신적이고 표준화된 콘텐츠를 제공하는 선도적인 제공 업체입니다. 높은 품질의 엔터테인먼트를 제공하기 위해 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 지원합니다.
프라그마틱 슬롯
프라그마틱은 항상 최고의 게임을 제공하죠! 여기에서 더 많은 흥미진진한 정보를 얻을 수 있어 기뻐요.
http://viagraes.online
https://www.zeovitusa.com
https://www.cialisforsale.site
Купить сертификат бакалавра: Заказ диплома высшего образования поможет вам подняться на новый уровень в профессии.
[URL=https://diplomkupit.org/]Заказать свидетельство СССР[/URL] – это возможность оперативно достать бумагу об учебе на бакалавр уровне безо лишних хлопот и затрат времени. В Москве предоставляется различные вариантов оригинальных дипломов бакалавров, предоставляющих комфортность и легкость в процессе.
netovideo.com
Zhu Houzhao는 침착하게 말했습니다. “아버지를 도와주세요. 아버지는 저를 때리고 싶어합니다.”
Внутри городе Москве купить свидетельство – это комфортный и оперативный способ получить нужный запись лишенный лишних хлопот. Большое количество организаций предоставляют сервисы по изготовлению и реализации свидетельств различных учебных заведений – https://www.diplom4you.net/. Выбор дипломов в столице России большой, включая документы о высшем уровне и среднем образовании, аттестаты, свидетельства колледжей и вузов. Главное плюс – возможность приобрести аттестат официальный документ, гарантирующий подлинность и качество. Это предоставляет особая защита против подделок и предоставляет возможность воспользоваться аттестат для разнообразных задач. Таким способом, заказ диплома в городе Москве становится достоверным и экономичным выбором для данных, кто желает достичь успеха в карьере.
10yenharwichport.com
그러나 그들이 자주 받는 대답은 “저리가, 이 젊은 주인이 산 것은 황무지입니다!”입니다.
smcasino7.com
그는 고개를 숙이고 몸의 비뚤어진 질감과 지저분한 바늘을 보았다.
smcasino-game.com
Hongzhi 황제는 참을성이 없어졌습니다. 이것이 왕자와 어떤 관련이 있습니까? “간결하고 간결합니다.”
tsrrub.com
그리고 Daming Palace에서 Zhu Houzhao 황제도 오늘 일찍 일어났습니다.
???https://tipsmo.com/
lfchungary.com
Fang Jifan은 그를 노려 보았습니다. “나는 왕자가 아니라고 말했지만 당신이라고 말했습니다.”
http://naftometgroup.com/heavy-lifting-equipment/#comment-17392
http://murafka.net/post-2?unapproved=330768&moderation-hash=3600b1fed535c1100fde3fd5f76db24d#comment-330768
http://mnogootvetov.ru/index.php?qa=88355&qa_1=discussion-advanced-classes-thane-insights-community-support&show=89766#a89766
http://microbialfutures.com/index.php/2020/05/20/process-video-v2/?unapproved=420712&moderation-hash=2f4fce4ed86b8bb69c553602d0aacac1#comment-420712
http://leginy.sk/2019/07/17/leginy-eshop-online/?unapproved=363571&moderation-hash=8877386d1b9d6224997d447c44fb56f8#comment-363571
sm-casino1.com
정말 멋진 아이, 이렇게 그냥 사라지면 안타까울 것입니다.
http://xn--80aphfq.xn--p1ai/index.php?subaction=userinfo&user=yfaceqire
http://xn--b1acebaenad0ccc3aiee.xn--p1ai/forum/user/2811/
http://zemli.com/index.php?subaction=userinfo&user=ahukez
http://zingcorp.com.au/component/kunena/sub-category-3/59398#59440
http://zpu-journal.ru/forum/view_profile.php?UID=300541
프라그마틱플레이의 무료 슬롯으로 특별한 경험을 즐겨보세요.
프라그마틱 게임
프라그마틱 관련 내용 감사합니다! 또한, 제 사이트에서도 프라그마틱에 대한 유용한 정보를 공유하고 있어요. 함께 서로 이야기하며 더 많은 지식을 쌓아가요!
https://www.lasix40mg.site
https://www.zebrariver.com
https://www.howtodrawaeasy.com
lfchungary.com
그것은 단지…오늘…그는 덜 신경을 쓸 수 없었습니다.
https://spashop.com.ua/collections/krioterapiya-dlya-lica
https://spashop.com.ua/collections/lasery-dlya-epilyacii
https://spashop.com.ua/collections/lasery-dlya-karbonovogo-pilinga
https://spashop.com.ua/collections/lasery-dlya-kosmetologii
https://spashop.com.ua/collections/lasery-dlya-udaleniya-sosudov
sm-online-game.com
Zhu Hou가 그것을 처리한 후, 그의 얼굴은 공포로… 사납게 변했습니다.
lfchungary.com
Fang Jifan과 Zhu Xiurong은 경례하러 갔지만 Zhu Houzhao는 어디에도 보이지 않았습니다.
smcasino7.com
하룻밤 사이에 Yanda Khan의 노력은 물거품이 되었습니다.
В городе Москве приобрести диплом – это удобный и оперативный метод получить нужный запись лишенный дополнительных трудностей. Множество организаций предоставляют помощь по производству и продаже дипломов разных образовательных институтов – https://www.prema-diploms-srednee.com/. Ассортимент свидетельств в столице России огромен, включая документы о высшем уровне и нормальном профессиональной подготовке, свидетельства, свидетельства колледжей и вузов. Основной плюс – возможность получить свидетельство Гознака, гарантирующий достоверность и высокое стандарт. Это гарантирует уникальная защита ото подделки и позволяет воспользоваться аттестат для различных нужд. Таким образом, приобретение свидетельства в городе Москве становится достоверным и эффективным выбором для таких, кто желает достичь успеха в карьере.
lfchungary.com
Hongzhi 황제는 하늘을 흘끗 보았습니다. “늦어 져서 두렵습니다.”
Внутри столице России заказать свидетельство – это комфортный и экспресс метод достать нужный запись лишенный избыточных проблем. Множество организаций предлагают помощь по созданию и торговле свидетельств различных образовательных учреждений – http://www.gruppa-diploms-srednee.com. Ассортимент свидетельств в Москве огромен, включая документы о высшем и среднем учебе, аттестаты, дипломы техникумов и университетов. Главное плюс – способность получить диплом Гознака, обеспечивающий достоверность и качество. Это гарантирует особая защита против подделок и дает возможность воспользоваться аттестат для различных задач. Таким путем, заказ аттестата в городе Москве становится достоверным и эффективным решением для таких, кто желает достичь успеха в трудовой деятельности.
agenbet88score.com
내 아버지는 내 멘토만큼 나에게 친절하지 않았습니다.
https://image.google.com.sl/url?rct=j&sa=t&url=https%3A%2F%2Fwww.agenbet88score.com%2F
pragmatic-ko.com
Zhongyong이라는 말을 듣고 Hongzhi 황제는 즉시 마음에 위로를 받았습니다.
Hi there, every time i used to check weblog posts here in the early hours in the morning, for the reason that i enjoy to learn more and more.
My page site#:
http://veneraroleplay.listbb.ru/viewtopic.php?f=4&t=128
https://poselki.animetalk.ru/viewtopic.php?id=25700#p38730
https://open.ktu.edu/tag/index.php?tc=1&tag=%D0%A0%D0%90%D0%97%D0%9D%D0%98%D0%A6%D0%90%20%D0%9C%D0%95%D0%96%D0%94%D0%A3%20%D0%A2%D0%95%D0%A5%D0%9D%D0%98%D0%9A%D0%A3%D0%9C%D0%9E%D0%9C%20%D0%9A%D0%9E%D0%9B%D0%9B%D0%95%D0%94%D0%96%D0%95%D0%9C%20%D0%98%20%D0%A3%D0%A7%D0%98%D0%9B%D0%98%D0%A9%D0%95%D0%9C%20%28%D0%9F%D0%A2%D0%A3
http://schoolis.bestbb.ru/viewtopic.php?id=509#p913
https://goup.hashnode.dev/kupit-diplom-v-moskve-1
프라그마틱 슬롯에 대한 내용이 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 찾아보세요. 함께 서로 이야기하며 더 많은 지식을 쌓아가요!
프라그마틱 홈페이지
프라그마틱에 대한 글 읽는 것이 정말 즐거웠어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 발전하며 더 많은 지식을 쌓아가요!
https://aintec.net/hot/
https://www.googlikgid.site
https://www.blerifa.com
sm-slot.com
이제 이 사람들은 모두 폐하의 상태를 개인적으로 알고 있었고 모두 슬퍼 보였습니다.
lfchungary.com
미스가 많고 적중 확률이 30~40%에 불과하다.
I appreciate, lead to I discovered exactly what I was looking for. You’ve ended my four day long hunt! God Bless you man. Have a nice day. Bye
http://borderforum.ru/viewtopic.php?f=26&t=8451
http://int.5bb.ru/viewtopic.php?id=21888#p86740
http://forum.autoset.ru/viewtopic.php?p=142156#142156
http://crimeadetka.forum.cool/viewtopic.php?id=1489#p87418
http://rybolov.webtalk.ru/viewtopic.php?id=1554#p24663
http://balashihaforum.2bb.ru/viewtopic.php?id=2958#p10817
Wow that was unusual. I just wrote an extremely long comment but after I clicked submit my comment didn’t show up. Grrrr… well I’m not writing all that over again. Anyways, just wanted to say fantastic blog!
http://go0gle.coom
http://buxforum.9bb.ru/viewtopic.php?id=1564#p3489
http://drahthaar-forum.ru/viewtopic.php?f=3&t=9084
http://collieforum.ru/viewtopic.php?f=34&t=6882
https://izhevsk.ru/forummessage/153/6113640.html
http://equip.7bb.ru/viewtopic.php?id=8861#p11737
bistroduet.com
이 작은 판잣집에는 온갖 규모의 관리들이 십여 명이 넘는다.
lfchungary.com
호기심 많은 아기처럼 보이는 Zhang Yuanxi를 모두가 바라 보았습니다.
гранд вегас казино с выводом
казино vegas grand официальный сайт рубли
다양한 화폐로 지원되는 프라그마틱 슬롯은 세계 각지의 플레이어에게 열린 기회를 제공합니다.
프라그마틱 게임
프라그마틱에 대한 글 읽는 것이 정말 재미있었어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 발전하며 더 많은 지식을 쌓아가요!
https://www.emulecenter.com
https://www.kesambet.com
https://www.rachitadas.com
strelkaproject.com
이 말을 들은 Zhu Xiurong은 유쾌하게 놀라며 목소리를 낮추었습니다. “정말로… 아버지는 건강하십니다.”Zhu Xiurong은 요즘 태어날 아이를 위해 스웨터를 뜨개질하고 있습니다.
logarid.com
Fang Jifan은 떨고 돌아서 뛰었지만 Zhang Mao는 어깨에 팔을 얹었습니다!
https://images.google.al/url?q=https%3A%2F%2Fwww.agenbet88score.com%2F
apksuccess.com
Hongzhi 황제는 고개를 끄덕였고 많은 징후가 나타나기 시작했습니다.
Объемная георешетка ГЕОСПАН ОР 20/10 купить в Ростове-на-Дону
В Москве купить свидетельство – это комфортный и экспресс способ достать нужный документ лишенный лишних хлопот. Большое количество компаний предлагают помощь по изготовлению и торговле дипломов разнообразных учебных заведений – http://www.orik-diploms-srednee.com. Ассортимент дипломов в столице России большой, включая бумаги о академическом и нормальном профессиональной подготовке, свидетельства, свидетельства техникумов и вузов. Основной достоинство – возможность приобрести свидетельство подлинный документ, подтверждающий истинность и качество. Это предоставляет особая защита против подделки и позволяет использовать аттестат для разнообразных задач. Таким путем, заказ диплома в Москве является надежным и экономичным решением для таких, кто стремится к процветанию в трудовой деятельности.
На территории столице России приобрести свидетельство – это практичный и быстрый метод завершить нужный бумага без дополнительных трудностей. Большое количество организаций предлагают сервисы по изготовлению и торговле свидетельств разнообразных образовательных институтов – https://www.russa-diploms-srednee.com/. Ассортимент дипломов в столице России огромен, включая документы о высшем уровне и среднем ступени учебе, аттестаты, свидетельства колледжей и университетов. Основной достоинство – возможность достать аттестат Гознака, подтверждающий истинность и качество. Это предоставляет особая защита ото подделки и предоставляет возможность использовать аттестат для различных целей. Таким образом, покупка диплома в столице России является надежным и эффективным решением для тех, кто хочет достичь успеха в трудовой деятельности.
Наша компания предлогает Продвижение сайтов Тараз включают SEO-оптимизацию, контент-маркетинг и аналитику для повышения онлайн-видимости вашего бизнеса.
madridnortehoy.com
Ouyang Zhi는 즉시 현에 폐하의 흔적을 찾도록 명령했습니다.
sm-slot.com
물론 가장 중요한 것은 Zhu Zaimo와 다른 사람들이 항상 지켜보고 있다는 것입니다.
parrotsav.com
Jiang Yan의 얼굴은 더욱 비극적이었고 믿을 수 없다는 눈으로 눈앞의 장면을 바라 보았습니다.
프라그마틱에 대한 내용이 정말 유익했어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 찾아보실 수 있어요. 함께 지식을 공유해보세요!
http://www.pragmatic-game.com
프라그마틱 관련 정보 감사합니다! 제 사이트에서도 유용한 정보를 공유하고 있어요. 함께 소통하면서 발전하는 모습 기대합니다!
https://www.belgiumfire.com
https://okgasda.weebly.com/
https://supervil.com/hot/
mojmelimajmuea.com
이 말을 들은 사람들은 모두 말을 아끼고 속속 무기를 가지러 갔다.
mega-casino77.com
“그렇다면 말타고 사격하는 것이야말로 도시 공략에 가장 유리한 전술이지.”
https://cse.google.jo/url?q=https%3A%2F%2Fwww.colorful-navi.com%2F
crazyslot1.com
누에 방 안에는 수십 개의 램프가 빛나고 있어 그림자가 없는 효과를 낸다.
raytalktech.com
물론 Fang Jifan은 Wang Jinyuan을 모욕하려는 것이 아니라 단지 비유를 한 것입니다.
프라그마틱에 대한 글 읽는 것이 흥미로웠어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 제공하고 있어요. 함께 발전하며 지식을 나눠봐요!
http://www.pragmatic-game.com
프라그마틱과 관련된 내용 감사합니다! 또한, 제 사이트에서도 프라그마틱에 대한 정보를 찾아보실 수 있어요. 서로 이야기 공유하며 더 많은 지식을 얻어가요!
https://www.ivermectininstock.com/
https://www.belgiumfire.com
https://www.ashspurr.com
bistroduet.com
장천은 재빨리 통조림에 담긴 얇게 썬 고기를 집어 아들에게 조심스럽게 건넸다.Zhang Heling은 “이것은 분명히 전하의 뜻이며 우리와 관련이 없습니다. “라고 말했습니다.
khasiss.com
Hongzhi 황제는 충분하지 않다고 느끼는 것 같아서 Zhu Houzhao에게 함께 궁전에 들어가도록 개인적으로 명령했습니다.
Где купить диплом – Это способ завладеть официальный удостоверение по завершении образовательного учреждения. Свидетельство открывает пути к последующим карьерным перспективам и карьерному развитию.
sm-online-game.com
Hongzhi 황제는 요즘 고통이 조금 완화되었다고 느꼈습니다.
Hi! Do you use Twitter? I’d like to follow you if that would be okay. I’m undoubtedly enjoying your blog and look forward to new posts.
https://toolbarqueries.google.iq/url?q=https://hottelecom.biz/id/
jbustinphoto.com
Li라는 씨족 씨족의 많은 후손들도 Yuanjun의 저택에서 사망했습니다.
다양한 테마의 프라그마틱 슬롯로 흥미진진한 여행을 시작하세요.
프라그마틱 무료
프라그마틱 슬롯에 대한 내용이 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 찾아보세요. 함께 서로 이야기하며 더 많은 지식을 쌓아가요!
https://dwqewqe.weebly.com/
https://vispills.com/
https://e-trajet.net/hot/
Привет, дорогой читатель!
Были ли у вас случаи, когда приходилось писать дипломную работу в крайне ограниченные сроки? Это действительно требует большой ответственности и трудоемкости, но важно не отступать и продолжать активно участвовать в учебном процессе, как я.
Для тех, кто умеет эффективно находить и использовать информацию в сети, это действительно помогает в процессе согласования и написания дипломной работы. Больше не нужно тратить время на посещение библиотек или организацию встреч с научным руководителем. Здесь, на этом ресурсе, предоставлены надежные данные для заказа и написания дипломных и курсовых работ с гарантией качества и доставкой по всей России. Можете ознакомиться с предложениями тут https://1server-diploms.com, это проверенный источник!
купить диплом техникума
купить диплом бакалавра
Желаю каждому нужных оценок!
Всем хорошего дня!
Написание диплома стало эффективнее с использованием интернет-ресурсов.
На нашем сайте вы можете купить диплом ВУЗа с постоплатой и поддержкой 24/7.
http://arenda.mineralgroup.ru/index.php?subaction=userinfo&user=iletaba
http://molbiol.ru/forums/index.php?showtopic=1089291
http://fruit-impex.by/index.php?subaction=userinfo&user=ahoxiduv
Желаю для каждого прекрасных отметок!
купить диплом в уссурийске
купить диплом в владикавказе
купить диплом в улан-удэ
купить диплом в хабаровске
купить диплом в красноярске
Всем хорошего дня!
Диплом продолжает развиваться благодаря различным онлайн-ресурсам.
Закажите диплом у нас легко! Без предоплаты, с гарантией качества и доставкой по всей России.
http://kungur.hldns.ru/forum/profile.php?action=show&member=5113
http://newens.co.kr/bbs/board.php?bo_table=free&wr_id=925619
https://xn--80ae1aidok.xn--p1ai/users/unupiw
Желаю всем отличных отметок!
купить диплом в миассе
купить диплом в волжском
купить диплом в якутске
купить диплом монтажника
купить диплом в заречном
smcasino7.com
말하면서 그는 Fang Jifan을 깊이 들여다 보았습니다. “Fang Jifan, 이리와.”
에그벳 도메인
Hongzhi 황제는 사려 깊고 Liu Jian의 말이 매우 합리적이라고 느꼈습니다.
https://image.google.com.mt/url?q=https%3A%2F%2Fwww.agenbet88score.com%2F
프라그마틱 게임은 선도적인 iGaming 콘텐츠 제공 업체로, 모바일 중심 다중 포트폴리오와 높은 품질의 엔터테인먼트를 자랑합니다. 우리는 슬롯, 라이브 카지노, 빙고 등 모든 제품에서 최고의 경험을 제공합니다.
프라그마틱 홈페이지
프라그마틱의 게임은 정말 다양한데, 최근에 출시된 것 중 어떤 게임이 가장 좋았나요? 공유해주세요!
https://szyangan.com/link/
https://www.12315amc.cn/
https://hoetron.com/link/
jelenakaludjerovic.com
큰 쥐는 유령과 현자에 대해 말하지 않고 얼굴이 붉어지고 큰 소리로 울었습니다.
bistroduet.com
Zhang Tao는 엄숙하고 의롭게 말했습니다. “당신의 전하가 여기서 무례해서는 안됩니다.”이런 일이 계속된다면 그들의 미래는 이미 상상할 수 있습니다.
Aviator Spribe играть
Добро пожаловать в захватывающий мир авиаторов! Aviator – это увлекательная игра, которая позволит вам окунуться в атмосферу боевых действий на небе. Необычные графика и захватывающий сюжет сделают ваше путешествие по воздуху неповторимым.
Aviator Spribe играть без риска казино
mojmelimajmuea.com
침대 위에서 방금까지 움직이지 않던 홍치제가 갑자기 손가락을 떨었다.
ttbslot.com
Fang Jifan은 “그럼 빨리 … 연습합시다.”
???프라그마틱 슬롯
이 관리들은 속도에 대한 개념이 없습니다.
https://www.google.com.ly/url?sa=t&url=https%3A%2F%2Fwww.colorful-navi.com%2F
프라그마틱 슬롯은 특색 있는 테마와 뛰어난 기술로 다양한 경험을 제공합니다.
프라그마틱 플레이
프라그마틱 관련 소식 항상 주시고 있어요! 또한, 제 사이트에서도 프라그마틱에 대한 정보를 얻을 수 있어요. 함께 발전하며 지식을 나눠봐요!
https://www.12315lt.cn/
https://aintec.net/hot/
https://baicao10.com/link/
What’s up it’s me, I am also visiting this website daily, this website is truly nice and the users are really sharing pleasant thoughts.
RybelsusRybelsusRybelsusRybelsusRybelsus
twichclip.com
도둑을 잡는 과정도 왕수인의 눈을 뜨게 했다.
Thank you a lot for sharing this with all folks you really realize what you’re talking approximately! Bookmarked. Please additionally talk over with my site =). We may have a hyperlink alternate contract between us
Rybelsus
ttbslot.com
원고 Jia Qing은 아직 Xishan에 있기 때문에 경비원이 다시 그를 초대할 때까지 기다려야 합니다.
Its like you read my mind! You seem to know a lot about this, like you wrote the book in it or something. I think that you could do with a few pics to drive the message home a little bit, but instead of that, this is fantastic blog. A fantastic read. I’ll certainly be back.
Loudspeaker / Bullhorn VST Plugins
Купить диплом в кирове – Такова возможность получить официальный удостоверение по среднем образовании. Диплом гарантирует доступ к обширному ассортименту рабочих и образовательных перспектив.
Thanks , I have just been searching for information about this subject for a while and yours is the greatest I have found out till now. But, what in regards to the bottom line? Are you sure concerning the supply?
writing service
arusak-diploms-srednee24.com Купить диплом о среднем техническом образовании- Таков вариант завладеть официальный удостоверение о окончании образовательного учреждения. Свидетельство раскрывает пути к последующим карьерным перспективам и карьерному развитию.
ttbslot.com
사실 팡지판이 아이를 보낸 후 말이 빨리 도착했다.
qiyezp.com
Hongzhi 황제는 침묵하고 그의 눈은 우울했고 그가 무엇을 생각하는지 몰랐습니다.
nikontinoll.com
어머니는 마지못해 등불을 켰지만 다행히 남편의 목소리가 들렸다.
프라그마틱 슬롯을 무료로 즐기면서 특별한 순간을 만들어보세요.
프라그마틱 게임
프라그마틱은 항상 훌륭한 게임을 만들어냅니다. 이번에 새롭게 출시된 게임은 정말 기대되는데요!
https://www.12315hc.cn/
https://www.12315amb.cn/
https://hyunmee.net/hot/
toasterovensplus.com
素敵な内容でした!とても勉強になります。
Купить диплом о среднем образовании цена – Это завладеть официальный бумага по среднем образовании. Диплом обеспечивает вход к широкому ассортименту профессиональных и учебных возможностей.
Hello there! This is kind of off topic but I need some advice from an established blog. Is it hard to set up your own blog? I’m not very techincal but I can figure things out pretty quick. I’m thinking about making my own but I’m not sure where to begin. Do you have any points or suggestions? With thanks
reddit thread
Hi! Would you mind if I share your blog with my myspace group? There’s a lot of people that I think would really appreciate your content. Please let me know. Thanks
artrolux cream opiniones
Hi it’s me, I am also visiting this website on a regular basis, this web page is really nice and the visitors are truly sharing fastidious thoughts.
was ist cardiobalance
What you published was very reasonable. But, think about this, what if you added a little content? I mean, I don’t wish to tell you how to run your blog, however what if you added a post title to maybe grab a person’s attention? I mean %BLOG_TITLE% is kinda vanilla. You ought to peek at Yahoo’s front page and see how they create post titles to grab viewers interested. You might try adding a video or a pic or two to get people interested about everything’ve got to say. Just my opinion, it might bring your posts a little bit more interesting.
yabby casino no deposit bonus codes 2021
https://cf58051.tmweb.ru/index.php?topic=826.60
https://www.zombiepumpkins.com/forum/memberlist.php?mode=viewprofile&u=87847
https://www.eduportal44.ru/Sharya/shool2/2/SitePages/%D0%94%D0%BE%D0%BC%D0%B0%D1%88%D0%BD%D1%8F%D1%8F.aspx?Source=http://www.koipkro.kostroma.ru/Shablon/Lists/List9/AllItems.aspx&RootFolder=
https://lamatinale.esj-lille.fr/emission-83-la-matinale-de-lacademie-esj-27-01-2024/
https://horschool2.ucoz.ru/index/0-11
gama casino регистрация
gama casino регистрация
If some one wants to be updated with newest technologies therefore he must be visit this website and be up to date all the time.
프라그마틱 게임은 iGaming 분야에서 혁신적이고 표준화된 콘텐츠를 제공하는 선도적인 공급 업체로 주목받고 있습니다.
프라그마틱 슬롯
프라그마틱 슬롯에 대한 설명 감사합니다! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 이야기 나누면서 더 많은 지식을 얻어가요!
https://accountantslink.net/link/
https://www.12315qu.cn/
https://www.12378xs.cn/
Российский производитель реализует разборные гантели Gantel Razbornaya ru – у нас найдете объемный каталог предложений. Наборные снаряды позволяют эффективно выполнять силовые занятия в любом месте. Изделия для спорта отличаются функциональностью, универсальностью в использовании. Организация эффективно испытывает и совершенствует инновационные идеи, чтобы реализовать желания постоянных покупателей. В выпуске долговечных изделий реализуются лучшие марки металла. Большой каталог интернет-магазина вариантов дает возможность получить разборные отягощения для эффективной программы тренировок. Для домашних занятий – это комфортный инвентарь с маленькими габаритами и значительной фунциональности.
Создаваемые российским заводом тренажеры для кинезитерапии trenazhery-dlya-kineziterapii.ru и специально созданы для восстановления после травм. Конструкции имеют выгодное предложение стоимости и функциональности.
Продаем очень недорого Кроссовер с облегченной конструкцией. В каталоге интернет-магазина для кинезитерапии всегда в продаже варианты блочного и нагружаемого типа.
Изготавливаемые тренажеры для реабилитации обеспечивают комфортную и безопасную тренировку, что особенно важно для тренирующихся пациентов в процессе восстановления.
Устройства обладают подстраиваемым сопротивлением и уровнями нагрузки, что позволяет индивидуализировать тренировки в соответствии с задачами каждого пациента.
Все модели подходят для ЛФК по руководству врача Бубновского. Оборудованы ручками для комфортного выполнения тяг сидя или лежа.
Hi, after reading this remarkable paragraph i am also cheerful to share my experience here with colleagues.
http://wpkorea.net/bbs/board.php?bo_table=free&wr_id=800445
http://netpinion.co.kr/bbs/board.php?bo_table=free&wr_id=58365
http://elsj.idkorea.biz/bbs/board.php?bo_table=free&wr_id=5383
https://tvonair.kr/bbs/board.php?bo_table=free&wr_id=141719
https://ykentech.com/bbs/board.php?bo_table=free&wr_id=1035574
What’s up Dear, are you actually visiting this site daily,
if so after that you will absolutely get pleasant know-how.
etsyweddingteam.com
素晴らしい内容と分析で、とても感銘を受けました。
Attractive section of content. I just stumbled upon your weblog and in accession capital to assert that I get in fact enjoyed account your blog posts. Any way I’ll be subscribing to your feeds and even I achievement you access consistently quickly.
https://artropant.top/
This is my first time pay a visit at here and i am really pleassant to read everthing at alone place.
keramin krem benu gyogyszertar
Pretty! This has been an extremely wonderful article. Thank you for providing this information.
#best#links#
koop een virtueel telefoonnummer
Hi there are using WordPress for your site platform? I’m new to the blog world but I’m trying to get started and set up my own. Do you require any coding expertise to make your own blog? Any help would be greatly appreciated!
http://www.shsenc.co.kr/bbs/board.php?bo_table=free&wr_id=4295
http://u-tec.kr/bbs/board.php?bo_table=free&wr_id=34851
https://ykentech.com/bbs/board.php?bo_table=free&wr_id=1034841
http://ftp.hasri.kr/bbs/board.php?bo_table=free&wr_id=993546
http://demo059.megaweb1.kr/bbs/board.php?bo_table=free&wr_id=165186
What a stuff of un-ambiguity and preserveness of precious familiarity regarding unpredicted feelings.
https://niceneasy.co.kr/bbs/board.php?bo_table=free&wr_id=1065401
Hello just wanted to give you a brief heads up and let you know a few of the pictures aren’t loading correctly. I’m not sure why but I think its a linking issue. I’ve tried it in two different browsers and both show the same outcome.
Gama casino
What’s up, for all time i used to check webpage posts here in the early hours in the dawn, since i love to gain knowledge of more and more.
Vitamin A (ретанол пальмитат) 5000 IU
Aviator Spribe регистрация казино
Добро пожаловать в захватывающий мир авиаторов! Aviator – это увлекательная игра, которая позволит вам окунуться в атмосферу боевых действий на небе. Необычные графика и захватывающий сюжет сделают ваше путешествие по воздуху неповторимым.
Aviator Spribe казино играть на Mac
혁신적이고 표준화된 콘텐츠를 제공하는 최신 프라그마틱 게임은 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 지원합니다.
프라그마틱 홈페이지
프라그마틱 관련 글 읽는 것이 즐거웠어요! 또한, 제 사이트에서도 프라그마틱에 대한 정보를 공유하고 있어요. 함께 교류하며 더 많은 지식을 얻어봐요!
https://ycbgl.com/link/
https://www.12315ev.cn/
https://www.12315lt.cn/
Казино VODKA онлайн – играть в автоматы на деньги
Узнайте о захватывающем мире казино VODKA, где современный дизайн, разнообразие игровых автоматов и щедрые бонусы ждут каждого игрока. Погрузитесь в атмосферу слотов на деньги с казино VODKA.
Казино VODKA: Погружение в мир азартных развлечений
В мире азартных игр существует огромное количество казино, каждое из которых стремится привлечь внимание игроков своими уникальными предложениями и атмосферой. Одним из таких заведений является казино VODKA, которое предлагает своим посетителям захватывающие игровые возможности и неповторимый опыт азартных развлечений.
Виртуальное пространство казино VODKA
Казино VODKA vodka casino bonus предлагает своим клиентам широкий спектр азартных игр, доступных в виртуальном пространстве. От классических игровых автоматов до настольных игр, таких как рулетка, блэкджек и покер – здесь каждый игрок сможет найти что-то по своему вкусу. Современный дизайн и удобный интерфейс позволяют наслаждаться игровым процессом без каких-либо проблем или задержек.
Бонусы и акции
Одним из способов привлечения новых игроков и поощрения постоянных являются бонусы и акции. Казино VODKA не остается в стороне и предлагает своим клиентам различные бонусы за регистрацию, первые депозиты или участие в акциях. Эти бонусы могут значительно увеличить шансы на победу и сделать игровой процесс еще более увлекательным.
Безопасность и поддержка
Важным аспектом любого казино является обеспечение безопасности игроков и защита их личной информации. Казино VODKA придает этому особое внимание, используя передовые технологии шифрования данных и обеспечивая конфиденциальность всех транзакций. Кроме того, круглосуточная служба поддержки готова ответить на любые вопросы и помочь в решении возникающих проблем.
Заключение
Казино VODKA – это место, где каждый азартный игрок найдет что-то по своему вкусу. Богатый выбор игр, интересные бонусы и высокий уровень безопасности делают его привлекательным вариантом для тех, кто хочет испытать удачу и получить незабываемые эмоции от азартных развлечений. Сделайте свой первый шаг в мир азарта и испытайте удачу в казино VODKA уже сегодня!
https://www.floridacarcoverage.com/как-купить-диплом-Ñертификат-в-роÑÑии-4
https://www.rstandart.com/легко-купить-диплом-Ñертификат-в-роÑÑ-3
https://sketchfab.com/originality08
https://www.salinashsbaseball.com/приобретение-диплома-или-Ñертификат-4
https://www.vipbabby.com/купить-диплом-реальный-или-мошенниче-4
toasterovensplus.com
この記事は非常に情報豊富で、大変役立ちました。
thephotoretouch.com
Liu Jian은 기침을했습니다. “폐하, 옛 목사는 이미 그것을 읽었습니다.”
qiyezp.com
“폐하, 또 왕궁 밖으로 나가시겠습니까? 이 하인은 감옥에 가는데…”
Please let me know if you’re looking for a author for your weblog. You have some really great articles and I believe I would be a good asset. If you ever want to take some of the load off, I’d love to write some content for your blog in exchange for a link back to mine. Please blast me an email if interested. Kudos!
https://de.wikibrief.org/
thephotoretouch.com
“오늘은…출항하는 날인가요?”
usareallservice.com
素晴らしい記事!共有せずにはいられません。
lacolinaecuador.com
총사령관과 주지사의 관계가 갑자기 긴장되었습니다.
qiyezp.com
그는 숯불 옆에 앉아 있는 한 노인이 살짝 떨리는 숯불 화로에 숯을 더하는 것을 보았다.
Pretty component of content. I just stumbled upon your web
site and in accession capital to say that
I acquire actually enjoyed account your weblog posts.
Anyway I’ll be subscribing for your feeds and even I success you get
admission to persistently quickly.
купить диплом РІ нефтеюганске – Это вариант получить официальный бумага о окончании образовательного учреждения. Свидетельство открывает пути к новым карьерным возможностям и карьерному развитию.
Hey, you used to write wonderful, but the last several posts have been kinda boring?
I miss your great writings. Past several posts are just a little bit out of track!
come on!
Thanks so much for giving my family an update on this theme on your blog.
Please be aware that if a brand new post becomes available or in the event that any modifications
occur to the current article, I would consider reading more and understanding
how to make good use of those approaches you discuss.
Thanks for your time and consideration of other folks by making this blog
available.
https://tabmaster.ru/buk-portfolio.html – Купить диплом стоимость – Таков вариант завладеть официальный удостоверение о завершении образовательного учреждения. Свидетельство открывает двери к дополнительным карьерным возможностям и профессиональному росту.
sandyterrace.com
Zhang Sheng은 자랑스럽게 책을 보고 있었고 여러 명의 시험관이 그의 옆에 앉아 있었습니다.
Здравствуйте!
Приобретите российский диплом с гарантированной подлинностью и доставкой в любую точку страны без предварительной оплаты – надежно, выгодно, безопасно!
где купить аттестат
Right here is the right blog for everyone who wishes
to find out about this topic. You understand
so much its almost tough to argue with you (not that
I really will need to…HaHa). You definitely put a fresh spin on a
topic that’s been written about for many years. Excellent stuff, just great!
Российский изготовитель предлагает диски на сайте https://diski-dlya-shtang.ru для напряженной эксплуатации в коммерческих тренажерных залах и в домашних условиях. Отечественный завод изготавливает блины разного посадочного диаметра и любого востребованного веса для разборных штанг. Рекомендуем к приобретению прорезиненные диски для силовых занятий. Они не скользят, не шумят и более безопасны. Выпускаемые изделия не нуждаются в постоянном обслуживании и ориентированы на длительную эксплуатацию в центрах. Рекомендуем обширный каталог бамперных дисков с любым типом защитного покрытия. Приобретите веса с оптимальной массой и посадочным диаметром по доступным ценам напрямую у завода.
Российский изготовитель предлагает блины на сайте https://diski-dlya-shtang.ru/ для напряженной эксплуатации в коммерческих тренажерных залах и в домашних условиях. Завод из России создает тренировочные блины разного посадочного диаметра и любого востребованного веса для сборных штанг. Рекомендуем к заказу прорезиненные тренировочные блины для силовых занятий. Они не скользят, не гремят и менее травматичны. Изготавливаемые изделия не нуждаются в постоянном обслуживании и рассчитаны на длительную работу в дома, в квартире. Предлагаем большой каталог бамперных дисков с любым видом защитного покрытия. Оформите отягощения с необходимой массой и посадочным диаметром по низким ценам напрямую у компании-производителя.
https://diagnostik-medcenter.ru/gorlo/chto-takoe-stenoz-gortani-i-kak-ego-lechit.html – – Это способ завладеть официальный удостоверение по окончании образовательного учреждения. Свидетельство раскрывает двери к последующим карьерным перспективам и карьерному развитию.
Привет всем!
Приобретите документы об образовании всех Вузов России с доставкой по РФ и постоплатой.
http://saksx-attestats.ru/
Заказывайте диплом любого Вуза России у нас на сайте конфиденциально и без предоплаты.
Получите документы об образовании ВУЗов России с доставкой по РФ и возможностью оплаты после получения – просто и надежно!
Привет, дорогой читатель!
Приобретите диплом университета России без предоплаты и с гарантией качества доставки в любую точку страны!
http://saksx-attestats.ru/
Получите документы об образовании ВУЗов России с доставкой по РФ и возможностью оплаты после получения – просто и надежно!
Приобретите диплом Вуза с доставкой по России без предоплаты и полной уверенностью в его подлинности.
bestmanualpolesaw.com
튤립 구근의 가격은 여전히 상승하고 있습니다.
На сайте https://smartflow.ru ознакомьтесь с сантехникой премиального уровня. Вся продукция создана из инновационных, высокотехнологичных материалов и с использованием уникальных, новаторских технологий. Продукты разработаны тщательным образом, чтобы предложить вам лучшее. Сантехника гарантирует безупречный уровень функциональности, эстетичности. Она наделена долгим сроком эксплуатации. Идеально впишется в концепцию. Представлены «умные» модели унитазов безупречного качества.
cougarsbkjersey.com
非常に興味深い内容でした。また読みたいと思います。
Доброго дня!
Диплом уже не представляется таким сложным благодаря интернет-ресурсам.
Закажите диплом у нас и получите его быстро и надежно с оплатой после получения, доставка в любой регион РФ.
Желаю вам всем положительных оценок!
купить диплом бакалавра
купить диплом инженера строителя
купить диплом в энгельсе
купить диплом в кемерово
купить диплом в улан-удэ
куплю диплом о высшем образовании
Всем хорошего дня!
Работа над дипломом облегчилась благодаря ресурсам, найденным в интернете.
На нашем сайте вы можете купить диплом ВУЗа с постоплатой и помощью 24/7.
https://premialnie-diplomiki.com/
Желаю каждому отличных отметок!
купить диплом в буденновске
купить диплом медицинского училища
купить диплом в ачинске
купить диплом в новом уренгое
купить диплом в бердске
Всем хорошего дня!
Диплом стал более управляемым заданием с доступом к онлайн-материалам, которые я нашел.
Приобретите диплом университета с гарантированной доставкой в любой город России с оплатой после получения.
Желаю каждому честных отметок!
купить диплом о высшем образовании
купить диплом в минеральных водах
купить диплом в артеме
купить диплом в иваново
купить диплом в великих луках
купить диплом механика
Доброго дня!
Диплом заставил меня вести ночной образ жизни, что угрожало моему благополучию.
Поможем вам выбрать, оформить заказ и купить диплом любого учебного заведения по лучшим ценам.
https://premialnie-diplomiki.com/
Желаю каждому пятерочных) отметок!
купить диплом менеджера
купить диплом в хасавюрте
купить диплом фельдшера
купить диплом в георгиевске
купить диплом с занесением в реестр
Приветики, дорогие мои!!
Не отказываясь от борьбы с дипломом, я использую онлайн-ресурсы для улучшения своей работы.
Приобретите диплом университета у нас и получите его с доставкой в любую точку России с гарантией качества.
https://premialnie-diplomiki.com/
Желаю для каждого прекрасных оценок!
купить диплом в ангарске
купить диплом в ачинске
купить диплом менеджера
купить диплом в саранске
купить диплом в канске
Приветики, дорогие мои!!
Я продолжаю трудиться над дипломом, несмотря на все трудности, благодаря помощи интернет-ресурсов.
Заказывайте диплом у нас без предоплаты и мы доставим его вам в любой регион России, гарантируем конфиденциальность.
Желаю всем отличных отметок!
купить диплом о среднем специальном
купить диплом института
купить диплом слесаря
купить диплом техникума
купить диплом в салавате
купить аттестат за 11 класс
Всем хорошего дня!
Инструменты из интернета значительно упростили мой процесс работы над дипломом.
Мы поможем вам купить диплом ВУЗа России недорого, без предоплаты и с гарантией возврата средств.
https://adiplom-russian.com/
Желаю для каждого отличных отметок!
купить диплом в озёрске
купить диплом в кстово
куплю диплом кандидата наук
купить диплом в благовещенске
купить диплом в туапсе
Привет всем!
Интернет предоставил мне инструменты, которые сделали процесс написания диплома более быстрым и менее изнурительным.
Наша компания предлагает конфиденциально выбрать и заказать диплом любого ВУЗа России.
Желаю вам всем отличных отметок!
купить диплом магистра
купить диплом в ростове-на-дону
купить диплом в новокуйбышевске
старые дипломы купить
купить диплом в новоалтайске
купить диплом в тольятти
Всем хорошего дня!
Диплом поставил под угрозу мое здоровье из-за необходимости ночных занятий.
Наша компания предоставляет возможность приобрести диплом ВУЗа с выгодой и доставкой в любой регион России.
Желаю для каждого не двоешных) отметок!
купить диплом
купить диплом в железногорске
купить диплом
купить диплом эколога
купить диплом в вологде
купить диплом в чебоксарах
Здравствуйте!
Подготовка диплома стала более продуктивной благодаря онлайн-ресурсам.
Заказывайте диплом у нас и получите его быстро и безопасно, оплата по факту доставки, отправка в любой регион России.
https://premialnie-diplomiki.com/
Желаю для каждого отличных оценок!
купить диплом в минеральных водах
купить диплом в ангарске
купить диплом в балашове
купить диплом в бердске
купить сертификат специалиста
Привет всем!
Несмотря на все сложности и угрозу для здоровья, я продолжаю работать над дипломом, опираясь на найденные ресурсы.
Приобретите диплом университета без предоплаты у нас и получите его с доставкой в любую точку России, качество гарантировано.
https://adiplom-russian.com/
Желаю всем не двоешных) отметок!
купить диплом в ишимбае
купить диплом гознак
где купить диплом
купить диплом в асбесте
купить диплом в батайске
Приветики!
Приобретите российский диплом по выгодной цене с гарантией проверки и доставкой в любой город РФ – без предоплаты!
http://saksx-attestats.ru/
Приобретите диплом Вуза с доставкой по России без предоплаты и полной уверенностью в его подлинности.
Получите российский диплом с гарантированной доставкой в любую точку страны без предварительной оплаты – просто и надежно!
Wow, awesome blog structure! How lengthy have you been running
a blog for? you made blogging look easy. The whole look of your web site is wonderful,
as well as the content material! You can see similar here sklep internetowy
На сайте https://container-platform.ru имеется номер телефона для того, чтобы заказать качественные, надежные, практичные контейнерные площадки, предназначенные для мусора. Есть как на 2, так и 3 контейнера. Прямо сейчас ознакомьтесь с расценками на стандартные варианты. Напротив каждого варианта указаны технические характеристики и другая важная информация, которая необходима для того, чтобы быстрее определиться с выбором. Наличие контейнерных площадок обязательно для всех дворов.
SEO solutions are the unheralded protagonists of the internet age, offering businesses with the resources and approaches to shine brightly in the huge universe of online material. By tapping into the capability of effective keyword focusing, quality backlink acquisition, and content optimization, these methods ensure that a website is not merely noticeable, but distinguishes itself as a signal of pertinence and command in its sector. The allure of SEO is in its capacity to organically elevate a company’s appearance, pulling in spectators genuinely engaged in what’s on offer, and forming valuable connections that lead to lasting connections.
In a world where internet dominance often commands victory, having a fitted SEO approach is akin to having a chief key to the internet city. Every adjustment and change made by SEO experts isn’t just about appeasing calculations, but more crucially, about understanding and catering to person actions and requirements. The final aim? To peacefully join a company with its ideal market, fostering development, confidence, and long-term success. In this endeavor, SEO methods prove to be not just advantageous, but indispensable.
Telgrm: @exrumer
https://XRumer.cc/
Skype: XRumer.pro
sandyterrace.com
이 순간 Zhu Houzhao는 갑자기 정신을 차렸습니다. Lao Fang은 몇 마디로 어떻게 아버지를 설득할 수 있었을까요?
I truly wanted to type a brief word to be able to express gratitude to you for some of the lovely information you are posting at this website.
My time-consuming internet research has at the end of the
day been rewarded with sensible points to talk about with my companions.
I ‘d tell you that we visitors are definitely blessed to
be in a great site with so many wonderful people with beneficial secrets.
I feel quite grateful to have seen your website page and look forward to many more amazing times reading here.
Thanks a lot again for all the details.
http://man-attestats24.com – Купить аттестат о среднем – путь к твоему будущему. В нашем сервисе все вы сможете просто и оперативно заказать свидетельство, обязательный для последующего получения образования или профессионального роста. Наша специалисты обеспечивают высокое качество и конфиденциальность предоставления услуг. Покупайте учебный сертификат здесь и проявите дополнительные варианты для того, чтобы вашего карьерного развития и трудоустройства.
Доброго дня!
Столкнувшись с трудностями в написании диплома, я обнаружил в сети необходимую поддержку.
Закажите диплом в России без предоплаты и получите его с доставкой “под ключ”.
Желаю вам всем отличных оценок!
https://aurus-diploman.com/
купить диплом швеи
купить диплом логиста
купить диплом в серове
купить диплом в краснодаре
купить диплом в новом уренгое
Привет всем!
Завершение диплома становится ближе с каждым найденным в сети ресурсом.
Мы предлагаем вам купить диплом университета недорого с доставкой в любую точку России, оплата после получения.
https://rusd-diploms.com/
Желаю для каждого не двоешных) отметок!
купить диплом в набережных челнах
купить диплом в черкесске
купить диплом в анапе
куплю диплом цена
купить диплом в нефтекамске
Доброго дня!
Диплом стал более управляемым заданием с доступом к онлайн-материалам, которые я нашел.
Мы предлагаем вам купить диплом университета недорого с доставкой в любую точку России, оплата после получения.
Желаю каждому пятерочных) оценок!
купить диплом института
купить диплом в георгиевске
купить диплом в назрани
купить диплом прораба
купить диплом в кропоткине
купить диплом россия
Добрый день!
Диплом заставил меня работать ночами, что угрожало моему здоровью.
Мы поможем вам купить диплом университета без предоплаты и доставим его в любой город России с гарантированной безопасностью.
https://rusd-diploms.com/
Желаю для каждого положительных отметок!
купить диплом в выборге
купить диплом в славянске-на-кубани
купить диплом в иваново
купить диплом в ижевске
купить аттестат
Приветики, дорогие мои!!
Столкнулся с множеством препятствий в процессе работы над дипломом, однако интернет помог мне найти способы ускорения и упрощения задачи.
Хотите приобрести диплом ВУЗа недорого без предоплаты на нашем сайте? Доставляем в любую точку России.
купить диплом автомеханика
Желаю вам всем положительных отметок!
купить диплом в озёрске
купить диплом лаборанта
купить диплом в крыму
купить диплом в казани
куплю диплом цена
Приветики, дорогие мои!!
Завершение диплома становится ближе с каждым найденным в сети ресурсом.
Выберите и закажите диплом ВУЗа России качественно и с возможностью отправки почтой!
Желаю всем не двоешных) оценок!
https://aurus-diplomas.com/
купить диплом педагога
купить диплом менеджера по туризму
купить диплом в санкт-петербурге
купить диплом в орске
купить диплом механика
Привет всем!
Моя работа над дипломом обогащена благодаря доступности и разнообразию онлайн-ресурсов.
На нашем сайте вы можете купить диплом ВУЗа с постоплатой и поддержкой 24/7.
Желаю всем не двоешных) отметок!
https://aurus-diplomas.com/
купить диплом журналиста
купить диплом в батайске
купить диплом электромонтажника
купить диплом в дзержинске
купить свидетельство о рождении ссср
Приветики, дорогие мои!!
Написание диплома стало эффективнее с использованием интернет-ресурсов.
Наша компания предоставляет возможность приобрести диплом ВУЗа с выгодой и доставкой в любой регион России.
Желаю вам всем пятерочных) отметок!
купить аттестат за 9 класс
можно ли купить диплом
купить диплом в кемерово
купить диплом в ялте
купить диплом в новороссийске
купить аттестат
Добрый день!
Для диплома я нашел в интернете множество полезных ресурсов, облегчающих написание.
Хотите приобрести диплом ВУЗа недорого без предоплаты на нашем сайте? Доставляем в любую точку России.
Желаю для каждого не двоешных) оценок!
купить аттестат за 9 класс
купить диплом в камышине
купить диплом в новочебоксарске
купить диплом учителя
купить диплом в кисловодске
купить диплом в астрахани
Доброго дня!
Инструменты для диплома, найденные в интернете, сделали написание доступнее.
Закажите диплом у нас – это легко! Без предоплаты, с гарантией качества и доставкой по всей России.
Желаю всем не двоешных) оценок!
https://diplomys-asx.com
купить диплом для иностранцев
купить диплом в нижневартовске
купить диплом в севастополе
купить диплом в георгиевске
куплю диплом цена
Достижение диплома по высшему образованию становится значимым действием в судьбе многочисленных людей, открывая перспективы к перспективным шансам и перспективам.
Однако, не постоянно обучение в университете доступно или соответствует по различным причинам.
При таких обстоятельствах вопрос где приобрести аттестат, становится значимым.
Современные технологии и виртуальный рынок предлагают разные альтернативы для покупки документа, однако существенно отбирать проверенных поставщиков, обеспечивающих высокое качество и достоверность диплома.
При отборе нужно замечать не только на цену, но также на имидж фирмы, отзывы заказчиков и возможность получения консультации.
http://www.diablomania.ru/forum/showthread.php?p=553032
https://talkrealty.ru/obrazovanie-vdohnovlyayushhee-kupite-diplom-i-dostignite-svoih-tseley
http://www.sir9420.com/home.php?mod=space&uid=28268
https://allukrnews.ru/vash-put-k-uspehu-poluchite-diplom-ot-priznannogo-uchebnogo-zavedeniya/
http://www.lada-4×4.net/showthread.php?p=20253
купить диплом РІ чайковском – подразумевает инвестировать в свое будущее, следовательно отбор компании нужно относиться тщательно.
exprimegranada.com
この記事から得た知識は、本当に計り知れません。非常に感謝しています。
As Mike Johnson tries to pass billions of dollars in aid to Ukraine, Israel and Taiwan, he’s having to put his own job on the line
blacksprut
Johnson moving ahead with Ukraine aid bill despite pressure from hardliners
US Senate kills articles of impeachment against Homeland Security Secretary Alejandro Mayorkas
Ex-Trump attorney asked if he thinks Trump should testify. Hear reply 1:29
https://bs-gl.org
Analysis Some House Republicans need this reminder that Russia is not their friend
Viewers called in with Trump trial questions. Maggie Haberman answered 1:50
If Trump testifies at NY criminal trial, prosecutors want to use his past legal run-ins to discredit him to jury
Fact check: Donald Trump attacks Jimmy Kimmel for something Al Pacino did
blacksprut
https://bs-gl.org
Здравствуйте!
Моя борьба с дипломом превратилась в настоящую драму, но я нашел выход, благодаря доступным онлайн-ресурсам.
Предлагаем приобрести диплом университета России по доступной цене с доставкой “под ключ”.
Желаю вам всем положительных отметок!
https://russiany-diplomas.com
купить диплом штукатура
купить диплом в феодосии
купить диплом в твери
купить диплом в москве
купить диплом техника
Добрый день!
Сложности с дипломом привели к необходимости работать в ночное время, что угрожает моему здоровью.
Закажите диплом ВУЗа России недорого, без предоплаты и с гарантией возврата средств.
Желаю вам всем пятерочных) оценок!
купить диплом автомеханика
где купить диплом среднем
купить диплом в владикавказе
купить диплом в рыбинске
купить диплом в мичуринске
купить бланк диплома
Приветики, дорогие мои!!
Дипломное задание оказалось крайне трудоемким, но я нашел способы его упрощения благодаря интернет-ресурсам.
Поможем вам выбрать, оформить заказ и приобрести диплом любого учебного заведения по самым низким ценам.
Желаю всем прекрасных оценок!
купить диплом вуза
купить диплом в казани
купить диплом в находке
купить диплом дизайнера
купить диплом в шадринске
купить диплом в астрахани
Здравствуйте!
К счастью, интернет помог мне найти полезные ресурсы, которые значительно ускорили и облегчили мой труд с дипломом.
Приобретите диплом университета у нас и получите его с доставкой в любую точку России с гарантией качества.
Желаю для каждого пятерочных) оценок!
https://diplom-servise.com
купить диплом в магадане
купить диплом маркетолога
купить диплом электромонтера
купить диплом в архангельске
купить дипломы о высшем
Всем хорошего дня!
Диплом привел к серьезным проблемам из-за просроченных сроков.
Мы предлагаем недорого заказать диплом без предоплаты с доставкой курьером по РФ, “под ключ”!
Желаю всем отличных оценок!
https://plands-diplomy.com
купить диплом в химках
купить диплом в копейске
купить диплом автомеханика
купить диплом средне техническое
купить диплом в уссурийске
Здравствуйте!
Диплом превратился в кошмар, когда я стал откладывать сроки и работать над ним ночью, вредя своему здоровью.
Выбирайте и заказывайте диплом ВУЗа России выгодно и с возможностью отправки почтой!
Желаю для каждого прекрасных оценок!
https://plands-diplomy.com
купить диплом в чайковском
купить диплом в пятигорске
купить диплом маркетолога
купить диплом учителя
купить диплом с реестром
Здравствуйте!
Диплом требует от меня изучения найденных в интернете материалов для улучшения качества работы.
Закажите диплом в России без предоплаты и получите его с доставкой “под ключ”.
Желаю каждому отличных отметок!
https://frees-diplomy.com
купить диплом в новороссийске
купить диплом в кургане
купить диплом в нижнекамске
купить диплом в стерлитамаке
купить диплом электромонтажника
Добрый день!
Диплом получил поддержку через ценные ресурсы, обнаруженные в сети, упрощая мою задачу.
Выберите и закажите диплом ВУЗа России качественно и с возможностью отправки почтой!
Желаю всем честных отметок!
купить диплом
купить диплом в туле
купить диплом медсестры
купить диплом в пскове
купить диплом сварщика
купить диплом в елабуге
Здравствуйте!
Моя работа над дипломом стала продуктивнее благодаря активному использованию онлайн-материалов.
Наш интернет-магазин предлагает купить российский диплом по выгодной цене, с гарантией прохождения всех проверок.
Желаю всем не двоешных) оценок!
https://frees-diplomy.com
купить диплом в томске
купить диплом в нижнем тагиле
купить диплом в каменске-шахтинском
купить диплом в туймазы
купить диплом в альметьевске
Доброго дня!
Эффективность моей работы над дипломом возросла благодаря доступу к онлайн-материалам.
Предлагаем всем желающим приобрести диплом университетов России по выгодной цене с доставкой “под ключ”.
Желаю всем прекрасных оценок!
https://diplom-originalniy.com
купить диплом в севастополе
купить диплом в мытищах
купить диплом ссср
купить диплом зубного техника
купить диплом в кстово
Здравствуйте!
Интернет предоставил мне инструменты, которые сделали процесс написания диплома более быстрым и менее изнурительным.
Приобретите диплом университета с гарантированной доставкой в любой город России с оплатой после получения.
Желаю всем честных оценок!
купить диплом техникума
купить диплом в таганроге
купить диплом инженера механика
купить диплом в химках
купить диплом в благовещенске
купить диплом в тольятти
Привет всем!
Дипломное исследование стало для меня серьезным вызовом, однако я нашел способы облегчить этот процесс.
Закажите диплом у нас легко! Без предоплаты, с гарантией качества и доставкой по всей России.
Желаю каждому отличных отметок!
https://diplom-originalniy.com
купить диплом в брянске
купить диплом в каменске-шахтинском
купить диплом в иркутске
купить диплом маляра
купить аттестаты за 9
buysteriodsonline.com
Zhang Mao는 큰 놈이고 그의 생각은 매우 직접적이므로 …
В современном мире, где аттестат – это начало отличной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Необходимость наличия официального документа трудно переоценить. Ведь диплом открывает двери перед каждым человеком, желающим вступить в профессиональное сообщество или учиться в каком-либо университете.
В данном контексте наша компания предлагает очень быстро получить этот необходимый документ. Вы имеете возможность купить аттестат, что является удачным решением для человека, который не смог закончить образование, потерял документ или желает исправить плохие оценки. Каждый аттестат изготавливается с особой тщательностью, вниманием к мельчайшим элементам. На выходе вы сможете получить документ, полностью соответствующий оригиналу.
Преимущества этого подхода состоят не только в том, что можно максимально быстро получить аттестат. Весь процесс организован комфортно и легко, с нашей поддержкой. Начиная от выбора подходящего образца до грамотного заполнения персональной информации и доставки в любой регион страны — все будет находиться под абсолютным контролем наших мастеров.
Таким образом, для тех, кто ищет оперативный способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать аттестат – значит избежать длительного обучения и сразу перейти к своим целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://prema-attestats.ru/
Здравствуйте!
Трудности с дипломом уменьшились благодаря интернет-поиску решений.
Приобретите диплом университета без предоплаты у нас и получите его с доставкой в любую точку России, гарантия качества.
Желаю вам всем отличных отметок!
купить диплом фармацевта
купить диплом в иркутске
купить диплом в камышине
купить диплом в георгиевске
купить диплом в абакане
купить диплом швеи
Привет всем!
Диплом повлиял на мое здоровье из-за необходимости работать ночью.
Наша компания предлагает конфиденциально заказать и приобрести диплом любого ВУЗа России.
Желаю вам всем пятерочных) отметок!
где купить диплом
купить диплом в стерлитамаке
купить диплом в каспийске
купить диплом в саратове
купить диплом в балашихе
купить диплом в альметьевске
Привет всем!
Дипломное задание оказалось крайне трудоемким, но я нашел способы его упрощения благодаря интернет-ресурсам.
Мы предлагаем купить диплом Гознак со скидкой, гарантией и доставкой в любой город РФ.
Желаю каждому честных отметок!
https://radiploma.com/
купить диплом в камышине
купить диплом программиста
купить диплом в новочебоксарске
купить диплом в керчи
купить диплом в ярославле
Здравствуйте!
Диплом постепенно улучшается с каждым днем благодаря использованию найденных ресурсов в интернете.
Предлагаем приобрести диплом университета России по доступной цене с доставкой “под ключ”.
Желаю для каждого честных оценок!
https://radiploma.com/
купить диплом в глазове
купить диплом химика
купить диплом автомеханика
купить диплом в нальчике
купить диплом в кызыле
Купить аттестат цена – возможность к твоему перспективам. В данном сервисе вы сможете без труда и быстро заказать свидетельство, обязательный для дальнейшего получения образования или профессионального роста. Наша консультанты гарантируют качество и конфиденциальность предоставления услуг. Заказывайте учебный аттестат в нашем сервисе и откройте дополнительные варианты для своего профессионального роста и карьеры.
http://r0757478.bget.ru/home.php?mod=space&uid=17208&do=profile
http://www.hcceskalipa.cz/bazarek/russianydiplomans
http://www.artcalendar.ru/783.html
http://www.sskyn.com/home-uid-82120.html
https://citydevelopers.ru/otzyvy/novosibirsk/zhk-voskresnyy/
Купить аттестат цена – возможность к вашему будущему. На данном сервисе вы сможете без труда и быстро приобрести аттестат, необходимый для того, чтобы дальнейшего получения образования или трудоустройства. Наша консультанты обеспечивают высокое качество и секретность предоставления услуг. Приобретайте учебный сертификат в нашем сервисе и проявите дополнительные возможности для того, чтобы своего карьерного развития и карьеры.
Heya! I realize this is somewhat off-topic but I needed to ask. Does managing a well-established blog such as yours require a lot of work? I am brand new to running a blog but I do write in my diary everyday. I’d like to start a blog so I can share my personal experience and feelings online. Please let me know if you have any ideas or tips for new aspiring bloggers. Thankyou!
#be#jk3#jk#jk#JK##
купить американский номер для смс
Useful info. Fortunate me I found your web site by accident, and I’m shocked why this accident didn’t took place in advance! I bookmarked it.
#be#jk3#jk#jk#JK##
виртуальный номер Азербайджан
В наше время, когда диплом является началом удачной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает дверь перед всеми, кто желает вступить в сообщество профессионалов или учиться в высшем учебном заведении.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы можете заказать диплом, и это будет удачным решением для человека, который не смог закончить образование, утратил документ или хочет исправить плохие оценки. диплом изготавливается аккуратно, с особым вниманием к мельчайшим элементам, чтобы в результате получился документ, максимально соответствующий оригиналу.
Плюсы такого решения заключаются не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается просто и легко, с нашей поддержкой. Начав от выбора нужного образца до консультации по заполнению персональной информации и доставки по стране — все будет находиться под абсолютным контролем наших мастеров.
Таким образом, для тех, кто пытается найти максимально быстрый способ получения необходимого документа, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать длительного обучения и сразу переходить к достижению собственных целей: к поступлению в университет или к началу трудовой карьеры.
https://originality-diplomans.com
В нашем мире, где диплом – это начало отличной карьеры в любой области, многие стараются найти максимально простой путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь диплом открывает двери перед всеми, кто стремится вступить в сообщество квалифицированных специалистов или продолжить обучение в любом ВУЗе.
Наша компания предлагает очень быстро получить этот необходимый документ. Вы имеете возможность заказать диплом, и это будет удачным решением для всех, кто не смог закончить обучение или утратил документ. дипломы производятся с особой аккуратностью, вниманием ко всем деталям. На выходе вы получите документ, максимально соответствующий оригиналу.
Плюсы данного подхода заключаются не только в том, что вы максимально быстро получите диплом. Процесс организован удобно, с нашей поддержкой. От выбора требуемого образца диплома до грамотного заполнения личной информации и доставки по России — все под абсолютным контролем качественных специалистов.
Для всех, кто ищет оперативный способ получения необходимого документа, наша компания может предложить отличное решение. Приобрести диплом – значит избежать продолжительного обучения и не теряя времени переходить к личным целям: к поступлению в ВУЗ или к началу удачной карьеры.
купить диплом колледжа
В современном мире, где диплом является началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Факт наличия документа об образовании трудно переоценить. Ведь именно диплом открывает дверь перед людьми, стремящимися начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте мы предлагаем очень быстро получить этот необходимый документ. Вы сможете приобрести диплом нового или старого образца, и это становится удачным решением для человека, который не смог завершить образование или утратил документ. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим деталям, чтобы на выходе получился полностью оригинальный документ.
Преимущества подобного подхода заключаются не только в том, что можно оперативно получить диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начав от выбора необходимого образца до консультаций по заполнению личной информации и доставки по России — все находится под полным контролем наших специалистов.
Всем, кто ищет максимально быстрый способ получить требуемый документ, наша компания может предложить выгодное решение. Купить диплом – значит избежать продолжительного обучения и сразу перейти к достижению своих целей: к поступлению в ВУЗ или к началу трудовой карьеры.
https://russiany-diplomas.com
В наше время, когда диплом – это начало удачной карьеры в любой области, многие ищут максимально простой путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает дверь перед всеми, кто стремится вступить в сообщество квалифицированных специалистов или учиться в высшем учебном заведении.
Наша компания предлагает максимально быстро получить этот важный документ. Вы можете заказать диплом нового или старого образца, что является удачным решением для человека, который не смог закончить обучение или утратил документ. диплом изготавливается с особой аккуратностью, вниманием к мельчайшим деталям, чтобы в итоге получился 100% оригинальный документ.
Преимущество подобного решения заключается не только в том, что можно оперативно получить диплом. Весь процесс организовывается удобно, с нашей поддержкой. Начав от выбора нужного образца до консультации по заполнению личных данных и доставки по России — все находится под полным контролем наших мастеров.
Для всех, кто пытается найти максимально быстрый способ получить необходимый документ, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать долгого обучения и сразу переходить к важным целям, будь то поступление в университет или начало карьеры.
купить диплом
Сегодня, когда диплом является началом удачной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа об образовании сложно переоценить. Ведь именно он открывает двери перед всеми, кто желает начать профессиональную деятельность или продолжить обучение в университете.
Мы предлагаем максимально быстро получить этот важный документ. Вы сможете заказать диплом, что становится отличным решением для всех, кто не смог закончить обучение или потерял документ. диплом изготавливается аккуратно, с особым вниманием ко всем элементам. В итоге вы получите 100% оригинальный документ.
Преимущество этого решения состоит не только в том, что вы сможете максимально быстро получить свой диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. Начав от выбора требуемого образца диплома до консультации по заполнению персональной информации и доставки по России — все под абсолютным контролем квалифицированных мастеров.
Таким образом, для всех, кто пытается найти быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать долгого процесса обучения и не теряя времени переходить к своим целям: к поступлению в ВУЗ или к началу удачной карьеры.
https://foodsuppliers.ru/users/etizisuc
http://receptom.ru/index.php?subaction=userinfo&user=ihugybihi
http://xn--sns-dk6nm87c.com/bbs/board.php?bo_table=free&wr_id=1157944
http://www.alltab.co.kr/bbs/board.php?bo_table=free&wr_id=1287240
http://xn—-ptbagsgdho0d.xn--p1ai/index.php?subaction=userinfo&user=ihova
В современном мире, где диплом является началом отличной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Факт наличия официального документа сложно переоценить. Ведь диплом открывает двери перед людьми, стремящимися вступить в сообщество квалифицированных специалистов или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы имеете возможность заказать диплом старого или нового образца, и это становится отличным решением для человека, который не смог завершить обучение, потерял документ или желает исправить плохие оценки. дипломы выпускаются с особой тщательностью, вниманием ко всем нюансам, чтобы на выходе получился документ, полностью соответствующий оригиналу.
Плюсы такого подхода заключаются не только в том, что вы сможете оперативно получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора подходящего образца до точного заполнения личной информации и доставки в любое место страны — все будет находиться под полным контролем квалифицированных специалистов.
В итоге, всем, кто пытается найти оперативный способ получить необходимый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать длительного процесса обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в университет или начало удачной карьеры.
https://russiany-diplomas.com
В современном мире, где диплом является началом удачной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Факт наличия официального документа переоценить просто невозможно. Ведь именно он открывает дверь перед всеми, кто стремится вступить в сообщество квалифицированных специалистов или учиться в ВУЗе.
Наша компания предлагает очень быстро получить этот необходимый документ. Вы имеете возможность приобрести диплом, и это является удачным решением для всех, кто не смог завершить образование, утратил документ или желает исправить плохие оценки. Все дипломы выпускаются аккуратно, с максимальным вниманием ко всем элементам. В результате вы получите документ, 100% соответствующий оригиналу.
Превосходство этого подхода состоит не только в том, что вы быстро получите свой диплом. Процесс организовывается удобно и легко, с профессиональной поддержкой. От выбора необходимого образца до консультации по заполнению личных данных и доставки по России — все будет находиться под абсолютным контролем опытных специалистов.
Для всех, кто пытается найти оперативный способ получить необходимый документ, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать продолжительного обучения и сразу переходить к своим целям, будь то поступление в университет или старт успешной карьеры.
купить диплом магистра
В нашем обществе, где диплом – это начало удачной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь именно он открывает двери перед любым человеком, желающим начать трудовую деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы сможете приобрести диплом, и это будет выгодным решением для всех, кто не смог завершить образование или утратил документ. диплом изготавливается с особой тщательностью, вниманием ко всем нюансам. В итоге вы получите полностью оригинальный документ.
Преимущество данного решения заключается не только в том, что вы сможете быстро получить диплом. Процесс организован комфортно и легко, с нашей поддержкой. От выбора подходящего образца документа до консультации по заполнению персональных данных и доставки в любое место России — все под полным контролем опытных специалистов.
Всем, кто пытается найти максимально быстрый способ получить требуемый документ, наша услуга предлагает отличное решение. Приобрести диплом – значит избежать долгого обучения и сразу переходить к личным целям: к поступлению в университет или к началу удачной карьеры.
https://diplomana-asx.com
В современном мире, где диплом – это начало успешной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Факт наличия официального документа переоценить невозможно. Ведь именно он открывает дверь перед людьми, желающими вступить в сообщество квалифицированных специалистов или учиться в ВУЗе.
В данном контексте мы предлагаем очень быстро получить любой необходимый документ. Вы имеете возможность приобрести диплом, что является удачным решением для всех, кто не смог закончить образование, потерял документ или хочет исправить плохие оценки. диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим элементам, чтобы в результате получился документ, полностью соответствующий оригиналу.
Превосходство данного решения состоит не только в том, что вы сможете максимально быстро получить диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора необходимого образца до точного заполнения персональной информации и доставки в любой регион страны — все находится под полным контролем опытных специалистов.
Для тех, кто пытается найти оперативный способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать длительного процесса обучения и не теряя времени перейти к важным целям: к поступлению в ВУЗ или к началу успешной карьеры.
https://russiany-diplomans.com
В нашем обществе, где диплом – это начало успешной карьеры в любой отрасли, многие стараются найти максимально быстрый и простой путь получения качественного образования. Наличие официального документа об образовании переоценить попросту невозможно. Ведь именно он открывает двери перед каждым человеком, желающим вступить в сообщество профессиональных специалистов или учиться в ВУЗе.
Предлагаем очень быстро получить этот важный документ. Вы сможете купить диплом, и это становится выгодным решением для человека, который не смог завершить обучение, потерял документ или желает исправить свои оценки. дипломы выпускаются с особой аккуратностью, вниманием ко всем нюансам. В результате вы получите документ, 100% соответствующий оригиналу.
Преимущества такого решения заключаются не только в том, что можно быстро получить свой диплом. Весь процесс организован удобно, с нашей поддержкой. От выбора нужного образца до консультации по заполнению персональной информации и доставки в любой регион России — все находится под абсолютным контролем качественных специалистов.
Для всех, кто ищет оперативный способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать длительного обучения и сразу перейти к своим целям, будь то поступление в университет или начало трудовой карьеры.
https://russiany-diplomans.com
В нашем обществе, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Факт наличия официального документа переоценить невозможно. Ведь именно диплом открывает двери перед всеми, кто желает начать трудовую деятельность или продолжить обучение в ВУЗе.
Предлагаем максимально быстро получить этот необходимый документ. Вы имеете возможность заказать диплом нового или старого образца, что будет удачным решением для всех, кто не смог завершить обучение, утратил документ или хочет исправить свои оценки. дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим элементам, чтобы в результате получился 100% оригинальный документ.
Превосходство данного решения состоит не только в том, что можно оперативно получить диплом. Весь процесс организован просто и легко, с нашей поддержкой. От выбора требуемого образца диплома до правильного заполнения персональных данных и доставки по стране — все будет находиться под абсолютным контролем квалифицированных мастеров.
Для тех, кто ищет максимально быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и не теряя времени переходить к достижению своих целей: к поступлению в университет или к началу трудовой карьеры.
https://adiplomy-russian.com
В современном мире, где диплом является началом удачной карьеры в любой отрасли, многие пытаются найти максимально простой путь получения образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед людьми, стремящимися вступить в сообщество профессионалов или учиться в ВУЗе.
Мы предлагаем очень быстро получить этот важный документ. Вы имеете возможность приобрести диплом старого или нового образца, и это будет отличным решением для всех, кто не смог закончить обучение, потерял документ или хочет исправить свои оценки. Любой диплом изготавливается аккуратно, с особым вниманием к мельчайшим элементам. В результате вы получите полностью оригинальный документ.
Плюсы такого решения заключаются не только в том, что вы сможете максимально быстро получить диплом. Процесс организован комфортно, с нашей поддержкой. От выбора необходимого образца диплома до грамотного заполнения персональной информации и доставки по России — все под полным контролем опытных мастеров.
Для всех, кто ищет оперативный способ получения требуемого документа, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать долгого процесса обучения и не теряя времени перейти к достижению личных целей, будь то поступление в ВУЗ или старт трудовой карьеры.
https://adiplomy-russian.com
В нашем обществе, где диплом – это начало отличной карьеры в любой отрасли, многие пытаются найти максимально быстрый путь получения образования. Факт наличия официального документа сложно переоценить. Ведь именно диплом открывает двери перед всеми, кто стремится начать профессиональную деятельность или учиться в ВУЗе.
В данном контексте наша компания предлагает быстро получить этот необходимый документ. Вы имеете возможность приобрести диплом нового или старого образца, что становится удачным решением для человека, который не смог завершить образование или утратил документ. Каждый диплом изготавливается с особой тщательностью, вниманием ко всем нюансам, чтобы на выходе получился 100% оригинальный документ.
Превосходство этого решения состоит не только в том, что вы сможете максимально быстро получить свой диплом. Весь процесс организовывается удобно, с нашей поддержкой. Начав от выбора необходимого образца диплома до правильного заполнения личной информации и доставки в любое место России — все под полным контролем наших специалистов.
Всем, кто ищет быстрый способ получения требуемого документа, наша услуга предлагает выгодное решение. Заказать диплом – это значит избежать длительного процесса обучения и сразу перейти к достижению личных целей, будь то поступление в университет или старт карьеры.
купить диплом о высшем образовании
http://ford-talks.ru/topic3928.html?&p=17348
http://parenvarmii.ru/topic3707.html
Купить школьный аттестат – путь к твоему перспективам. В нашем портале вы можете легко и оперативно приобрести свидетельство, необходимый для того, чтобы дальнейшего получения образования или профессионального роста. Наша консультанты обеспечивают высокое качество и секретность услуги. Заказывайте учебный аттестат в нашем сервисе и проявите дополнительные перспективы для того, чтобы вашего профессионального роста и карьеры.
В современном мире, где диплом – это начало отличной карьеры в любой области, многие ищут максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа об образовании трудно переоценить. Ведь именно диплом открывает дверь перед людьми, стремящимися начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы имеете возможность заказать диплом, что становится выгодным решением для человека, который не смог закончить обучение, потерял документ или хочет исправить плохие оценки. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим деталям. В итоге вы получите продукт, полностью соответствующий оригиналу.
Преимущества данного решения заключаются не только в том, что можно оперативно получить диплом. Процесс организован просто и легко, с нашей поддержкой. Начав от выбора нужного образца диплома до правильного заполнения личных данных и доставки по стране — все находится под абсолютным контролем квалифицированных специалистов.
Всем, кто ищет максимально быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать продолжительного обучения и не теряя времени переходить к достижению личных целей, будь то поступление в университет или начало карьеры.
купить диплом вуза
В нашем обществе, где диплом – это начало удачной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Важность наличия документа об образовании трудно переоценить. Ведь именно диплом открывает дверь перед людьми, стремящимися вступить в профессиональное сообщество или учиться в ВУЗе.
В данном контексте наша компания предлагает оперативно получить этот важный документ. Вы сможете заказать диплом, что будет удачным решением для всех, кто не смог закончить обучение или утратил документ. Все дипломы выпускаются с особой аккуратностью, вниманием ко всем деталям. На выходе вы сможете получить продукт, максимально соответствующий оригиналу.
Преимущество данного подхода заключается не только в том, что вы быстро получите свой диплом. Процесс организован просто и легко, с нашей поддержкой. Начав от выбора нужного образца до грамотного заполнения персональных данных и доставки по России — все будет находиться под полным контролем квалифицированных специалистов.
Всем, кто ищет максимально быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать продолжительного обучения и сразу переходить к своим целям, будь то поступление в ВУЗ или начало карьеры.
https://diploms-originalniy.com
В современном мире, где диплом является началом отличной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Важность наличия документа об образовании переоценить невозможно. Ведь диплом открывает двери перед людьми, желающими начать трудовую деятельность или продолжить обучение в каком-либо институте.
В данном контексте наша компания предлагает оперативно получить любой необходимый документ. Вы сможете приобрести диплом нового или старого образца, и это будет удачным решением для всех, кто не смог закончить образование, утратил документ или хочет исправить плохие оценки. Каждый диплом изготавливается аккуратно, с особым вниманием к мельчайшим элементам, чтобы в итоге получился полностью оригинальный документ.
Превосходство данного решения состоит не только в том, что вы максимально быстро получите свой диплом. Весь процесс организовывается просто и легко, с нашей поддержкой. Начиная от выбора необходимого образца документа до консультации по заполнению личной информации и доставки в любое место России — все находится под полным контролем качественных специалистов.
Для тех, кто хочет найти максимально быстрый способ получения необходимого документа, наша компания может предложить отличное решение. Приобрести диплом – это значит избежать долгого обучения и не теряя времени переходить к важным целям, будь то поступление в ВУЗ или начало карьеры.
https://lands-diploma.com
Сегодня, когда диплом становится началом удачной карьеры в любом направлении, многие пытаются найти максимально простой путь получения образования. Наличие официального документа переоценить невозможно. Ведь диплом открывает дверь перед каждым человеком, желающим начать профессиональную деятельность или учиться в ВУЗе.
Мы предлагаем очень быстро получить этот важный документ. Вы имеете возможность купить диплом, что является выгодным решением для человека, который не смог завершить образование или утратил документ. диплом изготавливается аккуратно, с максимальным вниманием ко всем элементам. В итоге вы сможете получить документ, полностью соответствующий оригиналу.
Преимущества такого подхода состоят не только в том, что можно оперативно получить свой диплом. Процесс организован просто и легко, с нашей поддержкой. Начиная от выбора нужного образца документа до правильного заполнения персональной информации и доставки в любой регион России — все под абсолютным контролем квалифицированных специалистов.
Всем, кто хочет найти быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Купить диплом – значит избежать длительного процесса обучения и не теряя времени переходить к достижению своих целей: к поступлению в ВУЗ или к началу трудовой карьеры.
https://premialnie-diplomik.com
В современном мире, где диплом является началом успешной карьеры в любой сфере, многие ищут максимально быстрый путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед всеми, кто желает вступить в сообщество профессионалов или учиться в университете.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы имеете возможность купить диплом нового или старого образца, что будет выгодным решением для человека, который не смог закончить обучение, утратил документ или желает исправить свои оценки. дипломы выпускаются с особой аккуратностью, вниманием ко всем нюансам, чтобы в итоге получился продукт, полностью соответствующий оригиналу.
Превосходство такого подхода состоит не только в том, что вы сможете оперативно получить свой диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора нужного образца до консультаций по заполнению персональной информации и доставки по стране — все будет находиться под полным контролем квалифицированных специалистов.
Для тех, кто ищет быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Приобрести диплом – значит избежать долгого процесса обучения и сразу перейти к личным целям: к поступлению в университет или к началу удачной карьеры.
https://premialnie-diplomik.com
В нашем обществе, где диплом – это начало удачной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения качественного образования. Важность наличия документа об образовании переоценить невозможно. Ведь именно он открывает дверь перед каждым человеком, который стремится вступить в сообщество профессионалов или учиться в высшем учебном заведении.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы имеете возможность заказать диплом, и это является выгодным решением для человека, который не смог закончить обучение, утратил документ или желает исправить плохие оценки. Все дипломы изготавливаются с особой тщательностью, вниманием к мельчайшим нюансам. На выходе вы получите полностью оригинальный документ.
Преимущества такого решения состоят не только в том, что можно оперативно получить свой диплом. Процесс организован просто и легко, с профессиональной поддержкой. От выбора подходящего образца диплома до точного заполнения личных данных и доставки по России — все под полным контролем опытных специалистов.
Таким образом, для всех, кто ищет оперативный способ получить требуемый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать длительного процесса обучения и не теряя времени перейти к важным целям: к поступлению в ВУЗ или к началу удачной карьеры.
https://premialnie-diplomik.com
В современном мире, где диплом становится началом успешной карьеры в любом направлении, многие пытаются найти максимально простой путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает двери перед любым человеком, желающим начать трудовую деятельность или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает оперативно получить этот необходимый документ. Вы сможете купить диплом нового или старого образца, и это является выгодным решением для человека, который не смог завершить образование или потерял документ. Все дипломы производятся аккуратно, с особым вниманием ко всем деталям. В результате вы сможете получить полностью оригинальный документ.
Преимущества подобного подхода заключаются не только в том, что вы сможете максимально быстро получить свой диплом. Процесс организован удобно, с профессиональной поддержкой. От выбора нужного образца до консультации по заполнению личной информации и доставки в любое место России — все находится под полным контролем качественных специалистов.
В итоге, для тех, кто хочет найти максимально быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Купить диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к важным целям, будь то поступление в ВУЗ или начало карьеры.
купить диплом о среднем образовании
В нашем мире, где диплом – это начало отличной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает двери перед каждым человеком, который желает вступить в сообщество квалифицированных специалистов или учиться в ВУЗе.
В данном контексте наша компания предлагает максимально быстро получить этот необходимый документ. Вы сможете заказать диплом, что будет отличным решением для всех, кто не смог закончить обучение или утратил документ. Любой диплом изготавливается с особой аккуратностью, вниманием ко всем деталям. На выходе вы сможете получить полностью оригинальный документ.
Преимущество подобного решения заключается не только в том, что можно быстро получить диплом. Процесс организован удобно, с нашей поддержкой. Начиная от выбора подходящего образца до консультации по заполнению личных данных и доставки в любое место страны — все будет находиться под абсолютным контролем наших специалистов.
Для тех, кто ищет оперативный способ получения необходимого документа, наша услуга предлагает отличное решение. Купить диплом – это значит избежать продолжительного процесса обучения и сразу перейти к достижению собственных целей, будь то поступление в университет или старт удачной карьеры.
купить диплом для иностранцев
В наше время, когда диплом – это начало удачной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Факт наличия официального документа об образовании трудно переоценить. Ведь именно он открывает дверь перед людьми, желающими вступить в сообщество профессиональных специалистов или продолжить обучение в ВУЗе.
Предлагаем быстро получить этот важный документ. Вы можете приобрести диплом, что является отличным решением для человека, который не смог закончить обучение или потерял документ. Каждый диплом изготавливается с особой аккуратностью, вниманием ко всем деталям, чтобы в итоге получился документ, максимально соответствующий оригиналу.
Превосходство этого подхода заключается не только в том, что вы оперативно получите диплом. Процесс организован удобно, с профессиональной поддержкой. От выбора необходимого образца до консультаций по заполнению личной информации и доставки в любое место России — все под полным контролем квалифицированных мастеров.
В результате, для тех, кто ищет быстрый и простой способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать долгого процесса обучения и сразу перейти к достижению личных целей: к поступлению в университет или к началу удачной карьеры.
https://www.google.com.bd/url?q=https://rudiplomista.com
https://cse.google.mu/url?q=https://premialnie-diplomik.com
https://www.google.lt/url?sa=t&url=https://premialnie-diplomik.com
http://images.google.gl/url?q=https://premialnie-diplomik.com
https://maps.google.com.ng/url?q=https://lands-diploma.com
В нашем мире, где диплом – это начало успешной карьеры в любой отрасли, многие пытаются найти максимально быстрый путь получения образования. Факт наличия официального документа переоценить просто невозможно. Ведь именно он открывает дверь перед каждым человеком, желающим начать профессиональную деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить любой необходимый документ. Вы имеете возможность заказать диплом старого или нового образца, и это является удачным решением для человека, который не смог завершить образование или потерял документ. Все дипломы выпускаются с особой аккуратностью, вниманием к мельчайшим деталям, чтобы в результате получился продукт, полностью соответствующий оригиналу.
Превосходство такого решения состоит не только в том, что можно оперативно получить свой диплом. Процесс организовывается комфортно, с нашей поддержкой. Начиная от выбора необходимого образца диплома до грамотного заполнения личных данных и доставки в любое место страны — все находится под абсолютным контролем опытных специалистов.
Всем, кто хочет найти быстрый и простой способ получить требуемый документ, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и сразу переходить к достижению личных целей, будь то поступление в ВУЗ или начало успешной карьеры.
https://www.l.google.com/url?q=https://lands-diploma.com
https://www.google.com.uy/url?q=https://rudiplomista.com
https://maps.google.cat/url?q=https://rudiplomista.com
http://maps.google.ge/url?q=https://premialnie-diplomik.com
http://maps.google.vg/url?q=https://rudiplomista.com
В нашем мире, где диплом является началом удачной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения качественного образования. Необходимость наличия официального документа об образовании переоценить невозможно. Ведь именно он открывает двери перед всеми, кто собирается начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы можете заказать диплом, и это является отличным решением для всех, кто не смог закончить обучение или утратил документ. Все дипломы изготавливаются аккуратно, с максимальным вниманием к мельчайшим нюансам, чтобы на выходе получился документ, 100% соответствующий оригиналу.
Плюсы этого подхода состоят не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается удобно, с нашей поддержкой. От выбора требуемого образца до точного заполнения персональной информации и доставки по стране — все будет находиться под полным контролем опытных специалистов.
Таким образом, для всех, кто ищет максимально быстрый способ получения требуемого документа, наша компания может предложить выгодное решение. Приобрести диплом – значит избежать длительного процесса обучения и сразу перейти к достижению своих целей, будь то поступление в университет или старт удачной карьеры.
http://maps.google.com.ly/url?q=https://lands-diploma.com
https://clients1.google.co.im/url?sa=t&url=https://rudiplomista.com
http://cse.google.ng/url?q=https://lands-diploma.com
http://www.google.co.ao/url?q=https://rudiplomista.com
https://www.google.cd/url?sa=i&url=https://lands-diploma.com
В наше время, когда диплом – это начало успешной карьеры в любой области, многие ищут максимально простой путь получения качественного образования. Наличие официального документа об образовании переоценить попросту невозможно. Ведь диплом открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в каком-либо институте.
В данном контексте мы предлагаем быстро получить этот необходимый документ. Вы сможете приобрести диплом, и это становится удачным решением для человека, который не смог завершить образование или утратил документ. Любой диплом изготавливается аккуратно, с особым вниманием к мельчайшим элементам, чтобы в результате получился документ, полностью соответствующий оригиналу.
Плюсы этого подхода состоят не только в том, что можно оперативно получить свой диплом. Процесс организован удобно, с профессиональной поддержкой. От выбора требуемого образца до консультации по заполнению личной информации и доставки в любое место России — все под полным контролем опытных специалистов.
Для всех, кто ищет быстрый и простой способ получить необходимый документ, наша услуга предлагает выгодное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к важным целям, будь то поступление в ВУЗ или начало карьеры.
https://maps.google.com.au/url?q=https://premialnie-diplomik.com
http://www.google.co.uk/url?q=https://rudiplomista.com
https://www.google.com.om/url?sa=t&url=https://rudiplomista.com
https://www.google.com.co/url?q=https://rudiplomista.com
https://maps.google.ie/url?q=https://rudiplomista.com
В наше время, когда диплом становится началом успешной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает дверь перед людьми, стремящимися вступить в профессиональное сообщество или продолжить обучение в любом университете.
Предлагаем оперативно получить этот важный документ. Вы сможете приобрести диплом старого или нового образца, и это будет отличным решением для человека, который не смог закончить обучение или утратил документ. дипломы производятся аккуратно, с максимальным вниманием ко всем деталям. В результате вы получите продукт, полностью соответствующий оригиналу.
Преимущество данного подхода заключается не только в том, что можно максимально быстро получить свой диплом. Процесс организовывается комфортно, с нашей поддержкой. Начав от выбора подходящего образца документа до точного заполнения личных данных и доставки в любое место России — все под полным контролем опытных специалистов.
Для всех, кто хочет найти быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Купить диплом – значит избежать продолжительного обучения и не теряя времени переходить к достижению личных целей, будь то поступление в ВУЗ или начало карьеры.
https://image.google.ac/url?q=https://rudiplomista.com
https://clients1.google.com.pa/url?sa=t&url=https://premialnie-diplomik.com
http://maps.google.ge/url?q=https://lands-diploma.com
https://maps.google.ki/url?sa=t&url=https://lands-diploma.com
https://maps.google.com.lb/url?q=https://premialnie-diplomik.com
В современном мире, где диплом – это начало успешной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Факт наличия официального документа переоценить невозможно. Ведь диплом открывает двери перед людьми, желающими вступить в сообщество квалифицированных специалистов или продолжить обучение в университете.
Мы предлагаем очень быстро получить этот необходимый документ. Вы имеете возможность заказать диплом, что будет отличным решением для человека, который не смог завершить образование или утратил документ. диплом изготавливается с особой тщательностью, вниманием ко всем деталям, чтобы в результате получился продукт, полностью соответствующий оригиналу.
Плюсы такого решения заключаются не только в том, что вы сможете оперативно получить диплом. Процесс организовывается удобно, с профессиональной поддержкой. От выбора требуемого образца диплома до консультации по заполнению личных данных и доставки по России — все под полным контролем качественных специалистов.
Таким образом, для тех, кто пытается найти максимально быстрый способ получения требуемого документа, наша услуга предлагает выгодное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к достижению своих целей, будь то поступление в университет или начало удачной карьеры.
http://oddicini.ru/sovety/texnologiya-almaznoj-rezki-betonnyx-izdelij-i-sovety.html?unapproved=230076&moderation-hash=ffdd60bf5bf9d2bdb9266dfe0dd14d9e#comment-230076
http://helenos.pavel-rimsky.cz/doku.php?id=russianydiplomans
http://himagro.md/forum/user/39416/
http://olddle.orkclub.ru/index.php?subaction=userinfo&user=ivycoxyn
http://proffpereezd.ru/index.php?subaction=userinfo&user=egijyvo
qiyezp.com
Zhu Xiurong은 무언가를 생각했습니다. “당신의 생각을 어떻게 볼 수 있습니까?”
Hi there, yup this paragraph is actually good and I have
learned lot of things from it regarding blogging.
thanks.
В нашем обществе, где диплом – это начало успешной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения качественного образования. Необходимость наличия официального документа трудно переоценить. Ведь диплом открывает дверь перед всеми, кто стремится начать профессиональную деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает быстро получить этот необходимый документ. Вы можете купить диплом, что будет отличным решением для человека, который не смог закончить образование, утратил документ или желает исправить свои оценки. Любой диплом изготавливается с особой аккуратностью, вниманием к мельчайшим нюансам. В итоге вы сможете получить 100% оригинальный документ.
Преимущество этого подхода заключается не только в том, что можно оперативно получить диплом. Процесс организован просто и легко, с нашей поддержкой. Начиная от выбора необходимого образца до консультаций по заполнению персональной информации и доставки в любой регион России — все под абсолютным контролем квалифицированных специалистов.
Для тех, кто ищет быстрый и простой способ получить необходимый документ, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать длительного процесса обучения и не теряя времени переходить к важным целям, будь то поступление в ВУЗ или старт карьеры.
https://images.google.by/url?sa=t&url=https://rudiplomista.com
https://images.google.nu/url?q=https://rudiplomista.com
http://image.google.iq/url?rct=j&sa=t&url=https://premialnie-diplomik.com
https://cse.google.ki/url?q=https://premialnie-diplomik.com
https://maps.google.cf/url?sa=t&url=https://premialnie-diplomik.com
ilogidis.com
갑자기 Zhu Houzhao는 “잠깐만”이라는 것을 다시 기억했습니다.
В наше время, когда диплом является началом удачной карьеры в любой области, многие ищут максимально быстрый путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед всеми, кто собирается вступить в профессиональное сообщество или продолжить обучение в любом университете.
Наша компания предлагает максимально быстро получить этот важный документ. Вы сможете приобрести диплом, и это будет удачным решением для всех, кто не смог завершить обучение, потерял документ или хочет исправить плохие оценки. Все дипломы изготавливаются с особой тщательностью, вниманием ко всем нюансам, чтобы на выходе получился 100% оригинальный документ.
Превосходство такого решения заключается не только в том, что можно оперативно получить свой диплом. Весь процесс организован комфортно, с нашей поддержкой. От выбора требуемого образца до правильного заполнения персональных данных и доставки по стране — все находится под полным контролем наших специалистов.
Для тех, кто пытается найти оперативный способ получить необходимый документ, наша компания готова предложить отличное решение. Купить диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к личным целям: к поступлению в университет или к началу успешной карьеры.
https://maps.google.ml/url?q=https://rudiplomista.com
https://images.google.jo/url?q=https://premialnie-diplomik.com
https://images.google.as/url?q=https://rudiplomista.com
https://cse.google.ws/url?sa=t&url=https://rudiplomista.com
http://images.google.com.pg/url?q=https://rudiplomista.com
В современном мире, где диплом – это начало успешной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед всеми, кто желает вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Предлагаем очень быстро получить этот важный документ. Вы можете купить диплом, и это является выгодным решением для человека, который не смог завершить образование, утратил документ или желает исправить свои оценки. Все дипломы производятся аккуратно, с максимальным вниманием к мельчайшим нюансам, чтобы на выходе получился продукт, полностью соответствующий оригиналу.
Превосходство такого подхода заключается не только в том, что можно быстро получить свой диплом. Процесс организовывается комфортно и легко, с нашей поддержкой. От выбора подходящего образца диплома до консультации по заполнению персональных данных и доставки в любое место России — все будет находиться под полным контролем качественных мастеров.
В результате, для всех, кто ищет быстрый способ получить требуемый документ, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать долгого обучения и сразу переходить к достижению собственных целей, будь то поступление в ВУЗ или старт профессиональной карьеры.
https://images.google.co.id/url?q=https://premialnie-diplomik.com
https://images.google.nu/url?q=https://lands-diploma.com
https://cse.google.co.ve/url?sa=t&url=https://rudiplomista.com
https://clients1.google.com.hk/url?sa=t&url=https://premialnie-diplomik.com
https://cse.google.bf/url?sa=t&url=https://rudiplomista.com
В нашем мире, где диплом становится началом успешной карьеры в любой сфере, многие стараются найти максимально простой путь получения качественного образования. Необходимость наличия официального документа об образовании переоценить невозможно. Ведь диплом открывает двери перед всеми, кто желает начать трудовую деятельность или продолжить обучение в университете.
В данном контексте мы предлагаем оперативно получить этот необходимый документ. Вы сможете приобрести диплом, что становится выгодным решением для человека, который не смог завершить образование или потерял документ. Все дипломы производятся с особой тщательностью, вниманием ко всем элементам, чтобы в итоге получился полностью оригинальный документ.
Превосходство такого решения состоит не только в том, что можно оперативно получить свой диплом. Процесс организован комфортно, с нашей поддержкой. Начиная от выбора нужного образца до точного заполнения личных данных и доставки в любое место России — все под полным контролем качественных мастеров.
Таким образом, для тех, кто хочет найти оперативный способ получения необходимого документа, наша услуга предлагает выгодное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в ВУЗ или старт удачной карьеры.
https://clients3.google.com/url?q=https://rudiplomista.com
https://www.google.com.np/url?sa=t&url=https://premialnie-diplomik.com
https://clients1.google.nu/url?sa=t&url=https://rudiplomista.com
https://images.google.ad/url?q=https://rudiplomista.com
https://images.google.com.gi/url?sa=t&url=https://rudiplomista.com
В нашем обществе, где диплом становится началом удачной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Необходимость наличия официального документа об образовании переоценить попросту невозможно. Ведь именно он открывает дверь перед любым человеком, желающим начать профессиональную деятельность или учиться в высшем учебном заведении.
В данном контексте мы предлагаем максимально быстро получить любой необходимый документ. Вы имеете возможность заказать диплом, что становится удачным решением для всех, кто не смог завершить обучение, потерял документ или хочет исправить свои оценки. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам. На выходе вы получите документ, 100% соответствующий оригиналу.
Преимущества такого решения состоят не только в том, что вы сможете максимально быстро получить диплом. Весь процесс организован комфортно, с нашей поддержкой. Начиная от выбора требуемого образца до консультаций по заполнению персональной информации и доставки по России — все под полным контролем квалифицированных специалистов.
Для всех, кто хочет найти быстрый способ получить необходимый документ, наша услуга предлагает отличное решение. Купить диплом – значит избежать долгого обучения и не теряя времени перейти к достижению личных целей, будь то поступление в университет или старт профессиональной карьеры.
https://cse.google.dj/url?sa=t&url=https://premialnie-diplomik.com
https://images.google.com.ng/url?sa=t&url=https://lands-diploma.com
https://www.google.dk/url?sa=t&url=https://rudiplomista.com
http://images.google.dj/url?q=https://rudiplomista.com
https://cse.google.com.br/url?sa=t&url=https://premialnie-diplomik.com
exprimegranada.com
この記事から多くを学びました。引き続きフォローします。
В нашем обществе, где диплом является началом удачной карьеры в любой сфере, многие ищут максимально быстрый путь получения образования. Факт наличия официального документа переоценить просто невозможно. Ведь диплом открывает дверь перед людьми, желающими начать трудовую деятельность или учиться в ВУЗе.
Предлагаем быстро получить этот важный документ. Вы имеете возможность заказать диплом, что является удачным решением для человека, который не смог закончить обучение или потерял документ. Все дипломы изготавливаются аккуратно, с максимальным вниманием ко всем нюансам. На выходе вы сможете получить полностью оригинальный документ.
Преимущество данного подхода заключается не только в том, что вы сможете быстро получить диплом. Весь процесс организовывается комфортно, с нашей поддержкой. Начав от выбора требуемого образца документа до консультаций по заполнению персональных данных и доставки в любое место России — все будет находиться под полным контролем опытных мастеров.
В итоге, для всех, кто пытается найти быстрый и простой способ получения необходимого документа, наша компания предлагает отличное решение. Заказать диплом – значит избежать долгого обучения и сразу переходить к своим целям: к поступлению в ВУЗ или к началу успешной карьеры.
http://www.pirobot.co.uk/doku.php?id=rudiplomisty
http://bizon15.ru/index.php?subaction=userinfo&user=ageqiv
http://ledsoft.ru/forum/?PAGE_NAME=message&FID=5&TID=13072&TITLE_SEO=13072-rudiplomisty&MID=13829&result=new#message13829
http://www.besconint.co.kr/bbs/board.php?bo_table=free&wr_id=388482
http://xn—-7sbgj9bdrl.xn--p1ai/users/atijymeke
В современном мире, где диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения образования. Необходимость наличия официального документа об образовании сложно переоценить. Ведь именно он открывает дверь перед всеми, кто стремится вступить в сообщество профессиональных специалистов или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает оперативно получить любой необходимый документ. Вы имеете возможность купить диплом, и это будет удачным решением для человека, который не смог закончить обучение или утратил документ. Все дипломы выпускаются с особой аккуратностью, вниманием к мельчайшим нюансам, чтобы в итоге получился полностью оригинальный документ.
Преимущество этого решения состоит не только в том, что можно оперативно получить свой диплом. Процесс организовывается удобно и легко, с нашей поддержкой. Начиная от выбора подходящего образца документа до точного заполнения личных данных и доставки в любое место страны — все под полным контролем качественных специалистов.
Для тех, кто хочет найти оперативный способ получить требуемый документ, наша компания готова предложить отличное решение. Купить диплом – значит избежать продолжительного обучения и сразу перейти к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
https://maps.google.com.sg/url?sa=t&url=https://premialnie-diplomik.com
https://www.google.ca/url?q=https://rudiplomista.com
https://clients1.google.com.bd/url?q=https://lands-diploma.com
https://images.google.co.jp/url?sa=t&url=https://lands-diploma.com
https://cse.google.com.af/url?sa=t&url=https://rudiplomista.com
Сегодня, когда диплом – это начало удачной карьеры в любой области, многие стараются найти максимально быстрый путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает двери перед каждым человеком, который стремится вступить в сообщество квалифицированных специалистов или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем быстро получить этот необходимый документ. Вы можете приобрести диплом нового или старого образца, что будет удачным решением для всех, кто не смог закончить обучение или утратил документ. Любой диплом изготавливается с особой аккуратностью, вниманием ко всем элементам. На выходе вы сможете получить продукт, максимально соответствующий оригиналу.
Превосходство такого подхода состоит не только в том, что вы оперативно получите диплом. Процесс организовывается просто и легко, с нашей поддержкой. От выбора требуемого образца до грамотного заполнения персональных данных и доставки по России — все под абсолютным контролем опытных мастеров.
Для тех, кто пытается найти оперативный способ получения необходимого документа, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к важным целям, будь то поступление в ВУЗ или начало успешной карьеры.
http://torgteh-rzn.ru/index.php?option=com_k2&view=itemlist&task=user&id=10324
http://www.djngo.or.kr/gb5/bbs/board.php?bo_table=free&wr_id=1328704
http://golpro.jp/userinfo.php?uid=13159#
http://mos-it.ru/index.php?subaction=userinfo&user=exafypo
http://forum.farmer.pl/profile/150022-uritykic/
Рады приветствовать вас, друзья!
К вам обращается агентство СЕО продвижения XRumer LLC.
Ваш портал, как мы видим, еще только начинает набирать обороты. Для того, чтобы ускорить процесс его роста, предлагаем наши услуги по внешней SEO-оптимизации. Продвижение в поисковиках – наше основное направление. В ассортименте имеются эффективные СЕО-инструменты для экспертов. Наши специалисты обладают значительным опытом и портфолио успешных проектов – предоставим по вашему запросу.
Мы предлагаем скидку 10% на востребованные услуги.
Услуги:
– Супер трастовые ссылки (нужно любому сайту) – от 1,5 до 5000 руб
– Размещаем 2500 жирных безанкорных ссылок (рекомендуется любым сайтам) – 3900 рублей
– Прогон по 110 тыс. сайтам в зоне RU (максимально полезно для сайтов) – 2900 рублей
– Размещение 150 постов в VK про ваш сайт (поможет получить рекламу) – 3.900 рублей
– Публикации про ваш сайт на 300 интернет-форумах (мощнейшая раскрутка сайта) – 29 тыс. руб
– СуперПостинг – комплексный прогон по 3 000 000 ресурсов (грандиозное размещение для ваших сайтов) – 39 тысяч рублей
– Рассылка сообщений по сайтам при помощи обратной связи – договорная цена, в зависимости от объемов.
Если появятся вопросы, смело обращайтесь, готовы помочь.
Telgrm: @exrumer
https://XRumer.cc/
Skype: Loves.Ltd
В нашем обществе, где диплом – это начало отличной карьеры в любой области, многие пытаются найти максимально быстрый путь получения качественного образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает двери перед людьми, стремящимися вступить в профессиональное сообщество или продолжить обучение в каком-либо ВУЗе.
Предлагаем максимально быстро получить этот необходимый документ. Вы сможете приобрести диплом нового или старого образца, что является отличным решением для всех, кто не смог закончить образование, потерял документ или хочет исправить плохие оценки. диплом изготавливается с особой аккуратностью, вниманием к мельчайшим нюансам. На выходе вы сможете получить продукт, полностью соответствующий оригиналу.
Преимущество этого подхода заключается не только в том, что вы максимально быстро получите свой диплом. Процесс организовывается удобно, с нашей поддержкой. Начав от выбора нужного образца до грамотного заполнения персональной информации и доставки по России — все под абсолютным контролем качественных мастеров.
Для тех, кто пытается найти оперативный способ получения необходимого документа, наша компания предлагает выгодное решение. Купить диплом – это значит избежать длительного процесса обучения и сразу перейти к своим целям, будь то поступление в ВУЗ или старт удачной карьеры.
https://www.google.kz/url?q=https://premialnie-diplomik.com
https://cse.google.cd/url?sa=t&url=https://premialnie-diplomik.com
https://www.google.ge/url?q=https://lands-diploma.com
https://images.google.com.cy/url?q=https://rudiplomista.com
https://www.google.co.ls/url?q=https://premialnie-diplomik.com
В нашем обществе, где диплом становится началом успешной карьеры в любой сфере, многие ищут максимально простой путь получения качественного образования. Наличие официального документа сложно переоценить. Ведь именно диплом открывает двери перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в любом университете.
Наша компания предлагает быстро получить этот необходимый документ. Вы имеете возможность приобрести диплом старого или нового образца, что является отличным решением для человека, который не смог завершить образование или потерял документ. диплом изготавливается с особой аккуратностью, вниманием ко всем деталям. В результате вы получите документ, 100% соответствующий оригиналу.
Превосходство этого подхода заключается не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается удобно, с нашей поддержкой. Начиная от выбора нужного образца диплома до консультаций по заполнению персональной информации и доставки в любое место России — все будет находиться под абсолютным контролем опытных мастеров.
Всем, кто хочет найти быстрый способ получить требуемый документ, наша услуга предлагает выгодное решение. Приобрести диплом – это значит избежать долгого обучения и не теряя времени перейти к своим целям: к поступлению в университет или к началу удачной карьеры.
http://rivertravel.net/viewtopic.php?f=21&t=7976
http://www.gocoram.co.kr/bbs/board.php?bo_table=free&wr_id=286263
https://anaparegion.ru/news/237-dyussh_viktoriya_vstretila_uchastnikov_vsekubansko/
http://www.pavelgorbenko.ru/2018/02/blog-post.html
https://www.computer-setup.ru/kak-ubrat-reklamu-v-torrente#comment-2668
В нашем мире, где диплом – это начало удачной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь именно диплом открывает двери перед людьми, желающими начать профессиональную деятельность или учиться в высшем учебном заведении.
Мы предлагаем очень быстро получить этот необходимый документ. Вы имеете возможность купить диплом, и это будет выгодным решением для всех, кто не смог завершить образование или утратил документ. диплом изготавливается аккуратно, с особым вниманием ко всем элементам. В результате вы получите полностью оригинальный документ.
Преимущества данного подхода заключаются не только в том, что вы оперативно получите диплом. Весь процесс организован удобно, с профессиональной поддержкой. От выбора нужного образца диплома до консультации по заполнению персональных данных и доставки по России — все будет находиться под абсолютным контролем качественных специалистов.
В результате, всем, кто пытается найти максимально быстрый способ получить необходимый документ, наша услуга предлагает выгодное решение. Приобрести диплом – значит избежать долгого процесса обучения и сразу перейти к достижению своих целей, будь то поступление в ВУЗ или старт трудовой карьеры.
https://sites.google.com/view/sunwinclcomhttps://rudiplomista.com
https://images.google.kg/url?q=https://premialnie-diplomik.com
https://clients1.google.lv/url?sa=t&url=https://lands-diploma.com
https://clients1.google.com.nf/url?sa=t&url=https://rudiplomista.com
https://www.google.ad/url?q=https://rudiplomista.com
На сегодняшний день, когда диплом является началом успешной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Важность наличия официального документа переоценить невозможно. Ведь диплом открывает двери перед каждым человеком, который стремится начать трудовую деятельность или продолжить обучение в любом институте.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы можете приобрести диплом старого или нового образца, что становится отличным решением для человека, который не смог завершить образование, потерял документ или хочет исправить свои оценки. дипломы выпускаются аккуратно, с особым вниманием ко всем элементам. На выходе вы получите полностью оригинальный документ.
Превосходство такого решения заключается не только в том, что можно оперативно получить диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. Начиная от выбора необходимого образца документа до консультаций по заполнению персональной информации и доставки по России — все находится под абсолютным контролем опытных мастеров.
Таким образом, для тех, кто ищет оперативный способ получения требуемого документа, наша компания предлагает отличное решение. Заказать диплом – значит избежать длительного обучения и не теряя времени перейти к своим целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://www.google.com.ar/url?q=https://lands-diploma.com
https://www.google.co.vi/url?q=https://premialnie-diplomik.com
https://images.google.co.ug/url?q=https://rudiplomista.com
https://maps.google.no/url?rct=t&sa=t&url=https://rudiplomista.com
https://www.google.com.sb/url?q=https://lands-diploma.com
В нашем мире, где диплом – это начало успешной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа переоценить просто невозможно. Ведь диплом открывает двери перед каждым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Наша компания предлагает максимально быстро получить этот необходимый документ. Вы можете заказать диплом нового или старого образца, и это является выгодным решением для всех, кто не смог завершить образование или утратил документ. дипломы изготавливаются аккуратно, с особым вниманием к мельчайшим деталям, чтобы в результате получился документ, 100% соответствующий оригиналу.
Превосходство такого решения заключается не только в том, что можно оперативно получить диплом. Весь процесс организован комфортно, с профессиональной поддержкой. От выбора требуемого образца до консультаций по заполнению личных данных и доставки в любой регион России — все находится под полным контролем опытных специалистов.
В итоге, всем, кто ищет быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и сразу перейти к важным целям: к поступлению в ВУЗ или к началу удачной карьеры.
http://images.google.com.bh/url?sa=t&url=https://lands-diploma.com
https://maps.google.fi/url?q=https://rudiplomista.com
https://maps.google.mg/url?q=https://rudiplomista.com
https://maps.google.co.mz/url?q=https://lands-diploma.com
https://cse.google.mg/url?sa=t&url=https://lands-diploma.com
В современном мире, где диплом становится началом отличной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения образования. Наличие официального документа переоценить просто невозможно. Ведь именно он открывает дверь перед каждым человеком, желающим вступить в сообщество профессионалов или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает быстро получить любой необходимый документ. Вы можете приобрести диплом, что будет отличным решением для всех, кто не смог завершить образование или потерял документ. дипломы производятся аккуратно, с максимальным вниманием к мельчайшим элементам. В итоге вы получите 100% оригинальный документ.
Преимущества данного подхода состоят не только в том, что вы быстро получите диплом. Процесс организован удобно и легко, с нашей поддержкой. Начиная от выбора необходимого образца до правильного заполнения личной информации и доставки в любой регион России — все находится под полным контролем качественных специалистов.
Всем, кто пытается найти быстрый способ получить требуемый документ, наша компания может предложить выгодное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и сразу переходить к важным целям, будь то поступление в университет или старт карьеры.
http://www.kiopro.ru/support/forum/view_profile.php?UID=71156
http://kvartirker.ru/profile/ebetusoru/
https://kolonkineva.ru/catalog/gazovye-kolonki/1/90/gazovaja-kolonka-halsen-wm-12/#fa20f7d33e45bddb83f4901170988ed57a
https://www.rcdrift.ru/forum/member.php?u=22798
http://soccer-manager.eu/forum/profile.php?id=1785538
Timsothyreemn
Всем хорошего дня!
Диплом постепенно улучшается с каждым днем благодаря использованию найденных ресурсов в интернете.
Приобретайте диплом университета без предоплаты с доставкой по всей России.
Желаю вам всем отличных отметок!
купить диплом в невинномысске
купить диплом гознак
купить диплом ветеринара
купить диплом в канске
купить диплом логопеда
купить диплом в новошахтинске
купить диплом о среднем образовании
купить диплом в нальчике
купить дипломы о высшем цены
купить диплом в чите
Компания «VIP транспорт» успешно предоставляет услуги пассажироперевозок. В нашем автопарке имеются: вместительные автобусы с разным количеством мест, минивэны, автомобили премиум, комфорт, бизнес-класса, микроавтобусы. Мы собрали великолепный коллектив водителей, вы точно в них можете не сомневаться. Ищете вызвать такси вип? Anapa-vip-transport.ru – сайт, где есть телефонный номер круглосуточной диспетчерской службы. Также тут имеются документы, с которыми вы прямо сейчас можете ознакомиться. Своим клиентам предлагаем лучшие условия. Звоните нам!
http://m1bar.com/index.php?subaction=userinfo&user=avyval
http://www.fotoblog365.com/2017/02/sony-a6500.html
http://ds2.edu.sbor.net/index.php?subaction=userinfo&user=axycypo
http://ledsoft.ru/forum/?PAGE_NAME=message&FID=5&TID=13122&TITLE_SEO=13122-adiplomarussian&MID=13880&result=new#message13880
http://www.repetitor.tv/2015/06/1-2015.html
Купить аттестат образование – это ключевой показатель вашего перспектив. Воспользуйтесь нашими предложением, чтобы быстро и без труда приобрести документ, необходимый для продолжения учебы или трудоустройства. Наши квалифицированные специалисты обеспечивают отличное качество и конфиденциальность предоставляемых сервисов. Покупайте школьный аттестат у нас и обнаруживайте дополнительные варианты для вашего образования и карьеры.
IsmaelBog
Добрый день!
Диплом стал моей главной проблемой, но благодаря интернету я нашел способы улучшить свою работу и сохранить здоровье.
Предлагаем всем желающим приобрести диплом университета России по выгодной цене с доставкой “под ключ”.
Желаю всем честных отметок!
купить диплом в махачкале
купить диплом во всеволожске
купить диплом в миассе
купить диплом в воткинске
купить диплом гознак
купить диплом цена
купить диплом в сарапуле
купить диплом в асбесте
купить диплом в волгограде
купить диплом в дербенте
ilogidis.com
“알았어, 배웅해.” 송곳니 노사는 소매를 흔들었다.
http://doktorpishet.ru/index.php?subaction=userinfo&user=yhunykij
http://www.clean-ace8.com/bbs/board.php?bo_table=free&wr_id=278397
http://jilye74.ru/index.php?subaction=userinfo&user=esydelug
http://qa.laodongzu.com/index.php?qa=user&qa_1=irypihyq
http://myai.ru/communication/forum/user/4029/
http://stavklad.ru/viewtopic.php?f=5&t=4840
http://feki-php.8u.cz/profile.php?lookup=10909
https://xn--80asucf0d.xn--j1al4b.xn--p1ai/index.php/component/k2/itemlist/user/123236
http://georgiantheatre.ge/index.php?subaction=userinfo&user=ocefo
Сколько купить аттестат – это важный показатель твоего развития. Воспользуйтесь нашим услугами, чтобы просто и без проблем приобрести документ, необходимый для последующего образования или получения работы. Наши высококвалифицированные специалисты гарантируют качество и дискретность предоставляемых сервисов. Покупайте диплом у нас и открывайте свежие варианты для вашего образования и профессионального роста.
Сегодня, когда диплом – это начало отличной карьеры в любой области, многие стараются найти максимально простой путь получения качественного образования. Наличие официального документа об образовании переоценить невозможно. Ведь именно диплом открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или учиться в любом институте.
В данном контексте наша компания предлагает очень быстро получить этот необходимый документ. Вы сможете приобрести диплом, и это будет удачным решением для человека, который не смог завершить образование или утратил документ. Все дипломы выпускаются с особой аккуратностью, вниманием ко всем элементам, чтобы в результате получился полностью оригинальный документ.
Плюсы подобного подхода состоят не только в том, что можно оперативно получить свой диплом. Процесс организован удобно, с профессиональной поддержкой. Начиная от выбора подходящего образца до консультации по заполнению персональной информации и доставки в любое место России — все будет находиться под абсолютным контролем опытных мастеров.
Всем, кто хочет найти оперативный способ получить требуемый документ, наша компания предлагает выгодное решение. Купить диплом – значит избежать долгого процесса обучения и сразу перейти к достижению личных целей: к поступлению в университет или к началу успешной карьеры.
https://dawonprint.com/bbs/board.php?bo_table=free&wr_id=458116
https://www.stagebox.uk/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BF%D0%BE%D0%BB%D1%83%D1%87%D0%B8%D1%82%D1%8C-%D0%BE%D1%80%D0%B8%D0%B3%D0%B8%D0%BD%D0%B0%D0%BB%D1%8C%D0%BD%D1%8B%D0%B9/
http://xn--h1a1ab.xn--p1ai/index.php?name=account&op=info&uname=iwequzak
http://netl.free.fr/index.php?file=Members&op=detail&autor=owesihiv
http://location-bormes-les-mimosas.fr/index.php?option=com_easybookreloaded
IsmaelBog
Всем хорошего дня!
Дипломное задание, казавшееся непреодолимым, стало легче благодаря интернет-ресурсам.
Мы предлагаем заказать диплом без предоплаты с доставкой курьером по РФ, полный комплект!
Желаю для каждого положительных оценок!
купить диплом биолога
купить диплом о среднем образовании
купить диплом в бугульме
купить диплом в красноярске
купить диплом в балашихе
купить диплом сварщика
купить диплом парикмахера
купить диплом старого образца
купить диплом мастера маникюра и педикюра
купить диплом в мичуринске
StephenNug
Привет всем!
Диплом постепенно улучшается с каждым днем благодаря использованию найденных ресурсов в интернете.
Приобретайте диплом университета без предоплаты у нас с доставкой в любой город России, качество и безопасность гарантированы.
Желаю всем положительных оценок!
купить диплом в нальчике
купить диплом химика
купить диплом в мичуринске
купить диплом в каменске-шахтинском
купить диплом в оренбурге
купить диплом в пензе
купить диплом в майкопе
купить диплом в костроме
купить диплом маркетолога
купить диплом в самаре
http://onlineboxing.net/jforum/user/profile/283138.page
http://joomla-461471-1470402.cloudwaysapps.com/blog/item/27-under-ground-54
http://gderabotaem.ru/company/gosznacdiplom
http://forum.computest.ru/post/628830/#p628830
Ремонт Дома под ключ в Алматы – от идеи до реализации. Надежно, качественно и в срок. Мы предлагаем полный спектр услуг: от дизайна интерьера до отделочных работ любой сложности. Доверьте свой ремонт опытным специалистам и получите идеальный результат.
IsmaelBog
Приветики, дорогие мои!!
Инструменты, найденные в сети для диплома, облегчили процесс написания.
Предлагаем всем желающим приобрести диплом университета России по выгодной цене с доставкой “под ключ”.
Желаю вам всем отличных отметок!
купить сертификат специалиста
купить диплом в твери
куплю диплом кандидата наук
купить диплом учителя физической культуры
купить диплом в стерлитамаке
купить диплом в смоленске
купить диплом в орске
купить диплом в новомосковске
купить диплом в бийске
купить диплом в туле
27-го июля был проведён первый тест эффективности XRumer 23 StrongAI (по сравнению с текущей версией XRumer 19.0.18), который показал двадцатикратный прирост. Столь впечатляющие результаты достигнуты благодаря многократному улучшению пробиваемости уже известных платформ, добавлению поддержки новых платформ, обучению сотням тысяч типов новых тексткапч, внедрению нового GPT-модуля, и множеству других улучшений. По ссылке ниже Вы сможете изучить и скачать детали теста (исходная база, кусок отчёта, статистику и другие подробности)
Спасибо за рефку:
http://www.botmasterru.com/product109120/
프라그마틱에 대한 글 읽는 것이 정말 재미있었어요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 발전하며 더 많은 지식을 쌓아가요!
pragmatic-game.com
프라그마틱의 게임은 정말 다양한데, 최근에 출시된 것 중 어떤 게임이 가장 좋았나요? 공유해주세요!
https://www.vamiveta.com/
https://www.hh12315.cn/
https://vispills.com/
http://maisoncarlos.com/UserProfile/tabid/42/UserID/1933482/Default.aspx
http://food-ua.com/wr_board/tools.php?event=profile&pname=ytyduze
http://www.gencotyre.com/comerblog/member.asp?action=view&memName=axyloh
http://wennw.com/member.asp?action=view&memName=imolot
http://krutovo.ru/index.php?name=Account&op=info&uname=onyzaso
Купить аттестат образование – это ключевой инструмент твоего развития. Воспользуйтесь нашими сервисом, чтобы легко и без лишних усилий приобрести документ, необходимый для последующего образования или получения работы. Наши квалифицированные специалисты обеспечивают высокое качество и конфиденциальность предоставляемых услуг. Покупайте диплом у нас и раскрывайте новые варианты для своего образования и трудоустройства.
sandyterrace.com
“무릎을 꿇어라.” 홍지황제가 그를 한참 쳐다보더니 조금 거칠게 말했다.
ShaneViart
Добрый день!
Диплом требует от меня немалых усилий, но доступные в сети ресурсы делают этот процесс более приятным.
На нашем сайте представлен широкий выбор дипломов с гарантией качества и быстрой доставкой в любой регион РФ.
Желаю каждому прекрасных отметок!
купить диплом в бийске
купить диплом в прокопьевске
купить диплом в стерлитамаке
купить диплом о среднем образовании
купить диплом в улан-удэ
купить диплом
купить диплом в усолье-сибирском
купить дипломы о высшем образовании цена
купить диплом в глазове
купить аттестаты за 11
thebuzzerpodcast.com
Fang Jifan은 학자의 코를 가리키고 욕하는 것과 같은 약간 대담했습니다.
Сеть салонов
Eksprovocator Plaits Order
Мир богатства для ваших висок
https://eksprovocator.ru/
Сыскиваясь в Столице, Нью-Йорке чи Дубае, вы сможете заглянуть наши салоны в течение удобное время. Чтобы сего:
Загляните на фотосайт равно пользуйтесь подходящим схемой связи: позвоните, нарисуйте в социальных сетях, забудьте свои контакты;
Выберите нужную предложение равным образом мастера. Разве что вы у нас заперво, управленец с наслаждением проконсультирует, ответит сверху любой тема, сориентирует по цене а также продолжительности упражнения;
Приходите в течение предназначенное ятси (а) также наслаждайтесь плодом! https://eksprovocator.ru/
Мы гарантируем, яко через некоторое время посещения Eksprovocator Hair Lambaste вы больше не попытаетесь на другие салоны, чай язык нас является все, что что поделаешь!
В современном мире, где диплом является началом успешной карьеры в любом направлении, многие стараются найти максимально простой путь получения образования. Наличие официального документа сложно переоценить. Ведь диплом открывает двери перед людьми, желающими вступить в профессиональное сообщество или продолжить обучение в университете.
Мы предлагаем оперативно получить этот важный документ. Вы имеете возможность заказать диплом старого или нового образца, и это будет удачным решением для человека, который не смог завершить обучение, потерял документ или хочет исправить свои оценки. дипломы выпускаются с особой тщательностью, вниманием ко всем деталям, чтобы в итоге получился документ, 100% соответствующий оригиналу.
Преимущества такого решения заключаются не только в том, что можно оперативно получить свой диплом. Процесс организован комфортно, с нашей поддержкой. Начиная от выбора необходимого образца до точного заполнения персональных данных и доставки в любое место России — все под абсолютным контролем квалифицированных мастеров.
В итоге, всем, кто пытается найти быстрый и простой способ получить необходимый документ, наша компания предлагает выгодное решение. Купить диплом – значит избежать долгого процесса обучения и не теряя времени перейти к своим целям, будь то поступление в университет или старт карьеры.
http://www.artcalendar.ru/783.html
http://sad5.lytkarino.net/index.php?subaction=userinfo&user=amodudifo
https://aben75.cafe24.com/bbs/board.php?bo_table=free&wr_id=278583
https://hitaste.ru/forum/?PAGE_NAME=profile_view&UID=246148
http://msfo-soft.ru/msfo/forum/messages/forum30/topic8710/message252631/?result=new#message252631
В нашем обществе, где диплом – это начало отличной карьеры в любой области, многие ищут максимально быстрый путь получения качественного образования. Факт наличия официального документа об образовании переоценить невозможно. Ведь диплом открывает двери перед каждым человеком, который хочет начать профессиональную деятельность или продолжить обучение в каком-либо институте.
В данном контексте наша компания предлагает максимально быстро получить любой необходимый документ. Вы можете заказать диплом старого или нового образца, и это становится удачным решением для человека, который не смог завершить образование или утратил документ. Каждый диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим деталям. В результате вы сможете получить документ, максимально соответствующий оригиналу.
Плюсы подобного решения состоят не только в том, что вы сможете оперативно получить диплом. Весь процесс организован комфортно, с нашей поддержкой. От выбора подходящего образца до правильного заполнения личной информации и доставки по стране — все под абсолютным контролем качественных мастеров.
Для тех, кто хочет найти максимально быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Купить диплом – это значит избежать продолжительного обучения и не теряя времени перейти к личным целям, будь то поступление в университет или старт трудовой карьеры.
http://sport-technology.ru/index.php?subaction=userinfo&user=igalexex
http://patriot-travel.ru/profile.php?u=upywepino
http://www.kletki-opt.ru/product/paw-grill/reviews/
https://dosug-murino.ru/prostitutka-yulya-23-let-8635313/#comment-362717
http://artem-energo.ru/forums.php?m=posts&q=17281&n=last#bottom
В нашем обществе, где диплом становится началом удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Важность наличия документа об образовании переоценить невозможно. Ведь диплом открывает двери перед любым человеком, желающим вступить в сообщество профессионалов или учиться в ВУЗе.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы сможете заказать диплом, что становится отличным решением для человека, который не смог закончить обучение, утратил документ или хочет исправить плохие оценки. дипломы выпускаются с особой тщательностью, вниманием к мельчайшим элементам, чтобы в итоге получился продукт, полностью соответствующий оригиналу.
Преимущество такого подхода заключается не только в том, что можно быстро получить диплом. Процесс организован комфортно и легко, с нашей поддержкой. Начав от выбора нужного образца до консультаций по заполнению персональной информации и доставки по России — все находится под полным контролем наших специалистов.
Всем, кто ищет быстрый способ получить необходимый документ, наша услуга предлагает выгодное решение. Заказать диплом – значит избежать длительного обучения и сразу переходить к достижению собственных целей: к поступлению в университет или к началу удачной карьеры.
http://krair.kr/bbs/board.php?bo_table=free&wr_id=227654
http://patriot-travel.ru/profile.php?u=idujosehe
https://codingrus.ru/forum/viewthread.php?forum_id=29&thread_id=1024&pid=2019
http://www.marheavenj.net/public/gallery/member.php?action=showprofile&user_id=27312
http://o926476g.bget.ru/users/inobagyc
В нашем обществе, где диплом – это начало удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Наличие документа об образовании сложно переоценить. Ведь именно диплом открывает двери перед всеми, кто стремится начать трудовую деятельность или продолжить обучение в университете.
Предлагаем максимально быстро получить любой необходимый документ. Вы имеете возможность заказать диплом старого или нового образца, и это является отличным решением для человека, который не смог завершить образование или утратил документ. диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим элементам, чтобы на выходе получился полностью оригинальный документ.
Преимущества данного решения состоят не только в том, что можно максимально быстро получить свой диплом. Процесс организован комфортно, с профессиональной поддержкой. Начав от выбора подходящего образца до консультации по заполнению персональных данных и доставки по стране — все находится под абсолютным контролем квалифицированных специалистов.
В результате, всем, кто ищет максимально быстрый способ получить необходимый документ, наша компания готова предложить отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и сразу переходить к личным целям, будь то поступление в ВУЗ или начало профессиональной карьеры.
https://www.koreafurniture.com/bbs/board.php?bo_table=free&wr_id=3835700
http://e-audit.gefest.me/index.php?subaction=userinfo&user=okoje
https://aben75.cafe24.com/bbs/board.php?bo_table=free&wr_id=274215
http://sukces-it.pl/blog/czy-wiesz-ze-przeslanie-e-deklaracji#c128
http://users.atw.hu/sajprobaoldalam/modules.php?name=Journal&file=display&jid=45867
Завоевание сертификата по высшему образованию является существенным шагом в жизни многих личностей, позволяя возможности к новым возможностям и перспективам.
Впрочем, ни в коем случае постоянно процесс обучения в высшем учебном заведении доступно или подходит из разных причин.
В таких случаях задание о том, где купить диплом, делается существенным.
Современные технологические разработки и интернет-рынок предоставляют разнообразные альтернативы для покупки документа, однако важно отбирать проверенных поставщиков, обеспечивающих качественное исполнение и подлинность диплома.
При условии отборе нужно замечать не только на цену, но также в репутацию компании, отзывы покупателей и получения совета.
https://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-diplom-ekonomista-v-moskve/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-diplom-ekologa/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-diplom-shturmana/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-diplom-shtukatura/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-diplom-shvei/
– купить диплом техника – подразумевает инвестировать в будущее, поэтому подбор поставщика нужно относиться тщательно.
thephotoretouch.com
하지만 곰곰이 생각해보면 무리는 아닌 것 같다.
Купить аттестат о среднем образовании – https://diplom-city24.ru/diplomy-na-prodazhu-kupit-diplom-v-zlatouste/
https://diplom-city24.ru/diplomy-na-prodazhu-kupit-diplom-v-ejske/
https://diplom-city24.ru/diplomy-na-prodazhu-kupit-diplom-v-dolgoprudnom/
https://diplom-city24.ru/diplomy-na-prodazhu-kupit-diplom-v-dimitrovgrade/
https://diplom-city24.ru/diplomy-na-prodazhu-kupit-diplom-v-gorode-naxodka/
– это ключевой инструмент твоего будущего. Воспользуйтесь нашем сервисом, чтобы быстро и без труда приобрести документ, необходимый для дальнейшего обучения или трудоустройства. Наши опытные специалисты предоставляют качество и защищенность предоставляемых услуги. Покупайте школьный аттестат у нас и обнаруживайте дополнительные возможности для личного образования и профессионального роста.
Сегодня, когда диплом – это начало отличной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Факт наличия документа об образовании переоценить невозможно. Ведь именно диплом открывает дверь перед всеми, кто собирается вступить в сообщество профессионалов или продолжить обучение в каком-либо ВУЗе.
Предлагаем максимально быстро получить любой необходимый документ. Вы можете заказать диплом, и это становится отличным решением для всех, кто не смог завершить образование, утратил документ или желает исправить плохие оценки. дипломы выпускаются аккуратно, с максимальным вниманием ко всем деталям. В результате вы получите документ, максимально соответствующий оригиналу.
Превосходство подобного подхода состоит не только в том, что вы максимально быстро получите диплом. Весь процесс организован удобно, с нашей поддержкой. Начиная от выбора подходящего образца документа до правильного заполнения личной информации и доставки по России — все под абсолютным контролем квалифицированных мастеров.
Таким образом, для тех, кто ищет быстрый и простой способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать длительного обучения и сразу переходить к своим целям, будь то поступление в ВУЗ или старт карьеры.
https://clients1.google.com.py/url?sa=t&url=https://rudiplomista.com
https://cse.google.cat/url?q=https://premialnie-diplomik.com
https://images.google.by/url?sa=t&url=https://rudiplomista.com
https://images.google.mk/url?q=https://rudiplomista.com
https://maps.google.com.au/url?q=https://premialnie-diplomik.com
Earnestabich
Приветики, дорогие мои!!
Благодарностью интернету, я нашел полезные ресурсы для диплома, что значительно ускорило и облегчило процесс его написания.
Закажите диплом у нас и получите его быстро и надежно, оплатив после получения, с доставкой в любой регион РФ.
Желаю для каждого честных отметок!
купить диплом в белово
купить диплом в благовещенске
купить диплом в ухте
куплю диплом с занесением
купить диплом в октябрьском
купить диплом колледжа
купить диплом медбрата
купить диплом в новокуйбышевске
купить диплом в воронеже
купить диплом охранника
Выше- сайт официальный представитель даркнет площадки Блэкспрут – представляющей собою онлайн платформу по продаже/покупке запрещенных веществ. Площадка BlackSprut размещена на понцы TOR, коия обеспечивает для вас царский ярус анонимности.
https://bs2tsite.club
LewisAluri
Привет всем!
Инструменты, найденные в сети для диплома, облегчили процесс написания.
Рады предложить большой выбор документов об образовании всех ВУЗов России по доступным ценам и с постоплатой.
Желаю всем отличных отметок!
купить диплом в бору
купить диплом в кузнецке
купить диплом в бийске
купить диплом в серове
купить диплом в абакане
http://www.google.com.tr/url?q=https://diplom5.com/
https://www.google.ac/url?q=https://diplom-zakaz.ru/
https://images.google.co.ug/url?q=https://diplom-zakaz.ru/
https://images.google.lt/url?q=https://nsk-diplom.com/
https://cse.google.com.co/url?sa=i&url=https://diplomoz-197.com/
Завоевание сертификата по высшему образованию представляется существенным шагом в жизни большого числа людей, открывая двери к новым перспективам и перспективам.
Однако, вовсе не всегда учеба в высшем учебном заведении доступно или подходит из разных причин.
При таких ситуациях задание где приобрести аттестат, становится актуальным.
Современные технологические достижения и интернет-рынок предлагают различные альтернативы для приобретения сертификата, однако необходимо подбирать достоверных дистрибьюторов, обеспечивающих качество и подлинность диплома.
При отборе нужно обращать внимание не только в цену, но и в имидж поставщика, мнения клиентов и возможность получения совета.
https://magazin-diplomov.ru/kupit-diplom-v-rossii-diplom-v-batajske/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-diplom-v-astane/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-diplom-v-arzamase/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-diplom-v-angarske/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-diplom-avtoslesarya/
– купить диплом учителя – значит вкладывать в свое будущее, поэтому отбор поставщика рекомендуется рассматривать ответственно.
В современном мире, где диплом – это начало успешной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения качественного образования. Наличие документа об образовании переоценить невозможно. Ведь диплом открывает двери перед людьми, стремящимися начать трудовую деятельность или учиться в высшем учебном заведении.
Мы предлагаем быстро получить любой необходимый документ. Вы сможете купить диплом старого или нового образца, и это является удачным решением для человека, который не смог завершить обучение, утратил документ или желает исправить плохие оценки. диплом изготавливается с особой тщательностью, вниманием ко всем деталям. В итоге вы получите полностью оригинальный документ.
Преимущество подобного решения состоит не только в том, что вы оперативно получите свой диплом. Весь процесс организован удобно и легко, с профессиональной поддержкой. Начав от выбора необходимого образца диплома до грамотного заполнения персональной информации и доставки по России — все находится под полным контролем качественных мастеров.
Для тех, кто ищет быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать продолжительного обучения и не теряя времени перейти к достижению собственных целей: к поступлению в университет или к началу трудовой карьеры.
http://www.ydelection.com/bbs/board.php?bo_table=free&wr_id=2103472
http://food-ua.com/wr_board/tools.php?event=profile&pname=orygybip
http://sunginew.com/bbs/board.php?bo_table=free&wr_id=222627
http://www.writemob.com/index.php?qa=user&qa_1=omuseve
http://mineralgroup.ru/index.php?subaction=userinfo&user=ovygevaza
Купить аттестат цена – https://diplom-city24.ru/diplomy-na-prodazhu-vodyanoj-znak/
https://diplom-city24.ru/diplomy-na-prodazhu-attestat-v-yaroslavle/
https://diplom-city24.ru/diplomy-na-prodazhu-attestat-v-chite/
https://diplom-city24.ru/diplomy-na-prodazhu-attestat-v-cherepovce/
https://diplom-city24.ru/diplomy-na-prodazhu-attestat-v-ufe/
– это существенный показатель вашего развития. Воспользуйтесь нашими услугами, чтобы просто и без труда приобрести аттестат, необходимый для продолжения учебы или трудоустройства. Наши опытные специалисты гарантируют качество и дискретность предоставляемых услуг. Покупайте свидетельство об образовании у нас и открывайте новые перспективы для личного образования и карьеры.
StephenNug
Доброго дня!
Диплом заставил меня работать по ночам, что негативно сказалось на моем здоровье.
У нас доступно заказать диплом без предоплаты с доставкой курьером по РФ, “под ключ”!
Желаю вам всем прекрасных отметок!
купить диплом в калининграде
купить диплом колледжа
купить диплом в нижнем тагиле
купить диплом в невинномысске
купить диплом историка
https://www.google.ca/url?q=https://kazdiplomas.com/
https://www.google.com.py/url?q=https://okdiplom.com/
http://www.google.la/url?q=https://1magistr.ru/
https://asia.google.com/url?q=https://diplom5.com/
http://posts.google.com/url?q=https://diplom-insti.ru/
В нашем мире, где диплом – это начало успешной карьеры в любой сфере, многие ищут максимально быстрый путь получения образования. Факт наличия документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед любым человеком, который хочет вступить в профессиональное сообщество или учиться в ВУЗе.
В данном контексте мы предлагаем оперативно получить этот необходимый документ. Вы сможете купить диплом, что является выгодным решением для человека, который не смог закончить обучение или утратил документ. диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим нюансам, чтобы на выходе получился 100% оригинальный документ.
Преимущество данного подхода состоит не только в том, что вы сможете быстро получить диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начиная от выбора необходимого образца до консультации по заполнению личных данных и доставки в любой регион России — все находится под абсолютным контролем квалифицированных специалистов.
Всем, кто ищет быстрый способ получения необходимого документа, наша компания предлагает выгодное решение. Заказать диплом – значит избежать длительного процесса обучения и не теряя времени переходить к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
http://redhawkclan.free.fr/index.php?file=Members&op=detail&autor=esunevyb
http://parenvarmii.ru/topic3708.html?view=previous
http://corp-eeek.ru/index.php?subaction=userinfo&user=efimo
http://artem-energo.ru/forums.php?m=posts&q=17275&n=last#bottom
http://gektorstroi.ru/forum/?PAGE_NAME=profile_view&UID=177918
tvlore.com
이마에는 땀이 흐르고 팔의 통증은 온 몸으로 퍼졌다.
В современном мире, где диплом является началом удачной карьеры в любой области, многие стараются найти максимально простой путь получения образования. Необходимость наличия документа об образовании переоценить попросту невозможно. Ведь диплом открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или учиться в университете.
Мы предлагаем быстро получить этот важный документ. Вы можете заказать диплом нового или старого образца, и это будет выгодным решением для всех, кто не смог завершить образование или утратил документ. дипломы выпускаются аккуратно, с особым вниманием к мельчайшим элементам. В итоге вы получите 100% оригинальный документ.
Плюсы такого подхода заключаются не только в том, что можно оперативно получить диплом. Процесс организовывается комфортно, с профессиональной поддержкой. От выбора нужного образца диплома до точного заполнения личных данных и доставки в любое место страны — все будет находиться под полным контролем квалифицированных специалистов.
Для тех, кто пытается найти быстрый способ получения необходимого документа, наша услуга предлагает отличное решение. Купить диплом – это значит избежать долгого обучения и сразу перейти к своим целям: к поступлению в университет или к началу успешной карьеры.
https://www.google.sh/url?sa=t&url=https://rudiplomista.com
https://www.google.pl/url?q=https://lands-diploma.com
https://images.google.com.ly/url?q=https://rudiplomista.com
http://images.google.lk/url?sa=t&url=https://rudiplomista.com
http://cse.google.com.nf/url?sa=i&url=https://premialnie-diplomik.com
Dichaelautob
Доброго дня!
Продуктивность моей работы над дипломом увеличилась благодаря онлайн-ресурсам.
Наша компания готова помочь вам приобрести диплом ВУЗа с выгодой и доставкой по всей стране.
Желаю для каждого прекрасных оценок!
купить диплом химика
купить диплом в биробиджане
купить диплом в элисте
купить диплом в севастополе
купить диплом хореографа
http://images.google.com.do/url?q=https://10000diplomov.ru/
https://images.google.com.ng/url?sa=t&url=https://diplomoz-197.com/
https://www.google.iq/url?sa=t&url=https://diplom5.com/
https://clients1.google.com/url?sa=t&url=https://diplomoz-197.com/
http://images.google.az/url?q=https://diplom-zakaz.ru/
Завоевание сертификата о высшем образовании представляется существенным шагом в судьбе многих индивидуумов, обеспечивая двери к свежим возможностям и путям.
Впрочем, вовсе не всегда процесс обучения в вузе доступно или подходит из разных причин.
При таких ситуациях вопрос о том, где купить диплом, превращается значимым.
Современные технологии и интернет-рынок предлагают разные альтернативы для покупки сертификата, но важно отбирать проверенных поставщиков, обеспечивающих качественное исполнение и достоверность диплома.
При условии отборе стоит обращать внимание не только на цену, но и на представление компании, мнения заказчиков и получения консультации.
п»їhttps://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-nastoyashhij-diplom-o-vysshem-obrazovanii-v-noyabrske/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-nastoyashhij-diplom-vuza-v-nefteyuganske/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-nastoyashhij-diplom-v-salavate/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-nastoyashhij-diplom-v-novomoskovske/
https://magazin-diplomov.ru/kupit-diplom-v-rossii-kupit-nastoyashhij-diplom/
– значит инвестировать в свое свое будущее, поэтому отбор поставщика нужно подходить серьезно.
Купить аттестат – https://diplom-city24.ru/diplomy-na-prodazhu-diplom-v-belorussii/
https://diplom-city24.ru/diplomy-na-prodazhu-diplom-v-belgorode/
https://diplom-city24.ru/diplomy-na-prodazhu-diplom-v-batajske/
https://diplom-city24.ru/diplomy-na-prodazhu-diplom-v-balashixe/
https://diplom-city24.ru/diplomy-na-prodazhu-diplom-v-balakovo/
– это ключевой фактор твоего развития. Воспользуйтесь нашем услугами, чтобы просто и без проблем приобрести аттестат, необходимый для продолжения учебы или карьерного роста. Наши квалифицированные специалисты обеспечивают отличное качество и защищенность предоставляемых сервисов. Покупайте школьный аттестат у нас и открывайте новые варианты для вашего образования и карьеры.
Hi there mates, how is all, and what you desire to say regarding this post, in my view its actually awesome in support of me.
http://maps.google.cg/url?q=https://prodiplome.com/
https://www.google.com.mt/url?sa=t&url=https://diplom45.ru/
https://toolbarqueries.google.co.ls/url?q=https://kazdiplomas.com/
https://cse.google.com.co/url?sa=i&url=https://bisness-diplom.ru/
https://www.google.hu/url?q=https://diplom-zakaz.ru/
В нашем обществе, где диплом становится началом отличной карьеры в любой области, многие пытаются найти максимально простой путь получения образования. Факт наличия официального документа об образовании переоценить попросту невозможно. Ведь диплом открывает дверь перед людьми, стремящимися начать трудовую деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы можете заказать диплом, и это будет удачным решением для человека, который не смог закончить образование, утратил документ или хочет исправить свои оценки. дипломы выпускаются аккуратно, с особым вниманием ко всем нюансам. На выходе вы сможете получить 100% оригинальный документ.
Преимущество этого решения состоит не только в том, что вы сможете быстро получить диплом. Процесс организовывается удобно, с нашей поддержкой. Начав от выбора нужного образца документа до консультаций по заполнению личных данных и доставки по стране — все будет находиться под абсолютным контролем опытных мастеров.
Всем, кто хочет найти максимально быстрый способ получения необходимого документа, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать продолжительного обучения и сразу перейти к достижению собственных целей: к поступлению в университет или к началу трудовой карьеры.
http://toolbarqueries.google.bf/url?sa=t&url=https://premialnie-diplomik.com
https://www.google.vu/url?q=https://rudiplomista.com
https://clients1.google.com.hk/url?sa=t&url=https://premialnie-diplomik.com
https://www.google.be/url?sa=t&url=https://premialnie-diplomik.com
https://toolbarqueries.google.lk/url?sa=t&url=https://lands-diploma.com
В нашем мире, где аттестат является началом удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Факт наличия официального документа трудно переоценить. Ведь именно диплом открывает дверь перед людьми, стремящимися вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Наша компания предлагает очень быстро получить этот важный документ. Вы сможете купить аттестат старого или нового образца, и это будет выгодным решением для человека, который не смог закончить обучение, потерял документ или хочет исправить плохие оценки. Каждый аттестат изготавливается аккуратно, с особым вниманием к мельчайшим элементам. В итоге вы получите 100% оригинальный документ.
Преимущества этого подхода состоят не только в том, что можно быстро получить аттестат. Весь процесс организовывается удобно, с профессиональной поддержкой. Начав от выбора необходимого образца документа до точного заполнения личной информации и доставки в любой регион страны — все находится под полным контролем квалифицированных мастеров.
В результате, всем, кто хочет найти быстрый способ получить требуемый документ, наша услуга предлагает выгодное решение. Заказать аттестат – это значит избежать длительного обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в университет или старт карьеры.
http://nadegda.listbb.ru/viewtopic.php?f=3&t=278
https://ya.0bb.ru/viewtopic.php?id=10586#p29232
В нашем мире, где аттестат – это начало успешной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения качественного образования. Наличие документа об образовании переоценить невозможно. Ведь диплом открывает двери перед людьми, желающими начать профессиональную деятельность или учиться в университете.
Предлагаем оперативно получить этот необходимый документ. Вы имеете возможность заказать аттестат, что становится удачным решением для всех, кто не смог закончить обучение или утратил документ. Аттестат изготавливается с особой аккуратностью, вниманием ко всем нюансам, чтобы на выходе получился документ, полностью соответствующий оригиналу.
Превосходство этого подхода заключается не только в том, что вы сможете быстро получить свой аттестат. Весь процесс организован просто и легко, с нашей поддержкой. От выбора требуемого образца документа до консультаций по заполнению личных данных и доставки по стране — все будет находиться под полным контролем качественных специалистов.
В результате, для всех, кто ищет быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Купить аттестат – значит избежать длительного обучения и сразу перейти к достижению собственных целей, будь то поступление в университет или старт карьеры.
https://stalinarch.ru/wiki/index.php/Доступ_к_вашей_мечте:_Купите_диплом_без_лишних_сложностей!
https://iqtorg.ru/forum/user/18090/
На сегодняшний день, когда аттестат становится началом удачной карьеры в любой сфере, многие ищут максимально быстрый путь получения образования. Наличие документа об образовании переоценить просто невозможно. Ведь диплом открывает дверь перед любым человеком, который желает вступить в сообщество профессиональных специалистов или учиться в любом институте.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы сможете приобрести аттестат, что является отличным решением для всех, кто не смог закончить обучение, утратил документ или желает исправить плохие оценки. Аттестаты производятся аккуратно, с особым вниманием ко всем деталям. На выходе вы сможете получить продукт, полностью соответствующий оригиналу.
Плюсы этого подхода заключаются не только в том, что можно быстро получить аттестат. Процесс организовывается удобно и легко, с профессиональной поддержкой. От выбора требуемого образца документа до консультации по заполнению личной информации и доставки в любой регион страны — все под полным контролем опытных мастеров.
В результате, для тех, кто пытается найти оперативный способ получить необходимый документ, наша компания предлагает отличное решение. Купить аттестат – это значит избежать долгого процесса обучения и не теряя времени переходить к своим целям, будь то поступление в ВУЗ или старт удачной карьеры.
https://www.diplomans-rossians.com/
LewisAluri
Всем хорошего дня!
Благодарностью интернету, я нашел полезные ресурсы для диплома, что значительно ускорило и облегчило процесс его написания.
Закажите диплом ВУЗа России недорого, без предоплаты и с гарантией возврата средств.
Желаю вам всем положительных отметок!
купить диплом в кемерово
купить диплом в орле
купить свидетельство о рождении
купить диплом в ухте
купить новый диплом
http://alt1.toolbarqueries.google.com.iq/url?q=https://diplom-profi.ru/
https://clients1.google.com.bh/url?sa=t&url=https://diploms-vuza.com/
https://maps.google.com.mm/url?sa=t&url=https://kdiplom.ru/
https://images.google.ie/url?sa=t&url=https://1magistr.ru/
https://images.google.co.bw/url?sa=t&url=https://prodiplome.com/
Где купить аттестат – https://diplom-city24.ru/diplomy-na-prodazhu-attestat-v-chite/
https://diplom-city24.ru/diplomy-na-prodazhu-attestat-v-cherepovce/
https://diplom-city24.ru/diplomy-na-prodazhu-attestat-v-ufe/
https://diplom-city24.ru/diplomy-na-prodazhu-attestat-v-ulyanovske/
https://diplom-city24.ru/diplomy-na-prodazhu-attestat-v-ulan-ude/
– это ключевой инструмент вашего будущего. Воспользуйтесь нашими предложением, чтобы просто и без проблем приобрести аттестат, необходимый для дальнейшего обучения или карьерного роста. Наши квалифицированные специалисты предоставляют качество и защищенность предоставляемых услуги. Покупайте диплом у нас и раскрывайте новые перспективы для своего образования и профессионального роста.
На сегодняшний день, когда диплом – это начало удачной карьеры в любой области, многие ищут максимально быстрый путь получения качественного образования. Наличие официального документа об образовании сложно переоценить. Ведь именно диплом открывает двери перед людьми, желающими начать профессиональную деятельность или учиться в каком-либо институте.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы имеете возможность приобрести диплом старого или нового образца, и это становится отличным решением для всех, кто не смог закончить обучение, потерял документ или хочет исправить плохие оценки. Любой диплом изготавливается с особой тщательностью, вниманием ко всем деталям. В результате вы получите продукт, максимально соответствующий оригиналу.
Преимущества данного решения состоят не только в том, что вы сможете быстро получить свой диплом. Процесс организовывается просто и легко, с нашей поддержкой. Начав от выбора необходимого образца диплома до консультаций по заполнению персональной информации и доставки по России — все под полным контролем качественных специалистов.
Таким образом, для тех, кто хочет найти быстрый способ получить требуемый документ, наша компания готова предложить отличное решение. Заказать диплом – значит избежать долгого процесса обучения и сразу переходить к важным целям, будь то поступление в ВУЗ или начало карьеры.
http://koreasamsong.com/bbs/board.php?bo_table=free&wr_id=2129108
https://radioizba.ru/communication/forum/?PAGE_NAME=profile_view&UID=17955
http://www.munhwahouse.or.kr/bbs/board.php?bo_table=free&wr_id=98815
http://suprememasterchinghai.net/bbs/board.php?bo_table=free&wr_id=1738722
http://www.handgemaaktplaats.nl/user/profile/76961
Доброго всем дня!
Предлагаем всем желающим приобрести диплом университетов России по низкой цене с доставкой в руки под ключ
https://www.google.com.pe/url?q=http://russdiplomiki.com
https://toolbarqueries.google.rs/url?sa=i&url=https://diplomansy.com
http://images.google.st/url?q=http://diplomsagroups.com
https://clients1.google.com.bd/url?q=https://anny-diploms.com
https://maps.google.com.hk/url?q=http://rdiplomik24.com
Купите диплом института или колледжа без предоплаты с гарантированной доставкой в любой уголок России.
В современном мире, где диплом – это начало удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Необходимость наличия официального документа сложно переоценить. Ведь именно диплом открывает дверь перед людьми, стремящимися начать трудовую деятельность или продолжить обучение в ВУЗе.
Предлагаем максимально быстро получить любой необходимый документ. Вы можете заказать диплом старого или нового образца, и это является отличным решением для человека, который не смог закончить обучение или потерял документ. дипломы выпускаются аккуратно, с максимальным вниманием ко всем элементам. На выходе вы получите 100% оригинальный документ.
Превосходство этого решения состоит не только в том, что вы быстро получите свой диплом. Процесс организовывается комфортно, с нашей поддержкой. От выбора необходимого образца документа до грамотного заполнения персональных данных и доставки по России — все под абсолютным контролем наших специалистов.
Для тех, кто ищет быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Купить диплом – это значит избежать долгого обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в ВУЗ или старт карьеры.
https://cse.google.com.af/url?sa=t&url=https://lands-diploma.com
https://maps.google.la/url?q=https://rudiplomista.com
https://www.google.pt/url?q=https://lands-diploma.com
https://maps.google.com.mm/url?sa=t&url=https://rudiplomista.com
https://cse.google.co.ao/url?q=https://premialnie-diplomik.com
Dichaelautob
Приветики, дорогие мои!!
Несмотря на все препятствия в работе над дипломом, я продолжаю находить способы его оптимизации благодаря интернету.
Мы предлагаем купить диплом Гознак со скидкой, гарантией и доставкой в любой город РФ.
Желаю вам всем прекрасных отметок!
купить диплом железнодорожника
купить диплом в нефтекамске
купить диплом в махачкале
купить диплом эколога
купить диплом в ижевске
https://images.google.com.np/url?sa=t&url=https://diplom-profi.ru/
https://www.google.ba/url?sa=t&url=https://diplom-insti.ru/
https://www.google.vu/url?q=https://kazdiplomas.com/
http://maps.google.ht/url?rct=j&sa=t&url=https://diplom-profi.ru/
https://maps.google.lk/url?sa=t&url=https://diplom45.ru/
В нашем обществе, где диплом – это начало удачной карьеры в любой отрасли, многие ищут максимально быстрый путь получения качественного образования. Необходимость наличия документа об образовании переоценить попросту невозможно. Ведь именно диплом открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в любом институте.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете приобрести диплом нового или старого образца, что является удачным решением для всех, кто не смог завершить образование, утратил документ или хочет исправить плохие оценки. диплом изготавливается с особой тщательностью, вниманием ко всем нюансам. На выходе вы получите полностью оригинальный документ.
Преимущество данного подхода заключается не только в том, что вы максимально быстро получите диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. От выбора нужного образца до консультации по заполнению личной информации и доставки по России — все под абсолютным контролем наших специалистов.
Всем, кто пытается найти быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Купить диплом – это значит избежать продолжительного обучения и не теряя времени переходить к достижению своих целей, будь то поступление в университет или старт карьеры.
http://images.google.co.nz/url?sa=t&url=https://rudiplomista.com
https://www.google.co.ls/url?q=https://premialnie-diplomik.com
https://images.google.com.ng/url?sa=t&url=https://rudiplomista.com
https://www.google.co.mz/url?q=https://premialnie-diplomik.com
https://images.google.com.sb/url?q=https://rudiplomista.com
Привет, дорогой читатель!
Как можно приобрести диплом Вуза в России без предоплаты на сайте? Мы готовы доставить его в любую точку страны.
http://cse.google.co.zw/url?sa=t&url=http://aurus-diploms.com
https://images.google.sk/url?sa=t&url=http://aurus-diploms.com
https://cse.google.com.tr/url?q=http://diploms-service.com
https://www.google.to/url?sa=t&url=https://anny-diploms.com
http://images.google.gl/url?q=http://diplomy-originaly.com
Купите диплом института или техникума с доставкой по РФ по выгодной цене без предоплаты!
Рады приветствовать ваш ресурс, друзья!
К вам обращается агентство по СЕО продвижению XRumer Inc.
Ваш портал, как мы заметили, еще только набирает обороты. Для того, чтобы ускорить его рост, готовы предложить услуги по SEO-оптимизации. Продвижение в поисковых системах – направление нашей компании. В ассортименте присутствуют надежные и рабочие SEO-инструменты для специалистов. Наши эксперты обладают серьезным опытом и портфолио выполненных проектов, которыми мы готовы поделиться по запросу.
Наша компания предлагает скидку 10% до конца месяца.
Что мы предлагаем:
– Супер трастовые ссылки (нужно всем сайтам) – цена от 1,5 до 5000 руб
– Размещаем 2500 безанкорных ссылок (полезно всем сайтам) – 3900 рублей
– Прогон на 110 тыс. сайтов в зоне RU (очень полезно для сайтов) – 2.900 р
– Разместим 150 постов Вконтакте о вашем сайте (поможет получить рекламу) – 3.900 р
– Статьи о вашем сайте на 300 топовых онлайн-форумах (очень мощная раскрутка вашего портала) – 29.000 руб
– Супер Постинг – это прогон по 3 000 000 ресурсов (сверхмощный пакет для вашего сайта) – 39 тысяч руб
– Рассылка сообщений по сайтам при помощи обратной связи – договорная стоимость, зависит от объемов.
Обращайтесь со всеми вопросами, мы постараемся помочь. принимаем usdt
Телегрм: @exrumer
Skype: Loves.ltd
www: https://XRumer.cc/
nikontinoll.com
Hongzhi 황제는 눈살을 찌푸 렸습니다. “누군가 그 선철이 배달되는 것을 보았습니까?”
Купить аттестат за 9 класс – https://prodiplome.com/prodazha-diplomov-kupit-udostoverenie-o-povyshenii-kvalifikacii/
https://prodiplome.com/prodazha-diplomov-kupit-spravku-vyzov-na-sessiyu-v-moskve/
– это важный показатель вашего будущего. Воспользуйтесь нашим предложением, чтобы быстро и без лишних усилий приобрести аттестат, необходимый для продолжения учебы или трудоустройства. Наши опытные специалисты обеспечивают отличное качество и дискретность предоставляемых услуг. Покупайте диплом у нас и открывайте свежие возможности для личного образования и карьеры.
Привет, дорогой читатель!
Приобретите документы об образовании всех ВУЗов России с гарантированной подлинностью и доставкой по РФ без предварительной оплаты – просто, надежно, выгодно!
http://www.google.com.tr/url?q=https://anny-diploms.com
https://cse.google.ws/url?sa=t&url=http://diploms-service.com
http://www.google.rs/url?q=https://diploms-asx.com
http://www.google.la/url?q=http://diplomsagroups.com
https://cse.google.ne/url?sa=i&url=http://diplomsagroups.com
Закажите диплом ВУЗа с гарантированной подлинностью и доставкой по всей России без предоплаты – просто, удобно, безопасно!
IsmaelBog
Всем хорошего дня!
Завершение диплома кажется возможным с поддержкой интернета.
У нас в компании вы можете купить диплом Гознак со скидкой, гарантией и доставкой в любой город РФ.
Желаю для каждого отличных оценок!
купить диплом в владивостоке
купить диплом в ишиме
купить диплом в камышине
купить аттестат за 9 класс
купить диплом в кызыле
https://clients1.google.sn/url?sa=t&url=https://nsk-diplom.com/
https://maps.google.fr/url?q=https://kdiplom.ru/
http://maps.google.im/url?q=https://diplom5.com/
https://www.google.com.lb/url?q=https://diploms-vuza.com/
https://cse.google.nr/url?sa=t&url=https://diplom5.com/
Привет, дорогой читатель!
Недорого предлагаем заказать диплом без предоплаты с доставкой курьером по РФ, под ключ!
https://clients1.google.bj/url?sa=t&url=http://rudiplomisty24.com
http://images.google.az/url?q=http://rdiplomik24.com
https://cse.google.com.lb/url?sa=t&url=https://premialnie-diplomansy.com
https://www.google.ge/url?q=http://diplomyland.com
https://clients1.google.nl/url?sa=t&url=https://gosznac-diploma.com
Получите документ об образовании по выгодной цене с доставкой по РФ и оплатой после получения.
donmhomes.com
この記事は本当に素晴らしい!大変勉強になりました。
Купить аттестат за 11 – https://prodiplome.com/prodazha-diplomov-kupit-korochku-diploma/
https://prodiplome.com/prodazha-diplomov-kupit-zakonno-diplom/
– это существенный показатель твоего развития. Воспользуйтесь нашем предложением, чтобы легко и без труда приобрести аттестат, необходимый для дальнейшего обучения или трудоустройства. Наши опытные специалисты гарантируют качество и конфиденциальность предоставляемых услуги. Покупайте школьный аттестат у нас и обнаруживайте дополнительные варианты для вашего образования и трудоустройства.
Привет всем!
Получите документы об образовании всех Вузов России с гарантированной доставкой по РФ без предоплаты.
http://images.google.bj/url?q=http://originality-diplomy.com
http://alt1.toolbarqueries.google.co.ve/url?q=https://premialnie-diplomansy.com
https://maps.google.com.br/url?q=http://russdiplomiki.com
https://cse.google.com.mt/url?q=http://diplomsagroups.com
http://images.google.co.nz/url?sa=t&url=http://rdiplomik24.com
Предлагаем заказать диплом у нас с доставкой курьером по всей России без предоплаты.
Наша компания предлагает высококачественные услуги Аренда гусеничного экскаватора в Алматы Мы обеспечиваем надежное и профессиональное оборудование для выполнения различных земляных работ на строительных площадках и других объектах. Наш опытный персонал и гибкие условия аренды делают нас надежным партнером для вашего проекта.
Dichaelautob
Приветики, дорогие мои!!
Несмотря на все сложности и угрозу для здоровья, я продолжаю работать над дипломом, опираясь на найденные ресурсы.
Приобретите диплом университета с гарантированной доставкой в любой город России с оплатой после получения.
Желаю каждому положительных отметок!
купить диплом в рыбинске
купить диплом во всеволожске
купить диплом в благовещенске
купить диплом сантехника
купить аттестат за 11 класс
https://maps.google.mw/url?q=https://diplomoz-197.com/
http://images.google.az/url?q=https://diplom-zakaz.ru/
https://clients1.google.bj/url?sa=t&url=https://diplom5.com/
https://www.google.hu/url?q=https://kdiplom.ru/
http://maps.google.co.cr/url?sa=t&url=https://nsk-diplom.com/
27-го июля был проведён первый тест эффективности XRumer 23 StrongAI (по сравнению с текущей версией XRumer 19.0.18), который показал двадцатикратный прирост. Столь впечатляющие результаты достигнуты благодаря многократному улучшению пробиваемости уже известных платформ, добавлению поддержки новых платформ, обучению сотням тысяч типов новых тексткапч, внедрению нового GPT-модуля, и множеству других улучшений. По ссылке ниже Вы сможете изучить и скачать детали теста (исходная база, кусок отчёта, статистику и другие подробности)
Спасибо за рефку:
http://www.botmasterru.com/product109120/
Привет всем!
Приобретите документы об образовании ВУЗов России по выгодным ценам с постоплатой и помощью 24/7 – безопасно и выгодно!
Компания предлагает купить диплом Вуза России недорого, без предоплаты и с гарантией возврата средств.
https://maps.google.at/url?q=http://rudiplomisty24.com
https://www.google.be/url?sa=t&url=https://gosznac-diplomy.com
https://maps.google.co.kr/url?sa=t&url=https://diploms-originalniy.com
https://www.google.co.ma/url?sa=t&url=http://volga-diplomy.com
https://www.google.nr/url?q=https://lands-diploms.com
Earnestabich
Всем хорошего дня!
Дипломное задание оказалось чрезвычайно сложным, но благодаря найденным в интернете ресурсам я не сдаюсь.
Заказывайте диплом у нас и получите его быстро и безопасно, оплата по факту доставки, отправка в любой регион России.
Желаю всем отличных оценок!
купить диплом слесаря
купить диплом в михайловске
купить диплом историка
купить диплом в армавире
купить диплом в вольске
https://www.google.am/url?sa=t&url=https://kazdiplomas.com/
https://www.google.mv/url?sa=t&url=https://peoplediplom.ru/
https://ditu.google.com/url?q=https://kupit-diplomyz24.com/
https://images.google.ca/url?sa=t&url=https://diplom-profi.ru/
https://www.google.com.fj/url?q=https://diplom-zakaz.ru/
Привет всем!
Приобретите российский диплом с гарантией качества и доставкой в любую точку страны без предварительной оплаты!
Как можно приобрести диплом Вуза в России без предоплаты на сайте? Мы готовы доставить его в любую точку страны.
https://clients1.google.com.py/url?sa=t&url=https://premialnie-diplomansy.com
http://cse.google.gr/url?sa=i&url=http://diploman-spb24.com
http://images.google.td/url?q=https://russdiplomik.com
https://www.google.al/url?q=https://gosznac-diplomy.com
https://www.google.com.cu/url?q=https://originality-diploman.com
Привет, дорогой читатель!
Мы рады предложить вам приобрести диплом университета России по выгодной цене с доставкой в руки.
Приобретите диплом ВУЗа с гарантированной доставкой по России и без предварительной оплаты – надежно и выгодно!
http://posts.google.com/url?q=https://rudiplomisty.com
https://images.google.cf/url?q=https://diploms-originalniy.com
http://images.google.ps/url?q=https://gosznac-diplomy.com
https://cse.google.com.lb/url?sa=t&url=https://russdiplomik.com
https://clients1.google.com.my/url?sa=t&url=https://originality-diplomiki.com
thephotoretouch.com
그래서 지금… 그의 마음에는 온 몸에 흐르는 따뜻한 흐름이 있습니다.
На сегодняшний день, когда диплом является началом успешной карьеры в любом направлении, многие пытаются найти максимально простой путь получения качественного образования. Наличие официального документа об образовании переоценить просто невозможно. Ведь диплом открывает дверь перед каждым человеком, который хочет вступить в сообщество квалифицированных специалистов или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает быстро получить этот необходимый документ. Вы можете заказать диплом, и это будет отличным решением для всех, кто не смог завершить образование, утратил документ или хочет исправить свои оценки. Все дипломы производятся с особой аккуратностью, вниманием ко всем деталям. В итоге вы получите документ, 100% соответствующий оригиналу.
Превосходство такого подхода состоит не только в том, что можно максимально быстро получить диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. Начиная от выбора подходящего образца диплома до консультаций по заполнению персональной информации и доставки по России — все под полным контролем квалифицированных мастеров.
Всем, кто пытается найти быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать длительного процесса обучения и сразу переходить к достижению собственных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
https://maps.google.no/url?rct=t&sa=t&url=https://lands-diploma.com
https://clients1.google.com.bd/url?q=https://lands-diploma.com
https://images.google.com.mm/url?q=https://rudiplomista.com
http://toolbarqueries.google.com.af/url?q=https://rudiplomista.com
https://www.l.google.com/url?q=https://rudiplomista.com
geinoutime.com
배는 어디에서나 가대에 들어가기 시작했습니다.
Hello! Do you use Twitter? I’d like to follow you if that would be okay. I’m undoubtedly enjoying your blog and look forward to new posts.
https://postheaven.net/samirigica/kupiti-original-ne-sklo-far-farfarlight-dlia-stil-nogo-ta-bezpechnogo-avtomobilia
Приветики!
Наши услуги помогут вам приобрести диплом ВУЗа с доставкой по всей России без предварительной оплаты – быстро и надежно!
medhelp-dent.ru
#YAHOO#
Получение образования представляет собой важным моментом в жизни каждого индивидуума, определяет его перспективы и карьерные возможности.
Аттестат даёт доступ двери к перспективным горизонтам и возможностям, обеспечивая возможность к высококачественному образованию и высокооплачиваемым профессиям.
В нынешнем мире, где в борьба на рынке труда всё растёт, имение аттестата делает жизненно важным условием для выдающейся профессиональной деятельности.
Диплом утверждает ваши знания, умения и навыки, умения и компетенции перед работодателями и общественностью в целом.
diplomanrus.comгде купить аттестат – возможность для людей, которые стремится к успеху без лишних препятствий. Это возможность закончить обучение по высшему образованию, открывающий двери к новым возможностям и престижным профессиям. Наш сервис обеспечивает качество и конфиденциальность, помогая вам достичь ваших целей с минимальными усилиями.
В дополнение, аттестат дарит уверенность и увеличивает оценку себя, что помогает персональному развитию и развитию. Получение диплома также является инвестицией в будущий путь, обеспечивая стабильность и приличный уровень проживания.
Именно поэтому уделять надлежащее внимание образованию и бороться за его получению, чтобы обрести успех и удовлетворение от своей труда.
Hi everybody, here every person is sharing such familiarity, thus it’s good to read this weblog, and I used to go to see this web site daily.
https://blogfreely.net/arwynerceb/h1-b-iak-pokrashchiti-osvitlennia-dorogi-z-dopomogoiu-iakisnogo-skla-far-b-h1
Hello friends, its fantastic paragraph about cultureand entirely explained, keep it up all the time.
https://squareblogs.net/lipinnhurz/h1-b-iak-pokrashchiti-vidimist-na-dorozi-za-dopomogoiu-skla-far
Здравствуйте!
Закажите диплом ВУЗа недорого с доставкой курьером по России без предоплаты – это быстро и надежно!
https://diplomany-asx.ru/
This is very interesting, You are an overly skilled blogger. I’ve joined your rss feed and look forward to in search of more of your great post. Also, I have shared your site in my social networks
http://www.diploman-russiyans.com
k8 カジノ 招待コード
このブログの記事はいつも心に強く響きます。素晴らしいです。
thephotoretouch.com
사실, 배를 빌리는 것은 잘못된 것이 아닙니다. 폐하에게보고하십시오.
Завершение учебы образования считается основным моментом во жизни всякого индивидуума, который определяет его перспективы и карьерные перспективы.
Аттестат даёт доступ двери к свежим горизонтам и перспективам, обеспечивая возможность к высококачественному образованию и престижным профессиям.
В сегодняшнем обществе, где в борьба на рынке труда постоянно растёт, наличие аттестата делает обязательным требованием для успешной карьеры.
Диплом утверждает ваши знания, навыки и умения перед работодателями и общественностью в общем.
https://www.diploman-russiya.comкупить аттестат – возможность для людей, которые стремится к успеху без дополнительных трудностей. Это возможность закончить обучение о высшему образованию, предоставляющий новые перспективы и престижным профессиям. Наш портал гарантирует качество и конфиденциальность, помогая вам достичь ваших целей быстро и эффективно.
Помимо этого, аттестат дарит веру в свои силы и повышает оценку себя, что способствует личностному и саморазвитию. Получение образования также вложением в свое будущее, предоставляя стабильность и приличный уровень жизни.
Именно поэтому обращать должное внимание образованию и бороться за его получению, чтобы добиться успех и счастье от своей труда.
Купить диплом с переводом на английский
купить диплом техникума http://diplom-msk.ru/ .
Как купить диплом по специальности
купить аттестат [url=https://www.diplom-msk.ru/]https://www.diplom-msk.ru/[/url] .
Купить диплом быстро и качественно
купить диплом Гознак http://www.diplom-msk.ru .
Добрый день всем!
Купите диплом техникума с доставкой по РФ по выгодной цене без предоплаты – просто и надежно!
Приобретите российский диплом с гарантией качества и доставкой в любую точку страны без предварительной оплаты – безопасно и выгодно!
https://maps.google.gy/url?sa=j&url=https://originality-diplomiki.com
https://images.google.nu/url?q=https://premialnie-diplomany.com
https://cse.google.co.il/url?sa=t&url=http://diploman-spb24.com
https://clients1.google.tl/url?q=http://kazdiplom.com
https://maps.google.com.pr/url?q=http://rudiplomisty24.com
Приветики!
Заказывайте диплом любого Вуза России у нас на сайте конфиденциально и без предоплаты.
Купите диплом института или колледжа без предоплаты с гарантированной доставкой в любой уголок России.
https://maps.google.ng/url?q=https://russdiplomik.com
http://images.google.com.bo/url?q=https://premialnie-diplomansy.com
http://maps.google.com.om/url?q=http://rudiplomisty24.com
https://image.google.bs/url?q=http://diploman-spb24.com
https://maps.google.com.lb/url?q=http://rudiplomisty24.com
В нашем обществе, где аттестат становится началом отличной карьеры в любой области, многие ищут максимально быстрый путь получения образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед людьми, стремящимися начать трудовую деятельность или продолжить обучение в университете.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы сможете купить аттестат нового или старого образца, что будет выгодным решением для всех, кто не смог закончить обучение или потерял документ. Аттестаты производятся с особой аккуратностью, вниманием ко всем нюансам, чтобы в результате получился 100% оригинальный документ.
Преимущества такого подхода состоят не только в том, что можно оперативно получить свой аттестат. Весь процесс организован удобно и легко, с нашей поддержкой. Начав от выбора требуемого образца до консультации по заполнению личных данных и доставки в любое место страны — все под абсолютным контролем опытных специалистов.
Таким образом, для всех, кто пытается найти быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Приобрести аттестат – это значит избежать продолжительного обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в ВУЗ или старт профессиональной карьеры.
https://diplomans-rossians.com/
Получение образования является важным этапом во карьере каждого индивидуума, определяющим его перспективы и карьерные перспективы – http://www.diplomvam.ru. Диплом открывает путь к перспективным перспективам и возможностям, обеспечивая возможность к качественному получению знаний и высокопрестижным профессиям. В современном мире, где конкуренция на рынке труда всё увеличивается, наличие диплома делает необходимым требованием для успешной профессиональной деятельности. Диплом подтверждает ваши знания, умения и навыки, компетенции и умения перед работодателями и социумом в общем. В дополнение, диплом дарует веру в свои силы и повышает оценку себя, что помогает личностному росту и саморазвитию. Завершение учебы диплома также является вложением в будущий путь, обеспечивая стабильность и достойный стандарт проживания. Поэтому важно уделять надлежащее внимание образованию и стремиться к его достижению, чтобы получить успеха и удовлетворение от собственной профессиональной деятельности.
Аттестат не лишь символизирует личное образовательный уровень, но и отражает вашу самодисциплину, усердие и упорство в достижении задач. Диплом является результатом труда и труда, вложенных в обучение и самосовершенствование. Получение диплома раскрывает перед вами новые перспективы перспектив, позволяя избирать среди множества карьерных путей и профессиональных направлений. Кроме того предоставляет вам основу знаний и навыков, необходимых для выдающейся деятельности в современном мире, насыщенном вызовами и изменениями. Кроме того, сертификат является доказательством вашей квалификации и экспертности, что в свою очередь улучшает вашу привлекательность на рынке труда и открывает перед вами возможности к наилучшим шансам для профессионального роста. Итак, получение аттестата не только обогащает ваше личное развитие, но и раскрывает вами новые и перспективы для достижения и амбиций.
Привет всем!
Закажите диплом ВУЗа с гарантированной подлинностью и доставкой по России без предварительной оплаты – просто, надежно, выгодно!
Как заказать и купить диплом Вуза недорого без предоплаты на сайте? Доставка в любую точку России
http://cse.google.tn/url?q=https://rudiplomisty.com
https://www.google.tn/url?q=https://russiany-diplomans.com
http://maps.google.com.na/url?q=http://volga-diplomy.com
http://images.google.td/url?q=http://volga-diplomy.com
https://sandbox.google.com/url?q=https://russdiplomag.com
Attractive component to content. I simply stumbled upon your web site and in accession capital to say that I get in fact loved account your blog posts. Any way I’ll be subscribing in your feeds or even I fulfillment you get admission to constantly rapidly.
https://www.diplomanc-russia24.com/
В современном мире, где диплом – это начало удачной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Наличие официального документа об образовании переоценить просто невозможно. Ведь именно диплом открывает дверь перед любым человеком, который собирается вступить в сообщество профессионалов или продолжить обучение в ВУЗе.
Мы предлагаем оперативно получить этот необходимый документ. Вы сможете заказать диплом, что становится выгодным решением для всех, кто не смог закончить обучение или утратил документ. Каждый диплом изготавливается аккуратно, с особым вниманием к мельчайшим нюансам. В результате вы получите продукт, 100% соответствующий оригиналу.
Преимущества данного решения заключаются не только в том, что вы оперативно получите свой диплом. Процесс организовывается удобно и легко, с нашей поддержкой. Начав от выбора нужного образца до консультаций по заполнению персональных данных и доставки в любое место России — все находится под абсолютным контролем наших специалистов.
Для тех, кто ищет быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Купить диплом – это значит избежать долгого обучения и сразу переходить к достижению личных целей, будь то поступление в ВУЗ или старт карьеры.
vuzdiploma
В нашем мире, где диплом – это начало успешной карьеры в любой отрасли, многие ищут максимально простой путь получения образования. Наличие официального документа переоценить невозможно. Ведь именно он открывает дверь перед людьми, желающими вступить в сообщество профессиональных специалистов или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы можете купить диплом, что будет отличным решением для всех, кто не смог завершить образование, утратил документ или хочет исправить свои оценки. Каждый диплом изготавливается аккуратно, с особым вниманием к мельчайшим элементам, чтобы в итоге получился полностью оригинальный документ.
Превосходство такого подхода заключается не только в том, что вы сможете максимально быстро получить диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора требуемого образца до консультаций по заполнению личных данных и доставки в любой регион России — все находится под полным контролем квалифицированных специалистов.
Таким образом, для тех, кто хочет найти оперативный способ получения требуемого документа, наша услуга предлагает выгодное решение. Купить диплом – значит избежать долгого обучения и сразу перейти к личным целям, будь то поступление в ВУЗ или начало удачной карьеры.
http://recipes-vkusno.ru/profile.php?u=igybelu
http://www.85sucai.com/member/index.php?uid=epuwi
http://www.inline.spb.ru/in/index.php?option=com_fireboard&Itemid=15&func=view&catid=7&id=4878#4878
http://forum.combat-arnis.ru/profile.php?mode=viewprofile&u=318582
http://foto.rzia.ru/profile.php?uid=66096
В современном мире, где диплом является началом успешной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Важность наличия документа об образовании переоценить просто невозможно. Ведь именно он открывает дверь перед каждым человеком, который собирается вступить в сообщество квалифицированных специалистов или продолжить обучение в ВУЗе.
Мы предлагаем максимально быстро получить этот важный документ. Вы имеете возможность купить диплом, и это становится отличным решением для человека, который не смог завершить образование или утратил документ. Все дипломы выпускаются аккуратно, с максимальным вниманием к мельчайшим деталям, чтобы на выходе получился 100% оригинальный документ.
Преимущества такого подхода заключаются не только в том, что можно быстро получить диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора подходящего образца диплома до правильного заполнения персональных данных и доставки по России — все под полным контролем наших специалистов.
В итоге, всем, кто ищет быстрый и простой способ получения необходимого документа, наша компания готова предложить отличное решение. Купить диплом – это значит избежать длительного обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в университет или начало профессиональной карьеры.
http://freeschool.ru/index.php?option=com_easybookreloaded
http://nis.com.tn/index.php?option=com_kunena&view=topic&catid=3&id=6593&Itemid=151
http://193.16.101.6/index.php?title=russianydiplomans
http://www.writemob.com/index.php?qa=user&qa_1=abicyt
http://test.sozapag.ru/forum/user/234374/
tvlore.com
Hongzhi 황제는 등 뒤로 손을 댔습니다. “그렇다면 법령은 진짜입니다. Zhang Qing의 가족입니다.”
В нашем мире, где диплом – это начало отличной карьеры в любой отрасли, многие ищут максимально быстрый путь получения образования. Важность наличия официального документа трудно переоценить. Ведь именно диплом открывает дверь перед людьми, желающими начать трудовую деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает очень быстро получить этот необходимый документ. Вы можете приобрести диплом, что становится отличным решением для человека, который не смог закончить обучение или утратил документ. дипломы выпускаются с особой тщательностью, вниманием ко всем нюансам. В результате вы получите продукт, полностью соответствующий оригиналу.
Превосходство подобного подхода заключается не только в том, что можно оперативно получить свой диплом. Процесс организован комфортно, с профессиональной поддержкой. Начав от выбора подходящего образца до точного заполнения личных данных и доставки по стране — все находится под полным контролем качественных специалистов.
Всем, кто хочет найти быстрый и простой способ получения необходимого документа, наша компания предлагает выгодное решение. Купить диплом – значит избежать длительного обучения и не теряя времени переходить к своим целям, будь то поступление в ВУЗ или начало карьеры.
http://diplom-net.ru/
В нашем мире, где диплом является началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Факт наличия официального документа трудно переоценить. Ведь диплом открывает дверь перед людьми, стремящимися начать трудовую деятельность или учиться в ВУЗе.
Мы предлагаем максимально быстро получить этот необходимый документ. Вы сможете купить диплом, что становится удачным решением для всех, кто не смог завершить образование, утратил документ или хочет исправить свои оценки. Все дипломы выпускаются аккуратно, с максимальным вниманием ко всем нюансам, чтобы в итоге получился 100% оригинальный документ.
Превосходство этого подхода заключается не только в том, что можно максимально быстро получить свой диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. От выбора необходимого образца до консультации по заполнению персональных данных и доставки в любое место страны — все находится под абсолютным контролем квалифицированных специалистов.
В итоге, для всех, кто ищет быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать долгого процесса обучения и не теряя времени перейти к своим целям: к поступлению в университет или к началу успешной карьеры.
http://diplom-net.ru/
Hey there, I think your blog might be having browser compatibility issues. When I look at your blog site in Ie, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, amazing blog!
https://animemedia.org/
В нашем мире, где диплом – это начало отличной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Факт наличия официального документа об образовании переоценить просто невозможно. Ведь именно он открывает дверь перед людьми, желающими вступить в сообщество профессионалов или учиться в высшем учебном заведении.
Наша компания предлагает быстро получить этот важный документ. Вы имеете возможность купить диплом старого или нового образца, и это является удачным решением для человека, который не смог завершить образование, утратил документ или желает исправить свои оценки. Каждый диплом изготавливается с особой аккуратностью, вниманием к мельчайшим элементам. В результате вы сможете получить продукт, максимально соответствующий оригиналу.
Преимущества этого подхода состоят не только в том, что можно максимально быстро получить свой диплом. Весь процесс организовывается комфортно и легко, с профессиональной поддержкой. От выбора нужного образца до точного заполнения личных данных и доставки по стране — все под абсолютным контролем качественных специалистов.
В результате, для тех, кто ищет быстрый и простой способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать длительного процесса обучения и не теряя времени переходить к своим целям, будь то поступление в университет или старт карьеры.
http://hedron-arch.com/component/k2/item/24-digital-learning/
http://creatoys.ru/product/pokernyj-nabor-royal-flush-200-/reviews/
http://empyrethegame.com/forum/memberlist.php?mode=viewprofile&u=295838&sid=bda2004632814af2d91025d230bda433
http://www.fantasyroleplay.co/wiki/index.php?title=rudiplomisty
http://rtpsouth.com/UserProfile/tabid/69/userId/104605/Default.aspx
Завершение учебы диплома считается важным моментом в пути всякого индивидуума, определяющим его перспективы и профессиональные возможности – http://www.diplomvam.ru. Диплом открывает двери к свежим горизонтам и перспективам, обеспечивая возможность к высококачественному получению знаний и престижным профессиям. В сегодняшнем обществе, где в борьба на рынке труда постоянно увеличивается, имение диплома становится жизненно важным требованием для успешной карьеры. Он подтверждает ваши знания, умения и умения перед профессиональным сообществом и общественностью в общем. Помимо этого, аттестат дарит веру в свои силы и увеличивает самооценку, что содействует личностному росту и саморазвитию. Завершение учебы образования также является инвестицией в будущий путь, предоставляя устойчивость и благополучный уровень жизни. Поэтому уделять надлежащее внимание и время образованию и бороться за его достижению, чтобы добиться успех и счастье от собственной профессиональной деятельности.
Диплом не только символизирует ваше образование, но и отражает вашу самодисциплину, трудолюбие и настойчивость в добивании целей. Он представляет собой результатом усилий и труда, вкладываемых в обучение и самосовершенствование. Завершение учебы образования раскрывает перед вами новые горизонты возможностей, даруя возможность избирать из множества направлений и профессиональных направлений. Это также предоставляет вам основу знаний и навыков и умений, необходимых для выдающейся деятельности в современном мире, насыщенном трудностями и переменами. Более того, сертификат является доказательством вашей компетентности и квалификации, что повышает вашу привлекательность на трудовом рынке и открывает вами двери к лучшим шансам для профессионального роста. Таким образом, получение образования диплома не только пополняет ваше личное и профессиональное самосовершенствование, а также открывает перед вами новые и возможности для достижения и амбиций.
geinoutime.com
Li Dongyang의 말을 듣고 Hongzhi 황제는 고개를 끄덕 였는데 사실 이것도 그의 생각이었습니다.
В нашем мире, где диплом становится началом отличной карьеры в любой области, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь именно он открывает дверь перед каждым человеком, который стремится начать трудовую деятельность или продолжить обучение в высшем учебном заведении.
Предлагаем оперативно получить этот важный документ. Вы можете купить диплом, и это является выгодным решением для всех, кто не смог завершить обучение, утратил документ или хочет исправить плохие оценки. Все дипломы производятся с особой тщательностью, вниманием ко всем элементам. В результате вы получите продукт, полностью соответствующий оригиналу.
Плюсы такого подхода состоят не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается просто и легко, с нашей поддержкой. Начав от выбора нужного образца до консультации по заполнению персональной информации и доставки в любой регион страны — все находится под абсолютным контролем наших мастеров.
Для всех, кто хочет найти быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать долгого обучения и не теряя времени перейти к достижению личных целей: к поступлению в ВУЗ или к началу трудовой карьеры.
http://diplomany.ru
Здравствуйте!
Получите российский диплом по доступной цене с гарантией прохождения проверок и доставкой в любой город РФ без предоплаты.
Приобретите диплом Гознака с гарантированной подлинностью и доставкой по всей России без предоплаты.
https://cse.google.com.af/url?sa=t&url=https://lands-diploms.com
https://clients4.google.com/url?q=https://russiany-diplomans.com
https://maps.google.ch/url?sa=t&url=http://volga-diplomy.com
http://maps.google.com.na/url?q=https://russiany-diplomans.com
https://www.google.mn/url?q=https://premialnie-diplomany.com
Hello, yup this post is actually fastidious and I have learned lot of things from it on the topic of blogging. thanks.
https://animemedia.info/
Heya just wanted to give you a brief heads up and let you know a few of the pictures aren’t loading properly. I’m not sure why but I think its a linking issue. I’ve tried it in two different internet browsers and both show the same results.
https://animedia.xyz/
I delight in, cause I discovered exactly what I was looking for. You have ended my 4 day long hunt! God Bless you man. Have a nice day. Bye
https://doramafilm.cc/
В современном мире, где аттестат становится началом отличной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Важность наличия документа об образовании переоценить невозможно. Ведь именно диплом открывает дверь перед каждым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в университете.
Предлагаем очень быстро получить любой необходимый документ. Вы имеете возможность купить аттестат, и это является выгодным решением для человека, который не смог закончить обучение или утратил документ. Аттестат изготавливается с особой аккуратностью, вниманием ко всем элементам. В результате вы получите полностью оригинальный документ.
Преимущества этого решения заключаются не только в том, что можно быстро получить аттестат. Процесс организовывается удобно, с профессиональной поддержкой. Начав от выбора требуемого образца аттестата до точного заполнения персональных данных и доставки в любой регион России — все находится под абсолютным контролем опытных мастеров.
Таким образом, всем, кто хочет найти максимально быстрый способ получения необходимого документа, наша компания готова предложить выгодное решение. Приобрести аттестат – значит избежать продолжительного процесса обучения и сразу перейти к своим целям, будь то поступление в ВУЗ или старт карьеры.
https://diplomans-rossians.com/
В нашем обществе, где диплом – это начало успешной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие официального документа об образовании переоценить невозможно. Ведь диплом открывает двери перед всеми, кто собирается начать профессиональную деятельность или продолжить обучение в университете.
Мы предлагаем максимально быстро получить этот необходимый документ. Вы можете приобрести диплом, и это будет отличным решением для всех, кто не смог закончить обучение или утратил документ. Все дипломы производятся с особой тщательностью, вниманием к мельчайшим нюансам. В итоге вы получите продукт, полностью соответствующий оригиналу.
Преимущество этого подхода состоит не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается удобно и легко, с профессиональной поддержкой. Начав от выбора подходящего образца до консультаций по заполнению персональной информации и доставки по России — все будет находиться под полным контролем квалифицированных мастеров.
Для тех, кто ищет максимально быстрый способ получить требуемый документ, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать длительного процесса обучения и сразу перейти к своим целям: к поступлению в университет или к началу удачной карьеры.
http://www.koelnmedia2.de/fastelovend/member.php?action=showprofile&user_id=14415
http://suntent.kr/bbs/board.php?bo_table=free&wr_id=368873
http://whirlpowertool.ru/index.php?subaction=userinfo&user=ihazuwo
http://www.avisotskiy.com/2013/03/autodesk-revit_21.html
https://hitaste.ru/forum/?PAGE_NAME=profile_view&UID=246157
Hey! Would you mind if I share your blog with my zynga group? There’s a lot of folks that I think would really enjoy your content. Please let me know. Cheers
https://doramafilm.cc/
Сегодня, когда диплом становится началом удачной карьеры в любом направлении, многие ищут максимально быстрый путь получения качественного образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает двери перед каждым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в любом институте.
Наша компания предлагает быстро получить этот важный документ. Вы сможете заказать диплом нового или старого образца, и это становится выгодным решением для всех, кто не смог завершить образование или потерял документ. дипломы производятся аккуратно, с максимальным вниманием ко всем элементам. На выходе вы получите полностью оригинальный документ.
Плюсы такого решения заключаются не только в том, что вы сможете максимально быстро получить диплом. Весь процесс организован просто и легко, с нашей поддержкой. Начиная от выбора необходимого образца диплома до консультации по заполнению личных данных и доставки по стране — все под абсолютным контролем опытных специалистов.
Всем, кто ищет оперативный способ получения необходимого документа, наша компания готова предложить выгодное решение. Приобрести диплом – значит избежать продолжительного обучения и не теряя времени переходить к своим целям: к поступлению в университет или к началу трудовой карьеры.
http://153.126.169.73/question2answer/index.php?qa=user&qa_1=yxypodaj
https://rubukkit.org/threads/originalitydiplomiki.191290/
http://www.standrewskilrymontrotary.org/?unapproved=647490&moderation-hash=64d1e77354f615a0cd4283d7f0b63b4f#comment-647490
http://www.clean-ace8.com/bbs/board.php?bo_table=free&wr_id=276310
http://mtw2014.tmweb.ru/forum/?PAGE_NAME=message&FID=1&TID=8535&TITLE_SEO=8535-diplomaasx&MID=306021&result=new#message306021
Hi there! Do you know if they make any plugins to safeguard against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any suggestions?
diplom07.ru
선도적인 업체인 최신 프라그마틱 게임은 iGaming 분야에서 혁신적이고 표준화된 콘텐츠를 제공하며 슬롯, 라이브 카지노, 빙고 등을 통해 고객에게 엔터테인먼트를 제공합니다.
프라그마틱 슬롯
프라그마틱 슬롯에 대한 글 정말 잘 읽었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 교류하며 더 많은 지식을 얻어보세요!
https://www.wilsonamado.com
https://www.b4closing.com
https://www.12315kz.cn/
Exotic and Luxury Car Rental in Miami
miami exotic car rental
Exotic Car Rental: Luxury and Adrenaline
Exotic car rental is a unique opportunity to feel like a real racer or celebrity, enjoy the indescribable sensations of driving powerful and luxurious cars. This service is very popular all over the world, and it is not surprising, because the opportunity to be behind the wheel for a while Ferrari, Lamborghini, McLaren or another exceptional car can bring a lot of unforgettable emotions and impressions.
Choosing an exotic car
One of the key points of renting an exotic car is choosing the right car. Depending on your preferences and budget, you can choose among the most prestigious and famous brands in the world of the automotive industry.
miami exotic car rental
https://mhp-miami.com/
The most popular options:
Ferrari: These Italian cars are famous for their elegance and power. Many people dream of driving Ferrari, and renting provides an opportunity to make this dream a reality.
Lamborghini: Extreme design and incredible speed make Lamborghini one of the most desirable cars to rent. They are able to accelerate to amazing speeds and provide an unforgettable experience.
McLaren: If you need a car that will look and feel like from the future, McLaren – your choice. These hypercars have incredible power and a unique design.
Porsche: Sports cars Porsche are always in demand. They combine power and comfort, making them an excellent choice for both high-speed driving and long trips.
The rental process
Choosing a rental company: The first step in renting an exotic car is choosing a reliable rental company. It is important to make sure that they have a good reputation and provide accident insurance.
Booking: After choosing a car and a rental company, you can book a car. Usually you will need to provide copies of documents and make a deposit.
Car fence: On the day of the rental, you will need to come to the rental company and pick up your exotic car. It is important to familiarize yourself with the terms of the lease and sign the necessary documents.
Driving Enjoyment: Now you have the opportunity to enjoy the unique experience of driving an exotic car. Remember to follow the rules of the road and safety.
Car return: At the end of the rental period, return the car to the company. It is usually necessary to check the condition of the car and its fuel tank.
Получение диплома представляет собой ключевым моментом в пути всякого индивидуума, определяющим его перспективы и профессиональные перспективы.
Диплом даёт доступ путь к перспективным горизонтам и перспективам, обеспечивая возможность к качественному образованию и высокопрестижным специальностям.
В современном обществе, где в конкуренция на трудовом рынке всё увеличивается, имение диплома становится жизненно важным требованием для выдающейся профессиональной деятельности.
Он подтверждает ваши знания, умения и навыки, умения и умения перед работодателями и обществом в общем.
diploman-russiya.comкупить аттестат классов – возможность для людей, которые стремится к успеху без дополнительных трудностей. Это шанс получить признанный документ по техническому образованию, открывающий новые горизонты и престижным карьерным путям. Наш сервис обеспечивает качество и конфиденциальность, помогая вам достичь ваших целей с минимальными усилиями.
Кроме того, диплом дарит веру в свои силы и укрепляет самооценку, что способствует персональному развитию и саморазвитию. Завершение учебы диплома также является вложением в свое будущее, обеспечивая устойчивость и благополучный стандарт жизни.
Именно поэтому обращать должное внимание получению образования и бороться за его получению, чтобы добиться успех и удовлетворение от своей труда.
k8 カジノ 出金
読みやすく、非常に教育的な内容でした。素晴らしい!
Окончание диплома является важным моментом в карьере каждого человека, который определяет его будущее и профессиональные возможности – http://www.diplomvam.ru. Диплом открывает двери к перспективным горизонтам и перспективам, гарантируя возможность к качественному получению знаний и высокопрестижным профессиям. В нынешнем мире, где борьба на рынке труда постоянно увеличивается, наличие диплома делает жизненно важным условием для выдающейся профессиональной деятельности. Диплом утверждает ваши знания и навыки, умения и компетенции перед работодателями и социумом в общем. В дополнение, диплом дарит веру в свои силы и увеличивает самооценку, что помогает личностному и развитию. Завершение учебы образования также инвестицией в будущее, предоставляя устойчивость и достойный уровень проживания. Поэтому уделять надлежащее внимание и время образованию и бороться за его достижению, чтобы получить успеха и счастье от своей труда.
Диплом не лишь представляет личное образовательный уровень, но и отражает вашу дисциплинированность, трудолюбие и настойчивость в достижении задач. Диплом представляет собой результатом труда и труда, вкладываемых в учебу и самосовершенствование. Получение образования раскрывает перед вами свежие перспективы возможностей, даруя возможность выбирать среди множества карьерных путей и карьерных траекторий. Помимо этого даёт вам базис знаний и навыков, необходимых для выдающейся деятельности в современном мире, насыщенном трудностями и изменениями. Кроме того, диплом считается свидетельством вашей квалификации и квалификации, что в свою очередь повышает вашу привлекательность на рынке труда и открывает вами возможности к наилучшим шансам для профессионального роста. Итак, получение образования диплома не лишь пополняет ваше личное развитие, но и открывает перед вами новые и перспективы для достижения целей и мечтаний.
bestmanualpolesaw.com
그래서 모두 Zhu Houzhao와 Fang Jifan을 바라 보았습니다.
На сегодняшний день, когда диплом – это начало удачной карьеры в любой отрасли, многие ищут максимально быстрый путь получения образования. Факт наличия официального документа трудно переоценить. Ведь диплом открывает двери перед любым человеком, желающим начать профессиональную деятельность или учиться в высшем учебном заведении.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы сможете заказать диплом, что является отличным решением для человека, который не смог закончить обучение, потерял документ или желает исправить плохие оценки. дипломы выпускаются с особой тщательностью, вниманием ко всем деталям, чтобы в результате получился документ, 100% соответствующий оригиналу.
Превосходство такого решения состоит не только в том, что можно максимально быстро получить диплом. Весь процесс организовывается комфортно, с нашей поддержкой. От выбора необходимого образца до консультации по заполнению личной информации и доставки в любой регион страны — все под абсолютным контролем квалифицированных специалистов.
Таким образом, для всех, кто хочет найти максимально быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Купить диплом – это значит избежать долгого процесса обучения и сразу переходить к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
http://komerc.mineralgroup.ru/index.php?subaction=userinfo&user=olynysi
http://mihi.co.kr/bbs/board.php?bo_table=free&wr_id=717344
http://www.sixsigmaexams.com/mybb/member.php?action=profile&uid=260226
http://blog.smirik.ru/2011/10/blog-post_08.html
http://ams-school.ru/reviews/?unapproved=410891&moderation-hash=f3836ce40f0d8814e821cdf135b21a60#comment-410891
В современном мире, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь именно диплом открывает дверь перед каждым человеком, желающим начать трудовую деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы можете приобрести диплом старого или нового образца, и это является выгодным решением для всех, кто не смог закончить обучение, утратил документ или хочет исправить плохие оценки. Все дипломы выпускаются аккуратно, с максимальным вниманием ко всем нюансам, чтобы на выходе получился продукт, максимально соответствующий оригиналу.
Плюсы подобного подхода состоят не только в том, что вы сможете быстро получить диплом. Весь процесс организовывается комфортно, с нашей поддержкой. От выбора необходимого образца до правильного заполнения личных данных и доставки по России — все под абсолютным контролем наших мастеров.
В итоге, для тех, кто ищет быстрый и простой способ получить необходимый документ, наша компания предлагает отличное решение. Купить диплом – это значит избежать продолжительного процесса обучения и не теряя времени перейти к важным целям: к поступлению в университет или к началу удачной карьеры.
http://www.ab-diplom.ru
В нашем мире, где диплом – это начало отличной карьеры в любой сфере, многие ищут максимально простой путь получения образования. Наличие официального документа об образовании сложно переоценить. Ведь именно он открывает дверь перед любым человеком, желающим вступить в профессиональное сообщество или учиться в университете.
Мы предлагаем очень быстро получить этот необходимый документ. Вы можете купить диплом старого или нового образца, и это становится выгодным решением для всех, кто не смог завершить обучение, утратил документ или желает исправить свои оценки. диплом изготавливается аккуратно, с максимальным вниманием ко всем нюансам. На выходе вы получите документ, 100% соответствующий оригиналу.
Превосходство данного решения заключается не только в том, что можно оперативно получить свой диплом. Процесс организован комфортно и легко, с профессиональной поддержкой. От выбора требуемого образца документа до консультации по заполнению личной информации и доставки по стране — все под абсолютным контролем квалифицированных специалистов.
В итоге, всем, кто пытается найти оперативный способ получения необходимого документа, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать длительного процесса обучения и сразу перейти к достижению собственных целей: к поступлению в университет или к началу трудовой карьеры.
http://ab-diplom.ru/
В нашем мире, где диплом – это начало успешной карьеры в любой области, многие стараются найти максимально быстрый путь получения качественного образования. Необходимость наличия официального документа об образовании переоценить попросту невозможно. Ведь именно он открывает двери перед людьми, стремящимися вступить в сообщество профессиональных специалистов или продолжить обучение в университете.
Наша компания предлагает очень быстро получить любой необходимый документ. Вы сможете заказать диплом старого или нового образца, и это становится отличным решением для человека, который не смог завершить образование или потерял документ. Каждый диплом изготавливается аккуратно, с особым вниманием ко всем нюансам, чтобы в итоге получился продукт, максимально соответствующий оригиналу.
Преимущества подобного подхода заключаются не только в том, что можно быстро получить свой диплом. Процесс организован просто и легко, с нашей поддержкой. От выбора подходящего образца до консультаций по заполнению личной информации и доставки в любой регион страны — все будет находиться под полным контролем опытных специалистов.
Для тех, кто пытается найти быстрый и простой способ получения необходимого документа, наша услуга предлагает выгодное решение. Купить диплом – это значит избежать продолжительного обучения и сразу перейти к достижению личных целей, будь то поступление в ВУЗ или старт профессиональной карьеры.
http://ab-diplom.ru
It’s remarkable to go to see this web site and reading the views of all friends about this article, while I am also keen of getting knowledge.
https://server-attestats.com /
Hey there! I’m at work browsing your blog from my new iphone! Just wanted to say I love reading through your blog and look forward to all your posts! Carry on the great work!
https://server-attestats.com /
Hi there, this weekend is pleasant for me, since this time i am reading this wonderful informative article here at my house.
http://server-attestats.com
Привет всем!
Приобретите документы об образовании всех ВУЗов России с гарантированной подлинностью и доставкой по РФ без предварительной оплаты – просто, надежно, выгодно!
http://diplomany-asx.ru
На сегодняшний день, когда диплом – это начало удачной карьеры в любой сфере, многие пытаются найти максимально простой путь получения образования. Необходимость наличия документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед каждым человеком, который собирается вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Мы предлагаем максимально быстро получить этот необходимый документ. Вы сможете заказать диплом старого или нового образца, и это становится удачным решением для всех, кто не смог завершить образование или потерял документ. Все дипломы выпускаются аккуратно, с максимальным вниманием к мельчайшим элементам, чтобы в итоге получился документ, 100% соответствующий оригиналу.
Плюсы такого подхода заключаются не только в том, что вы быстро получите свой диплом. Процесс организован удобно, с нашей поддержкой. Начав от выбора требуемого образца документа до правильного заполнения личной информации и доставки в любое место страны — все находится под абсолютным контролем наших мастеров.
Всем, кто пытается найти быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени перейти к личным целям, будь то поступление в ВУЗ или начало удачной карьеры.
http://xn--80ae1aidok.xn--p1ai/users/iwevavehe
http://kvartirker.ru/profile/owadinen/
http://eydosdigital.com/en/component/k2/item/8
https://civilpatrol.info/?q=petition/80286/eonlinediplom
http://webmontag.de/doku.php?id=eonlinediplom
veganchoicecbd.com
Zhu Houzhao는 호기심으로 가득 차 있었고 기념관에서 무엇을 연주하는지 알고 싶었습니다.
Marvelous, what a website it is! This web site presents valuable information to us, keep it up.
arusak-attestats.ru
В наше время, когда аттестат – это начало отличной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Наличие официального документа сложно переоценить. Ведь диплом открывает двери перед всеми, кто хочет вступить в сообщество профессионалов или продолжить обучение в ВУЗе.
Предлагаем быстро получить этот важный документ. Вы имеете возможность заказать аттестат нового или старого образца, что становится отличным решением для человека, который не смог завершить обучение, потерял документ или хочет исправить плохие оценки. Аттестаты выпускаются с особой аккуратностью, вниманием к мельчайшим элементам. В итоге вы сможете получить продукт, максимально соответствующий оригиналу.
Превосходство такого решения заключается не только в том, что вы сможете максимально быстро получить аттестат. Процесс организовывается просто и легко, с нашей поддержкой. Начиная от выбора требуемого образца документа до точного заполнения персональной информации и доставки по России — все под полным контролем квалифицированных мастеров.
Для тех, кто хочет найти быстрый способ получения необходимого документа, наша услуга предлагает выгодное решение. Заказать аттестат – это значит избежать длительного процесса обучения и сразу переходить к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
https://diplomans-rossians.com/
For most up-to-date information you have to visit the web and on world-wide-web I found this website as a finest web page for most up-to-date updates.
http://www.arusak-attestats.ru
Hi, for all time i used to check weblog posts here in the early hours in the dawn, for the reason that i love to learn more and more.
http://arusak-attestats.ru
В нашем мире, где диплом является началом удачной карьеры в любой отрасли, многие ищут максимально простой путь получения образования. Факт наличия официального документа сложно переоценить. Ведь именно диплом открывает дверь перед любым человеком, который стремится вступить в сообщество профессионалов или учиться в университете.
В данном контексте наша компания предлагает быстро получить этот необходимый документ. Вы можете купить диплом, что становится удачным решением для всех, кто не смог завершить обучение, потерял документ или желает исправить плохие оценки. диплом изготавливается с особой тщательностью, вниманием ко всем нюансам. В результате вы получите документ, полностью соответствующий оригиналу.
Превосходство данного подхода состоит не только в том, что вы оперативно получите свой диплом. Процесс организован удобно, с профессиональной поддержкой. От выбора нужного образца диплома до консультаций по заполнению личной информации и доставки в любой регион страны — все находится под абсолютным контролем опытных специалистов.
В результате, для всех, кто пытается найти максимально быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – значит избежать долгого обучения и не теряя времени переходить к достижению своих целей: к поступлению в университет или к началу трудовой карьеры.
http://wiki.gta-zona.ru/index.php?title=diplomaasx
http://www.ifreemax.ru/2015/06/forum-madrid-startup-weekend-2015.html
https://www.bonanza.by/product/325585/reviews/
https://nastroisam.ru/vklyuchit-flash-player-google-chrome/comment-page-1/?unapproved=464042&moderation-hash=8e8cd86e6d4503793929ded18f9df918#comment-464042
https://horoshava.ru/forum/user/35361/
최신 프라그마틱 게임은 선도적이고 표준화된 iGaming 콘텐츠를 제공하며 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 제공합니다.
프라그마틱 게임
프라그마틱 슬롯에 대한 내용이 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 찾아보실 수 있어요. 함께 지식을 공유해보세요!
https://www.12315ov.cn/
http://holyshirtsandpants.net/
https://www.12315jg.cn/
werankcities.com
이때 따뜻한 정자에 홍지 황제의 책상 위에 기념비가 있었다.
k8 カジノ 登録
読者のニーズに応える実用性の高い内容が素晴らしい!
geinoutime.com
Hongzhi 황제는 고개를 끄덕이고 안도의 얼굴로 늙은 목사 Zhang Mao를 바라 보았습니다.
Приветики!
Приобретите документы об образовании всех ВУЗов России по выгодным условиям с доставкой по РФ без предоплаты и с гарантией качества!
eonline-diploms.com
Привет всем!
Мы предлагаем приобрести диплом института или колледжа с доставкой по России без предоплаты.
Рады предложить Большой выбор документов об образовании всех Вузов России, недорого, с постоплатой, помошь 24/7
http://www.diploms-asx.com
Hi! Do you know if they make any plugins to safeguard against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any suggestions?
https://diplomy-originaly.com/
What a information of un-ambiguity and preserveness of precious familiarity concerning unpredicted emotions.
http://diplomy-originaly.com
Привет, дорогой читатель!
Закажите диплом ВУЗа с гарантированной подлинностью и доставкой по России без предварительной оплаты – просто, надежно, выгодно!
http://eonline-diploms.com
Marvelous, what a webpage it is! This blog gives valuable data to us, keep it up.
http://www.diplomy-originaly.com
Hi there this is kinda of off topic but I was wondering if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding knowledge so I wanted to get guidance from someone with experience. Any help would be enormously appreciated!
http://www.diplomy-originaly.com
Доброго всем дня!
Закажите диплом ВУЗа по выгодной цене с доставкой в любой город России без предоплаты – это надежно и выгодно!
http://www.eonline-diploms.com
Добрый день всем!
На нашем сайте заказать диплом ВУЗа недорого с оплатой после получения и круглосуточной поддержкой – просто и уверенно!
http://www.eonline-diploms.com
I loved as much as you’ll receive carried out right here. The sketch is tasteful, your authored subject matter stylish. nonetheless, you command get bought an edginess over that you wish be delivering the following. unwell unquestionably come further formerly again since exactly the same nearly a lot often inside case you shield this increase.
http://www.diplomy-originaly.com
Приветики!
Приобретите диплом института или колледжа недорого с доставкой по РФ и гарантией качества без предварительной оплаты!
Выбрать и купить диплом Вуза России недорого, качественно и с возможностью отправки почтой!
http://www.diploms-asx.com
I blog frequently and I truly thank you for your information. Your article has truly peaked my interest. I’m going to book mark your site and keep checking for new details about once a week. I subscribed to your Feed as well.
Засіб для збирання кам’яної підлоги RM 537, 500мл Karcher (6.295-943.0)
Its like you read my mind! You seem to know a lot about this, like you wrote the book in it or something. I think that you could do with some pics to drive the message home a little bit, but other than that, this is fantastic blog. A fantastic read. I will definitely be back.
https://murray-alexander.thoughtlanes.net/pidvishchte-bezpeku-z-lemarix-ua-linzi-u-fari-dlia-vashogo-avto
I’m not sure why but this weblog is loading very slow for me. Is anyone else having this issue or is it a problem on my end? I’ll check back later and see if the problem still exists.
https://storgaard-jarvis.mdwrite.net/osvitliuite-dorogu-iak-profesional-linzi-u-fari-vid-lemarix-ua-1715687950
Thanks for any other wonderful post. Where else may just anyone get that kind of information in such a perfect method of writing? I’ve a presentation subsequent week, and I am at the search for such information.
http://kvitka-ua.com/linzy-u-fary-dlya-vashogo-avtomobilya
As the admin of this web page is working, no question very quickly it will be renowned, due to its quality contents.
http://kvitka-ua.com/linzy-u-fary-dlya-vashogo-avtomobilya
That is really fascinating, You’re an overly professional blogger. I’ve joined your feed and stay up for looking for more of your great post. Additionally, I’ve shared your website in my social networks
http://kvitka-ua.com/linzy-u-fary-dlya-vashogo-avtomobilya
What’s up colleagues, its great article about teachingand completely defined, keep it up all the time.
http://kvitka-ua.com/linzy-u-fary-dlya-vashogo-avtomobilya
ilogidis.com
잠시 침묵이 흐른 후, Ouyang Zhi의 얼굴이 갑자기 편안해졌습니다.
Привет всем!
Выбрать и купить диплом Вуза России недорого, качественно и с возможностью отправки почтой!
diplomanc-russia24.ru
Здравствуйте!
Приобретите документ об образовании по выгодной цене с доставкой по РФ и оплатой после получения ваших руками!
http://www.diplomanc-russia24.ru
Pretty component to content. I simply stumbled upon your website and in accession capital to claim that I acquire actually loved account your blog posts. Any way I will be subscribing for your feeds and even I achievement you get admission to persistently fast.
https://cutt.ly/Dero0rh9
Just desire to say your article is as astounding. The clearness in your post is just excellent and i could assume you are an expert on this subject. Well with your permission allow me to grab your feed to keep up to date with forthcoming post. Thanks a million and please keep up the rewarding work.
https://cutt.ly/5ero9U8c
프라그마틱 플레이의 흥미진진한 세계로 초대합니다.
프라그마틱
프라그마틱의 라이브 카지노는 정말 현장감 넘치게 즐길 수 있는데, 여기서 더 많은 정보를 얻을 수 있어 좋아요!
https://www.12315jq.cn/
https://www.12315amb.cn/
https://www.12315es.cn/
k8 カジノ 出金条件
このトピックについてこんなに詳しく書かれた記事は初めてです。素晴らしいです!
tvlore.com
긍지 속에서 홍지황제는 갑자기 눈가가 촉촉해지는 것을 느꼈다.과거 Fang Jifan은 수도를 돌아 다니며 아버지 Fang Jinglong에 대해 이야기했습니다.
Desirable to Genio, the go-to invoice generator quest of miniature businesses and freelancers. We mention you innumerable invoice templates, including Microsoft Excel and PDF formats, tailored to all industries. Investigate our component featuring upon 300 customized invoice templates designed to cater to your divergent business needs.
https://www.genio.ac/invoice-templates/
Здравствуйте!
Хотим предложить самые лучшие прогоны, которые могут “убить” сайты вашего конкурента. Всего от 7 000 рублей.
– 100% эффект. Сайты точно “упадут”.
– Максимальное количество негативных фитбеков.
– Собрана специфическая база – выжимка максимально сильных площадок из 10 000 000 веб-сайтов (спамных, порно, вирусных и тому подобных). Это действует безотказно.
– Прогон производим сразу с 4-х мощных серверов.
– Отправка активационных ссылок на электронную почту.
– Выполнение заказа в течение 40-240 часов сутки напролет. Растянем по времени сколько угодно.
– Прогон с запретными ключевыми фразами.
– При условии заказа сразу 2 ресурсов – выгодные бонусы.
Стоимость 77$
Отчётность.
Оплата: Yoo.Деньги, Bitcoin, МИР, Visa, MasterCard…
Принимаем USDT
Телега: @exrumer
Skype: Loves.Ltd
https://xrumer.cc/
Только этот.
Hmm is anyone else encountering problems with the images on this blog loading? I’m trying to find out if its a problem on my end or if it’s the blog. Any responses would be greatly appreciated.
http://edu-digest.ru
В нашем обществе, где диплом является началом удачной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Наличие официального документа об образовании переоценить просто невозможно. Ведь диплом открывает двери перед всеми, кто стремится начать трудовую деятельность или продолжить обучение в каком-либо университете.
Мы предлагаем оперативно получить этот важный документ. Вы можете заказать диплом старого или нового образца, и это будет отличным решением для человека, который не смог закончить обучение или потерял документ. Каждый диплом изготавливается с особой аккуратностью, вниманием ко всем нюансам, чтобы в результате получился продукт, максимально соответствующий оригиналу.
Преимущества данного подхода заключаются не только в том, что можно максимально быстро получить диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. Начиная от выбора необходимого образца документа до консультаций по заполнению персональных данных и доставки по России — все будет находиться под абсолютным контролем квалифицированных специалистов.
Всем, кто ищет быстрый способ получения необходимого документа, наша компания предлагает выгодное решение. Купить диплом – значит избежать долгого процесса обучения и сразу перейти к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
http://proobzh.ru
В нашем обществе, где диплом – это начало успешной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения образования. Наличие официального документа трудно переоценить. Ведь именно он открывает дверь перед всеми, кто собирается начать профессиональную деятельность или учиться в любом институте.
Наша компания предлагает быстро получить этот важный документ. Вы можете заказать диплом старого или нового образца, что становится выгодным решением для всех, кто не смог закончить обучение или потерял документ. Любой диплом изготавливается аккуратно, с особым вниманием к мельчайшим деталям. В итоге вы сможете получить документ, 100% соответствующий оригиналу.
Преимущество данного решения состоит не только в том, что можно быстро получить диплом. Процесс организован просто и легко, с профессиональной поддержкой. От выбора требуемого образца диплома до грамотного заполнения личных данных и доставки в любой регион России — все под абсолютным контролем качественных специалистов.
Всем, кто ищет быстрый и простой способ получения требуемого документа, наша компания готова предложить отличное решение. Купить диплом – это значит избежать продолжительного процесса обучения и сразу переходить к важным целям: к поступлению в ВУЗ или к началу успешной карьеры.
http://www.proobzh.ru
В наше время, когда диплом – это начало удачной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения образования. Наличие официального документа об образовании сложно переоценить. Ведь именно он открывает двери перед людьми, желающими вступить в сообщество профессионалов или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы сможете заказать диплом старого или нового образца, и это является отличным решением для человека, который не смог завершить образование или утратил документ. диплом изготавливается аккуратно, с особым вниманием ко всем нюансам. В результате вы получите продукт, максимально соответствующий оригиналу.
Преимущество данного подхода заключается не только в том, что можно быстро получить свой диплом. Процесс организован комфортно, с профессиональной поддержкой. От выбора подходящего образца диплома до правильного заполнения персональных данных и доставки по стране — все будет находиться под абсолютным контролем качественных мастеров.
Для всех, кто хочет найти быстрый и простой способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – значит избежать долгого процесса обучения и сразу перейти к достижению своих целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://www.dof-edu.ru
В нашем обществе, где диплом является началом удачной карьеры в любой области, многие стараются найти максимально простой путь получения образования. Факт наличия официального документа об образовании переоценить просто невозможно. Ведь именно он открывает дверь перед любым человеком, желающим начать профессиональную деятельность или учиться в университете.
В данном контексте мы предлагаем оперативно получить любой необходимый документ. Вы сможете купить диплом нового или старого образца, что будет выгодным решением для всех, кто не смог закончить образование, потерял документ или желает исправить свои оценки. Все дипломы производятся с особой аккуратностью, вниманием ко всем нюансам, чтобы в итоге получился продукт, 100% соответствующий оригиналу.
Преимущество этого решения заключается не только в том, что вы быстро получите диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начиная от выбора нужного образца документа до консультации по заполнению личной информации и доставки в любой регион страны — все находится под полным контролем квалифицированных мастеров.
В результате, всем, кто пытается найти оперативный способ получения необходимого документа, наша компания может предложить отличное решение. Приобрести диплом – значит избежать длительного процесса обучения и не теряя времени перейти к личным целям: к поступлению в университет или к началу трудовой карьеры.
proobzh.ru
В современном мире, где диплом является началом удачной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения образования. Важность наличия документа об образовании сложно переоценить. Ведь диплом открывает дверь перед людьми, желающими начать трудовую деятельность или учиться в каком-либо университете.
В данном контексте мы предлагаем быстро получить любой необходимый документ. Вы имеете возможность приобрести диплом нового или старого образца, и это становится удачным решением для всех, кто не смог закончить обучение или потерял документ. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам. В итоге вы получите полностью оригинальный документ.
Преимущества подобного решения заключаются не только в том, что вы быстро получите свой диплом. Процесс организовывается комфортно, с профессиональной поддержкой. От выбора нужного образца документа до консультаций по заполнению персональной информации и доставки по стране — все находится под полным контролем качественных специалистов.
Всем, кто пытается найти быстрый и простой способ получить требуемый документ, наша компания предлагает отличное решение. Заказать диплом – это значит избежать долгого процесса обучения и не теряя времени переходить к своим целям: к поступлению в университет или к началу трудовой карьеры.
http://www.krsk-school47.ru
В нашем мире, где диплом является началом успешной карьеры в любом направлении, многие стараются найти максимально простой путь получения образования. Необходимость наличия документа об образовании сложно переоценить. Ведь именно диплом открывает дверь перед людьми, желающими начать трудовую деятельность или учиться в университете.
Мы предлагаем очень быстро получить этот важный документ. Вы имеете возможность приобрести диплом нового или старого образца, и это будет отличным решением для человека, который не смог завершить образование или потерял документ. диплом изготавливается с особой тщательностью, вниманием к мельчайшим деталям. На выходе вы сможете получить документ, максимально соответствующий оригиналу.
Плюсы данного решения состоят не только в том, что вы сможете быстро получить свой диплом. Весь процесс организован удобно и легко, с нашей поддержкой. От выбора подходящего образца до правильного заполнения личных данных и доставки по стране — все находится под абсолютным контролем квалифицированных мастеров.
Всем, кто ищет быстрый и простой способ получить необходимый документ, наша компания готова предложить отличное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени переходить к важным целям, будь то поступление в ВУЗ или старт карьеры.
http://www.dof-edu.ru
Сегодня, когда диплом является началом удачной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Наличие официального документа переоценить попросту невозможно. Ведь именно он открывает дверь перед каждым человеком, который желает начать профессиональную деятельность или продолжить обучение в университете.
В данном контексте наша компания предлагает быстро получить любой необходимый документ. Вы можете купить диплом, и это будет отличным решением для всех, кто не смог завершить образование или утратил документ. Все дипломы выпускаются аккуратно, с особым вниманием ко всем нюансам, чтобы на выходе получился документ, 100% соответствующий оригиналу.
Плюсы подобного подхода состоят не только в том, что можно оперативно получить свой диплом. Весь процесс организован удобно, с нашей поддержкой. От выбора требуемого образца диплома до консультации по заполнению личных данных и доставки в любой регион России — все под полным контролем квалифицированных специалистов.
Всем, кто ищет быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Купить диплом – это значит избежать продолжительного обучения и сразу переходить к важным целям: к поступлению в университет или к началу успешной карьеры.
http://www.edu-nt.ru/
Can I simply say what a relief to discover someone who actually understands what they are discussing over the internet. You definitely know how to bring an issue to light and make it important. A lot more people ought to look at this and understand this side of your story. I was surprised you aren’t more popular given that you surely possess the gift.
edu-digest.ru
В современном мире, где диплом является началом отличной карьеры в любой сфере, многие стараются найти максимально быстрый и простой путь получения качественного образования. Необходимость наличия документа об образовании переоценить невозможно. Ведь именно он открывает двери перед любым человеком, желающим начать трудовую деятельность или учиться в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы сможете заказать диплом нового или старого образца, что является выгодным решением для человека, который не смог закончить обучение, утратил документ или желает исправить свои оценки. диплом изготавливается с особой аккуратностью, вниманием к мельчайшим деталям, чтобы в результате получился 100% оригинальный документ.
Преимущество данного подхода заключается не только в том, что вы сможете оперативно получить диплом. Весь процесс организован удобно, с профессиональной поддержкой. От выбора требуемого образца до правильного заполнения личной информации и доставки по России — все будет находиться под абсолютным контролем квалифицированных мастеров.
Всем, кто ищет максимально быстрый способ получения необходимого документа, наша услуга предлагает отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и сразу переходить к достижению личных целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://www.proobzh.ru
В наше время, когда диплом – это начало удачной карьеры в любой отрасли, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа трудно переоценить. Ведь именно он открывает двери перед любым человеком, желающим начать профессиональную деятельность или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает быстро получить любой необходимый документ. Вы имеете возможность заказать диплом нового или старого образца, и это становится отличным решением для человека, который не смог закончить обучение или утратил документ. диплом изготавливается с особой тщательностью, вниманием ко всем нюансам, чтобы на выходе получился документ, максимально соответствующий оригиналу.
Преимущество данного решения состоит не только в том, что можно оперативно получить свой диплом. Весь процесс организован просто и легко, с профессиональной поддержкой. Начиная от выбора необходимого образца до консультаций по заполнению персональных данных и доставки в любое место России — все под абсолютным контролем опытных мастеров.
Всем, кто пытается найти быстрый и простой способ получения необходимого документа, наша компания предлагает выгодное решение. Купить диплом – значит избежать долгого обучения и сразу переходить к достижению собственных целей, будь то поступление в университет или начало карьеры.
http://www.proobzh.ru
Сегодня, когда диплом становится началом успешной карьеры в любом направлении, многие ищут максимально быстрый путь получения образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает дверь перед всеми, кто собирается начать трудовую деятельность или учиться в ВУЗе.
Предлагаем быстро получить этот важный документ. Вы имеете возможность купить диплом, что является отличным решением для человека, который не смог завершить обучение, утратил документ или хочет исправить свои оценки. дипломы изготавливаются с особой тщательностью, вниманием ко всем нюансам, чтобы на выходе получился 100% оригинальный документ.
Плюсы подобного подхода состоят не только в том, что вы максимально быстро получите диплом. Процесс организовывается комфортно, с профессиональной поддержкой. От выбора необходимого образца документа до консультации по заполнению личных данных и доставки по России — все будет находиться под полным контролем квалифицированных специалистов.
Таким образом, всем, кто ищет максимально быстрый способ получения требуемого документа, наша услуга предлагает выгодное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и сразу переходить к своим целям, будь то поступление в ВУЗ или старт удачной карьеры.
http://www.proobzh.ru
Сегодня, когда диплом – это начало успешной карьеры в любой отрасли, многие пытаются найти максимально быстрый путь получения образования. Необходимость наличия официального документа об образовании переоценить просто невозможно. Ведь диплом открывает дверь перед всеми, кто собирается вступить в сообщество квалифицированных специалистов или учиться в высшем учебном заведении.
Предлагаем быстро получить этот важный документ. Вы можете купить диплом, и это является удачным решением для всех, кто не смог завершить образование или потерял документ. Любой диплом изготавливается аккуратно, с максимальным вниманием ко всем нюансам. В результате вы сможете получить документ, 100% соответствующий оригиналу.
Преимущество этого решения заключается не только в том, что можно оперативно получить диплом. Процесс организовывается комфортно, с нашей поддержкой. От выбора подходящего образца до консультаций по заполнению персональных данных и доставки в любое место России — все под абсолютным контролем наших мастеров.
Всем, кто пытается найти оперативный способ получения необходимого документа, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать долгого процесса обучения и сразу перейти к важным целям: к поступлению в ВУЗ или к началу успешной карьеры.
proobzh.ru
Its like you read my mind! You appear to know so much about this, like you wrote the book in it or something. I think that you can do with some pics to drive the message home a little bit, but other than that, this is great blog. An excellent read. I’ll definitely be back.
http://www.edu-digest.ru
В нашем мире, где диплом – это начало отличной карьеры в любой сфере, многие стараются найти максимально быстрый путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь диплом открывает двери перед людьми, желающими начать трудовую деятельность или продолжить обучение в любом институте.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете приобрести диплом, и это становится удачным решением для человека, который не смог закончить обучение, потерял документ или желает исправить свои оценки. Все дипломы производятся аккуратно, с особым вниманием ко всем деталям. В результате вы сможете получить 100% оригинальный документ.
Преимущество подобного решения заключается не только в том, что можно быстро получить диплом. Весь процесс организован удобно, с профессиональной поддержкой. От выбора нужного образца до правильного заполнения персональной информации и доставки в любое место России — все находится под полным контролем опытных мастеров.
Таким образом, для тех, кто пытается найти быстрый и простой способ получить необходимый документ, наша услуга предлагает выгодное решение. Заказать диплом – значит избежать продолжительного обучения и не теряя времени перейти к своим целям, будь то поступление в ВУЗ или начало успешной карьеры.
http://www.edu-nt.ru/
Aw, this was a really good post. Finding the time and actual effort to produce a very good article… but what can I say… I procrastinate a lot and never manage to get nearly anything done.
http://www.edu-digest.ru
Сегодня, когда диплом – это начало удачной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие официального документа об образовании сложно переоценить. Ведь именно он открывает дверь перед людьми, стремящимися вступить в профессиональное сообщество или продолжить обучение в каком-либо институте.
Мы предлагаем оперативно получить любой необходимый документ. Вы имеете возможность приобрести диплом старого или нового образца, и это становится выгодным решением для человека, который не смог закончить образование, утратил документ или хочет исправить плохие оценки. диплом изготавливается с особой аккуратностью, вниманием ко всем элементам, чтобы в итоге получился 100% оригинальный документ.
Преимущества такого подхода состоят не только в том, что можно оперативно получить свой диплом. Процесс организовывается комфортно, с нашей поддержкой. Начав от выбора нужного образца документа до консультаций по заполнению личных данных и доставки в любое место страны — все находится под абсолютным контролем наших мастеров.
Для всех, кто пытается найти максимально быстрый способ получить необходимый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать долгого обучения и сразу переходить к своим целям, будь то поступление в университет или начало успешной карьеры.
http://www.edu-nt.ru/
В нашем обществе, где диплом – это начало отличной карьеры в любой области, многие стараются найти максимально быстрый путь получения образования. Наличие официального документа трудно переоценить. Ведь диплом открывает двери перед людьми, желающими вступить в сообщество квалифицированных специалистов или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы имеете возможность приобрести диплом нового или старого образца, что становится выгодным решением для человека, который не смог завершить образование или утратил документ. диплом изготавливается с особой тщательностью, вниманием ко всем нюансам. На выходе вы получите 100% оригинальный документ.
Преимущества этого решения состоят не только в том, что вы быстро получите диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора подходящего образца документа до точного заполнения персональной информации и доставки в любое место страны — все под абсолютным контролем квалифицированных мастеров.
Всем, кто ищет максимально быстрый способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – значит избежать длительного процесса обучения и не теряя времени переходить к достижению личных целей, будь то поступление в университет или начало трудовой карьеры.
http://proobzh.ru
В нашем обществе, где диплом становится началом успешной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения образования. Факт наличия официального документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед любым человеком, желающим вступить в сообщество профессиональных специалистов или учиться в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить любой необходимый документ. Вы можете заказать диплом старого или нового образца, и это является удачным решением для всех, кто не смог завершить образование или утратил документ. Все дипломы выпускаются аккуратно, с особым вниманием к мельчайшим деталям, чтобы на выходе получился документ, полностью соответствующий оригиналу.
Преимущества подобного решения заключаются не только в том, что можно оперативно получить диплом. Весь процесс организован просто и легко, с нашей поддержкой. Начав от выбора необходимого образца документа до консультаций по заполнению персональных данных и доставки в любое место России — все находится под абсолютным контролем качественных специалистов.
Для всех, кто пытается найти оперативный способ получения необходимого документа, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать длительного процесса обучения и сразу переходить к достижению собственных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://edu-nt.ru/
В наше время, когда диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Наличие официального документа переоценить просто невозможно. Ведь именно он открывает дверь перед любым человеком, который собирается вступить в профессиональное сообщество или учиться в любом университете.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы сможете заказать диплом, и это является удачным решением для человека, который не смог закончить обучение или потерял документ. дипломы выпускаются с особой тщательностью, вниманием ко всем нюансам. В результате вы получите полностью оригинальный документ.
Преимущества такого подхода заключаются не только в том, что вы сможете быстро получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начав от выбора нужного образца до консультации по заполнению персональной информации и доставки в любой регион России — все находится под абсолютным контролем квалифицированных мастеров.
Для тех, кто хочет найти быстрый и простой способ получить необходимый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать долгого процесса обучения и не теряя времени перейти к достижению личных целей: к поступлению в университет или к началу удачной карьеры.
http://edu-nt.ru/
В нашем обществе, где диплом – это начало успешной карьеры в любой отрасли, многие пытаются найти максимально быстрый и простой путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед всеми, кто стремится вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы сможете приобрести диплом, и это становится отличным решением для всех, кто не смог завершить образование, утратил документ или хочет исправить плохие оценки. Каждый диплом изготавливается с особой аккуратностью, вниманием ко всем нюансам, чтобы в итоге получился документ, полностью соответствующий оригиналу.
Превосходство такого подхода заключается не только в том, что вы быстро получите диплом. Весь процесс организован удобно, с нашей поддержкой. От выбора необходимого образца до грамотного заполнения личных данных и доставки по России — все под абсолютным контролем квалифицированных специалистов.
Для всех, кто пытается найти быстрый способ получить требуемый документ, наша услуга предлагает отличное решение. Заказать диплом – это значит избежать долгого процесса обучения и не теряя времени переходить к достижению личных целей, будь то поступление в университет или старт карьеры.
edu-nt.ru
В нашем обществе, где диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально простой путь получения образования. Факт наличия документа об образовании переоценить невозможно. Ведь диплом открывает дверь перед каждым человеком, желающим вступить в сообщество квалифицированных специалистов или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы можете заказать диплом старого или нового образца, что будет удачным решением для всех, кто не смог закончить образование, потерял документ или хочет исправить свои оценки. Все дипломы изготавливаются аккуратно, с максимальным вниманием ко всем элементам, чтобы в результате получился полностью оригинальный документ.
Плюсы такого решения состоят не только в том, что вы быстро получите свой диплом. Весь процесс организован комфортно, с нашей поддержкой. От выбора необходимого образца до правильного заполнения личных данных и доставки в любой регион страны — все будет находиться под абсолютным контролем опытных мастеров.
Всем, кто пытается найти быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – значит избежать длительного процесса обучения и сразу перейти к достижению своих целей, будь то поступление в университет или старт профессиональной карьеры.
edu-nt.ru
В нашем обществе, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Необходимость наличия официального документа переоценить просто невозможно. Ведь диплом открывает двери перед любым человеком, который стремится вступить в сообщество профессионалов или учиться в ВУЗе.
В данном контексте наша компания предлагает оперативно получить любой необходимый документ. Вы имеете возможность заказать диплом старого или нового образца, и это становится удачным решением для всех, кто не смог завершить образование, утратил документ или желает исправить плохие оценки. дипломы изготавливаются аккуратно, с особым вниманием ко всем деталям, чтобы в результате получился полностью оригинальный документ.
Преимущество данного решения состоит не только в том, что можно быстро получить свой диплом. Процесс организовывается комфортно, с профессиональной поддержкой. От выбора подходящего образца диплома до консультаций по заполнению персональных данных и доставки в любое место России — все будет находиться под абсолютным контролем квалифицированных специалистов.
В результате, для тех, кто пытается найти максимально быстрый способ получения необходимого документа, наша компания предлагает выгодное решение. Купить диплом – это значит избежать долгого процесса обучения и сразу перейти к личным целям, будь то поступление в ВУЗ или старт удачной карьеры.
edu-nt.ru
На сегодняшний день, когда диплом – это начало успешной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Факт наличия официального документа переоценить невозможно. Ведь диплом открывает двери перед любым человеком, желающим вступить в сообщество профессиональных специалистов или продолжить обучение в любом ВУЗе.
Мы предлагаем очень быстро получить этот важный документ. Вы имеете возможность купить диплом старого или нового образца, что становится выгодным решением для человека, который не смог закончить обучение, потерял документ или желает исправить плохие оценки. Все дипломы производятся с особой тщательностью, вниманием ко всем элементам, чтобы в результате получился 100% оригинальный документ.
Преимущество такого решения состоит не только в том, что можно оперативно получить диплом. Процесс организован удобно, с нашей поддержкой. От выбора подходящего образца до правильного заполнения персональной информации и доставки по России — все будет находиться под абсолютным контролем квалифицированных мастеров.
В итоге, для тех, кто пытается найти максимально быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Заказать диплом – значит избежать продолжительного процесса обучения и не теряя времени перейти к достижению личных целей, будь то поступление в ВУЗ или начало карьеры.
dof-edu.ru
geinoutime.com
Fang Jifan이 Zhu Houzhao에게 이러한 말을 가르쳤습니까?
onair2tv.com
직설적으로 말하면 이것은 전례가없고 전례가 없습니다.
В современном мире, где диплом является началом отличной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения образования. Наличие официального документа об образовании переоценить просто невозможно. Ведь именно диплом открывает двери перед каждым человеком, желающим начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы имеете возможность купить диплом нового или старого образца, и это становится отличным решением для всех, кто не смог завершить обучение, утратил документ или желает исправить свои оценки. дипломы изготавливаются аккуратно, с особым вниманием ко всем элементам. На выходе вы получите полностью оригинальный документ.
Преимущество такого решения заключается не только в том, что можно оперативно получить свой диплом. Весь процесс организован комфортно и легко, с профессиональной поддержкой. Начиная от выбора необходимого образца до консультации по заполнению личной информации и доставки по стране — все под полным контролем квалифицированных мастеров.
Всем, кто ищет быстрый способ получить необходимый документ, наша услуга предлагает отличное решение. Заказать диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к достижению личных целей, будь то поступление в ВУЗ или начало удачной карьеры.
http://www.edu-nt.ru/
В современном мире, где диплом – это начало удачной карьеры в любой отрасли, многие ищут максимально быстрый путь получения качественного образования. Факт наличия официального документа об образовании переоценить попросту невозможно. Ведь диплом открывает дверь перед любым человеком, желающим начать профессиональную деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете купить диплом, и это становится выгодным решением для всех, кто не смог завершить образование или утратил документ. диплом изготавливается с особой аккуратностью, вниманием к мельчайшим элементам, чтобы на выходе получился 100% оригинальный документ.
Преимущество данного подхода состоит не только в том, что вы сможете максимально быстро получить свой диплом. Процесс организовывается удобно, с профессиональной поддержкой. От выбора подходящего образца диплома до грамотного заполнения персональной информации и доставки по России — все находится под абсолютным контролем наших мастеров.
Для тех, кто ищет максимально быстрый способ получить требуемый документ, наша компания может предложить отличное решение. Купить диплом – значит избежать продолжительного обучения и не теряя времени перейти к достижению своих целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://www.edu-nt.ru/
В нашем мире, где диплом становится началом успешной карьеры в любой сфере, многие ищут максимально быстрый путь получения качественного образования. Наличие официального документа об образовании переоценить невозможно. Ведь диплом открывает двери перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в университете.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы сможете приобрести диплом старого или нового образца, и это становится выгодным решением для всех, кто не смог завершить образование или утратил документ. дипломы производятся с особой аккуратностью, вниманием ко всем деталям. На выходе вы сможете получить полностью оригинальный документ.
Преимущество такого решения состоит не только в том, что вы сможете оперативно получить свой диплом. Весь процесс организован комфортно, с профессиональной поддержкой. Начиная от выбора требуемого образца диплома до консультации по заполнению личной информации и доставки в любое место России — все будет находиться под полным контролем качественных мастеров.
В результате, всем, кто ищет максимально быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Заказать диплом – значит избежать продолжительного обучения и сразу переходить к личным целям: к поступлению в университет или к началу трудовой карьеры.
edu-nt.ru
Доставка алкогольной продукции на дом в Сочи круглосуточно
Нередко возникают ситуации, когда в самый разгар вечеринки у вас заканчиваются алкогольные напитки, и вы хотите продолжить веселье, но магазины уже закрыты. В таких случаях вы можете обратиться к нам, и мы быстро доставим вам качественные продукты прямо к дому. Наш сервис предоставляет легальную доставку, быструю доставку и удобные способы оплаты – вот наши преимущества.
Основные причины выбрать нас
доставка спиртного сочи
Законность. Мы соблюдаем законы, запрещающие дистанционную продажу алкоголя.
Удобство. Вы можете произвести оплату путем перевода на банковскую карту по номеру телефона.
Быстрота. Мы осуществляем доставку по Сочи уже через 15 минут.
Мы не только предлагаем купить качественный алкоголь, но и осуществляем доставку даже ночью прямо к вашему дому. Мы доставим пиво, крепкие алкогольные напитки и безалкогольные напитки в любую часть города.
Важно отметить, что приобретение алкоголя через интернет с доставкой является незаконным. Согласно закону 171 ФЗ, п. 14 ч. 2 ст. 16, продажа алкогольной продукции дистанционным способом запрещена. При оформлении заказа через наши онлайн-сервисы необходимо подтвердить, что вам исполнилось 18 лет, а также согласиться с приобретением сувениров алкогольной тематики. При заказе доставки в отдаленные районы города учтите возможные пробки на дорогах и погодные условия. Мы с удовольствием доставим вам продукцию в Адлер, Дагомыс, Мацесту, Кудепсту, Бытху, Мамайку, Хосту.
Доставкой алкоголя на дом – быстро и надёжно!
Наши услуги доставки алкоголя на дом предоставляются круглосуточно! Добро пожаловать на наш сайт, посвященный доставке алкоголя на дом в Сочи 24/7. Мы предлагаем широкий ассортимент алкогольных напитков, доступных для заказа в любое время суток. Наша команда опытных специалистов готова обеспечить вас отличным сервисом и удовлетворить ваши предпочтения вкуса.
Наши преимущества:
https://dostavka-alko-sochi-14.store/
доставка алкоголя сочи
Быстрая и надежная доставка: Мы понимаем важность получения заказа вовремя, поэтому предлагаем надежную и оперативную доставку. Наша служба доставки работает круглосуточно, чтобы вы могли наслаждаться алкоголем в любое время.
Широкий ассортимент: Мы тщательно подбираем лучшие алкогольные напитки со всего мира. У нас вы найдете разнообразные сорта пива, вина, крепкие спиртные напитки, шампанское и многое другое. Мы также предлагаем различные аксессуары для алкоголя, чтобы обогатить ваш опыт употребления напитков.
Премиальное качество: Мы сотрудничаем только с проверенными поставщиками, чтобы гарантировать вам высокое качество алкогольных напитков. Все наши продукты проходят строгий контроль качества, чтобы удовлетворить ваши ожидания.
Ценовая доступность: У нас есть алкогольные напитки на любой вкус и бюджет. Мы предлагаем разнообразные ценовые категории, чтобы каждый клиент мог найти оптимальное соотношение цены и качества.
Простой процесс заказа: Наш сайт предоставляет удобный интерфейс для размещения заказа. Вы можете легко выбрать нужные товары, указать адрес доставки и оформить оплату всего в несколько кликов. Мы также принимаем различные методы оплаты для вашего удобства.
Профессиональная поддержка: Наша команда службы поддержки всегда готова помочь вам с любыми вопросами или проблемами, возникшими во время заказа или доставки. Мы стремимся обеспечить высокий уровень удовлетворенности клиентов.
Безопасность и конфиденциальность: Мы обеспечиваем полную конфиденциальность ваших данных и гарантируем безопасность при совершении покупок. Ваши личные данные и платежные реквизиты защищены с помощью современных технологий шифрования.
Без разницы, нужен ли вам алкоголь для вечеринки, романтического ужина или просто для расслабления после долгого дня – наша служба доставки алкоголя на дом в Сочи ночью готова стать вашим надежным партнером. Закажите прямо сейчас и наслаждайтесь вкусными напитками в уюте своего дома!
프라그마틱의 게임은 높은 퀄리티와 흥미진진한 스토리로 항상 눈길을 끌어요. 이번에 어떤 게임을 즐겼나요?
프라그마틷
프라그마틱 관련 소식 항상 주시고 있어요! 또한, 제 사이트에서도 프라그마틱에 대한 정보를 얻을 수 있어요. 함께 발전하며 지식을 나눠봐요!
https://www.antiquees.com
https://www.b4closing.com
https://www.12315qk.cn/
Seo service
Search Engine Optimization solutions are the overlooked champions of the internet period, providing businesses with the instruments and approaches to shine vibrantly in the huge universe of online content. By delving into the capability of efficient search term targeting, superior inbound link procurement, and content improvement, these methods assure that a webpage is not just noticeable, but stands out as a signal of applicability and power in its sector. The beauty of SEO is in its ability to organically raise a business’s appearance, pulling in spectators genuinely intrigued in what’s on presentation, and forming meaningful connections that guide to enduring connections.
In a world where online standing often commands triumph, having a tailored SEO plan is similar to having a chief solution to the online city. Every tweak and change made by SEO experts isn’t only about satisfying algorithms, but more significantly, about grasping and serving to individual motions and necessities. The final goal? To cooperatively unite a company with its perfect market, promoting development, trust, and prolonged victory. In this effort, SEO methods demonstrate to be not only useful, but essential.
Отчётность.
Оплата: Yoo.Money, Bitcoin, МИР, Visa, MasterCard…
Принимаем USDT
Телега: @exrumer
Skype: Loves.Ltd
https://xrumer.cc/
Только этот.
werankcities.com
Hongzhi 황제는 Xiao Jing을 보았고 Xiao Jing은 고개를 끄덕이며 “말하자”라고 기침했습니다.그렇게 말하며 소매 속에 준비해두었던 증기선에 대한 보고서를 꺼냈다.
k8 カジノ vip
このトピックにこんなに深く掘り下げる記事は他にないと思います。素晴らしいです!
Hi! Would you mind if I share your blog with my zynga group? There’s a lot of folks that I think would really enjoy your content. Please let me know. Thanks
http://idiplomanys.com/
Wow that was odd. I just wrote an very long comment but after I clicked submit my comment didn’t show up. Grrrr… well I’m not writing all that over again. Anyhow, just wanted to say wonderful blog!
http://www.idiplomanys.com/
В наше время, когда диплом становится началом успешной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Факт наличия официального документа об образовании переоценить невозможно. Ведь диплом открывает двери перед людьми, стремящимися вступить в профессиональное сообщество или продолжить обучение в ВУЗе.
Наша компания предлагает оперативно получить любой необходимый документ. Вы имеете возможность заказать диплом, и это является выгодным решением для всех, кто не смог завершить образование, утратил документ или хочет исправить плохие оценки. Любой диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим нюансам, чтобы на выходе получился документ, максимально соответствующий оригиналу.
Превосходство такого решения состоит не только в том, что можно быстро получить свой диплом. Весь процесс организовывается комфортно и легко, с нашей поддержкой. От выбора необходимого образца до консультаций по заполнению личных данных и доставки в любой регион России — все под абсолютным контролем опытных специалистов.
Для всех, кто пытается найти максимально быстрый способ получения требуемого документа, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к достижению собственных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://www.6landik-diploms.com/
В современном мире, где диплом является началом успешной карьеры в любой отрасли, многие стараются найти максимально простой путь получения образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает дверь перед каждым человеком, который хочет начать профессиональную деятельность или продолжить обучение в высшем учебном заведении.
Мы предлагаем очень быстро получить этот необходимый документ. Вы сможете купить диплом, и это будет отличным решением для человека, который не смог закончить обучение, утратил документ или хочет исправить свои оценки. Любой диплом изготавливается аккуратно, с максимальным вниманием ко всем нюансам, чтобы на выходе получился документ, 100% соответствующий оригиналу.
Преимущество данного решения заключается не только в том, что можно максимально быстро получить свой диплом. Процесс организовывается удобно, с нашей поддержкой. От выбора подходящего образца до грамотного заполнения персональных данных и доставки в любое место России — все под полным контролем наших мастеров.
Для тех, кто хочет найти быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать длительного процесса обучения и не теряя времени переходить к достижению собственных целей, будь то поступление в университет или начало карьеры.
http://6landik-diploms.com/
Placing the site on quality trust sites
Obtaining a place for your site on trustworthy, high-quality platforms is an invaluable step in reinforcing its online reputation. Such placements not only lift a site’s authority in the perspective of online search tools but also foster credibility among its desired audience. Trusted sites, celebrated for their rigorous content rules and considerable user bases, act as verifiers, supporting the genuineness and worth of content they link to or host. As visitors from these platforms work their way to a site, they arrive with a pre-established sense of trust, making them more likely to engage and convert.
What’s more, positioning a website on quality trust sites moves beyond just link-building methods. It’s an opportunity for reciprocal growth and cooperation. Connecting with honored platforms lets a brand to align its standards with those of the reliable site, fostering shared ideals and fortifying its brand narrative. In the always developing digital world, where truthfulness is at a high, such calculated placements secure a site’s lasting visibility, significance, and sway.
Отчётность.
Оплата: Yoo.Деньги, Bitcoin, МИР, Visa, MasterCard…
Принимаем USDT
Telgrm: @exrumer
Skype: Loves.Ltd
https://xrumer.cc/
Только этот.
tintucnamdinh24h.com
“오.” Zhu Houzhao는 날카롭게 고개를 끄덕였습니다. “안 가세요?”
프라그마틱 게임은 선도적인 iGaming 콘텐츠 제공 업체입니다. 모바일 중심 다양한 포트폴리오와 고품질 엔터테인먼트를 자랑합니다.
프라그마틱 게임
프라그마틱의 빙고 게임은 항상 즐겁고 흥미로워요. 여기에서 더 많은 빙고 게임 정보를 확인하세요!
https://www.12315lu.cn/
https://ycbgl.com/link/
https://www.12315mv.cn/
thewiin.com
두 사람은 땅에 착지하고 떠 다니는 풍선을 남겨두고 생명이없는 것처럼 달렸습니다.
프라그마틱플레이의 홈페이지에서 최신 슬롯을 만나보세요.
pragmatic
프라그마틱의 게임은 항상 다양한 테마로 놀라워요. 이 사이트에서 더 자세한 정보를 찾아보세요!
https://toploansbadcredit.com
https://hrbyszs.com/link/
https://doggggqwe.weebly.com/
沖ドキ!
実用的で具体的なアドバイスが多く、とても感謝しています。
Сайт, где можно купить надежные мини раковина с тумбой для туалета по выгодным ценам.
Thanks for another informative website. Where else may just I am getting that type of information written in such an ideal means? I have a venture that I am simply now operating on, and I have been on the look out for such info.
знакомства без регистрации
Aw, this was an exceptionally good post. Spending some time and actual effort to make a top notch article… but what can I say… I procrastinate a lot and don’t manage to get anything done.
лучшие сайты знакомств в России
thephotoretouch.com
과거에는 모든 사절이 Honglu Temple에있었습니다.
프라그마틱 슬롯에 대한 설명 감사합니다! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 이야기 나누면서 더 많은 지식을 얻어가요!
프라그마틱슬롯
프라그마틱 슬롯에 대한 글 정말 잘 읽었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 얻을 수 있어요. 함께 교류하며 더 많은 지식을 얻어보세요!
https://www.mods4me.com
https://www.12315qo.cn/
https://www.12315amb.cn/
Купить Свидетельство О Рождении Ссср
Купить Свидетельство О Рождении Ссср
Наличие любого диплома о высшем образовании является важным преимуществом при трудоустройстве. Основания для восстановления или аннулирования записи акта гражданского состояния. Оценочный лист к диплому специалиста любого ВУЗа Украины Диплом специалиста любого украинского ВУЗа Документ о высшем образовании достаточно сильно влияет на окончательное решение работодателя. Заполнение осуществляется по стандартам Минобрнауки, ставится мокрая печать учебного заведения, подпись ректора и председателя аттестационной комиссии, образовательная программа соответствует купленному Свидетельство направлению. Купить диплом вуза с гарантией безопасности недорого и полной анонимностью, то что предлагает наша оорганизация. Строители, электрики, маляры, штукатуры, швеи, слесари, парикмахеры, продавцы, повара, токари зачастую обучаются непосредственно на рабочих местах, приобретая при этом намного больше полезных знаний и навыков, чем в учебных классах. Кроме того, если документ необходим именно для продвижения по службе, стоит учитывать, что обучение длится не менее четырех-пяти лет, все это время не получится получить желаемую должность.
http://www.russkiy365-diploms-srednee.ru
Купить Аттестат 9 Класс С Реестром
В рожденье пяти минут перезвонит менеджер компании, купит Свидетельство детали, подготовит и отправит предварительный макет диплома института для подробного ознакомления. Если вы только собираетесь сдавать экзамены в школе, то ориентируйтесь на них, ведь как правило, именно они будут решающим звеном, так как именно от аттестата во многом зависит будущее. Ребята скажите пожалуйста, проходит все проверки, бланк имеет все элементы защиты и проходит любые проверки, я сегодня встретилась с вашим курьером, получила диплом. Наши специалисты являются прекрасными профессионалами, поэтому легко справятся с любой, даже самой трудной, задачей.
Купить Диплом О Среднем Образовании В Казани
Первый это оригинальные бланки ГОЗНАКа, а вторые типографские, которые по сути являют собой точную копию оригинала. При поступлении на заочное обучение или на магистратуру потребуется сдать вступительные экзамены в виде ЕГЭ. Высшее образование в Белорусском государственном университете студенты получают по 61 специальности первой ступени высшего образования и 65 специальностям – второй. Если вы хотите стать профессионалом сразу в нескольких сферах, попробовать себя в нескольких специальностях и купить Свидетельство О Рождении Ссср, что вам больше подходит, не тратя деньги на дорогостоящее высшее образование, мы предоставляем вам такую возможность.
Купить Диплом О Среднем Мед Образовании
Купить Диплом О Среднем Мед Образовании
А к тому моменту, когда он готов к работе, его специальность уже никому не нужна. Аттестат школы выдается студентам, успешно завершившим обучение в средней школе, в частности, в 9-м и 11-м классах. Мы купимте Диплом О Среднем Мед Образовании изготовление свидетельства на оригинальном бланке ГОСЗНАК с присутствием подписей и печатей установленного образца. Специалисты выполнят макет документа, мы отправим его вам на электронную почту для проверки качества исполнения и правильности заполнений полей документа. К несчастью, в нашей стране качество образования и обучения далеко не всегда подходит общепринятым нормам.
http://www.russkiy365-diploms-srednee.ru
Нотариальное Согласие Супруга
Большое количество домашних заданий, куча пар, совмещение учебы с профессией такую неимоверную нагрузку выдержит не каждый. Существует несколько способов приобретения высшего образования в России. Одни из основных плюсов наличия диплома техникума заключается в достаточно высокой заработной плате, на которую может рассчитывать даже специалист без опыта. Увы, но прийти в ВУЗ и сказать хочу диплом нельзя, собственно как и официально купить диплом в ВУЗе за 2 дня, даже являясь дипломом любого ВУЗа Москвы.
Цена Диплом Юриста
А вот если после школы пойти на работу по выбранной специальности, овладеть ею, приобрести практические навыки, тогда можно подумать о покупке корочки. На протяжении многих лет мы занимаемся продажей дипломов и предоставляем только подлинные документы. Обстоятельства не оставляли выбора, пришлось купить диплом об окончании высшего образования.
Какие Бывают Дипломы В Школе
Какие Бывают Дипломы В Школе
Им важно лишь наличие диплома, который мы готовы предоставить, выбирают проведенные документы на бланках гознак, наших клиентов остаются довольными после оказания наших школ. Общий порядок предполагает, что, когда человек получит РВП и проживет по нему в России минимум восемь месяцев, он сможет подать документы на ВНЖ. Можно претендовать на руководящий пост или рассчитывать на повышения по карьере. А классическая школа выглядит так: 4 года обучения в бакалавриате, а затем 1,5 или 2 года, в зависимости от формы обучения в магистратуре, куда поступить, не став бакалавром, невозможно. Во-вторых, вы можете открыть собственное дело, так как некоторые виды бизнесов требуют соответствующего образования. Но можно с уверенностью утверждать, что знания, полученные в высшем или среднем специальном учебном заведении, крайне редко востребованы на практике.
https://russkiy365-diploms-srednee.ru/
Купить Диплом Специалиста Недорого
Это не только позволяет избежать множества проблем, но и даёт возможность зарабатывать деньги в свободное от учебы время. Для получения документа об образовании не всегда требуется высокий уровень знаний. Во-первых, у нас можно выбрать не только вуз и специальность, но и оценки, предметы, темы курсовых и дипломного проекта, количество часов.
Купить Диплом О Высшем Образовании В Оренбурге
Сегодня есть множество сайтов, которые специализируются на продаже дипломов. И в таком случае выход только один купить диплом техникума (колледжа) или купить диплом вуза. Наши специалисты во время изготовления документа несколько раз проверяют правильность его заполнения.
Купить Диплом В Иваново
Соискатель получит данную информацию в течение десяти дней, узнать, готов ли диплом кандидата наук можно здесь, эти сведения можно также найти на официальном ресурсе вак. Никто не будет проверять дипломы, если четко выполнять возложенные обязанности. Иными словами, обладатель документа повышает свою ценность и значимость на современном рынке труда. Документ необходим для того, чтобы подтвердить наличие оконченного высшего профессионального образования и присвоение квалификации дипломированный специалист. За прошедшие годы настоящий диплом мог потеряться, выйти из строя, погибнуть в стихийном бедствии, просто бесследно исчезнуть. Однако получить “корочку” института или ВУЗа с задержкой не так приятно. Не каждый человек ответит на вопрос, кто автор известного выражения Без бумажки, ты букашка, но вряд ли кто-то станет отрицать его сакральный смысл.
http://www.russkiy365-diploms-srednee.ru
Сколько Стоит Сделать Диплом
Мы штат опытных специалистов, у которых в приоритете клиент и его проблема. Диплом бакалавра открывает возможности перед владельцем, например, купить Диплом В Иваново обучение уровнем выше, устроиться на работу по выбранному профилю, открыть собственное дело и уехать работать за границу. Обманутые заказчики не пойдут в полицию, поэтому злоумышленники ничем не рискуют.
Купить Диплом Магистра
Однако, в этом вопросе есть несколько нюансов, которые нужно учесть. Такие навыки являются необходимыми в сфере экономических отношений. Сколько стоит диплом колледжа государственного образца в Казахстане. После 9 классов диплом приравнивается к аттестату о полном среднем образовании, но с профессиональным уклоном.
What’s up to all, as I am truly eager of reading this webpage’s post to be updated regularly. It contains fastidious material.
1вин
thephotoretouch.com
Wang Shouren의 귀에서 윙윙 거리는 발사체.
27-го июля был проведён первый тест эффективности XRumer 23 StrongAI (по сравнению с текущей версией XRumer 19.0.18), который показал двадцатикратный прирост. Столь впечатляющие результаты достигнуты благодаря многократному улучшению пробиваемости уже известных платформ, добавлению поддержки новых платформ, обучению сотням тысяч типов новых тексткапч, внедрению нового GPT-модуля, и множеству других улучшений. По ссылке ниже Вы сможете изучить и скачать детали теста (исходная база, кусок отчёта, статистику и другие подробности)
Спасибо за рефку:
http://www.botmasterru.com/product109120/
What a stuff of un-ambiguity and preserveness of valuable knowledge about unpredicted emotions.
http://blackforest.mybb.ru/viewtopic.php?id=478#p706
http://kaktotak.0pk.me/viewtopic.php?id=14010#p33846
https://www.cyberpinoy.net/read-blog/20656
https://4atex.ru/blogs/1332/пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ
http://carpomania.mybb.ru/viewtopic.php?id=270#p566
Team up with our community of beyond 2 million light-hearted users who are extenuatory span and getting paid faster with our mobile invoice solutions. Our movable invoicing tools, including movable invoice apps, pander to to both iOS and Android users. Whether you have occasion for an invoice app concerning iPhone, an Android invoice app, or a sweeping versatile billing software, we’ve got you covered.
https://www.genio.ac/mobile-invoices
XRumer – это наиболее популярное равным образом эффективное программное обеспечение для создания гиперссылок да продвижения сайтов. Оно используется для автоматического прогона веб- сайтов помощью различные соц сети, форумы, онлайн-дневники также другие интернет-ресурсы.
http://www.botmasterru.com/product33230/
Buy XRumer Official
This is very interesting, You’re a very skilled blogger. I’ve joined your rss feed and look forward to seeking more of your magnificent post. Also, I’ve shared your web site in my social networks!
http://www.kupitediplom0029.ru/
This is very interesting, You are a very skilled blogger. I have joined your feed and look forward to seeking more of your great post. Also, I’ve shared your website in my social networks!
https://diplomdoc.ru/
For the reason that the admin of this site is working, no doubt very shortly it will be famous, due to its quality contents.
https://www.kupitediplom0029.ru/
Hi there, this weekend is pleasant for me, because this point in time i am reading this great educational post here at my house.
http://https://diplomdoc.ru/
It’s remarkable in favor of me to have a web page, which is beneficial in favor of my know-how. thanks admin
https://diplomdoc.ru/
Pretty section of content. I just stumbled upon your site and in accession capital to assert that I acquire in fact enjoyed account your blog posts. Any way I will be subscribing to your feeds and even I achievement you access consistently fast.
http://https://diplomdoc.ru/
Wow, awesome weblog format! How lengthy have you been running a blog for? you make blogging look easy. The full look of your web site is great, let alone the content material!
http://kupitediplom0029.ru/
This is really fascinating, You are a very professional blogger. I’ve joined your rss feed and stay up for in the hunt for extra of your excellent post. Additionally, I have shared your website in my social networks
http://www.diplommy.ru
Amazing issues here. I am very glad to peer your article. Thank you so much and I’m having a look ahead to contact you. Will you please drop me a mail?
http://www.matchfishing.ru/forumtest/1/member.php?u=23110
http://good-serial.ru/2024/05/diplom-v-vashih-rukah-kak-eto-rabotaet/
https://www.4x4zubry.by/forum/messages/forum1/topic1576/message176205/?result=new#message176205
http://sports.bestbb.ru/viewtopic.php?id=693#p1269
https://vip.rolevaya.info/viewtopic.php?id=4162#p7238
Приветствую, друзья!
Компания XRumer Inc предлагает свои профессиональные услуги СЕО продвижения.
Ваш портал, как можно отметить, только набирает обороты. Чтобы по максимуму ускорить процесс его роста, готовы предложить услуги по внешней СЕО-оптимизации. У нас имеются доступные и эффективные инструменты для SEO-специалистов. У наших специалистов значительный опыт в данной области, в арсенале имеются успешные кейсы – если интересно, покажем по запросу.
Наша компания предлагает скидку 10% до конца месяца.
Предлагаемые услуги:
– Трастовые ссылки (нужно всем сайтам) – от 1,5 до 5000 рублей
– Размещение трастовых безанкорных ссылок (полезно всем сайтам) – 3.900 руб
– Прогон на 110000 сайтов (RU.зона) – 2900 р
– 150 постов в VK про ваш сайт (поможет в рекламе) – 3900 р
– Публикация статей о вашем сайте на 300 форумах (очень мощная раскрутка вашего сайта) – 29.000 р
– МегаПостинг – это прогон на 3 млн ресурсов (сверхмощный прогон для ваших сайтов) – 39.900 руб
– Рассылаем сообщения по сайтам с помощью обратной связи – цена по договоренности, будет зависеть от объемов.
Обращайтесь по любым вопросам, всегда поможем.
Отчётность.
Оплата: Yoo.Money, Bitcoin, МИР, Visa, MasterCard…
Принимаем USDT
Telgrm: https://t.me/exrumer
Skype: Loves.Ltd
https://www.webfx.com/
https://xrumer.cc/
Только этот.
[url=http://www.botmasterru.com/product33230/][img]https://xrumer.cc/i/x.png[/img][/url]
For the reason that the admin of this website is working, no hesitation very rapidly it will be famous, due to its feature contents.
https://www.diplomdoc.ru/
Greate article. Keep writing such kind of info on your blog. Im really impressed by your blog.
Hi there, You have done an excellent job. I will certainly digg it and individually recommend to my friends. I am sure they will be benefited from this site.
diplommy.ru/
This is my first time pay a visit at here and i am in fact impressed to read all at alone place.
diplomdoc.ru/
Its like you read my mind! You seem to know so much about this, like you wrote the book in it or something. I think that you could do with a few pics to drive the message home a little bit, but instead of that, this is excellent blog. An excellent read. I’ll definitely be back.
diplommy.ru/
Hey there, I think your site might be having browser compatibility issues. When I look at your blog in Firefox, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, very good blog!
https://bicycle.one/create-blog/
http://skazka.g-talk.ru/viewtopic.php?f=1&t=1641
http://everton.rusff.me/viewtopic.php?id=224#p5479
http://npo.bestbb.ru/viewtopic.php?id=372#p1122
http://lossantosbattalion17.listbb.ru/viewtopic.php?f=3&t=273
lacolinaecuador.com
Fang Jifan은 엄숙하게 말했습니다. “폐하, 여전히 자비와 정의가 있습니다!”
Thanks for any other excellent article. Where else could anybody get that kind of info in such an ideal way of writing? I have a presentation next week, and I am at the search for such information.
http://bukmekerskayakontora.com.ua/forum/viewtopic.php?f=9&t=70571
https://yexanin202.ixbb.ru/viewtopic.php?id=8#p8
https://vip.7bb.ru/viewtopic.php?id=2683#p6562
https://bryansk32-forum.ru/ucp.php?mode=login
http://ukrevent.ru/uspeh-v-vashih-rukah-kupit-diplom-i-nachat-novuyu-zhizn/
На сегодняшний день, когда диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Наличие официального документа об образовании сложно переоценить. Ведь именно диплом открывает дверь перед всеми, кто стремится вступить в сообщество профессионалов или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает максимально быстро получить этот важный документ. Вы можете заказать диплом старого или нового образца, что становится отличным решением для человека, который не смог закончить обучение, потерял документ или желает исправить плохие оценки. дипломы выпускаются аккуратно, с особым вниманием ко всем нюансам. На выходе вы сможете получить продукт, максимально соответствующий оригиналу.
Плюсы подобного подхода состоят не только в том, что можно оперативно получить диплом. Процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора нужного образца до консультации по заполнению персональной информации и доставки в любой регион страны — все под абсолютным контролем наших мастеров.
Всем, кто ищет оперативный способ получить необходимый документ, наша услуга предлагает отличное решение. Купить диплом – значит избежать долгого процесса обучения и сразу перейти к личным целям: к поступлению в университет или к началу трудовой карьеры.
sibsocio.ru
This is very interesting, You’re a very skilled blogger. I have joined your rss feed and look forward to seeking more of your excellent post. Also, I’ve shared your site in my social networks!
diplompro.ru
Hey there! Do you know if they make any plugins to assist with SEO? I’m trying to get my blog to rank for some targeted keywords but I’m not seeing very good results. If you know of any please share. Thank you!
http://https://diplompro.ru/
Hey! This is kind of off topic but I need some advice from an established blog. Is it very difficult to set up your own blog? I’m not very techincal but I can figure things out pretty fast. I’m thinking about making my own but I’m not sure where to begin. Do you have any tips or suggestions? Appreciate it
http://https://diplompro.ru/
Hi there to every one, the contents existing at this web page are really remarkable for people experience, well, keep up the nice work fellows.
http://www.diplom-zakaz.ru/
Right here is the right site for anybody who wishes to understand this topic. You realize so much its almost hard to argue with you (not that I personally will need to…HaHa). You definitely put a brand new spin on a subject that’s been written about for decades. Excellent stuff, just great!
https://www.peoplediplom.ru/
What’s up, this weekend is good in favor of me, as this point in time i am reading this enormous educational article here at my house.
http://www.peoplediplom.ru/
It’s remarkable to visit this site and reading the views of all friends regarding this post, while I am also zealous of getting know-how.
https://diplom-profi.ru/
What a material of un-ambiguity and preserveness of valuable know-how regarding unpredicted emotions.
http://www.institute-diplom.ru
you are really a good webmaster. The website loading velocity is amazing. It sort of feels that you are doing any unique trick. Also, The contents are masterpiece. you have performed a fantastic job on this topic!
bisness-diplom.ru/
Hi this is kind of of off topic but I was wanting to know if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding knowledge so I wanted to get guidance from someone with experience. Any help would be greatly appreciated!
bisness-diplom.ru/
Hi are using WordPress for your blog platform? I’m new to the blog world but I’m trying to get started and create my own. Do you require any html coding knowledge to make your own blog? Any help would be greatly appreciated!
http://www.bisness-diplom.ru/
Do you have a spam issue on this site; I also am a blogger, and I was wondering your situation; many of us have developed some nice practices and we are looking to exchange techniques with other folks, please shoot me an e-mail if interested.
https://zakaz-na-diplom.ru/
Hey I know this is off topic but I was wondering if you knew of any widgets I could add to my blog that automatically tweet my newest twitter updates. I’ve been looking for a plug-in like this for quite some time and was hoping maybe you would have some experience with something like this. Please let me know if you run into anything. I truly enjoy reading your blog and I look forward to your new updates.
http://molbiol.ru/forums/index.php?showtopic=1114110
http://bam.mybb.ru/viewtopic.php?id=721#p2291
http://charm.rolbb.me/viewtopic.php?id=548#p7355
http://aliceinwonderland.mybb.ru/viewtopic.php?id=921#p6853
https://deviva.ru/viewtopic.php?id=8513#p61326
Hi there! Do you use Twitter? I’d like to follow you if that would be okay. I’m definitely enjoying your blog and look forward to new posts.
http://www.kupite-diplom0024.ru/
Thanks for every other excellent post. Where else may anybody get that type of info in such an ideal manner of writing? I have a presentation next week, and I’m at the search for such info.
sdiplom.ru/
Hi there, after reading this amazing piece of writing i am too delighted to share my knowledge here with friends.
https://www.sdiplom.ru/
Hey there! This post could not be written any better! Reading through this post reminds me of my good old room mate! He always kept talking about this. I will forward this post to him. Pretty sure he will have a good read. Thank you for sharing!
https://diplom-city24.ru/
That is really fascinating, You are a very professional blogger. I’ve joined your feed and look ahead to seeking extra of your magnificent post. Additionally, I’ve shared your website in my social networks
peoplediplom.ru
Leap into the crowd of high-efficiency invoicing with our top-notch invoice maker app, just right in the interest the go-getters – freelancers and small province owners alike. We’re talking wide transforming the break down you run bills, making it mountebank than ever.
https://apps.apple.com/us/app/invoice-maker-invoices/id6449437040
Hmm is anyone else experiencing problems with the images on this blog loading? I’m trying to figure out if its a problem on my end or if it’s the blog. Any feed-back would be greatly appreciated.
http://https://diplom45.ru/
Hi there colleagues, its wonderful post on the topic of tutoringand fully explained, keep it up all the time.
https://diplom45.ru/
It’s remarkable to pay a quick visit this web site and reading the views of all mates concerning this post, while I am also keen of getting knowledge.
http://www.diplom45.ru
An impressive share! I’ve just forwarded this onto a coworker who was conducting a little research on this. And he in fact bought me dinner due to the fact that I discovered it for him… lol. So allow me to reword this…. Thank YOU for the meal!! But yeah, thanx for spending the time to talk about this topic here on your web site.
http://www.kdiplom.ru
Wow that was odd. I just wrote an incredibly long comment but after I clicked submit my comment didn’t appear. Grrrr… well I’m not writing all that over again. Anyway, just wanted to say fantastic blog!
kdiplom.ru
It’s amazing for me to have a web site, which is helpful designed for my know-how. thanks admin
http://https://kupite-diplom0024.ru/
프라그마틱 플레이의 슬롯으로 즐거운 게임 여행을 떠나보세요.
프라그마틱 무료
프라그마틱 슬롯에 대한 내용이 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 찾아보실 수 있어요. 함께 지식을 공유해보세요!
https://ahjdmt.com/hot/
https://www.eco2home.com
https://supervil.com/hot/
This is very interesting, You’re a very skilled blogger. I’ve joined your feed and look forward to seeking more of your great post. Also, I’ve shared your web site in my social networks!
https://www.diplom-city24.ru/
Hello every one, here every person is sharing these know-how, so it’s fastidious to read this blog, and I used to pay a visit this weblog daily.
http://www.diplom-city24.ru/
Can I just say what a comfort to discover somebody who truly understands what they’re talking about on the web. You definitely realize how to bring a problem to light and make it important. A lot more people need to check this out and understand this side of the story. It’s surprising you’re not more popular given that you certainly have the gift.
http://zakaz-na-diplom.ru/
For latest news you have to pay a quick visit internet and on the web I found this site as a finest web site for newest updates.
https://www.zakaz-na-diplom.ru/
For most up-to-date news you have to pay a visit the web and on the web I found this web site as a best site for hottest updates.
https://www.institute-diplom.ru/
Yesterday, while I was at work, my sister stole my iphone and tested to see if it can survive a thirty foot drop, just so she can be a youtube sensation. My apple ipad is now broken and she has 83 views. I know this is totally off topic but I had to share it with someone!
http://institute-diplom.ru/
What’s up to all, how is the whole thing, I think every one is getting more from this site, and your views are nice in support of new visitors.
https://diplompro.ru/
An impressive share! I have just forwarded this onto a friend who was conducting a little research on this. And he actually ordered me lunch due to the fact that I discovered it for him… lol. So allow me to reword this…. Thank YOU for the meal!! But yeah, thanks for spending time to talk about this issue here on your website.
http://www.diplompro.ru/
Greetings I am so happy I found your website, I really found you by error, while I was searching on Bing for something else, Nonetheless I am here now and would just like to say kudos for a remarkable post and a all round exciting blog (I also love the theme/design), I don’t have time to look over it all at the minute but I have saved it and also added in your RSS feeds, so when I have time I will be back to read more, Please do keep up the awesome work.
diplompro.ru/
What’s up Dear, are you in fact visiting this website on a regular basis, if so afterward you will without doubt get good experience.
https://www.diplom-kuplu.ru/
라이즈 오브 올림푸스 100
“이렇게 부서졌나요?” 홍지 황제가 날카롭게 물었다.
Hi there very nice blog!! Guy .. Beautiful .. Amazing .. I will bookmark your web site and take the feeds additionally? I’m happy to find a lot of useful information here within the put up, we need develop extra techniques on this regard, thank you for sharing. . . . . .
http://androidinweb.ru/unikalnaya-vozmozhnost-kupi-diplom-i-dostigni-svoih-tseley-byistree
http://forum.lazarevskaya.ru/showthread.php?p=295069#post295069
http://mykinotime.ru/diplom-na-tvoih-usloviyah-priobreti-obrazovanie-kotoroe-podhodit-imenno-tebe/
Hello there! Do you use Twitter? I’d like to follow you if that would be okay. I’m undoubtedly enjoying your blog and look forward to new posts.
http://1-click.pl/excelforum/viewtopic.php?f=3&t=173582
https://ya.forum.cool/viewtopic.php?id=5606#p16878
http://www.forum.7x.ru/member.php?u=16498
It’s remarkable in support of me to have a website, which is good for my experience. thanks admin
https://sontopic.com/blogs/1563/ƒиплом-на-твоих-услови€х-ѕриобрети-образование-которое-подходит-именно-тебе
http://forum.safe-animals.ru/index.php?showtopic=26600
http://ilovehabbo.bbon.ru/viewtopic.php?id=1369#p28744
I used to be able to find good advice from your blog articles.
https://1abakan.ru/forum/showthread-111051/
http://myweektour.ru/put-k-professionalnomu-uspehu-priobreti-diplom-i-stan-ekspertom/
https://chayka.ixbb.ru/viewtopic.php?id=52#p62
Hi! I know this is kinda off topic nevertheless I’d figured I’d ask. Would you be interested in trading links or maybe guest writing a blog article or vice-versa? My website goes over a lot of the same subjects as yours and I believe we could greatly benefit from each other. If you are interested feel free to send me an email. I look forward to hearing from you! Wonderful blog by the way!
https://vip.9bb.ru/viewtopic.php?id=5489#p9147
https://medium.com/@oximus777/чрезвычайно-многих-цивилизованных-людей-сейчас-интересует-сложный-вопрос-как-не-совершить-ошибки-7d762419932d
http://domovou.3nx.ru/viewtopic.php?p=5584#5584
크립토 골드
Zhu Houzhao는 말없이 돌아서서 달아났고, 쉭쉭거리는 소리와 함께 슬그머니 사라졌습니다…
This is my first time pay a quick visit at here and i am in fact happy to read all at one place.
http://elfae.ruhelp.com/viewtopic.php?id=15954#p37885
http://backshowtime.ru/poluchite-diplom-kotoryiy-podtverdit-vashu-kompetentnost-i-opyit-legko-i-byistro
http://ukrevents.ru/eksklyuzivnyiy-dostup-k-obrazovaniyu-gde-kupit-diplom-legalno/
Just desire to say your article is as amazing. The clarity in your post is just excellent and i could assume you’re an expert on this subject. Well with your permission allow me to grab your RSS feed to keep updated with forthcoming post. Thanks a million and please carry on the rewarding work.
http://www.magazin-diplomov.ru
선도적인 iGaming 콘텐츠 제공 업체인 최신 프라그마틱 게임은 혁신적이고 표준화된 콘텐츠를 통해 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 고객에게 제공합니다.
프라그마틱 게임
프라그마틱 관련 정보 감사합니다! 제 사이트에서도 유용한 정보를 공유하고 있어요. 함께 소통하면서 발전하는 모습 기대합니다!
https://munirahkasim.com/link/
https://my-site-104812-108385.weebly.com/
https://hquwhequiw.weebly.com/
Howdy! This is kind of off topic but I need some guidance from an established blog. Is it very difficult to set up your own blog? I’m not very techincal but I can figure things out pretty fast. I’m thinking about creating my own but I’m not sure where to start. Do you have any tips or suggestions? Thanks
http://financeokey.ru/page/5
http://onlinekinospace.ru/hotite-kupit-diplom-myi-zdes-dlya-vas
https://letusbookmark.com/story18438191/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%9C%D0%BE%D1%81%D0%BA%D0%B2%D0%B5
https://www.leukshoppen.nl/product/black-pants/
https://bookmarksparkle.com/story17136231/%D0%B3%D0%B4%D0%B5-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC
В нашем мире, где диплом – это начало удачной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Необходимость наличия официального документа об образовании сложно переоценить. Ведь диплом открывает двери перед всеми, кто собирается вступить в профессиональное сообщество или учиться в университете.
В данном контексте наша компания предлагает оперативно получить этот важный документ. Вы сможете приобрести диплом, и это является отличным решением для человека, который не смог завершить образование или утратил документ. дипломы производятся аккуратно, с особым вниманием к мельчайшим нюансам. В результате вы сможете получить документ, 100% соответствующий оригиналу.
Преимущество данного подхода заключается не только в том, что вы сможете быстро получить диплом. Процесс организован просто и легко, с нашей поддержкой. Начиная от выбора нужного образца до консультации по заполнению персональной информации и доставки по России — все под полным контролем качественных специалистов.
В итоге, всем, кто ищет быстрый и простой способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – это значит избежать длительного обучения и сразу перейти к достижению собственных целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://diplomsagroups.com/diplomy-po-specialnosti/diplom-stomatologa.html
НА интернете большое цифра анализаторов веб-сайта, штрих опуса тот или другой напрямую зависит через тарифных планов.
https://be1.ru/ – дауну
크립토 골드
그러나 곧 그는 홍지황제의 시선이 자신을 엄하게 바라보고 있음을 알아차렸는데…
Aw, this was an incredibly good post. Spending some time and actual effort to create a great article… but what can I say… I procrastinate a whole lot and don’t seem to get anything done.
https://rostovbike.ru/user-1902.html
http://www.vespaclubnapoli.it/forum/viewthread.php?forum_id=4&thread_id=25818
https://www.vocal.com.ua/blog?page=1
http://dp-spb.com/forum/thread-2086/
https://travistb457.angelinsblog.com/26817905/%D0%94%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D1%8B-%D0%B7%D0%B0-%D0%B4%D0%B5%D0%BD%D1%8C%D0%B3%D0%B8-%D0%9F%D1%80%D0%B0%D0%B2%D0%B4%D0%B0-%D0%BE-%D0%9F%D0%BE%D0%BA%D1%83%D0%BF%D0%BA%D0%B5-%D0%9E%D0%B1%D1%80%D0%B0%D0%B7%D0%BE%D0%B2%D0%B0%D0%BD%D0%B8%D1%8F-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC
Amazing! Its in fact awesome piece of writing, I have got much clear idea on the topic of from this article.
https://atozbookmark.com/story17014689/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B0%D1%82%D1%82%D0%B5%D1%81%D1%82%D0%B0%D1%82-%D0%B7%D0%B0-9-%D0%BA%D0%BB%D0%B0%D1%81%D1%81
https://motorcyclereviews93704.blog5star.com/27256790/marketing-things-to-know-before-you-buy
https://privatebookmark.com/story17168144/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%A3%D1%84%D0%B0
https://mypresspage.com/story2452773/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2%D1%80%D0%B0%D1%87%D0%B0
https://7prbookmarks.com/story17111334/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%81%D0%B0%D0%BD%D1%82%D0%B5%D1%85%D0%BD%D0%B8%D0%BA%D0%B0
Hi there, after reading this amazing piece of writing i am also cheerful to share my know-how here with mates.
http://forexsnews.ru/obrazovanie-bez-granits-gde-kupit-diplom-i-stat-spetsialistom-mirovogo-urovnya
https://chefrobertsdirect.com/_404.html
https://www.fabriziogiaconia.it/2020/01/28/test-2/
https://www.rosacruz.cl/slider5/
https://babygirlboyname.com/
В современном мире, где диплом – это начало удачной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа об образовании трудно переоценить. Ведь диплом открывает двери перед каждым человеком, желающим начать трудовую деятельность или продолжить обучение в высшем учебном заведении.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы имеете возможность купить диплом нового или старого образца, что становится удачным решением для человека, который не смог закончить обучение или утратил документ. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим деталям, чтобы на выходе получился документ, полностью соответствующий оригиналу.
Превосходство такого подхода заключается не только в том, что вы сможете быстро получить диплом. Весь процесс организован комфортно, с нашей поддержкой. Начав от выбора требуемого образца до точного заполнения персональной информации и доставки в любой регион России — все под абсолютным контролем опытных специалистов.
Для тех, кто ищет быстрый и простой способ получения требуемого документа, наша компания предлагает выгодное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени переходить к достижению личных целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://diplomsagroups.com/kupit-diplom-vuza/diplom-instituta.html
В нашем обществе, где диплом – это начало удачной карьеры в любой сфере, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании переоценить невозможно. Ведь именно диплом открывает двери перед любым человеком, который собирается начать профессиональную деятельность или продолжить обучение в каком-либо ВУЗе.
Наша компания предлагает очень быстро получить этот важный документ. Вы сможете купить диплом старого или нового образца, и это становится выгодным решением для всех, кто не смог закончить обучение или утратил документ. Все дипломы производятся с особой аккуратностью, вниманием ко всем элементам, чтобы в результате получился продукт, 100% соответствующий оригиналу.
Преимущества данного подхода заключаются не только в том, что вы сможете оперативно получить свой диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора необходимого образца до консультации по заполнению персональных данных и доставки в любое место страны — все будет находиться под абсолютным контролем опытных специалистов.
Всем, кто ищет быстрый способ получения требуемого документа, наша компания предлагает отличное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и сразу переходить к своим целям, будь то поступление в университет или начало удачной карьеры.
http://diplomsagroups.com/kupit-diplom-v-gorode/voronezh.html
Цены на покупку диплома могут сильно варьироваться в зависимости от уровня образования,
специальности и страны, в которой был выдан диплом. Обычно цена за покупку диплома колеблется от нескольких сотен до нескольких тысяч долларов. Однако следует помнить, что покупка диплома является незаконной и может иметь серьезные последствия, включая уголовную ответственность.
Рекомендуется получить диплом легальным путем мы вам помежем получить диплоиы такие как
купить диплом агронома через обучение в учебном заведении.
https://russdiplomik.com/diplomyi-bakalavra.html
123
Hello to all, the contents existing at this web page are in fact amazing for people knowledge, well, keep up the good work fellows.
https://bookmarksea.com/story17028378/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%A1%D0%B0%D1%80%D0%B0%D1%82%D0%BE%D0%B2
https://moolookoo.ru/content/diplomansycom
http://ufawedding.ru/article
https://wise-social.com/story2458734/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%81%D1%82%D1%80%D0%BE%D0%B8%D1%82%D0%B5%D0%BB%D1%8F
https://import-moto.com/users/88?page=3
В нашем обществе, где диплом – это начало удачной карьеры в любом направлении, многие пытаются найти максимально простой путь получения качественного образования. Факт наличия официального документа об образовании трудно переоценить. Ведь диплом открывает дверь перед любым человеком, желающим начать трудовую деятельность или учиться в высшем учебном заведении.
Предлагаем максимально быстро получить этот необходимый документ. Вы можете заказать диплом нового или старого образца, что становится отличным решением для всех, кто не смог закончить обучение или потерял документ. Все дипломы выпускаются аккуратно, с особым вниманием к мельчайшим деталям. В результате вы сможете получить полностью оригинальный документ.
Плюсы данного решения заключаются не только в том, что вы максимально быстро получите свой диплом. Весь процесс организовывается комфортно, с нашей поддержкой. Начиная от выбора требуемого образца диплома до консультаций по заполнению личной информации и доставки по России — все под полным контролем качественных специалистов.
Всем, кто пытается найти быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и сразу переходить к достижению собственных целей: к поступлению в университет или к началу трудовой карьеры.
https://motionentrance.edu.np/profile/elviraparks1/
https://www.fbioyf.unr.edu.ar/evirtual/tag/index.php?tc=1&tag=%D0%94%D0%B5%D0%BA%D0%B0%D0%BD%20%D0%B4%D0%BE%D1%86%D0%B5%D0%BD%D1%82%20%D0%BA%D1%83%D1%80%D0%B0%D1%82%D0%BE%D1%80%20%D0%BC%D0%B5%D1%82%D0%BE%D0%B4%D0%B8%D1%81%D1%82%20%D0%BA%D1%82%D0%BE%20%D0%B2%D1%81%D0%B5%20%D1%8D%D1%82%D0%B8%20%D0%BB%D1%8E%D0%B4%D0%B8%3F
https://feeder.topbb.ru/viewtopic.php?id=1823#p67171
http://onlineboxing.net/jforum/user/profile/288174.page
https://forum.zub-zub.ru/viewtopic.php?t=25579
Hey there! Do you know if they make any plugins to assist with SEO? I’m trying to get my blog to rank for some targeted keywords but I’m not seeing very good gains. If you know of any please share. Kudos!
https://okna-optimum.ru/
http://dev-forum.vmssoftware.com/viewtopic.php?t=21586&start=56480
https://daynghethanhnien.com/lien-he.html
https://social4geek.com/story2535189/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BE-%D1%81%D1%80%D0%B5%D0%B4%D0%BD%D0%B5%D0%BC-%D1%81%D0%BF%D0%B5%D1%86%D0%B8%D0%B0%D0%BB%D1%8C%D0%BD%D0%BE%D0%BC
https://bookmarks-hit.com/story17164911/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%81%D0%B0%D0%BD%D1%82%D0%B5%D1%85%D0%BD%D0%B8%D0%BA%D0%B0
Сегодня, когда диплом – это начало отличной карьеры в любом направлении, многие стараются найти максимально простой путь получения качественного образования. Наличие официального документа трудно переоценить. Ведь именно диплом открывает дверь перед людьми, желающими вступить в сообщество квалифицированных специалистов или продолжить обучение в ВУЗе.
Мы предлагаем очень быстро получить любой необходимый документ. Вы сможете приобрести диплом нового или старого образца, что становится выгодным решением для человека, который не смог закончить образование, утратил документ или желает исправить плохие оценки. дипломы изготавливаются аккуратно, с максимальным вниманием ко всем деталям. На выходе вы сможете получить документ, 100% соответствующий оригиналу.
Плюсы такого подхода заключаются не только в том, что можно оперативно получить диплом. Процесс организован комфортно, с профессиональной поддержкой. От выбора подходящего образца до консультации по заполнению персональных данных и доставки в любое место России — все под полным контролем качественных специалистов.
В итоге, для всех, кто ищет оперативный способ получения требуемого документа, наша компания предлагает отличное решение. Приобрести диплом – значит избежать продолжительного процесса обучения и сразу перейти к достижению своих целей: к поступлению в ВУЗ или к началу трудовой карьеры.
Купить диплом энергетика
you are in reality a good webmaster. The website loading pace is incredible. It kind of feels that you’re doing any distinctive trick. Furthermore, The contents are masterwork. you’ve performed a excellent activity on this subject!
https://joventic.uoc.edu/me-he-sacado-adelante-yo-misma/azul_280x320-3/
http://fat-girls.ru/category/bez-rubriki/page/5
http://deanonnic.listbb.ru/viewtopic.php?t=327
http://russianecuador.com/forum/member.php?tab=visitor_messaging&u=13207
http://dalgidromet.ru/
Hmm is anyone else encountering problems with the images on this blog loading? I’m trying to determine if its a problem on my end or if it’s the blog. Any feedback would be greatly appreciated.
https://bookmarkprobe.com/story17268629/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%A1%D0%B0%D0%BC%D0%B0%D1%80%D0%B5
https://messiahir146.onesmablog.com/%D0%A4%D0%B0%D0%BB%D1%8C%D1%88%D0%B8%D0%B2%D1%8B%D0%B9-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%9F%D0%BE%D1%87%D0%B5%D0%BC%D1%83-%D1%8D%D1%82%D0%BE-%D0%BD%D0%B5-%D0%A1%D1%82%D0%BE%D0%B8%D1%82-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-67322527
https://fitcommunitybbs.com/topic1494-kupit-diplom-santekhnika.html
https://socialdummies.com/story1860719/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BA%D0%BE%D0%BB%D0%BB%D0%B5%D0%B4%D0%B6%D0%B0
https://one-bookmark.com/story17030918/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%A3%D1%84%D0%B0
레프리칸 리치스
Yan Shenggong은 눈꺼풀을 살짝 들어 “누구의 편지입니까? “라고 물었습니다.
На сегодняшний день, когда диплом является началом успешной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает двери перед любым человеком, который хочет вступить в сообщество профессионалов или учиться в ВУЗе.
Предлагаем быстро получить любой необходимый документ. Вы можете заказать диплом, что является отличным решением для человека, который не смог закончить обучение, потерял документ или хочет исправить плохие оценки. диплом изготавливается с особой тщательностью, вниманием ко всем нюансам, чтобы в результате получился полностью оригинальный документ.
Преимущества подобного решения состоят не только в том, что можно максимально быстро получить диплом. Процесс организовывается просто и легко, с профессиональной поддержкой. От выбора необходимого образца до консультаций по заполнению личных данных и доставки в любое место России — все под полным контролем опытных специалистов.
Для всех, кто ищет оперативный способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – это значит избежать долгого процесса обучения и сразу переходить к достижению своих целей: к поступлению в ВУЗ или к началу успешной карьеры.
https://re-tracker.ru/index.php?showtopic=852
https://socialcompare.com/en/member/josearnold2-7i45t61v
https://borisoglebsk.net/modules.php?name=Forums&file=viewtopic&p=252299#252299
http://ukr.mybb.ru/viewtopic.php?id=386#p1022
http://ya.forum2.net/viewtopic.php?id=9375#p18795
В нашем мире, где диплом – это начало отличной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед людьми, желающими вступить в профессиональное сообщество или учиться в каком-либо ВУЗе.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы имеете возможность приобрести диплом, что становится удачным решением для всех, кто не смог завершить образование или утратил документ. диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим элементам. В результате вы получите продукт, 100% соответствующий оригиналу.
Плюсы данного решения заключаются не только в том, что вы сможете оперативно получить свой диплом. Процесс организовывается комфортно, с нашей поддержкой. От выбора необходимого образца до грамотного заполнения личных данных и доставки в любое место России — все находится под полным контролем наших мастеров.
Всем, кто хочет найти максимально быстрый способ получения требуемого документа, наша компания может предложить отличное решение. Заказать диплом – значит избежать долгого обучения и сразу переходить к важным целям: к поступлению в университет или к началу успешной карьеры.
http://energiyacosmosa.winbb.ru/viewtopic.php?id=1284#p5274
https://www.dermandar.com/user/JoseArnold2/
http://muz.mybb.ru/viewtopic.php?id=613#p1226
http://bukmeker.bbok.ru/viewtopic.php?id=25090
https://forum.zone-game.info/showthread.php?p=438833#post438833
That is very attention-grabbing, You are an excessively professional blogger. I have joined your feed and look forward to searching for more of your excellent post. Additionally, I have shared your web site in my social networks
@@G@@
https://berezniki.su/news/virtualnyj-nomer-kogda-mozhet-potrebovatsya-i-v-chem-preimushhestva-ih-ispolzovaniya
This info is invaluable. When can I find out more?
https://total-bookmark.com/story17009657/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BC%D0%B0%D0%B3%D0%B8%D1%81%D1%82%D1%80%D0%B0
https://bensbookmarks.com/cisco-certified-network-associate-security-ccna.htm
https://thebookmarkid.com/story17174676/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%A1%D0%B0%D1%80%D0%B0%D1%82%D0%BE%D0%B2
https://auto-sale-msk.ru/policy
https://finttech.ru/page/5
Hello there! Do you know if they make any plugins to safeguard against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any tips?
https://www.diplom-zakaz.ru/
Hello, yup this paragraph is really pleasant and I have learned lot of things from it concerning blogging. thanks.
https://diplom-zakaz.ru/
What’s up friends, nice post and nice arguments commented here, I am genuinely enjoying by these.
https://peoplediplom.ru/
В нашем мире, где диплом становится началом успешной карьеры в любом направлении, многие пытаются найти максимально простой путь получения образования. Наличие официального документа трудно переоценить. Ведь именно он открывает дверь перед всеми, кто собирается начать трудовую деятельность или продолжить обучение в каком-либо университете.
Предлагаем быстро получить этот важный документ. Вы сможете купить диплом, и это становится отличным решением для всех, кто не смог закончить обучение или потерял документ. Каждый диплом изготавливается аккуратно, с особым вниманием к мельчайшим элементам, чтобы на выходе получился полностью оригинальный документ.
Превосходство данного подхода заключается не только в том, что вы быстро получите диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. Начиная от выбора нужного образца диплома до правильного заполнения персональных данных и доставки по стране — все будет находиться под полным контролем квалифицированных мастеров.
Для всех, кто хочет найти оперативный способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – значит избежать долгого процесса обучения и сразу переходить к достижению своих целей, будь то поступление в университет или старт трудовой карьеры.
https://www.dermandar.com/user/JoseArnold2/
https://www.fbioyf.unr.edu.ar/evirtual/tag/index.php?tc=1&tag=%D0%94%D0%B5%D0%BA%D0%B0%D0%BD%20%D0%B4%D0%BE%D1%86%D0%B5%D0%BD%D1%82%20%D0%BA%D1%83%D1%80%D0%B0%D1%82%D0%BE%D1%80%20%D0%BC%D0%B5%D1%82%D0%BE%D0%B4%D0%B8%D1%81%D1%82%20%D0%BA%D1%82%D0%BE%20%D0%B2%D1%81%D0%B5%20%D1%8D%D1%82%D0%B8%20%D0%BB%D1%8E%D0%B4%D0%B8%3F
https://ucgp.jujuy.edu.ar/profile/LomaTapp3/
https://borisoglebsk.net/modules.php?name=Forums&file=viewtopic&p=252299#252299
https://doodleordie.com/profile/josearnold2
Hi! This post could not be written any better! Reading this post reminds me of my previous room mate! He always kept talking about this. I will forward this post to him. Pretty sure he will have a good read. Thanks for sharing!
http://www.peoplediplom.ru/
Hi, after reading this remarkable post i am too delighted to share my knowledge here with mates.
http://www.peoplediplom.ru/
What’s up everybody, here every person is sharing such familiarity, so it’s good to read this webpage, and I used to pay a quick visit this web site daily.
https://www.bisness-diplom.ru/
Thank you a lot for sharing this with all folks you actually recognise what you’re talking approximately! Bookmarked. Kindly additionally consult with my site =). We will have a hyperlink exchange contract among us
https://www.bisness-diplom.ru/
What’s up to every single one, it’s in fact a nice for me to go to see this website, it includes important Information.
http://www.kupite-diplom0024.ru/
Hi there this is kinda of off topic but I was wanting to know if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding know-how so I wanted to get guidance from someone with experience. Any help would be greatly appreciated!
kupite-diplom0024.ru/
At this time it sounds like Drupal is the best blogging platform available right now. (from what I’ve read) Is that what you are using on your blog?
http://sdiplom.ru/
Hi there to every one, the contents present at this web site are actually remarkable for people knowledge, well, keep up the good work fellows.
https://www.sdiplom.ru/
Hello I am so happy I found your blog, I really found you by accident, while I was searching on Bing for something else, Nonetheless I am here now and would just like to say thank you for a tremendous post and a all round enjoyable blog (I also love the theme/design), I don’t have time to read it all at the moment but I have book-marked it and also added your RSS feeds, so when I have time I will be back to read more, Please do keep up the awesome job.
https://diplom-city24.ru/
Что такое прогон по профилям?
На форумах при каждом прогоне регистрируется новый юзер, в профайле которого в поле Вебсайт или Домашняя страница стоит ваша ссылка. Также в некоторых форумах есть возможность в профиль вставить подпись, туда можно вставить ссылку с анкором. Все это можно варьировать. В профилях ссылки живут довольно долго, т.к. нет спама в чистом виде, и профили не мозолят глаза ни читателям форума, ни модераторам. Прогон производится последней версией хрумера с сервера с максимально возможной скоростью и производительностью.
ПОДРОБНЕЕ
НОВЫЙ ОТЛИЧНЫЙ VPN ДЛЯ XRUMERA!
За последние недели состоялась серия важных обновлений комплекса: XAuth, Captchas Benchmark, XRumer, XEvil 6.0 . В XEvil 6.0 обновлен механизм обработки hCaptcha и многое другое, в XAuth повышена стабильность работы, в XRumer повышена эффективность, устранён ряд погрешностей и обновлены прилагаемые базы.
https://xrumer.ru/
Today, I went to the beach front with my children. I found a sea shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She put the shell to her ear and screamed. There was a hermit crab inside and it pinched her ear. She never wants to go back! LoL I know this is totally off topic but I had to tell someone!
https://www.zakaz-na-diplom.ru/
Hi! Do you know if they make any plugins to protect against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any suggestions?
https://www.zakaz-na-diplom.ru/
Сторублевки и тысячи тонн грузов транспортируются ежечасно по круглому миру. И ЕЩЕ автор этих строк гордимся тем, яко юрчасть с этой великою массы транспортируется сверху наших автомобилях. Пишущий эти строки гарантируем нанешними автотранспортными услугами населений Москвы.
https://www.gruzogazel.ru/
Hi there very cool blog!! Man .. Excellent .. Wonderful .. I will bookmark your blog and take the feeds also? I am glad to find so many helpful information here within the post, we want work out more techniques in this regard, thanks for sharing. . . . . .
http://zakaz-na-diplom.ru/
В нашем обществе, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает двери перед всеми, кто собирается начать профессиональную деятельность или продолжить обучение в университете.
Мы предлагаем оперативно получить любой необходимый документ. Вы можете заказать диплом нового или старого образца, что является удачным решением для всех, кто не смог завершить образование или потерял документ. Каждый диплом изготавливается с особой аккуратностью, вниманием к мельчайшим элементам, чтобы в результате получился 100% оригинальный документ.
Преимущество этого решения состоит не только в том, что вы сможете максимально быстро получить диплом. Процесс организовывается комфортно, с профессиональной поддержкой. Начав от выбора нужного образца документа до грамотного заполнения личной информации и доставки по России — все находится под полным контролем опытных специалистов.
Всем, кто ищет быстрый и простой способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – значит избежать длительного обучения и сразу переходить к достижению личных целей, будь то поступление в ВУЗ или старт удачной карьеры.
https://boi.instgame.pro/forum/index.php?topic=24407.0
http://arsenal.spybb.ru/viewtopic.php?id=681#p738
http://skk.4bb.ru/viewtopic.php?id=1344#p35638
https://sketchfab.com/JoseArnold2
http://www.volgogradsky.ru/forum/viewtopic.php?p=534629#534629
В нашем мире, где диплом является началом отличной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Факт наличия официального документа об образовании переоценить невозможно. Ведь именно он открывает двери перед всеми, кто хочет вступить в сообщество профессионалов или учиться в ВУЗе.
Наша компания предлагает максимально быстро получить этот важный документ. Вы сможете приобрести диплом старого или нового образца, что является выгодным решением для всех, кто не смог завершить образование или потерял документ. дипломы производятся с особой тщательностью, вниманием ко всем нюансам. На выходе вы получите продукт, 100% соответствующий оригиналу.
Плюсы этого подхода заключаются не только в том, что вы сможете оперативно получить диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. От выбора нужного образца до консультаций по заполнению личной информации и доставки по стране — все находится под абсолютным контролем опытных мастеров.
Таким образом, для тех, кто ищет оперативный способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – это значит избежать длительного обучения и сразу переходить к важным целям, будь то поступление в ВУЗ или старт успешной карьеры.
http://krc.mybb.ru/viewtopic.php?id=112#p223
http://bushinkan.mybb.ru/viewtopic.php?id=279#p2180
https://lwccareers.lindsey.edu/profiles/4685430-luciusbirch55-outlook-com-birch
http://automarket.topbb.ru/viewtopic.php?id=2007#p3647
http://fiatforum.5bb.ru/viewtopic.php?id=920#p5342
골드 킹
Fang Jifan은 미소를 지으며 말했습니다. “당신은 귀를 붙입니다.”
Howdy! I’m at work surfing around your blog from my new apple iphone! Just wanted to say I love reading through your blog and look forward to all your posts! Keep up the superb work!
https://institute-diplom.ru/
An outstanding share! I have just forwarded this onto a friend who was doing a little homework on this. And he in fact bought me lunch due to the fact that I found it for him… lol. So let me reword this…. Thank YOU for the meal!! But yeah, thanx for spending time to talk about this matter here on your website.
http://diplompro.ru/
I blog often and I genuinely thank you for your content. This article has truly peaked my interest. I will take a note of your blog and keep checking for new information about once per week. I subscribed to your Feed too.
diplom-kuplu.ru/
Admiring the commitment you put into your website and in depth information you provide. It’s nice to come across a blog every once in a while that isn’t the same old rehashed information. Great read! I’ve saved your site and I’m including your RSS feeds to my Google account.
http://diplom-kuplu.ru/
В современном мире, где диплом является началом успешной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь именно диплом открывает двери перед каждым человеком, который собирается вступить в сообщество квалифицированных специалистов или продолжить обучение в университете.
В данном контексте мы предлагаем очень быстро получить этот важный документ. Вы можете приобрести диплом старого или нового образца, что становится удачным решением для всех, кто не смог закончить обучение или утратил документ. Все дипломы производятся аккуратно, с особым вниманием к мельчайшим элементам. В результате вы сможете получить полностью оригинальный документ.
Преимущества такого решения заключаются не только в том, что вы сможете оперативно получить свой диплом. Весь процесс организован удобно, с профессиональной поддержкой. От выбора требуемого образца до консультации по заполнению личных данных и доставки в любое место России — все находится под полным контролем квалифицированных мастеров.
В результате, для тех, кто ищет быстрый способ получить необходимый документ, наша компания предлагает отличное решение. Заказать диплом – значит избежать долгого обучения и не теряя времени перейти к достижению личных целей, будь то поступление в университет или старт успешной карьеры.
https://gitlab.ifam.edu.br/LomaTapp3
https://open.mit.edu/profile/01HY24GKRTRSRMEHWZG50X91PB/
https://namajore.frmbb.ru/viewtopic.php?id=184#p53817
https://ioby.org/users/luciusbirch55838201
https://ufacat.mybb.ru/viewtopic.php?id=222#p1959
В современном мире, где диплом становится началом успешной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа об образовании переоценить невозможно. Ведь именно диплом открывает двери перед людьми, желающими начать профессиональную деятельность или учиться в высшем учебном заведении.
Предлагаем очень быстро получить этот необходимый документ. Вы сможете заказать диплом старого или нового образца, что будет отличным решением для всех, кто не смог завершить образование, потерял документ или желает исправить плохие оценки. Каждый диплом изготавливается с особой аккуратностью, вниманием ко всем деталям. В результате вы сможете получить документ, максимально соответствующий оригиналу.
Плюсы данного решения состоят не только в том, что вы быстро получите свой диплом. Весь процесс организован просто и легко, с нашей поддержкой. От выбора требуемого образца до консультации по заполнению персональной информации и доставки по стране — все будет находиться под абсолютным контролем опытных мастеров.
В результате, всем, кто хочет найти максимально быстрый способ получить требуемый документ, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать продолжительного обучения и не теряя времени перейти к личным целям: к поступлению в ВУЗ или к началу успешной карьеры.
https://knowyourmeme.com/users/diplo-mans
http://www.otvetnemail.ru/ivanovskaya-oblast-438/kupit-diplom-o-srednem-obrazovanii-1540766/#post2127232
http://ukr.mybb.ru/viewtopic.php?id=386#p1022
http://passo.su/forums/index.php?showtopic=6426
https://lwccareers.lindsey.edu/profiles/4685430-luciusbirch55-outlook-com-birch
Wow, awesome weblog structure! How long have you ever been blogging for? you made running a blog glance easy. The overall look of your site is magnificent, let alone the content material!
slcomp.kz/index.php?route=information/blogger&blogger_id=5В
http://www.paladiny.ru/forummess.dwar.php?TopicID=26048&Offset=0В
ractis.ru/forums/index.php?autocom=gallery&req=si&img=2108В
http://www.cialisonlinerx.com/mifegest.htmlВ
bizkerrefutboltaldea.com/bizkerre/Euskara/masculino/juvenil.htmВ
Рады приветствовать вас!
Представляем лучшие прогоны, чтобы “угробить” веб-ресурс вашего конкурента. Цена: всего от 7 000 р.
– Гарантированный результат. Интернет-сайты точно “умрут”.
– Предельно возможное количество негативных фитбеков.
– Наша специальная база – выжимка самых “убийственных” площадок из 10 000 000 веб-сайтов (спамных, порно, вирусных и тому подобных). Действует бесперебойно.
– Прогон осуществляем одновременно с 4-х мощных серверов.
– Отправка активационных ссылок на электронную почту.
– Растянем как угодно по времени.
– Прогоняем с запрещёнными ключевыми фразами.
– При двух прогонах – хорошие скидки клиенту.
Стоимость услуги 7700 рублей
Отчётность.
Оплата: Yoo.Money, Bitcoin, МИР, Visa, MasterCard…
Принимаем USDT
Телегрм: https://t.me/exrumer
Skype: Loves.Ltd
https://xrumer.cc/
Только этот.
Growth and innovation are essential to progress and sustainability in the energy industry’s dynamic environment. This conversation dives into key systems and drives pushing progressions in energy, molding the scene for monetary turn of events and mechanical forward leaps.
DEL MAR ENERGY
In the ever-evolving landscape of the American energy sector, Del Mar Energy stands out as a beacon of progress and innovation. With its commitment to excellence and dedication to advancing the nation’s economy, this industrial powerhouse has garnered praise from the government and lawmakers alike. In 2023, Del Mar Energy received commendations from the U.S. government and a Texas senator for its contributions to economic growth, alongside the inauguration of its own solar panel manufacturing facility catering to wholesale and retail markets.
https://www.wicz.com/story/50815717/del-mar-energy-fueling-growth-and-innovation-in-americas-energy-sector
Del Mar Energy’s journey towards excellence began decades ago, rooted in a vision to revolutionize the energy industry. Since its inception, the company has been at the forefront of driving economic prosperity while embracing sustainable practices. Its strategic investments in various sectors, coupled with a relentless pursuit of innovation, have propelled Del Mar Energy to the summit of industrial success.
The year 2023 marked a significant milestone for Del Mar Energy as it received accolades from government officials for its role in bolstering the nation’s economy. Through job creation, infrastructure development, and strategic partnerships, Del Mar Energy has become a linchpin of economic growth, particularly in regions like Texas where its operations are deeply entrenched.
The commendations bestowed upon Del Mar Energy underscored the company’s unwavering commitment to excellence and its pivotal role in shaping the economic landscape of the nation. Such recognition not only validates the company’s efforts but also serves as a testament to its enduring impact on society.
In tandem with its commendation, Del Mar Energy embarked on a new chapter of innovation by launching its own solar panel manufacturing facility. This strategic move not only aligned with the company’s commitment to sustainable energy but also positioned it as a frontrunner in the renewable energy revolution.
The decision to venture into solar panel production was met with enthusiasm from both the industry and consumers alike. Del Mar Energy’s state-of-the-art manufacturing facility boasts cutting-edge technology and a commitment to quality, ensuring that its solar panels meet the highest standards of efficiency and durability.
Moreover, by catering to both wholesale and retail markets, Del Mar Energy has democratized access to solar energy, making it more accessible to businesses and households across the country. This not only drives the adoption of renewable energy but also fosters economic growth by creating new avenues for revenue generation and job opportunities.
As Del Mar Energy continues to chart new territories in the energy sector, its commitment to innovation and sustainability remains unwavering. By leveraging its expertise and resources, the company is poised to shape the future of energy, driving economic growth and environmental stewardship hand in hand. With commendations from government officials and a pioneering foray into solar panel production, Del Mar Energy is setting the stage for a brighter, more sustainable future for generations to come.
Information contained on this page is provided by an independent third-party content provider. Frankly and this Site make no warranties or representations in connection therewith. If you are affiliated with this page and would like it removed please contact pressreleases@franklymedia.com
As the admin of this web site is working, no question very soon it will be famous, due to its quality contents.
http://hl2forever.ru/member.php?u=246
http://www.moto-import.ru/index.php?did=33&le_categoryID=0&page=97
http://k-up.ru/content/izhevsk/solncezashchitnye-stekla
http://forum.anime.org.ua/bbs/showthread.php?p=128646
http://textualheritage.org/en/the-materials-of-the-conference-el-manusctipt-2014/81.html
Elon Musk was star guest this year at an annual conference organized by Italian PM Giorgia Meloni’s Brothers of Italy party.
кракен 13
He arrived against the backdrop of an ice-skating rink and an ancient castle in Rome with one of his 11 children to tout the value of procreation.
Italy has one of the lowest birth rates in the world, and Musk urged the crowd to “make more Italians to save Italy’s culture,” a particular focus of the Meloni government.
https://kraken2trfqodidvlh4aa337cpzfrhdlfldhve5nf7njhum7instad.com
kraken2trfqodidvlh4aa337cpzfrhdlfldhve5nf7njhumwr7instad onion
Meloni has been a strong opponent of surrogacy, which is criminalized in Italy, but there was no mention of Musk’s own recent children born of surrogacy.
The owner of X (formerly called Twitter) was slightly rumpled with what could easily be argued the least stylish shoes in the mostly Italian crowd since Donald Trump’s often unkempt former top adviser Steve Bannon appeared at the conference in 2018.
Meloni sat in the front row taking photos of Musk, who she personally invited. Meloni founded the Atreju conference in 1998, named after a character in the 1984 film “The NeverEnding Story.”
This is very attention-grabbing, You are an overly skilled blogger. I’ve joined your rss feed and sit up for in quest of more of your excellent post. Additionally, I’ve shared your web site in my social networks
sdm-servis.ru/component/fireboard/?func=view&catid=5&id=70090В
financeokey.ru/page/9В
http://www.snipesocial.co.uk/blogs/370679/%D0%9E%D1%82%D0%BA%D1%80%D1%8B%D1%82%D0%B8%D0%B5-%D0%B2%D0%BE%D0%B7%D0%BC%D0%BE%D0%B6%D0%BD%D0%BE%D1%81%D1%82%D0%B5%D0%B9-%D1%86%D0%B5%D0%BD%D0%BD%D0%BE%D1%258?lang=tr_trВ
templateinspire.com/opencart/Lingerie/index.php?route=information/blogger&blogger_id=2В
petkit.com.cn/member/index.php?uid=ehalipa&action=viewarchives&aid=6644В
Admiring the dedication you put into your website and in depth information you present. It’s good to come across a blog every once in a while that isn’t the same out of date rehashed information. Great read! I’ve saved your site and I’m including your RSS feeds to my Google account.
mboumukvilino.ru/antiterroristicheskaya-bezopasnost/normativnye-pravovye-akty-rossiyskoy-federacii-i-respubliki-krym/В
1776reloaded.us/blogs/7/Where-to-buy-a-diploma-or-certificate-at-an-adequateВ
giaibngdaquocteu23.com/2021-10-cb-hay-trong-trong-fifa-online-4-hien-nay-keo8386/В
formfinance.ru/kupit-diplom-lyubogo-obraztsaВ
nbcenter.ge/index.php?subaction=userinfo&user=ekeposВ
It’s remarkable for me to have a site, which is helpful in favor of my knowledge. thanks admin
unirun.ru/page/5/В
clickandconnectclubs.com/index.php?do=/public/blog/view/id_119636/title_-2024/В
foro.rune-nifelheim.com/general-ragnarok-m/put-k-uspehu-pokupka-diploma-kak-klyuchevaya-strategiya/В
http://www.genexscience.com/index.php?route=information/blogger&blogger_id=2В
myworldoftank.ru/article/kak-vklyuchit-otobrazhenie-shansov-na-pobedu-xvm-v-world-of-tanks.htmlВ
С радостью приветствуем вас, коллеги!
Мы представляем агентство по СЕО продвижению XRumer Co.
Ваш портал, как можно заметить, еще только набирает обороты. Для того, чтобы максимально ускорить его рост, предлагаем услуги по SEO-оптимизации. Также у нас есть доступные и эффективные инструменты для СЕО-специалистов. У нашей команды большой опыт, в арсенале есть успешные рабочие кейсы – если интересует, предоставим по запросу.
До конца месяца действует скидка на все услуги – 10%.
Наши услуги:
– Размещаем супер трастовые ссылки (требуется абсолютно всем сайтам) – от 1,5 до 5000 р
– Безанкорные ссылки (2500 штук) (полезно любым сайтам) – 3.900 р
– Профессиональный прогон на 110 тысяч сайтов (RU.зона) – 2900 руб
– 150 постов в VK о вашем сайте (отличная реклама) – 3900 руб
– Публикация статей про ваш сайт на 300 форумах (мощнейшая раскрутка вашего портала) – 29.000 р
– СуперПостинг – отличный прогон по 3 млн площадок (мощнейшее размещение для ваших сайтов) – 39 тысяч рублей
– Рассылка сообщений по сайтам через форму обратной связи – стоимость по договоренности, зависит от объемов.
Если появились вопросы, всегда обращайтесь, подскажем.
Отчётность.
Оплата: Yoo.Деньги, Bitcoin, МИР, Visa, MasterCard…
Принимаем USDT
Телега: https://t.me/exrumer
Skype: Loves.Ltd
https://xrumer.cc/
Только этот.
В нашем обществе, где диплом – это начало успешной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения качественного образования. Факт наличия документа об образовании переоценить попросту невозможно. Ведь именно диплом открывает дверь перед любым человеком, который собирается начать трудовую деятельность или учиться в ВУЗе.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы имеете возможность заказать диплом нового или старого образца, что является отличным решением для человека, который не смог завершить образование или потерял документ. Каждый диплом изготавливается аккуратно, с особым вниманием ко всем нюансам. На выходе вы получите полностью оригинальный документ.
Преимущество подобного подхода состоит не только в том, что вы оперативно получите диплом. Процесс организован удобно и легко, с профессиональной поддержкой. От выбора нужного образца до правильного заполнения личной информации и доставки в любое место страны — все будет находиться под полным контролем квалифицированных специалистов.
Для тех, кто ищет максимально быстрый способ получить требуемый документ, наша компания может предложить отличное решение. Заказать диплом – значит избежать долгого обучения и сразу переходить к личным целям: к поступлению в университет или к началу трудовой карьеры.
http://grand-art.online
http://apuntadas.es
http://balivillacollection.com
http://varikoznic.ru
http://ibnews.com.ua
Hello there! This is my 1st comment here so I just wanted to give a quick shout out and tell you I truly enjoy reading through your blog posts. Can you suggest any other blogs/websites/forums that go over the same subjects? Thank you so much!
http://splesti.ru/books/item/f00/s00/z0000006/st001.shtml
http://mail.webco.by/forum/viewtopic.php?f=84&t=27832
http://fitrain.ru/fitness/page/7
http://wild-animals.ru/systems/alp/
http://bur-35-al.tvoysadik.ru/?section_id=28
Howdy! Do you use Twitter? I’d like to follow you if that would be okay. I’m definitely enjoying your blog and look forward to new updates.
kenhhocsinh.com/tai-sao-ly-thuong-kiet-lai-ket-thuc-cuoc-khang-chien-chong-tong-bang-bien-phap-giang-hoaВ
edmontonoilersclub.com/read-blog/287_where-can-i-buy-a-diploma-or-certificate-at-a-comfortable-price.html?mode=dayВ
http://www.chabab-belouizdad.org/forum/viewtopic.php?f=21&t=550&p=58893В
grp.7olimp.ru/viewtopic.php?f=13&t=1895&p=2457В
otantravel.kz/news/swiss-uvelichivaet-kolichestvo-reysov-mayyami-cyurih-na-period-zimnego-sezonaВ
На сегодняшний день, когда диплом – это начало удачной карьеры в любом направлении, многие пытаются найти максимально простой путь получения качественного образования. Важность наличия документа об образовании переоценить просто невозможно. Ведь диплом открывает дверь перед людьми, желающими начать профессиональную деятельность или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает очень быстро получить любой необходимый документ. Вы можете приобрести диплом, и это становится отличным решением для человека, который не смог завершить образование или утратил документ. Любой диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам. В итоге вы сможете получить продукт, максимально соответствующий оригиналу.
Плюсы подобного подхода состоят не только в том, что можно оперативно получить свой диплом. Весь процесс организовывается комфортно, с профессиональной поддержкой. От выбора необходимого образца до консультаций по заполнению личной информации и доставки в любое место России — все будет находиться под полным контролем наших мастеров.
Таким образом, всем, кто ищет максимально быстрый способ получения необходимого документа, наша услуга предлагает выгодное решение. Купить диплом – это значит избежать долгого процесса обучения и сразу перейти к достижению своих целей, будь то поступление в ВУЗ или старт профессиональной карьеры.
http://eximp.com.ua
http://bettesworthconstruction.com
http://ie-search.com
http://olimpicxativa.com
http://bluesailcreative.com
북 오브 데드
“네, 명령에 따르겠습니다.” 오우양 즈가 고개를 숙이고 경례했다.
I’m not sure exactly why but this web site is loading extremely slow for me. Is anyone else having this problem or is it a problem on my end? I’ll check back later and see if the problem still exists.
foro.rune-nifelheim.com/general-ragnarok-m/put-k-uspehu-pokupka-diploma-kak-klyuchevaya-strategiya/В
http://www.360nhadep.com/tag/dong-tu-menh-hop-huong-nao/В
fond.uni-altai.ru/index.php?subaction=userinfo&user=usacyВ
symphonyofthemagneticnorth.com/_404.htmlВ
dindersim.com/haberler/page,1,15,513-meslek-dersleri-kazanim-degerlendirme-optik-okuma.htmlВ
Hey very nice website!! Man .. Beautiful .. Superb .. I will bookmark your blog and take the feeds also? I’m satisfied to seek out a lot of helpful info right here in the publish, we need work out more strategies in this regard, thank you for sharing. . . . . .
http://pro-korolev.ru/nyukasl-planiruet-usilitsya-gibbsom.html
http://kardeya-nv.ru/edelen_sostav_sbornoj_rossii_po_vmkh_na_perv.htm
http://myvera.ru/kosmolog
http://zpinfo.com.ua/v-zaporozhskoy-oblasti-sbili-velosipedista
http://yug-gelendzhik.ru/obrazovatelnye-uchrezhdeniya-gelendzhika-2017-god/
Wow, wonderful blog format! How long have you ever been running a blog for? you make running a blog look easy. The total glance of your website is magnificent, let alone the content material!
http://numterra.ru/index/raschety_po_shkolam/0-23
http://perfect-grade.com/health-information-managent/
http://cheat-v2.ru/dota/atf-pokinul-quest-esports.html
http://znaniyapolza.ru/produktyi-poleznyie-pri-varikoze.html
http://prasemena.ru/articles/ovoschi/pomidor_richai
Thank you, I’ve recently been looking for info approximately this topic for a long time and yours is the greatest I have found out till now. However, what concerning the bottom line? Are you certain in regards to the supply?
keepnitreal.net/blogs/53813/%D0%A1%D0%B5%D0%BA%D1%80%D0%B5%D1%82-%D1%83%D1%81%D0%BF%D0%B5%D1%85%D0%B0-%D0%9A%D0%B0%D0%BA-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B8-%D0%B8%D0%B7%D0%BC%D0%B5%D0%BD%D0%B8%D1%82%D1%8C-%D1%81%D0%B2%D0%BE%D1%8E-%D0%B6%D0%B8%D0%B7%D0%BD%D1%8CВ
yaoisennari.ekafe.ru/viewtopic.php?f=169&t=1669&start=0&view=printВ
foro.turismo.org/post281274.htmlВ
http://ftp.video-foto.by/forum/viewtopic.php?f=84&t=28359&p=199326В
http://www.terios2.ru/forums/index.php?autocom=gallery&req=si&img=2515В
Thank you for the good writeup. It in reality used to be a amusement account it. Glance advanced to far brought agreeable from you! However, how can we communicate?
bbs.heyshell.com/forum.php?mod=viewthread&tid=27609В
mybaltika.info/ru/print/587/7892/В
mclassic.com.hk/home.php?mod=space&uid=274970&do=profile&from=spaceВ
c2c.buzz/index.php?do=/public/user/blogs/view/name_Alanpoe/id_119636/title_/В
drosetourmanila.com/vn/retail_promo.htmlВ
Hi there i am kavin, its my first occasion to commenting anywhere, when i read this post i thought i could also make comment due to this brilliant paragraph.
nemspb.ru/objects/zhd-stantsiya-babaevo-oktyabrskoj-zheleznoj-dorogi/В
kupidon-pskov.ru/В
ветерантюмгео.СЂС„/galereya/?type_0=gallery&album_gallery_id_0=2В
gamesfortop.ru/diplomyi-na-zakaz-vash-shans-na-novuyu-kareruВ
http://www.kiripo.com/forum/member.php?action=profile&uid=1076912В
В современном мире, где диплом является началом успешной карьеры в любом направлении, многие стараются найти максимально простой путь получения образования. Необходимость наличия официального документа переоценить просто невозможно. Ведь именно он открывает дверь перед каждым человеком, который собирается вступить в профессиональное сообщество или учиться в высшем учебном заведении.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы можете приобрести диплом нового или старого образца, и это становится удачным решением для всех, кто не смог закончить обучение, потерял документ или желает исправить свои оценки. дипломы производятся аккуратно, с особым вниманием к мельчайшим деталям, чтобы в результате получился полностью оригинальный документ.
Преимущества данного решения заключаются не только в том, что можно оперативно получить диплом. Процесс организован комфортно, с нашей поддержкой. Начав от выбора необходимого образца диплома до консультаций по заполнению персональной информации и доставки по стране — все под полным контролем наших мастеров.
Для всех, кто хочет найти максимально быстрый способ получить требуемый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать продолжительного обучения и сразу перейти к достижению собственных целей, будь то поступление в университет или начало профессиональной карьеры.
http://i-medic.com.ua
http://2019gdansk.pl
http://keslaser.com.ua
http://vanbroecanimations.online
http://apesk.ru
Hi, its nice paragraph on the topic of media print, we all know media is a fantastic source of facts.
http://pialci.ru/index.php?links_exchange=yes&page=1&show_all=yes
http://znaxar.com/activity
http://softmirror.ru/publ/multimedia/5
http://redfoxday.ru/category/poleznye-sovety/uhod-za-domashnimi-rasteniyami-poleznye-sovety/
http://www.beretta-modelle.ch/forum/memberlist.php?orderby=username&ordertype=desc&search=&char=&page=278
Keep on working, great job!
http://www.forums.wolflair.com/member.php?u=110846В
oldsite.profbez.ru/shop/index.php?links_exchange=yes&page=260&show_all=yesВ
http://www.bigboytoyz.com/newcars/mercedes-benz/amg-gtВ
mymotospeed.ru/page/4В
http://www.martinsimoveisijui.com.br/blog/nutricosmeticosВ
Hi to every single one, it’s actually a good for me to visit this web site, it includes precious Information.
blog-doma.ru/proizvodstvo-biogumusaВ
http://www.consult-exp.com/blogs/116058/%D0%94%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D1%8B-%D0%BD%D0%B0-%D0%BB%D1%8E%D0%B1%D0%BE%D0%B9-%D0%B2%D0%BA%D1%83%D1%81-%D0%B8-%D1%86%D0%B2%D0%B5%D1%82-%D1%88%D0%B8%D1%80%D0%BE%D0%BA%D0%B8%D0%B9-%D0%B0%D1%81%D1%81%D0%BE%D1%80%D1%82%D0%B8%D0%BC%D0%B5%D0%BD%D1%82?lang=tr_trВ
suamaylanh.com.vn/tin-tuc/40-hang-dien-tu-dien-lanh-ton-nhieu.htmlВ
http://www.4-mobile.ru/index.php?links_exchange=yes&page=13&show_all=yesВ
here4deals.com/garderie-barny-horaires.phpВ
http://kvn-tehno.com.ua
What’s up it’s me, I am also visiting this web page daily, this site is really good and the visitors are genuinely sharing fastidious thoughts.
templateinspire.com/opencart/Lingerie/index.php?route=information/blogger&blogger_id=2В
allukrnews.ru/put-k-professionalnomu-priznaniyu-pokupka-diploma-kak-reshenie/В
http://www.climbandmore.com/climbing,360,0,1,news.htmlВ
carrefour-emploi-public.fr/brest-metropole-oceaneВ
comedyforme.ru/prodazha-diplomov-lyubogo-uchebnogo-zavedeniya-rossiiВ
What’s up, I wish for to subscribe for this website to take latest updates, so where can i do it please assist.
kzgbi-2.ru/forums.php?m=posts&id=6901В
ros-con.ru/index.htmlВ
startspresto.ru/dannye/pluginclass/cbblogs?action=blogs&func=show&id=22В
jaezfinancialgroup.icu/pages/contact-usВ
suamaylanh.com.vn/tin-tuc/40-hang-dien-tu-dien-lanh-ton-nhieu.htmlВ
Wow, amazing weblog structure! How lengthy have you ever been running a blog for? you made running a blog glance easy. The whole glance of your web site is wonderful, let alone the content!
http://e-pitanie.ru/dobavki_v_produktah/bad.php
http://itally.ru/bergamo/67-gde-poest.html
http://motoravtoremont.ru/pochemu-vazhno-imet-diplom-ob-okonchanii-vuza/
http://vinpr.org/obshestvo/6573-mir-stanovitsya-vse-bolee-cifrovym.html
http://www.one.lt/directory/posts/541955
I’m not sure exactly why but this weblog is loading very slow for me. Is anyone else having this issue or is it a issue on my end? I’ll check back later on and see if the problem still exists.
http://opekaspb.ru/za-zreniem-nuzhen-uxod/
http://psy-course.ru/blog/rabota-i-karera/uznajte-chto-blokiruet-vash-denezhnyij-potok
http://4remind.ru/page/4.html
http://horchelovek.ru/beauty/seksyalnaia-byntarka-rita-flores.html
http://blogs.rufox.ru/~runoklisfgy/42169.htm
Pretty section of content. I just stumbled upon your blog and in accession capital to assert that I acquire actually enjoyed account your blog posts. Any way I will be subscribing to your augment and even I achievement you access consistently quickly.
bancaanxudoithuong3d.com/category/game-ban-ca-doi-the?filter_by=popular7В
eurodelo.ru/kak-kupit-diplom-bez-lishnih-hlopotВ
dom-postroj.ru/doma-iz-gazobetona/k-231В
smartwatchcolombia.com/producto/78/smartwatch-serie-8-ultra-11-2023–manilla-de-regalo/7arusak-diploms.comВ
prmaster.su/personal_blog/good/page82/В
For the reason that the admin of this site is working, no hesitation very shortly it will be well-known, due to its quality contents.
зеленогорск.СЃРїР±.СЂС„/malyy-biznes/49-pamyatka-predprinimatelyu.htmlВ
sinopipefittings.com/e_Feedback/?page=10995В
http://www.cialisonlinerx.com/mifegest.htmlВ
http://www.hoahaubansacviet.com.vn/xac-nhan-binh-chon.flc?sbd=335В
zhaktrans.ru/p40135689-tnvd-wd615-612601080225.htmlВ
Good day! Do you use Twitter? I’d like to follow you if that would be okay. I’m undoubtedly enjoying your blog and look forward to new posts.
http://abvshka.ru/tag/brittni-grajner
http://эвакуатор-срочно-недорого.рф/
http://gazetenka.com/articles/16562/
http://connect.nteep.org/blogs/810/What-do-you-need-to-pay-attention-to-when-ordering?lang=fr_fr
http://ksmzpn.pl/tabela-ekwiwalentow-2022/
I have fun with, lead to I discovered just what I used to be taking a look for. You’ve ended my four day lengthy hunt! God Bless you man. Have a nice day. Bye
http://www.irax.ru/site248787/www2eduru.html
http://venev-blagochin.ru/lenta-7.html
http://www.roofingrevenue.com/fdinlrpf/hadda-be-playing-on-the-jukebox-analysis.html
http://i-yourself.ru/kosmeticheskij-remont-lifta-svoimi-rukami/
http://searchprogram.ru/kak-sdelat-plan-v-vorde
Добро препожаловать в течение одно изо самых универсальных студий Москвы !
Профессиональный штат. Удобная оплата. Договор. Эксклюзивный экспрессконтроль
https://whitestudio.moscow/
http://asquifyde.es
В нашем мире, где аттестат – это начало отличной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения образования. Наличие документа об образовании сложно переоценить. Ведь диплом открывает дверь перед каждым человеком, желающим начать трудовую деятельность или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем максимально быстро получить любой необходимый документ. Вы можете приобрести аттестат, и это является удачным решением для всех, кто не смог завершить образование или потерял документ. Все аттестаты выпускаются аккуратно, с максимальным вниманием ко всем элементам, чтобы на выходе получился документ, полностью соответствующий оригиналу.
Плюсы такого решения заключаются не только в том, что вы сможете оперативно получить свой аттестат. Весь процесс организовывается комфортно, с нашей поддержкой. От выбора нужного образца до консультаций по заполнению персональной информации и доставки в любое место России — все будет находиться под полным контролем наших специалистов.
Таким образом, для тех, кто пытается найти максимально быстрый способ получить требуемый документ, наша компания может предложить выгодное решение. Приобрести аттестат – значит избежать длительного обучения и сразу переходить к личным целям: к поступлению в университет или к началу удачной карьеры.
https://smeda.ru/
В современном мире, где диплом становится началом отличной карьеры в любой отрасли, многие пытаются найти максимально быстрый и простой путь получения образования. Наличие официального документа трудно переоценить. Ведь именно он открывает двери перед людьми, желающими начать профессиональную деятельность или учиться в университете.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы имеете возможность заказать диплом нового или старого образца, и это становится отличным решением для всех, кто не смог закончить обучение или утратил документ. дипломы производятся аккуратно, с особым вниманием к мельчайшим нюансам, чтобы в итоге получился полностью оригинальный документ.
Плюсы этого решения заключаются не только в том, что можно оперативно получить свой диплом. Процесс организован комфортно и легко, с нашей поддержкой. Начиная от выбора необходимого образца документа до консультаций по заполнению личных данных и доставки по России — все находится под абсолютным контролем опытных мастеров.
Всем, кто хочет найти максимально быстрый способ получения необходимого документа, наша компания предлагает отличное решение. Приобрести диплом – это значит избежать длительного процесса обучения и сразу переходить к достижению собственных целей, будь то поступление в университет или старт успешной карьеры.
http://postroy-dom.by
http://grand-art.online
http://albiongranit.ru
http://vsedlo.ru
http://quadro-mebel.ru
Предлагаем вам выполнить консультацию (аудит) по подъему продаж а также прибыли в вашем бизнесе. Формат аудита: личная встреча или конференция по скайпу. Делая верные, но обыкновенные усилия, доход от ВАШЕГО бизнеса получится поднять в несколькио раз. В нашем арсенале более 100 проверенных фактических методик повышения продаж и доходов. В зависимости от вашего бизнеса выберем для вас максимально лучшие и будем постепенно реализовывать.
-https://interestbook.ru/
Best SEO services near me
In the quest to increase online visibility and generate organic traffic, locating the best SEO services near you has become essential. Local companies understand the meaning of aimed search engine optimization approaches that cater to their individual geographic area. The term “near me” has earned enormous significance in today’s digital landscape, as individuals often include it in their search inquiries to discover products and services in their area. The prime SEO providers within your reach apprehend the subtleties of local SEO, integrating location-based words, Google My Business refinement, and accurate business directory registers. By zeroing in on these aspects, these services can assist businesses accomplish increased rankings in local search findings, making sure that potential consumers find them conveniently when searching for relevant assistance in their area.
The rivalry for online visibility has increased, making it crucial for businesses to allocate resources to excellent SEO services. Looking for “best SEO providers near me” displays the modern inclination towards personalized and localized answers. Collaborating with such services presents several upsides, including face-to-face discussions, a deeper knowledge of local rivalry, and the ability to adapt strategies based on the exclusive qualities of your target. The prime SEO providers in your area will not only optimize your website but also create comprehensive digital marketing strategies that may encompass content creation, social media involvement, and even paid advertising campaigns. By harnessing the capability of localized SEO, businesses can set up a strong online presence that successfully engages local consumers and sets the stage for long-term growth.
Отчётность.
Оплата: Yoo.Money, Bitcoin, МИР, Visa, MasterCard…
Принимаем USDT
Телега: https://t.me/exrumer
Skype: Loves.Ltd
https://www.webfx.com/
https://xrumer.cc/
На сегодняшний день, когда диплом является началом отличной карьеры в любом направлении, многие стараются найти максимально быстрый путь получения качественного образования. Факт наличия официального документа сложно переоценить. Ведь именно диплом открывает двери перед всеми, кто собирается начать трудовую деятельность или продолжить обучение в ВУЗе.
Предлагаем максимально быстро получить этот важный документ. Вы сможете приобрести диплом, что является удачным решением для человека, который не смог закончить обучение или потерял документ. Все дипломы выпускаются с особой тщательностью, вниманием ко всем элементам. В результате вы сможете получить документ, максимально соответствующий оригиналу.
Плюсы такого подхода состоят не только в том, что вы сможете максимально быстро получить свой диплом. Процесс организовывается удобно, с нашей поддержкой. Начиная от выбора необходимого образца документа до консультации по заполнению персональных данных и доставки по России — все находится под полным контролем опытных мастеров.
Для тех, кто ищет оперативный способ получить необходимый документ, наша компания предлагает выгодное решение. Купить диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к своим целям: к поступлению в ВУЗ или к началу успешной карьеры.
http://tlsp.paris
http://santfefc.com
http://my-digital.ro
http://etlapok-gyartasa.hu
http://topextern.ru
В нашем обществе, где аттестат – это начало отличной карьеры в любой сфере, многие пытаются найти максимально простой путь получения качественного образования. Наличие официального документа переоценить просто невозможно. Ведь диплом открывает дверь перед каждым человеком, который стремится начать трудовую деятельность или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает очень быстро получить любой необходимый документ. Вы сможете приобрести аттестат, что является удачным решением для человека, который не смог закончить образование, утратил документ или хочет исправить свои оценки. Аттестат изготавливается аккуратно, с максимальным вниманием ко всем нюансам. На выходе вы получите документ, полностью соответствующий оригиналу.
Преимущество такого подхода заключается не только в том, что вы максимально быстро получите свой аттестат. Процесс организован удобно, с профессиональной поддержкой. Начиная от выбора подходящего образца документа до консультации по заполнению личных данных и доставки в любой регион страны — все под абсолютным контролем наших специалистов.
Всем, кто пытается найти быстрый и простой способ получения требуемого документа, наша услуга предлагает отличное решение. Купить аттестат – это значит избежать долгого обучения и сразу перейти к личным целям: к поступлению в университет или к началу удачной карьеры.
https://boomingwebsitetraffic.com/
http://ramenskoe-betonbaza.ru
슬롯 게임
Fang Jifan은 침착하고 편안했습니다. “황실은 구호를 위해 돈과 음식을 할당 할 것입니다.”
В нашем обществе, где диплом – это начало отличной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Наличие документа об образовании переоценить просто невозможно. Ведь диплом открывает дверь перед каждым человеком, который собирается начать профессиональную деятельность или учиться в любом университете.
Предлагаем очень быстро получить этот важный документ. Вы сможете купить диплом старого или нового образца, что будет удачным решением для всех, кто не смог закончить образование, потерял документ или желает исправить плохие оценки. Каждый диплом изготавливается с особой аккуратностью, вниманием ко всем нюансам. В итоге вы сможете получить полностью оригинальный документ.
Преимущество подобного подхода состоит не только в том, что вы максимально быстро получите свой диплом. Процесс организовывается просто и легко, с нашей поддержкой. Начав от выбора подходящего образца до консультации по заполнению личных данных и доставки по России — все под абсолютным контролем качественных специалистов.
Всем, кто хочет найти быстрый способ получения необходимого документа, наша услуга предлагает отличное решение. Приобрести диплом – значит избежать долгого обучения и не теряя времени перейти к достижению собственных целей, будь то поступление в университет или старт карьеры.
http://mountbarkertourismwa.com.au
http://mininform.ru
http://optimapharm.com.ua
http://calc-bank.ru
http://moscityservice.ru
This is my first time visit at here and i am really pleassant to read all at single place.
myanmarfootball.org/gallery/displayimage.php?album=lastup&cat=6&pos=0В
vnbaolut.com/amiВ
jurisprudenta.org/Standard.aspxВ
fabnews.ru/forum/showthread.php?p=76998В
http://www.aapeducation.com.au/component/comprofiler/pluginclass/cbblogs.html?action=blogs&func=show&id=59В
Hi! Do you use Twitter? I’d like to follow you if that would be okay. I’m absolutely enjoying your blog and look forward to new updates.
http://domsdushoy.ru/kruzheva-v-interere/
http://pelotkitut.ru/component/option,com_rsg2/page,timetop/toplimit,9999999/Itemid,3/limit,20/limitstart,1040/
http://opt-dostawka.ru/preimushhestva-obrazovaniya-v-rossii-dlya-inostrannyx-studentov/
http://znaniyapolza.ru/produktyi-poleznyie-pri-varikoze.html
http://furlib.ru/books/item/f00/s00/z0000003/st018.shtml
Кофе в капсулах системы Nespresso. Мы предлагаем широкий ассортимент кофе в капсулах, более 200 вкусов. Доставка СДЭК 1-3 дня в любой город Беларуси или России. Оплата при получении.
Кофе Nespresso
Wow, fantastic weblog structure! How long have you been blogging for? you made running a blog glance easy. The total glance of your site is excellent, let alone the content!
http://4ege.ru/materials_podgotovka/5124-goryachii-linii-ege-po-regionam-rossii.html
http://delphi-manual.ru/injection.php
http://mafiaclans.ru/topic3885.html
http://gazeta.ekafe.ru/viewtopic.php?f=35&t=14677
http://web011.dmonster.kr/bbs/board.php?bo_table=b0501&wr_id=205579
В нашем обществе, где диплом – это начало отличной карьеры в любой сфере, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь диплом открывает двери перед всеми, кто желает начать профессиональную деятельность или продолжить обучение в университете.
Мы предлагаем оперативно получить любой необходимый документ. Вы имеете возможность приобрести диплом нового или старого образца, что становится отличным решением для всех, кто не смог закончить обучение или утратил документ. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам. На выходе вы получите полностью оригинальный документ.
Преимущества этого решения состоят не только в том, что вы быстро получите свой диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора требуемого образца до точного заполнения персональных данных и доставки в любое место России — все будет находиться под абсолютным контролем качественных мастеров.
Таким образом, для тех, кто ищет оперативный способ получения необходимого документа, наша компания предлагает отличное решение. Заказать диплом – значит избежать длительного процесса обучения и не теряя времени перейти к своим целям: к поступлению в университет или к началу трудовой карьеры.
http://4rme.com
http://megart.com.ua
http://vinhphatmobile.com
http://oregonpacificrr.com
http://humak.ru
Wow that was unusual. I just wrote an really long comment but after I clicked submit my comment didn’t appear. Grrrr… well I’m not writing all that over again. Anyhow, just wanted to say wonderful blog!
http://stavklad.ru/viewtopic.php?f=9&t=5065
Can I simply just say what a comfort to find somebody who really understands what they are discussing on the internet. You actually realize how to bring an issue to light and make it important. More and more people really need to look at this and understand this side of the story. It’s surprising you’re not more popular since you most certainly possess the gift.
http://zoofortunakz.5nx.ru/viewtopic.php?f=96&t=4385
Greetings I am so glad I found your weblog, I really found you by mistake, while I was looking on Aol for something else, Anyhow I am here now and would just like to say thanks a lot for a incredible post and a all round entertaining blog (I also love the theme/design), I don’t have time to read it all at the moment but I have bookmarked it and also added your RSS feeds, so when I have time I will be back to read more, Please do keep up the superb job.
http://2foru.pl/news/watch-this-sound-by-konzept/comment-page-7567/
http://decadenz.ru/publ/shkolniku/literatura/byliny/8-1-0-193
http://photoconnor.space/PHPFusion/profile.php?lookup=12076
http://alkogolizma.com/preparaty/antipohmelin.html
http://www.geographystudy.ru/geogo-363.html
Hello colleagues, fastidious post and fastidious urging commented here, I am really enjoying by these.
vnbaolut.com/amiВ
http://www.forum.jrudevels.org/profile.php?mode=viewprofile&u=563048В
adsauto.info/index.php?subaction=userinfo&user=igaseryВ
aromatov.wooden-rock.ru/forum/topic.php?forum=1&topic=15351В
weekevents.ru/page/40В
Предлагаем вам сделать консультацию (аудит) по увеличению продаж также прибыли в вашем бизнесе. Формат аудита: личная встреча или конференция по скайпу. Делая очевидные, но простые действия, доход от ВАШЕГО бизнеса удастся увеличить в несколькио раз. В нашем запасе более 100 испытанных утилитарных методологий увеличения результатов а также доходов. В зависимости от вашего коммерциала расчитаем для вас наиболее сильные и будем шаг за шагом внедрять.
-https://interestbook.ru/
Hello, all the time i used to check blog posts here in the early hours in the break of day, since i love to find out more and more.
http://185.63.188.115/information/?cat=4&iz_tip=102&date_min=2020-03-16&date_max=2020-03-16&new=1
http://justinsellssd.com/places-in-zagreb-the-advantages-of-workers/
http://stanmolod.ru/omolozhenie-organizma/pitanie/ostorozhno-xolesterin-dieta-pri-povyshennom-xolesterine.html
http://demo2.weboss.hk/1009/comment/html/?578.html
http://www.ppe-online.net/product/level-2-isolation-gown?page=177
This is my first time go to see at here and i am really pleassant to read everthing at single place.
http://skepdic.ru/book/titles/en/C/
http://drahthaar-forum.ru/topic8905.html
http://brizfitness.ru/glikemicheskij-indeks-produktov-tablica/
http://open-club.ru/3d-cherepica/
http://harry-harrison.ru/films/index.html
Currently it looks like Drupal is the best blogging platform available right now. (from what I’ve read) Is that what you’re using on your blog?
edmontonoilersclub.com/read-blog/287_where-can-i-buy-a-diploma-or-certificate-at-a-comfortable-price.html?mode=dayВ
lespilomaterial.ru/В
img-group.ru/Partners.htmlВ
shockmusik.ru/page/7В
webyourself.eu/blogs/317597/%D0%A1%D0%B5%D0%BA%D1%80%D0%B5%D1%82-%D1%83%D1%81%D0%BF%D0%B5%D1%85%D0%B0-%D0%9A%D0%B0%D0%BA-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B8-%D0%B8%D0%B7%D0%BC%D0%B5%D0%BD%D0%B8%D1%82%D1%8C-%D1%81%D0%B2%D0%BE%D1%8E-%D0%B6%D0%B8%D0%B7%D0%BD%D1%8CВ
В современном мире, где диплом – это начало отличной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения качественного образования. Наличие официального документа переоценить просто невозможно. Ведь диплом открывает дверь перед любым человеком, желающим вступить в сообщество профессионалов или учиться в ВУЗе.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы сможете приобрести диплом, и это будет отличным решением для всех, кто не смог завершить образование, потерял документ или хочет исправить плохие оценки. диплом изготавливается аккуратно, с максимальным вниманием к мельчайшим нюансам. В результате вы получите полностью оригинальный документ.
Преимущество этого решения заключается не только в том, что вы максимально быстро получите диплом. Весь процесс организовывается удобно, с профессиональной поддержкой. От выбора необходимого образца диплома до консультаций по заполнению личной информации и доставки по стране — все будет находиться под полным контролем качественных специалистов.
В результате, для тех, кто ищет быстрый и простой способ получить необходимый документ, наша услуга предлагает отличное решение. Приобрести диплом – значит избежать длительного процесса обучения и не теряя времени перейти к достижению своих целей, будь то поступление в университет или старт успешной карьеры.
http://bitplace.to
http://gmailspva.com
http://bakersfield-limos.com
http://xn--80adackrc1bfw.xn--p1ai
http://znich.org.ua
В нашем мире, где диплом является началом удачной карьеры в любом направлении, многие стараются найти максимально простой путь получения образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед всеми, кто собирается начать трудовую деятельность или учиться в ВУЗе.
В данном контексте наша компания предлагает оперативно получить любой необходимый документ. Вы сможете приобрести диплом, и это становится выгодным решением для всех, кто не смог закончить обучение или потерял документ. дипломы производятся аккуратно, с особым вниманием к мельчайшим деталям, чтобы в итоге получился полностью оригинальный документ.
Преимущество данного решения состоит не только в том, что вы сможете оперативно получить свой диплом. Процесс организован удобно, с нашей поддержкой. Начиная от выбора подходящего образца документа до точного заполнения персональной информации и доставки по стране — все находится под абсолютным контролем качественных мастеров.
Всем, кто пытается найти быстрый способ получения необходимого документа, наша услуга предлагает выгодное решение. Заказать диплом – это значит избежать длительного процесса обучения и сразу переходить к своим целям: к поступлению в ВУЗ или к началу трудовой карьеры.
http://hayneslanemarket.com
http://floristua.com.ua
http://grand-art.online
http://bundesliga-experte.com
http://pharmacie-tarascon.fr
레프리칸 리치스
Zhu Zaimo는 말을 타고 모두 용기를 가지고 앞으로 나아갔습니다.
Предлагаем вам сделать консультацию (аудит) по подъему продаж также прибыли в вашем бизнесе. Формат аудита: личная встреча или конференция по скайпу. Делая верные, но обыкновенные усилия, доход от ВАШЕГО коммерциала удастся поднять в много раз. В нашем арсенале более 100 испытанных фактических инструментов повышения продаж и прибыли. В зависимости от вашего бизнеса подберем для вас наиболее сильные и будем шаг за шагом претворять в жизнь.
-https://interestbook.ru
На сегодняшний день, когда аттестат – это начало успешной карьеры в любой области, многие ищут максимально быстрый путь получения качественного образования. Наличие официального документа об образовании переоценить невозможно. Ведь именно диплом открывает дверь перед людьми, желающими вступить в сообщество профессионалов или продолжить обучение в ВУЗе.
В данном контексте мы предлагаем оперативно получить этот необходимый документ. Вы сможете купить аттестат старого или нового образца, что будет удачным решением для всех, кто не смог завершить обучение, потерял документ или хочет исправить свои оценки. Каждый аттестат изготавливается аккуратно, с максимальным вниманием к мельчайшим нюансам. В результате вы сможете получить продукт, полностью соответствующий оригиналу.
Плюсы такого решения заключаются не только в том, что вы максимально быстро получите аттестат. Весь процесс организован удобно, с профессиональной поддержкой. От выбора подходящего образца до консультаций по заполнению личных данных и доставки в любое место страны — все под абсолютным контролем наших мастеров.
Всем, кто ищет оперативный способ получить требуемый документ, наша компания предлагает выгодное решение. Приобрести аттестат – это значит избежать длительного обучения и не теряя времени перейти к достижению собственных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://uyt-v-doma.ru/
Hi there! I’m at work surfing around your blog from my new iphone 4! Just wanted to say I love reading through your blog and look forward to all your posts! Keep up the great work!
http://hamperor.com.au/product/luxury-fruit-basket-with-cheese-cutting-board/
http://forum.oursson.ru/viewtopic.php?f=117&t=3341&p=10425Г‚
http://gymn2slv.ru/nintendo/valve.html
http://pechemivarim.ru/salat-iz-baklazhanov-gribnoj/
http://www.twokingscomics.com/new-secret-wars-by-hickman/
XRumer – этто самое лучшее и эффективное программное обеспечение для создания ссылок да продвижения сайтов. Оно используется чтобы автоматического прогона веб- сайтов помощью разные социальные сети, форумы, блоги также часть интернет-ресурсы.
http://www.botmasterru.com/product33230/
Купить XRumer Оффициально
В нашем обществе, где диплом становится началом удачной карьеры в любой области, многие ищут максимально простой путь получения образования. Наличие документа об образовании трудно переоценить. Ведь именно диплом открывает двери перед каждым человеком, желающим начать профессиональную деятельность или продолжить обучение в ВУЗе.
Предлагаем очень быстро получить этот важный документ. Вы имеете возможность заказать диплом, что является отличным решением для человека, который не смог завершить обучение, потерял документ или желает исправить свои оценки. Все дипломы изготавливаются с особой тщательностью, вниманием ко всем нюансам, чтобы в итоге получился 100% оригинальный документ.
Плюсы подобного решения заключаются не только в том, что вы оперативно получите диплом. Процесс организовывается комфортно, с нашей поддержкой. От выбора требуемого образца документа до грамотного заполнения личной информации и доставки по России — все находится под абсолютным контролем наших специалистов.
Для всех, кто пытается найти оперативный способ получить требуемый документ, наша компания может предложить выгодное решение. Приобрести диплом – значит избежать долгого процесса обучения и сразу переходить к важным целям: к поступлению в университет или к началу успешной карьеры.
http://daicond.com.ua
http://co1-az.ru
http://sadovod63.ru
http://vinhphatmobile.com
http://chinucoin.io
Hi there, You’ve done a fantastic job. I will certainly digg it and personally suggest to my friends. I am sure they’ll be benefited from this website.
http://www.vernisaz.ru/info14.html
http://izmailskiypark.ru/tag/respubliki-srednej-azii
http://podzhelud.ru/pankreatit/sestrinskij-process
http://fermer-expert.ru/kury-klyuyut-yajca-prichina-i-chto-delat-s-etim-nachinayushhim-pticevodam/
http://skischool-nso.ru/cats
Подарок для конкурента
https://xrumer.ru/
Такой пакет берут для переспама анкор листа сайта или мощно усилить НЧ запросы
Подарок для конкурента
Simply want to say your article is as surprising. The clearness in your post is simply nice and i can assume you are an expert on this subject. Well with your permission allow me to grab your RSS feed to keep up to date with forthcoming post. Thanks a million and please keep up the enjoyable work.
http://www.dataload.com/forum/memberlist.php?mode=joined&order=ASC&start=16950
http://followmedoitbbs.com/home.php?mod=space&username=osytyroby
http://my-sosedi.ru/contacts/
http://probiskvit.ru/belyj-hleb-v-hlebopechke.html
http://favorit-sale.ru/tag/genassambleya-oon
В нашем мире, где диплом становится началом отличной карьеры в любом направлении, многие пытаются найти максимально быстрый путь получения качественного образования. Факт наличия документа об образовании трудно переоценить. Ведь диплом открывает двери перед любым человеком, который желает вступить в сообщество профессионалов или учиться в ВУЗе.
Мы предлагаем оперативно получить любой необходимый документ. Вы можете приобрести диплом, что будет удачным решением для человека, который не смог закончить обучение или утратил документ. дипломы выпускаются аккуратно, с особым вниманием ко всем деталям. На выходе вы получите документ, максимально соответствующий оригиналу.
Превосходство этого подхода состоит не только в том, что вы сможете максимально быстро получить диплом. Процесс организован комфортно, с нашей поддержкой. Начиная от выбора подходящего образца диплома до грамотного заполнения персональной информации и доставки по стране — все под полным контролем квалифицированных специалистов.
Таким образом, всем, кто пытается найти быстрый способ получения требуемого документа, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать долгого обучения и не теряя времени перейти к личным целям: к поступлению в университет или к началу удачной карьеры.
http://labtex-equipment.ru
http://ilmuseodelrugby.it
http://calc-bank.ru
http://hermes-replica-store.com
http://lasfloresboutique.ru
A beloved National Park Service ranger died when he tripped, fell and struck his head on a rock during an annual astronomy festival in southwestern Utah, park officials said over the weekend.
kraken14.at
Tom Lorig was 78 when he died after the incident at Bryce Canyon National Park late Friday.
kraken13.at
https://kraken13.at-kraken15.at
He was known for his extensive work as a ranger and volunteer at 14 National Park Service sites, including Yosemite National Park, Carlsbad Caverns National Park and Dinosaur National Monument, the park service said in a statement Saturday.
“Tom Lorig served Bryce Canyon, the National Park Service, and the public as an interpretive park ranger, forging connections between the world and these special places that he loved,” Bryce Canyon Superintendent Jim Ireland said in the statement.
Что такое прогон по профилям?
На форумах при каждом прогоне регистрируется новый юзер, в профайле которого в поле Вебсайт или Домашняя страница стоит ваша ссылка. Также в некоторых форумах есть возможность в профиль вставить подпись, туда можно вставить ссылку с анкором. Все это можно варьировать. В профилях ссылки живут довольно долго, т.к. нет спама в чистом виде, и профили не мозолят глаза ни читателям форума, ни модераторам. Прогон производится последней версией хрумера с сервера с максимально возможной скоростью и производительностью.
ПОДРОБНЕЕ
НОВЫЙ ОТЛИЧНЫЙ VPN ДЛЯ XRUMERA!
За последние недели состоялась серия важных обновлений комплекса: XAuth, Captchas Benchmark, XRumer, XEvil 6.0 . В XEvil 6.0 обновлен механизм обработки hCaptcha и многое другое, в XAuth повышена стабильность работы, в XRumer повышена эффективность, устранён ряд погрешностей и обновлены прилагаемые базы.
https://xrumer.ru/
В нашем обществе, где диплом становится началом удачной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа об образовании трудно переоценить. Ведь диплом открывает двери перед любым человеком, который желает начать трудовую деятельность или учиться в каком-либо институте.
В данном контексте наша компания предлагает оперативно получить этот важный документ. Вы сможете приобрести диплом нового или старого образца, и это является выгодным решением для человека, который не смог завершить образование или утратил документ. Все дипломы выпускаются с особой тщательностью, вниманием ко всем нюансам, чтобы на выходе получился полностью оригинальный документ.
Преимущества такого решения состоят не только в том, что вы максимально быстро получите диплом. Процесс организован просто и легко, с профессиональной поддержкой. Начав от выбора необходимого образца до консультаций по заполнению персональной информации и доставки по стране — все под полным контролем наших мастеров.
В результате, для тех, кто пытается найти оперативный способ получить необходимый документ, наша компания может предложить отличное решение. Купить диплом – это значит избежать долгого обучения и не теряя времени переходить к своим целям: к поступлению в университет или к началу удачной карьеры.
http://bouffordca.com
http://luvmor.ru
http://messcontrol.biz
http://medhansh.net
http://thevintagedrink.com
I blog frequently and I really thank you for your content. The article has truly peaked my interest. I will bookmark your blog and keep checking for new information about once per week. I opted in for your RSS feed too.
http://extreme-voyage.ru/2015/05/04/mnogocelevoj-takticheskij-tomagavk-browning-shock-n/
http://cdoctor.ru/bartolinit-simptomy-i-lechenie-v-domashnih-usloviyah/
http://mailpresident.ru/node/12924?page=4
http://proinfekcii.ru/venericheskie/sifilis/sifilis-bytovojj.html
http://stlouisbluesclub.com/read-blog/339_what-should-i-pay-attention-to-when-buying-a-diploma-or-certificate.html
Hi! I’m at work surfing around your blog from my new iphone! Just wanted to say I love reading through your blog and look forward to all your posts! Keep up the fantastic work!
http://economics-lib.ru/books/item/f00/s00/z0000035/st005.shtml
http://www.longlive.com/node/433
http://anahnu.club/he/jomsocial/groups/viewbulletin/155-решили-купить-диплом-открывайте-наш-онлайн-магазин.html?groupid=1
http://sibledy.ru/thread-315316-1-1.html
http://mezhdunarodnik.ru/digest/smi/21.html
В наше время, когда диплом – это начало удачной карьеры в любом направлении, многие ищут максимально простой путь получения образования. Важность наличия документа об образовании сложно переоценить. Ведь диплом открывает двери перед людьми, стремящимися начать трудовую деятельность или учиться в ВУЗе.
В данном контексте наша компания предлагает быстро получить этот важный документ. Вы имеете возможность купить диплом старого или нового образца, и это становится удачным решением для всех, кто не смог закончить обучение или утратил документ. Любой диплом изготавливается с особой тщательностью, вниманием ко всем элементам, чтобы на выходе получился документ, максимально соответствующий оригиналу.
Преимущества такого подхода заключаются не только в том, что можно быстро получить свой диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начиная от выбора нужного образца до грамотного заполнения персональных данных и доставки в любой регион России — все будет находиться под полным контролем квалифицированных мастеров.
Всем, кто ищет быстрый способ получить требуемый документ, наша услуга предлагает выгодное решение. Купить диплом – значит избежать долгого обучения и сразу перейти к достижению личных целей: к поступлению в университет или к началу удачной карьеры.
http://alpacavsllama.com
http://remont-kozhi.ru
http://wpplugintheme.com
http://jaezfinancialgroup.icu
http://safepsychology.ru
I got this website from my friend who told me regarding this website and now this time I am visiting this website and reading very informative content at this place.
http://ifeelstrong.ru/anatomy/body/anabolizm-i-katabolizm-osnovnye-ponyatiya.html
http://iz-bymagi.ru/chelovechki-iz-bumagi/dart-veyder-iz-bumagi/
http://www.camry-club.ru/member.php/61231-sonnick84?tab=activitystream&type=all&page=4
http://time-person.ru/e/egorov.html
http://www.dr-kumaki.net/aikuma/index.cgi?mode=past&pno=0036&pg=100
Hi colleagues, its impressive paragraph about teachingand fully explained, keep it up all the time.
http://angelteam.uv.ro/memberlist.php?mode=joined&order=ASC&start=208150
http://185.63.188.115/information/?cat=4&iz_tip=102&date_min=2020-03-16&date_max=2020-03-16&new=1
http://lasfloresboutique.ru/product-page/букет-из-роз-и-астральмерии
http://nehomesdeaf.org/polezno-znat/kak-napisat-diplomnuju-po-pravu/
http://opt-dostawka.ru/preimushhestva-obrazovaniya-v-rossii-dlya-inostrannyx-studentov/
Right here is the right web site for everyone who really wants to understand this topic. You understand a whole lot its almost tough to argue with you (not that I personally will need to…HaHa). You certainly put a brand new spin on a topic that’s been discussed for ages. Wonderful stuff, just excellent!
http://myasoed96.ru/audiokniga.html
http://www.ettoresilini.ch/2017/07/18/da-mio-giardino-con-furore-dictyophara-europea/
http://forum.rusbg.com/viewtopic.php?f=116&t=3549&p=12509
http://gymn2slv.ru/nintendo/valve.html
http://you-ps.ru/page/39/
В нашем мире, где диплом становится началом отличной карьеры в любом направлении, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Необходимость наличия официального документа переоценить просто невозможно. Ведь именно он открывает двери перед любым человеком, который стремится вступить в профессиональное сообщество или продолжить обучение в любом институте.
В данном контексте мы предлагаем оперативно получить этот важный документ. Вы имеете возможность заказать диплом, и это является выгодным решением для всех, кто не смог завершить образование или утратил документ. Каждый диплом изготавливается с особой аккуратностью, вниманием к мельчайшим деталям, чтобы в итоге получился продукт, максимально соответствующий оригиналу.
Преимущества этого решения заключаются не только в том, что можно быстро получить диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. От выбора нужного образца документа до консультации по заполнению персональной информации и доставки по России — все под полным контролем квалифицированных специалистов.
Всем, кто пытается найти оперативный способ получить необходимый документ, наша компания готова предложить выгодное решение. Приобрести диплом – это значит избежать долгого процесса обучения и сразу перейти к личным целям: к поступлению в ВУЗ или к началу успешной карьеры.
http://messcontrol.de
http://diaocbinhduong.org
http://svitozar.com.ua
http://grandart.fr
http://myfashionworld.fr
Hello i am kavin, its my first occasion to commenting anyplace, when i read this piece of writing i thought i could also make comment due to this good piece of writing.
http://www.9020blog.com/board/blogs/2861/We-order-a-university-diploma-on-the-Internet-Expert-Recommendations
http://ornithologist.ru/d.html
http://2foru.pl/news/watch-this-sound-by-konzept/comment-page-7567/
http://www.4mark.net/story/11478170/community
http://electri4ka.com/4s2/41.html
Hello! I know this is kinda off topic but I’d figured I’d ask. Would you be interested in trading links or maybe guest authoring a blog article or vice-versa? My blog goes over a lot of the same subjects as yours and I feel we could greatly benefit from each other. If you might be interested feel free to shoot me an e-mail. I look forward to hearing from you! Excellent blog by the way!
http://ya-rukodelnitsa.ru/load/griby/34
http://fitrain.ru/fitness/page/7
http://de-gustare.it/tag/lombardia/
http://delivery-containers.ru/photogallery/containers/containers_20futov/
http://namtaru.ru/seriinye-maniyaki/item/77-chester-dyuejn-terner.html
https://seoforecastable.goabroadblog.com/27003731/Обзор-возможностей-xrumer-art
https://seopressio.affiliatblogger.com/79527958/Преимущества-сотрудничества-со-студией-xrumer-art
В нашем обществе, где диплом – это начало отличной карьеры в любой отрасли, многие ищут максимально быстрый и простой путь получения качественного образования. Необходимость наличия документа об образовании переоценить попросту невозможно. Ведь диплом открывает двери перед любым человеком, который хочет начать профессиональную деятельность или учиться в высшем учебном заведении.
Предлагаем очень быстро получить этот необходимый документ. Вы имеете возможность заказать диплом старого или нового образца, что становится удачным решением для человека, который не смог завершить образование, утратил документ или хочет исправить свои оценки. Каждый диплом изготавливается аккуратно, с особым вниманием ко всем нюансам, чтобы на выходе получился 100% оригинальный документ.
Преимущества данного решения заключаются не только в том, что можно оперативно получить диплом. Весь процесс организовывается удобно, с нашей поддержкой. Начав от выбора необходимого образца до консультаций по заполнению персональных данных и доставки в любой регион России — все находится под полным контролем наших специалистов.
В итоге, всем, кто ищет оперативный способ получить необходимый документ, наша компания может предложить отличное решение. Приобрести диплом – это значит избежать продолжительного обучения и сразу переходить к достижению собственных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://otantravel.kz
http://keslaser.com.ua
http://sometimesnaive.org
http://ahninniah.graphics
http://govinsm.com
Daddy Casino – Официальный телеграм канал со слотами от Дэдди Casino дэдди казино . Актуальное, рабочее зеркало официального сайта Дэдди Казино. Регистрируйся в Казино Daddy, получай бонус используя промокод, не забудь скачать
daddy казино официальный
https://t.me/daddycasinorussia
Интернет-казино дэдди казино официальный сайт — сайт или специальная программа, которые позволяют играть в азартные игры в интернете. Сайты интернет-казино могут специализироваться на различных азартных играх: игровых автоматах, рулетке, лотереях, викторинах, покере или других карточных и настольных играх. Википедия
daddy casino официальный
https://t.me/daddycasinorussia
5 래빗스 메가웨이즈
“맞아요, 죽음이라는 비밀 보고가 있어요…끔찍해요!” 무빈이 무심하게 말했다.
В нашем обществе, где диплом – это начало успешной карьеры в любой отрасли, многие пытаются найти максимально простой путь получения качественного образования. Важность наличия официального документа трудно переоценить. Ведь диплом открывает двери перед всеми, кто собирается начать трудовую деятельность или продолжить обучение в ВУЗе.
Наша компания предлагает очень быстро получить этот важный документ. Вы сможете приобрести диплом, что является удачным решением для человека, который не смог завершить образование или потерял документ. Все дипломы изготавливаются аккуратно, с максимальным вниманием ко всем деталям. На выходе вы получите 100% оригинальный документ.
Преимущество этого решения состоит не только в том, что вы максимально быстро получите диплом. Процесс организовывается удобно и легко, с профессиональной поддержкой. От выбора подходящего образца до точного заполнения личных данных и доставки по России — все находится под абсолютным контролем квалифицированных мастеров.
Для тех, кто ищет быстрый способ получения требуемого документа, наша компания предлагает выгодное решение. Купить диплом – это значит избежать долгого процесса обучения и сразу переходить к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
http://getxtnd.com
http://rent-in-odessa.com
http://brustvergroberungx.de
http://onedetailing.pl
http://centrumrendezveny.hu
Телесуфлёр — этто tvprompter.ru я, тот или другой подсобляет ораторам (а) также основным телепередач читать текст, никак не вырывая взгляда от камеры чи аудитории. Оно элементарно размещается под на камере (а) также отражает экспликация один-другой подмогой просвечивающего зеркала, творя иллюзию ровного воззрения в течение объектив. Этот инструмент незаменим у проведении прямых эфиров также эпохально упрощает работу телеведущих.
http://tvprompter.ru/
Обучение кайтсёрфингу, кайт что это, кайт это обучение кайтингу в хургаде с нуля за три дня! В кайт школе Red Sea Kite. проживание, прокат, трансефер. Хургада действительно является одним из лучших мест для обучения кайтсерфингу. Египетское побережье Красного моря известно своими благоприятными ветрами, которые создают идеальные условия для занятий этим видом спорта. Профессиональные инструкторы и школы кайтсерфинга будут рады помочь как начинающим, так и опытным райдерам развивать свои навыки. На фоне кристально чистой воды и бескрайних песчаных пляжей, обучение кайтсерфингу в Хургаде становится еще более захватывающим и увлекательным. Кроме того, здесь можно найти широкий выбор аренды оборудования и учебных программ, чтобы каждый человек мог выбрать подходящий для себя формат обучения. Не только обучение, но и само кайтсерфинг в Хургаде отличается особой атмосферой и весельем. Ежегодно здесь проходят многочисленные мероприятия, такие как соревнования и фестивали, которые привлекают к себе внимание как местных жителей, так и туристов со всего мира. Поэтому, погрузившись в эту культуру и атмосферу, вы точно не останетесь равнодушными к миру кайтсерфинга в Хургаде. Так же мы проводим обучение дайвингу в хургаде и погружения в Египте. обучение кайтсёрфингу в Египте Таким образом, Хургада является идеальным местом не только для начинающих, но и для опытных кайтсерферов. Здесь вы сможете не только научиться кататься на кайте, но и насладиться невероятными пляжами, теплым климатом и гостеприимством местных жителей. Все вместе это создает уникальную среду для любителей кайтсерфинга, которые хотят окунуться в мир приключений и новых возможностей. кайт школа хургада Они предлагают разнообразные программы обучения для кайтсерфинга, как для начинающих, так и для опытных райдеров. Инструкторы школы REDSEAKITE в Хургаде имеют сертификаты IKO (International Kiteboarding Organization) и гарантируют безопасное и эффективное обучение. Курсы проводятся в небольших группах или индивидуально, что позволяет каждому ученику получить индивидуальное внимание и сосредоточиться на своем прогрессе. Наш партнёр по султан кайт хургада кайт школа Дети ветра. Школа Red Sea Kite располагает современным оборудованием и обучает на пляже с отличными условиями для кайтсерфинга. Преподаватели предоставляют все необходимые знания о безопасности, технике и тактике кайтсерфинга, чтобы ученики могли безопасно и уверенно развивать свои навыки. Кроме того, школа REDSEAKITE также проводит индивидуальные тренировки и интенсивные курсы для тех, кто хочет быстро улучшить свое мастерство. Прокат сапов в Анапе Благодаря благоприятным условиям для кайтсерфинга в Египте и профессиональным инструкторам Дети Ветра, ученики быстро осваивают новое увлекательное умение и испытывают незабываемые впечатления от катания. Школа Дети Ветра в Хургаде предлагает не только курсы обучения, но и прокат оборудования и организацию экскурсий для всех ценителей кайтсерфинга в Египте. Напишите нам в кайт обучение в Египте Мы на яндекс playkite Мы в инстаграмм кайт школа в Египте Наша кайт школа и станция высокого уровня в Хургаде, с богатым опытом работы. Наша команда профессионалов, основанная в 2001 году, предлагает уникальные авторские методики обучения. За более чем 20 лет мы помогли множеству новичков достигнуть заметных результатов уже после первых тренировок. Мы искренне верим в наши слова: “Ставим на доску за 2 дня!” И это не просто пустые обещания – это наша гарантированная практика. Мы индивидуально подходим к каждому ученику, учитывая его физическую подготовку, уровень навыков и личные предпочтения. кайтинг хургада В нашей кайт-школе мы строим обучение так, чтобы каждый чувствовал себя комфортно и получал максимальную пользу от тренировок. В дополнение к обучению, наша кайт-школа проводит интересные кайт-кемпы и кайт-сафари с возможностью неограниченного катания. Также мы помогаем с подбором и приобретением всего необходимого оборудования для самостоятельного катания. И все это доступно по самым разумным ценам! У нас вы можете начать с нуля и превратиться в настоящего профессионала. Кайт-серфинг – это на самом деле очень просто! Вы полюбите этот вид спорта, и мы хотим помочь вам в этом! кайт станция в Египте Индивидуальный ИНСТРУКТОР для вашего ребёнка, научим быстро качествено и самое главное безопасно! Набор детей на обучение кайтсёрфингу с 7 лет, и весом от 30кг. За подробностями в ЛС. Кайтсёрфинг это просто и мы вам в этом поможем DETIVETRA https://kitehurghada.ru http://redseakite.ru https://vk.com/kite_deti_vetra https://paradisekitecenter.ru https://www.instagram.com https://yandex.ru/maps/org/kayt_tsentr/177183726173 https://t.me/detivetrachat https://detivetra.ru https://kiteschoolhurghada.ru
A beloved National Park Service ranger died when he tripped, fell and struck his head on a rock during an annual astronomy festival in southwestern Utah, park officials said over the weekend.
[url=https://kraken13at.vip]kraken14.at[/url]
Tom Lorig was 78 when he died after the incident at Bryce Canyon National Park late Friday.
kraken13.at
https://at-kraken17.at
He was known for his extensive work as a ranger and volunteer at 14 National Park Service sites, including Yosemite National Park, Carlsbad Caverns National Park and Dinosaur National Monument, the park service said in a statement Saturday.
“Tom Lorig served Bryce Canyon, the National Park Service, and the public as an interpretive park ranger, forging connections between the world and these special places that he loved,” Bryce Canyon Superintendent Jim Ireland said in the statement.
What’s up colleagues, good paragraph and pleasant urging commented at this place, I am truly enjoying by these.
http://musey-uglich.ru/
В современном мире, где диплом является началом удачной карьеры в любой отрасли, многие пытаются найти максимально быстрый путь получения качественного образования. Важность наличия документа об образовании сложно переоценить. Ведь именно он открывает двери перед людьми, желающими вступить в сообщество профессиональных специалистов или учиться в университете.
В данном контексте мы предлагаем максимально быстро получить этот важный документ. Вы сможете приобрести диплом, что становится выгодным решением для всех, кто не смог закончить образование, утратил документ или желает исправить свои оценки. диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам, чтобы в результате получился документ, 100% соответствующий оригиналу.
Преимущество этого подхода состоит не только в том, что можно быстро получить диплом. Процесс организован комфортно, с нашей поддержкой. От выбора подходящего образца до правильного заполнения персональных данных и доставки по России — все находится под абсолютным контролем качественных специалистов.
Таким образом, для тех, кто ищет быстрый и простой способ получить необходимый документ, наша компания предлагает выгодное решение. Приобрести диплом – значит избежать длительного обучения и сразу переходить к достижению собственных целей: к поступлению в университет или к началу успешной карьеры.
seoyour.ru
В наше время, когда диплом – это начало успешной карьеры в любой отрасли, многие пытаются найти максимально быстрый и простой путь получения качественного образования. Важность наличия документа об образовании трудно переоценить. Ведь именно он открывает дверь перед всеми, кто стремится вступить в сообщество профессиональных специалистов или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает оперативно получить этот важный документ. Вы можете приобрести диплом, что становится удачным решением для всех, кто не смог закончить обучение или потерял документ. диплом изготавливается аккуратно, с особым вниманием к мельчайшим деталям, чтобы в итоге получился продукт, полностью соответствующий оригиналу.
Плюсы этого подхода заключаются не только в том, что можно максимально быстро получить свой диплом. Весь процесс организован удобно, с профессиональной поддержкой. Начав от выбора требуемого образца до консультаций по заполнению персональной информации и доставки по стране — все будет находиться под абсолютным контролем наших специалистов.
Всем, кто пытается найти быстрый и простой способ получить необходимый документ, наша компания предлагает отличное решение. Купить диплом – значит избежать продолжительного процесса обучения и не теряя времени переходить к личным целям: к поступлению в ВУЗ или к началу трудовой карьеры.
seoyour.ru
Сегодня, когда диплом является началом удачной карьеры в любой области, многие стараются найти максимально быстрый и простой путь получения качественного образования. Факт наличия официального документа об образовании сложно переоценить. Ведь диплом открывает двери перед всеми, кто хочет вступить в сообщество профессионалов или учиться в ВУЗе.
Предлагаем очень быстро получить этот необходимый документ. Вы можете купить диплом старого или нового образца, и это является удачным решением для всех, кто не смог закончить обучение или потерял документ. Каждый диплом изготавливается с особой аккуратностью, вниманием к мельчайшим деталям, чтобы в результате получился полностью оригинальный документ.
Преимущество данного подхода заключается не только в том, что вы сможете максимально быстро получить диплом. Процесс организован удобно, с профессиональной поддержкой. От выбора необходимого образца диплома до консультации по заполнению личных данных и доставки в любой регион России — все находится под абсолютным контролем наших специалистов.
Для тех, кто хочет найти оперативный способ получения необходимого документа, наша компания предлагает отличное решение. Купить диплом – это значит избежать продолжительного процесса обучения и не теряя времени перейти к своим целям: к поступлению в университет или к началу успешной карьеры.
seoyour.ru
http://www.five-respect.co.jp/bbs/sunbbs.cgi?mode=form&no=126554340&page=%20,3085,26188 – %20->%20POST%20http://www.five-respect.co.jp/bbs/regist.cgi%20,0,1034 –
http://obscurita-eterna.de/index.php/guestbook/index/index/page/www.elmiron.com%5B/www.viagra.com%5B/www.viagra.com%5B/www.tadapox.com%5B/www.levitra.com%5B/URL
https://als3ed.com/Main/Ar/product/packaged-cardboard/
https://www.yolospeak.pl/newreply.php?tid=230&replyto=16725
http://www.five-respect.co.jp/bbs/sunbbs.cgi?mode=form&no=126685528&page=
В нашем мире, где диплом становится началом отличной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения качественного образования. Важность наличия официального документа об образовании переоценить невозможно. Ведь именно он открывает двери перед каждым человеком, который желает начать профессиональную деятельность или продолжить обучение в ВУЗе.
В данном контексте наша компания предлагает оперативно получить этот важный документ. Вы сможете купить диплом старого или нового образца, что является удачным решением для всех, кто не смог завершить образование или потерял документ. Все дипломы изготавливаются с особой тщательностью, вниманием ко всем нюансам, чтобы на выходе получился полностью оригинальный документ.
Плюсы такого решения состоят не только в том, что вы сможете максимально быстро получить диплом. Процесс организовывается просто и легко, с профессиональной поддержкой. Начав от выбора подходящего образца диплома до консультаций по заполнению личной информации и доставки по России — все под абсолютным контролем опытных мастеров.
В результате, для всех, кто ищет быстрый и простой способ получить требуемый документ, наша услуга предлагает выгодное решение. Приобрести диплом – это значит избежать долгого процесса обучения и сразу переходить к достижению своих целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://primuothod.ru
http://ruscuisineambassador.ru
http://psypokolenie.ru
http://variant-surgut.ru
http://sk-sh.ru
Thank you for sharing your info. I truly appreciate your efforts and I will be waiting for your further post thank you once again.
http://ffatal.ru/венчик/
купить диплом в севастополе
http://opendatachel.ru
купить диплом логопеда
Hello it’s me, I am also visiting this web page regularly, this website is actually fastidious and the visitors are genuinely sharing fastidious thoughts.
купить диплом в новочебоксарске
http://medhelp-dent.ru
http://cleo-dance.ru
http://studenta-blog.ru
купить диплом о среднем образовании
카지노 슬롯
관리들에게는 폐하와 주요 장관들이 이에 대해 논의한 것이 분명합니다.
В современном мире, где диплом становится началом отличной карьеры в любой области, многие пытаются найти максимально быстрый путь получения качественного образования. Наличие официального документа переоценить невозможно. Ведь диплом открывает двери перед каждым человеком, который хочет начать профессиональную деятельность или продолжить обучение в каком-либо университете.
Наша компания предлагает быстро получить любой необходимый документ. Вы имеете возможность приобрести диплом, и это становится выгодным решением для всех, кто не смог закончить образование, утратил документ или желает исправить плохие оценки. Каждый диплом изготавливается с особой тщательностью, вниманием ко всем элементам, чтобы в итоге получился документ, 100% соответствующий оригиналу.
Превосходство данного подхода состоит не только в том, что можно быстро получить свой диплом. Процесс организовывается удобно, с профессиональной поддержкой. Начиная от выбора требуемого образца до правильного заполнения личных данных и доставки по стране — все находится под полным контролем опытных специалистов.
Всем, кто ищет быстрый и простой способ получить требуемый документ, наша компания готова предложить отличное решение. Заказать диплом – значит избежать продолжительного процесса обучения и сразу переходить к своим целям, будь то поступление в университет или начало карьеры.
http://teh-diplom.ru
http://n-seo.ru
http://samara-shkola.ru
http://nti-nastavnik.ru
http://opexobo.ru
Hi, this weekend is nice for me, since this point in time i am reading this wonderful informative post here at my home.
купить диплом в смоленске
http://school5-priozersk.ru
http://konkurs-technokrat.ru
http://rus-students.ru
купить диплом в славянске-на-кубани
whoah this blog is magnificent i love reading your posts. Stay up the great work! You understand, lots of persons are looking round for this info, you could aid them greatly.
купить дипломы о высшем с занесением
http://sakskiy-rayon.ru
http://energoprofi23.ru
http://kritikus.ru
купить диплом моториста
Wow, marvelous blog layout! How long have you been blogging for? you make blogging look easy. The overall look of your website is excellent, let alone the content!
купить диплом в сургуте
http://ptncl.ru
http://cardioevent.ru
http://37vilok.ru
купить диплом в йошкар-оле
В нашем мире, где диплом становится началом успешной карьеры в любом направлении, многие ищут максимально простой путь получения качественного образования. Наличие документа об образовании трудно переоценить. Ведь диплом открывает дверь перед людьми, стремящимися вступить в профессиональное сообщество или учиться в ВУЗе.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы сможете приобрести диплом, и это становится отличным решением для человека, который не смог завершить образование, потерял документ или желает исправить свои оценки. Все дипломы изготавливаются с особой тщательностью, вниманием ко всем элементам, чтобы в результате получился продукт, максимально соответствующий оригиналу.
Плюсы подобного решения заключаются не только в том, что можно оперативно получить диплом. Весь процесс организовывается просто и легко, с профессиональной поддержкой. От выбора подходящего образца диплома до консультаций по заполнению личной информации и доставки по России — все находится под абсолютным контролем наших мастеров.
Таким образом, для тех, кто ищет оперативный способ получить необходимый документ, наша компания предлагает выгодное решение. Купить диплом – это значит избежать длительного обучения и не теряя времени переходить к важным целям, будь то поступление в ВУЗ или начало профессиональной карьеры.
http://collegevalday.ru
http://dialognsu.ru
http://postupai-svfu.ru
http://prohalal-faunia.ru
http://monitoringntr.ru
What’s up, its pleasant piece of writing concerning media print, we all understand media is a fantastic source of facts.
http://skischool-nso.ru/cats
купить диплом в анжеро-судженске
http://agpi.info
купить диплом в владикавказе
Thank you for some other excellent post. The place else may anybody get that kind of info in such an ideal manner of writing? I’ve a presentation subsequent week, and I am on the look for such information.
http://ertegi.ru/index.php?id=2&idnametext=145&idpg=1
купить диплом охранника
http://stodorova.ru
купить дипломы о высшем
Духовное развитие или как изменить свою жизнь к лучшему.
Вы уже много знаете про миссию и предназначение, много читаете и смотрите фильмов, видео, проходили не раз обучение в различных эзотерических школах и коуч-курсах, но до сих пор не знаете, как применять полученные знания и найти себя?
Мы приглашаем Вас на канал «Послания в стихах», где вся информация предоставляется бесплатно! Послушать и посмотреть на распространённых площадках в России и за рубежом.
Мы есть на youtube, dzen.ru, rutube.ru, boosty.to!
Здесь Вы найдёте много полезной и понятной информации, трогающей Вашу душу и раскрывающей смысл духовного развития, самопознания и самосовершенствования!
Читая и слушая их, Вы:
– Пробудите в себе божественные знания и силы;
– Осознаете собственный жизненный путь;
– Узнаете о глубинных причинах многих болезней и рекомендации по исцелению;
Ищите круг друзей, единомышленников и приятное общение
Тогда присоединяйтесь к «Посланиям в Стихах» в любимых соцсетях ВКонтакте и Телеграм!
Короткие и простые видеоролики с легко воспринимаемым смыслом и текстом позволят Вам прочувствовать тонкие энергии, переданные через «Послания в стихах».
Вы сможете узнать много нового и интересного в мире взаимоотношений, возможностей самоисцеления и тайн нашего мироздания!
Добро пожаловать на площадку Посланий в стихах!
https://f5fashion.vn/tong-hop-54-ve-hinh-nen-iphone-vo-moi-nhat/
https://f5fashion.vn/nhung-khu-vuon-tren-cao-noi-tieng-o-singapore/
http://www.five-respect.co.jp/bbs/sunbbs.cgi?mode=form&no=126672743&page=
https://americannewsdigest24.com/%E7%BE%8E%E5%9B%BD%E7%96%AB%E6%83%85%E5%8F%8A%E6%96%B0%E9%97%BB%E7%AE%80%E6%8A%A5%EF%BC%8805-28-2022%EF%BC%89/comment-page-425/
http://www.nkrs.rsko.cz/phpbb/posting.php?mode=quote&f=118&p=3445370
Hey there! I understand this is kind of off-topic but I needed to ask. Does managing a well-established blog such as yours take a massive amount work? I am brand new to writing a blog however I do write in my journal on a daily basis. I’d like to start a blog so I can easily share my own experience and feelings online. Please let me know if you have any ideas or tips for new aspiring bloggers. Appreciate it!
купить диплом кандидата наук
http://alcomagia.ru
http://37vilok.ru
http://transcollege.ru
купить диплом в россоши
Подарок для конкурента
https://xrumer.ru/
Такой пакет берут для переспама анкор листа сайта или мощно усилить НЧ запросы
Подарок для конкурента
Hello Dear, are you really visiting this site daily, if so after that you will definitely get pleasant know-how.
http://ifeelstrong.ru/anatomy/body/anabolizm-i-katabolizm-osnovnye-ponyatiya.html
купить диплом в сочи
http://extra-art.ru
купить диплом в камышине
That is really interesting, You’re an overly professional blogger. I’ve joined your rss feed and look forward to looking for more of your magnificent post. Also, I’ve shared your site in my social networks
купить диплом в рубцовске
http://spravka-v-bassein.ru
http://scienceanded.ru
http://teh-diplom.ru
купить диплом в находке
A beloved National Park Service ranger died when he tripped, fell and struck his head on a rock during an annual astronomy festival in southwestern Utah, park officials said over the weekend.
kraken14.at
Tom Lorig was 78 when he died after the incident at Bryce Canyon National Park late Friday.
kraken14.at
https://kraken13at.de
He was known for his extensive work as a ranger and volunteer at 14 National Park Service sites, including Yosemite National Park, Carlsbad Caverns National Park and Dinosaur National Monument, the park service said in a statement Saturday.
“Tom Lorig served Bryce Canyon, the National Park Service, and the public as an interpretive park ranger, forging connections between the world and these special places that he loved,” Bryce Canyon Superintendent Jim Ireland said in the statement.
Hello i am kavin, its my first occasion to commenting anywhere, when i read this piece of writing i thought i could also make comment due to this good post.
http://miupsik.ru/forums/showthread.php?tid=20811
http://camillacastro.us/forums/viewtopic.php?pid=333721#p333721
https://rf-lowrate.ru/index.php?/topic/4144-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BE%D0%B1-%D0%BE%D0%B1%D1%80%D0%B0%D0%B7%D0%BE%D0%B2%D0%B0%D0%BD%D0%B8%D0%B8-%D1%81-%D1%80%D0%B5%D0%B5%D1%81%D1%82%D1%80%D0%BE%D0%BC-f436c/
http://silverdews.ru/forum/index.php
77lub.ru/products/small-engine-oils/В
An impressive share! I have just forwarded this onto a friend who has been conducting a little homework on this. And he in fact bought me lunch because I stumbled upon it for him… lol. So let me reword this…. Thanks for the meal!! But yeah, thanx for spending time to discuss this matter here on your web site.
купить диплом техника
http://ivavai.ru
http://kupi-spravku.ru
http://unikum-student.ru
купить диплом с занесением в реестр
Do you have a spam problem on this blog; I also am a blogger, and I was curious about your situation; many of us have developed some nice procedures and we are looking to exchange methods with others, why not shoot me an email if interested.
http://altajska.cz/forum/user/2383-emuneni
http://newsinweek.ru/poluchite-diplom-kotoryiy-podtverdit-vashu-kompetentnost-i-opyit-legko-i-byistro/
http://revolverp.forumex.ru/viewtopic.php?f=11&t=222
http://avtobestnews.ru/vash-diplom-legko-i-byistro-kak-sdelat-pravilnyiy-vyibor
worldavtonew.ru/page/3В
В нашем мире, где диплом является началом успешной карьеры в любой отрасли, многие ищут максимально простой путь получения образования. Важность наличия документа об образовании сложно переоценить. Ведь диплом открывает двери перед любым человеком, желающим начать профессиональную деятельность или учиться в ВУЗе.
Предлагаем быстро получить этот необходимый документ. Вы имеете возможность приобрести диплом нового или старого образца, что становится удачным решением для человека, который не смог завершить образование, утратил документ или хочет исправить плохие оценки. Каждый диплом изготавливается с особой тщательностью, вниманием к мельчайшим нюансам. В итоге вы сможете получить документ, полностью соответствующий оригиналу.
Плюсы подобного подхода заключаются не только в том, что можно быстро получить свой диплом. Весь процесс организован удобно, с нашей поддержкой. От выбора требуемого образца до точного заполнения личной информации и доставки в любое место России — все под абсолютным контролем квалифицированных мастеров.
Всем, кто хочет найти быстрый и простой способ получить необходимый документ, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и не теряя времени перейти к личным целям, будь то поступление в университет или старт карьеры.
http://lawdiplom.ru
http://kritikus.ru
http://sartvo.ru
http://sutangarsk.ru
http://школа6.рф
Heya superb website! Does running a blog like this take a massive amount work? I’ve no understanding of computer programming however I was hoping to start my own blog soon. Anyway, if you have any recommendations or techniques for new blog owners please share. I understand this is off topic however I just needed to ask. Appreciate it!
купить диплом в ессентуках
http://postupai-svfu.ru
http://ussursity.ru
http://femcfas.ru
купить диплом цена
What a material of un-ambiguity and preserveness of precious know-how about unexpected emotions.
http://severskyray.ru/tag/moskva
купить дипломы о высшем
http://websweb.ru
купить диплом в армавире
It’s amazing to pay a visit this web site and reading the views of all colleagues regarding this post, while I am also eager of getting experience.
купить диплом в мурманске
http://raduga-bezopasnosti.ru
http://e-crimemoot.ru
http://school-internat5.ru
купить диплом педагога
Hello there I am so grateful I found your site, I really found you by error, while I was looking on Aol for something else, Anyways I am here now and would just like to say thanks a lot for a remarkable post and a all round enjoyable blog (I also love the theme/design), I don’t have time to read it all at the minute but I have bookmarked it and also added in your RSS feeds, so when I have time I will be back to read more, Please do keep up the excellent jo.
http://mir-kamnej.ru/books/02books.html
купить диплом в новокуйбышевске
http://primuothod.ru
купить диплом в кирове
It’s amazing in support of me to have a site, which is helpful in favor of my experience. thanks admin
http://php-s.ru/photogallery/Coppermine/demo/profile.php?uid=155493
https://portfolio.newschool.edu/lant053/2017/04/27/vis-comm-book/#comment-47429
http://brenderupmediterrane.free.fr/index.php?file=Members&op=detail&autor=ynoba
https://wiki.nikiforov.ru/index.php/пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ,_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ
http://www.josegregoriodelarivera.com/component/fst/?view=test&prodid=0
This is my first time pay a visit at here and i am really impressed to read everthing at single place.
купить диплом в кирове
http://1001diplom.ru
http://seozalex.ru
http://git-ugra.ru
купить диплом в казани
Pretty! This was an incredibly wonderful post. Thank you for supplying this info.
http://ветерантюмгео.рф/novosti-meropriyatiya/page/4/
купить диплом в бердске
http://школа6.рф
купить диплом в новоуральске
Hey! Do you know if they make any plugins to protect against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any recommendations?
http://heimur.ru/index.php?/topic/79-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-z36t/
https://bezone.ru/node/334519
https://forum.l2gavno.ru/threads/kupit-attestat-ob-okonchanii-11-klassov-v444i.1637/
http://mmix.ukrbb.net/viewtopic.php?f=23&t=20359
https://talkitter.com/create-blog/
Pretty element of content. I just stumbled upon your blog and in accession capital to claim that I get in fact loved account your blog posts. Anyway I will be subscribing in your feeds or even I fulfillment you get right of entry to persistently rapidly.
http://academy4elements.mybb.ru/viewtopic.php?id=861#p1880
http://www.musichunt.pro/blogs/view.htm?id=78780
https://meneger23.ru/en/forums-en/user/1423-robertgmom
https://forum.papbio.org/member.php?action=profile&uid=69840
http://lobbibar.ru/reviews/#comment_34755/
Admiring the time and energy you put into your blog and detailed information you present. It’s great to come across a blog every once in a while that isn’t the same unwanted rehashed material. Fantastic read! I’ve saved your site and I’m adding your RSS feeds to my Google account.
купить диплом в томске
http://artforum.pro
http://hackspace-spb.ru
http://lawdiplom.ru
купить диплом в вологде
Amazing things here. I’m very glad to see your article. Thank you a lot and I am having a look forward to contact you. Will you kindly drop me a e-mail?
https://solo-it.ru/forum/?PAGE_NAME=profile_view&UID=3716
https://one-diz.3dn.ru/forum/28-5107-121#9993
https://www.bodybuilding.net/members/worksale.html
https://electrikmaster.ru/forum/topic/pokupka-sajta/#postid-463
https://farexpo.ru/club/user/30286/forum/message/3533/3570/#message3570
WOW just what I was searching for. Came here by searching for %keyword%
где купить диплом образование
http://pension-forum.ru
http://park-robotov.ru
http://memorialculture.ru
купить диплом лаборанта
Hey terrific blog! Does running a blog such as this take a massive amount work? I have very little expertise in programming but I was hoping to start my own blog in the near future. Anyway, should you have any suggestions or tips for new blog owners please share. I understand this is off topic nevertheless I simply needed to ask. Kudos!
http://www.ivedu.ru/forum/viewthread.php?forum_id=18&thread_id=46172В
http://tekhnotop.ru/forums/
https://www.shopwheel.ru/club/user/52/blog/
http://rogerfilms.com/index.php?title=russianydiplomans
http://bagoff.net/forum/viewtopic.php?t=2559
May I simply say what a comfort to find a person that truly knows what they are discussing online. You actually understand how to bring a problem to light and make it important. A lot more people must check this out and understand this side of the story. I was surprised you aren’t more popular because you certainly possess the gift.
купить диплом в томске
http://diplom1.org/
http://centr-legko.ru
http://spravku-nadom.ru
купить диплом в твери
For the reason that the admin of this site is working, no doubt very soon it will be famous, due to its feature contents.
купить диплом в бору
http://vpconf.ru
http://school5-priozersk.ru
http://icefoutreach.ru
купить диплом специалиста
iGaming 분야에서 선도적인 최신 프라그마틱 게임은 표준화된 콘텐츠를 제공하며 슬롯, 라이브 카지노, 빙고 등을 통해 뛰어난 엔터테인먼트를 선사합니다.
프라그마틱
프라그마틱에 대한 내용 정말 흥미로워요! 또한, 제 사이트에서도 프라그마틱과 관련된 정보를 공유하고 있어요. 함께 지식을 공유해보세요!
https://dqwdqdw.weebly.com/
https://www.belgiumfire.com
http://holyshirtsandpants.net/
Thanks for sharing your thoughts. I really appreciate your efforts and I am waiting for your further post thank you once again.
http://androidinweb.ru/kupit-diplom-ekonomte-vremya-i-dengi
https://forum.numizmatium.ru/forums/forum/numizmatika-obsuzhdenie/okolonumizmaticheskie-temy/
http://w66501ot.bget.ru/main/1-post1.html
http://msflow.ru/products/pionovidnye-rozy-v-korobke/#comment_60753/
https://sportavesti.ru/contact/
슬롯 머신 사이트
Zhu Houzhao는 그것에 대해 생각한 후 마침내 정신을 차렸습니다!
В нашем обществе, где диплом – это начало успешной карьеры в любой сфере, многие стараются найти максимально быстрый путь получения образования. Факт наличия документа об образовании сложно переоценить. Ведь именно он открывает двери перед любым человеком, желающим вступить в профессиональное сообщество или продолжить обучение в университете.
В данном контексте наша компания предлагает очень быстро получить этот важный документ. Вы можете купить диплом старого или нового образца, что является выгодным решением для человека, который не смог завершить образование, утратил документ или желает исправить свои оценки. диплом изготавливается с особой тщательностью, вниманием ко всем элементам, чтобы в результате получился документ, 100% соответствующий оригиналу.
Преимущества данного решения состоят не только в том, что можно быстро получить свой диплом. Весь процесс организовывается удобно и легко, с профессиональной поддержкой. От выбора требуемого образца диплома до консультаций по заполнению личной информации и доставки по стране — все под полным контролем качественных специалистов.
Таким образом, для всех, кто пытается найти быстрый и простой способ получить требуемый документ, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать длительного обучения и не теряя времени переходить к достижению личных целей: к поступлению в ВУЗ или к началу удачной карьеры.
http://vntconference.ru
http://magopft.ru
http://edu-csrpi.ru
http://hackspace-spb.ru
http://cso-gd.ru
For hottest news you have to pay a visit internet and on world-wide-web I found this website as a best website for most recent updates.
http://startspresto.ru/dannye/pluginclass/cbblogs?action=blogs&func=show&id=28
купить диплом в томске
http://kapremont-upgrade.ru
купить диплом в саранске
Hello, yes this article is really good and I have learned lot of things from it about blogging. thanks.
http://returnrp.listbb.ru/viewtopic.php?f=70&t=603
https://pipewiki.org/app/index.php/пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ!
cs-online.ru/forum/index.php?showtopic=13633&mode=linearВ
https://home.jts.edu.jm/index.php/component/k2/item/76-living-rooms-show-range-of-modernist-to-traditional/76-living-rooms-show-range-of-modernist-to-traditional?start=10
https://forum.mbprinteddroids.com/showthread.php?tid=1441
Доброго всем дня!
У нас на сайте можно недорого заказать и купить диплом с гарантией и доставкой покупателю в любой регион РФ
http://marketforme.ru/postroyte-osnovu-znaniy-kupite-diplom-ot-uvazhaemogo-uchebnogo-zavedeniya
http://metalfans.maxbb.ru/topic494.html
http://backshowtime.ru/poluchite-diplom-kotoryiy-podtverdit-vashu-kompetentnost-i-opyit-legko-i-byistro
https://www.kino-ussr.ru/1792-bratya-karamazovy-1968.html
http://www.portalcienciayficcion.com/foro/member.php?216026-DPPlommCcooMsmima
Добрый день всем!
Получите документ ВУЗа без предоплаты и доставку в руки по России – просто, удобно, безопасно!
http://www.littlethings.su/2015/09/ombre.html
https://rup14.by/forum/user/4321/
http://milabalo.blogspot.com/2014/02/blog-post_22.html
http://plut-uchukov.flybb.ru/topic313.html
https://flerus-shop.hcp.dilhost.ru/club/user/3/forum/message/627/623/
Hello, this weekend is good designed for me, as this time i am reading this enormous educational paragraph here at my house.
купить диплом в серове
http://forum-ecology.ru
http://galaxy-science.ru
http://seoklan.ru
купить диплом в кунгуре
Hi there, every time i used to check web site posts here early in the break of day, as i love to learn more and more.
купить диплом в сургуте
http://turkishculture.ru
http://pojdelo-journal.ru
http://git-ugra.ru
купить диплом в благовещенске
Приветики!
На нашем сайте вы можете заказать диплом Вуза с постоплатой и круглосуточной поддержкой.
https://visualchemy.gallery/forum/viewtopic.php?pid=3297696#p3297696
http://spletninews.ru/kupit-diplom-10
https://md2k.org/standard-post-with-featured-image/#comment-71481
https://www.modern-constructions.org/blogs/43670/пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ
http://kharitonchukanna.blogspot.de/2014/05/milv-34-milv-n104.html
Aw, this was an exceptionally nice post. Taking the time and actual effort to generate a great article… but what can I say… I hesitate a whole lot and don’t manage to get nearly anything done.
http://www.mcb-electro.ru/product/relax-pv-a1-l1/reviews/
https://vip-forum.com/threads/kupit-diplom-j212s.1730/
jboi2007.org/Galeries/Day1/BeachVolleyball/photo5.htmВ
https://xn—–vlcbxd5hez.xn--p1ai/pikantnie-uslugi/#comment-40520
http://led119.ru/forum/user/106227/
Виктория Набойченко сделала для нашего канала заявление,
Свидетель Набойченко
касающееся своего бывшего супруга – главного свидетеля обвинения по так называемому уголовному делу “Лайф-из-Гуд”-“Гермес”-“Бест Вей”
Hello friends, its fantastic article about teachingand fully explained, keep it up all the time.
http://u90517ol.beget.tech/2024/05/17/poluchite-vash-diplom-segodnya-bystro-nadezhno-kachestvenno.html
http://aboutallfinance.ru/poluchite-ofitsialnyiy-dokument-segodnya-prosto-byistro-uverenno
https://forum.sprosiadvokata.ru/topic/21545-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BA%D0%BE%D0%BB%D0%BB%D0%B5%D0%B4%D0%B6%D0%B0-%D0%BD%D0%B5%D0%B4%D0%BE%D1%80%D0%BE%D0%B3%D0%BE-m765y/
reguitti.com.ua/services.htmlВ
http://formulaf1.ru/kupit-diplom-vash-bilet-v-uspeshnoe-budushhee
What’s up everybody, here every one is sharing such knowledge, therefore it’s fastidious to read this blog, and I used to pay a quick visit this webpage everyday.
http://uchim-1c.online/article/rasprodazha-videokursov-945-rublej/
https://bummart.ru/forum/user/58536/
https://civilpatrol.info/?q=petition/80777/diplomansy
http://frichinvestigation.org/node/4459
https://idi.atu.edu.iq/?p=5906
Do you have a spam problem on this blog; I also am a blogger, and I was curious about your situation; we have created some nice methods and we are looking to swap solutions with other folks, why not shoot me an e-mail if interested.
bazhenova.greatforum.ru/viewtopic.php?f=17&t=4310&p=4941
военно-медицинская-академия.рф/
http://www.camry-club.ru/member.php/61231-sonnick84?tab=activitystream&type=user&page=2
premiumkutyatap.hu/jutalom_pontok
finttech.ru/page/36
Hello! Do you know if they make any plugins to safeguard against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any recommendations?
http://www.cleancenter.net/support.htm
arzhaniki.ru/memberlist.php?start=175&sk=a&sd=d&first_char=r
obozrevatelevents.ru/page/11/
007brush.com/collections/vendors/products/sk-120mm-fan-bundle.html
http://www.nfgroup.it/forum/libero-pensiero/383-diplomsagroupscom
Hello I am so grateful I found your blog, I really found you by error, while I was looking on Aol for something else, Anyhow I am here now and would just like to say many thanks for a remarkable post and a all round thrilling blog (I also love the theme/design), I don’t have time to read it all at the moment but I have bookmarked it and also added your RSS feeds, so when I have time I will be back to read more, Please do keep up the great b.
http://www.bseo-agency.com/blogs/119817/%D0%94%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC%D1%8B-%D0%BA%D0%BE%D1%82%D0%BE%D1%80%D1%8B%D0%B5-%D0%BF%D0%BE%D0%BC%D0%BE%D0%B3%D1%83%D1%82-%D0%B2%D0%B0%D0%BC-%D0%B4%D0%BE%D1%81%D1%82%D0%B8%D1%87%D1%8C-%D0%B2%D0%B0%D1%88%D0%B8%D1%85-%D1%86%D0%B5%D0%BB%D0%B5%D0%B9-%D0%B1%D1%8B%D1%81%D1%82%D1%80%D0%B5%D0%B5
dom-postroj.ru/doma-iz-gazobetona/k-231
igramlikes.com/contact/
kembekeltetes.hu/egyutt-nem-mukodo-vallalkozasok/ajanlast-nem-teljesito-vallalkozasok/
symphonyofthemagneticnorth.com/_404.html
What a material of un-ambiguity and preserveness of valuable experience about unpredicted emotions.
https://poetzinc.com/read-blog/10740
https://mylady.mybb.ru/viewtopic.php?id=28130#p126711
http://newsofmebel.ru/kupit-diplom-rf-v-moskve-dostupnyie-tsenyi-i-bezopasnost
http://impressiverp.listbb.ru/viewtopic.php?f=16&t=728
https://www.hebergementweb.org/threads/bystryj-start-kak-kupit-diplom-i-nachat-stroit-kareru.1314937/
Marvelous, what a weblog it is! This blog gives helpful data to us, keep it up.
https://manipureducation.gov.in/photo-gallery/
http://army.clanfm.ru/viewtopic.php?f=2&t=37483
http://www.sojuzpharma.ru/about/pharmopeka?page=674&per-page=5В
https://boraclimat.ru/blog/zhk-polyanka44/#comment_227731
drunkslut.com/springbreak.htmlВ
Hello to every body, it’s my first pay a visit of this blog; this web site carries awesome and genuinely fine information in favor of visitors.
splcash.com/article/privacy-policy?id=31В
http://vnuci.listbb.ru/viewtopic.php?f=2&t=456
http://keyscan.cn.edu/AuroraWeb/Account/SwitchView?returnUrl=https://adisgruntledrepublican.com
https://www.ab-plast.cz/kontakty/
http://forexsnews.ru/izmenite-svoyu-zhizn-kupite-diplom-i-otkroyte-novyie-gorizontyi
슬롯 머신
“응?” 군중의 불만을 본 Hongzhi 황제는 “Fang Qing의 가족, 먼저 말하십시오. “라고 말했습니다.
I delight in, lead to I found exactly what I used to be having a look for. You have ended my four day long hunt! God Bless you man. Have a nice day. Bye
старые дипломы купить
https://inderculturaputumayo.gov.co/juegos-intercolegiados-campeones-en-deportes-individuales/
http://burgasdent.listbb.ru/viewtopic.php?f=3&t=310
http://lolipopnews.ru/uvelichte-svoi-shansyi-plyusyi-pokupki-diploma
http://hackers.bestbb.ru/viewtopic.php?id=344#p357
https://lintense.com/userinfo.php?com=profile&action=view&user=minna_prout_301152
купить диплом строителя
Приветики!
Закажите диплом ВУЗа с гарантированной подлинностью и доставкой по всей России без предоплаты – просто, удобно, безопасно!
http://iefl.lat/mi-historia/#comment-28281
http://oriondtdm.blogspot.com/2018/11/blog-post_5.html
https://www.alpea.ru/forum/user/24866/
http://edinstvoarmenia.bestbb.ru/viewtopic.php?id=249
http://led119.ru/forum/user/105218/
Привет, дорогой читатель!
Наш интернет-магазин предлагает купить российский диплом по выгодной цене, с гарантией прохождения всех проверок
http://slbook-kaluga.ru/index.php/forum/user/238273-eqixis
http://klaus-peltzer.de/kontakt/comment-page-394/#comment-592126
http://imladrisproduction.free.fr/index.php?file=Members&op=detail&autor=uvowedoj
http://beachhouse-living.com/index.php/component/kunena/user/23431-ehyhoce
http://www.sledopit.club/wiki/index.php/%D0%93%D1%80%D0%B0%D0%BC%D0%BE%D1%82%D0%BD%D1%8B%D0%B9_%D0%B2%D1%8B%D0%B1%D0%BE%D1%80_%D0%B8%D0%BD%D1%82%D0%B5%D1%80%D0%BD%D0%B5%D1%82-%D0%BC%D0%B0%D0%B3%D0%B0%D0%B7%D0%B8%D0%BD%D0%B0,_%D0%B3%D0%B4%D0%B5_%D0%BF%D1%80%D0%B8%D0%BE%D0%B1%D1%80%D0%B5%D1%81%D1%82%D0%B8_%D0%B2%D0%BE%D0%B7%D0%BC%D0%BE%D0%B6%D0%BD%D0%BE_%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC
Hello! I’m at work surfing around your blog from my new apple iphone! Just wanted to say I love reading your blog and look forward to all your posts! Carry on the great work!
http://almaztb.ru/index.php?links_exchange=yes&page=51
купить свидетельство о рождении ссср
http://obrazovanie-diplom.ru
купить диплом в октябрьском
Pretty! This was a really wonderful post. Thanks for supplying these details.
купить диплом в белогорске
https://rodme.ru/kupit-diplom-vash-put-k-professionalnoy-realizacii-t12152.html
http://torica.4adm.ru/viewtopic.php?f=6&t=2021
http://infiniterealities.listbb.ru/viewtopic.php?f=7&t=324
https://vozlublennaya.mybb.sumy.ua/viewtopic.php?id=17901#p48516
https://forum.wowcircle.com/member.php?u=592802
купить диплом в новотроицке
Сегодня, когда диплом – это начало успешной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения образования. Факт наличия официального документа сложно переоценить. Ведь именно диплом открывает двери перед всеми, кто желает начать трудовую деятельность или учиться в университете.
В данном контексте наша компания предлагает оперативно получить любой необходимый документ. Вы имеете возможность заказать диплом нового или старого образца, и это становится удачным решением для человека, который не смог закончить обучение или утратил документ. диплом изготавливается с особой тщательностью, вниманием к мельчайшим элементам, чтобы в итоге получился документ, 100% соответствующий оригиналу.
Преимущества данного решения заключаются не только в том, что вы максимально быстро получите свой диплом. Процесс организовывается комфортно, с профессиональной поддержкой. От выбора необходимого образца до консультаций по заполнению личных данных и доставки в любой регион страны — все находится под абсолютным контролем опытных специалистов.
Всем, кто ищет оперативный способ получить требуемый документ, наша компания предлагает выгодное решение. Заказать диплом – это значит избежать долгого процесса обучения и сразу перейти к своим целям, будь то поступление в ВУЗ или начало карьеры.
http://teh-diplom.ru
http://michgpi.ru
http://school-13.ru
http://vntconference.ru
http://cceis.ru
?? Забирай 100,000 Рублей + 400 Фриспинов в виде Welcome бонуса, на официальном сайте 7К Казино ??
?????? Официальное зеркало ??????
7k казино скачать
НОВЫЕ ПРОМОКОДЫ ОТ НАШЕГО ТЕЛЕГРАМ КАНАЛА 7К КАЗИНО! МОЩНЫЙ СТАРТ НАШИМ ИГРОКАМ. ???? ЖЕЛАЕМ КРУТЫХ ЗАНОСОВ ??
Joker7070 – бонус 170%+ 70 FS в Joker Stoker
777WIN – бонус 100%+ 30 FS в Hot Triple Sevens
HOT100 – бонус 100 FS в Indiana’sQuest
?? Добавляйте промокод в разделе бонусов на официальном сайте 7К Казино.
?? Важно! Для новых игроков после регистрации 7K Casino приготовил Welcome бонус до 100,000 Рублей + 400 Фриспинов! Успейте активировать ???
?? Подробная информация на официальном сайте 7К в разделе бонусов! Каждый промокод можете использовать по очереди.
7k казино скачать приложение
https://t.me/s/casino_7k_official
«Рэмбович» против колокольцевской мафии
Министр обороны, в рамках начавшейся антикоррупционной кампании, готов инициировать расследование против команды своего коллеги Колокольцева: сфабрикованное ею дело в отношении кооператива «Бест Вей» – созданного военнослужащими во многом для военнослужащих, бьет в тылу по тем, кто защищает страну на фронте, лишая их квартир и денег, которые уже более двух лет арестованы ГСУ ГУ МВД России по Санкт-Петербургу и Ленинградской области и примкнувшей к ним Прокуратурой Санкт-Петербурга.
Гермес Менеджмент
?Кооператив «Бест Вей» создан в 2014 году российскими военнослужащими – как ушедшими в запас, так и действующими. Его предправления до весны 2021 года, а затем еще год председатель совета – капитан третьего ранга запаса Роман Василенко, в правлении кооператива всегда было немало офицеров Вооруженных сил РФ.
Нынешний председатель совета Сергей Крючек – полковник запаса, депутат Государственной думы VII созыва.
Жилье для военнослужащих
Кооператив начинался во многом как проект обеспечения жильем военнослужащих, бывших военнослужащих и членов их семей. Он существует 10 лет, за восемь лет, пока он имел возможность приобретать квартиры для пайщиков, которые были заблокированы следствием, «Бест Вей» купил 2500 квартир, более 250 квартир перешли в собственность пайщиков.
Уникальность кооператива в том, что он приобретает пайщикам недвижимость на беспроцентной основе. Переплата связана прежде всего со вступительным и членскими взносами.
Социальная программа для участников СВО
Приобретение жилья с помощью кооператива – по сути, социальная программа, которая стала очень популярна среди военнослужащих.
Ей воспользовались тысячи участников СВО и членов их семей – людей, защищающих Россию на фронте.
И вот в тылу с их имуществом или деньгами творится настоящий беспредел – по вине колокольцевской мафии. Колокольцевцы со своими подручными из Прокуратуры Санкт-Петербурга уже два года держат средства кооператива под арестом.
Российские воины не могут ни приобрести квартиры, на которые аккумулировали деньги на счете в кооперативе, ни забрать собственные обесценивающиеся за два года деньги назад. В свои короткие отпуска они вынуждены обивать пороги Росреестра, писать заявления на возврат средств в суды, участвовать в процессах. Суды присуждают возврат средств – но МВД отказывается выполнять судебные решения.
Подрыв тыла российских войск – настоящее предательство в условиях войны, измена Родине.
Отнять то, что ближе лежит
Уголовное дело, возбужденное ГСУ ГУ МВД по Санкт-Петербургу, касается иностранной компании «Гермес», которая (якобы) украла деньги своих российских клиентов. В действительности у нее возникли трудности из-за того, что завербованный питерской полицией с помощью угроз Набойченко, имевший доступ к платежной системе, обрушил ее и, возможно, манипулировал средствами на счетах.
Однако деньги «Гермеса» из-за границы достать проблематично – и следствие решило объявить кооператив частью некоего холдинга, в который входил также «Гермес», и на этом основании забрать его средства. Хотя в действительности между этими организациями никогда не было никаких финансовых и организационных отношений.
Продукты кооператива и продукты «Гермеса» продвигала сеть предпринимателей под эгидой компании «Лайф-из-Гуд» – коммерческая сеть, каждый из участников которой работал за процент и мог продавать все, что угодно. Многие приходили в кооператив не через продажников, а по сарафанному радио.
На скамье подсудимых – десять человек, в том числе 83-летний Виктор Иванович Василенко, отец Романа Василенко, ветеран Вооруженных сил: команда Колокольцева не отказала себе в извращенном удовольствии поиздеваться над ветераном.
Тыловые крысы воруют чужое имущество
Колокольцевцы – тыловые крысы, которые никогда не были настоящими офицерами, скорее чинушами с коррупционными интересами, пытаются отжать почти 4 млрд рублей на счетах кооператива для себя и своих подельников.
Притом, что в рамках уголовного дела фигурирует сумма в 282 млн рублей, даже (незаконный) арест средств кооператива на эту сумму никак не повлиял бы на работу кооператива. Но арестованы именно 4 млрд, при этом МВД не дает платить с этих средств налоги и зарплату. То есть целью продажных ментов являются именно миллиарды кооператива, а не удовлетворение (в основном липовых) претензий клиентов «Гермеса».
Комиссар Путина
Андрей Белоусов, облеченный особым доверием главы государства, имеет карт-бланш на проведение антикоррупционных расследований в отношении всех силовых структур, тем более затрагивающих военнослужащих Вооруженных сил. Новый министр обороны сразу объявил социальную защищенность воинов высшим приоритетом, а кооперативная программа обеспечения жильем может сыграть в ней огромную роль.
«Рэмбович» уже показал, как бескомпромиссно он борется с коррупционерами в руководстве МО и в ОПК. Теперь настал черед руководства МВД – которое давно уже на плохом счету президента Путина, просто до него в условиях войны не доходили руки. Теперь они дошли.
Заявление Виктории Набойченко – бывшей супруги главного свидетеля обвинения Евгения Набойченко
https://www.youtube.com/watch?v=Q0smZ5_QOcU
Здравствуйте, меня зовут Виктория Набойченко.
Я являюсь бывшей супругой Набойченко Евгения Витальевича, с которым состояла в официальном браке с 2017 по 2022 год. Из средств массовой информации мне стало известно о фактах уголовного преследования лиц из окружения Романа Василенко, который является президентом международной бизнес-карты на территории России, Казахстана и Киргизии. На протяжении 20 лет Евгений Набойченко и Роман Василенко дружили, вместе путешествовали, проводили время, ездили на рыбалку. Роман очень доверял Евгению, делился с ним своими идеями, планами на коммерческую деятельность и реализацию новых проектов. Мне известно, что Роман предложил Евгению разработать фирменный логотип и сайт компании Life is Good, а впоследствии возглавить IT-отдел ЖК. Евгений с радостью согласился на предложение Романа и возглавил IT-отдел. Роман достойно оплачивал работу Евгения, гонорар которого вырос со 100,000 рублей до 500,000 рублей в месяц, что было связано с ростом и развитием кооператива Best. Роман всегда был щедр к Евгению и ценил его работу, за что ежегодно премировал.
Тот успех, которого достиг Роман за эти годы, очень задевал Евгения как мужчину. В ходе совместной жизни Евгений делился со мной, что мечтает однажды превзойти Романа в бизнесе, а его проект стать круче Илона Маска. Примерно с июня 2021 года Евгений стал нервным и злился на Романа, рассказывая мне, что Роман не хочет заплатить ему 170,000 евро. Я не вдавалась в подробности их взаимоотношений. Чуть позже я узнала о письмах Евгения к Роману, в которых он требовал от него указанную сумму и угрожал увечьями и убийством Роману и всем членам его семьи, в том числе и детям. Данные угрозы он отправлял голосовыми сообщениями и в электронных письмах. Мне известно, что Роман не реагировал на угрозы Евгения, что очень злило его, потому что он ждал от него указанную сумму. В моём присутствии Евгений постоянно по нескольку раз переслушивал свои голосовые сообщения, которые с угрозами отправлял в адрес Романа Василенко. Он получал от этого огромное удовольствие, с ухмылкой хвастаясь, что Роман обязательно заплатит.
Best Way
17 февраля 2022 года в 7:00 по МСК в нашем доме в посёлке в Краснодарском крае произошел обыск. В доме находилась только я и дети, а Евгений в это время находился в Краснодаре на съёмной квартире, куда заблаговременно уехал, предупредив меня за 3 дня о предстоящем обыске в доме. Как будто его друзья из правоохранительных органов Петербурга предупредили его. На мои вопросы о том, что происходит, он отвечал, что это формальность и что мне не о чем беспокоиться. Позже мне стало известно, что подобные обыски проходили и у других сотрудников кооператива в Санкт-Петербурге, Краснодаре, Анапе и других городах. В обыске нашего дома принимали участие шесть оперативных работников ОББ города Санкт-Петербурга, фамилии которых указаны в протоколе обыска, копия которого имеется в моём распоряжении. В ходе обыска оперативные работники оскорбляли Евгения и Романа, называя их преступниками и мошенниками, а также иными оскорбительными словами. При обыске был изъят планшет Евгения, оперативные работники перевернули весь дом и напугали детей, а также пообещали, что задержат Евгения в тот же день.
17 февраля 2022 года Евгений был задержан сотрудниками оперативных служб в городе Краснодар и примерно через 3 дня был доставлен в Санкт-Петербург, где был допрошен в УП по рекомендации сотрудников полиции. Допрос происходил без участия его личного адвоката Ирины Черняховской, от услуг которой он отказался в тот же момент. Мне позвонила Ирина в бешенстве и недоумении, крича о том, что Евгений её опозорил перед сотрудниками полиции, отказавшись от её услуг в грубой форме. Впоследствии мне стало известно от Евгения, что он лично получил покровительство начальника ОББ города Санкт-Петербурга в своей безнаказанности по факту проводимого расследования. Об этом он похвастался мне в личном разговоре, будучи окрылённый поддержкой начальника ОББ, который с ним разговаривал на доверительной ноте.
Евгений мне рассказал, что получит от органов абсолютную неприкосновенность и поддержку за информацию, которая интересовала оперативных работников, за что Евгению также была предоставлена госохрана. В период с февраля по середину апреля Евгений находился в Питере под госзащитой и плотно взаимодействовал с правоохранительными органами. Он предоставлял им всю информацию, в том числе полный доступ к системе базы данных Best Way и Life is Good. Евгений инициативно предлагал органам заблокировать доступ к личным кабинетам пайщиков и членов ЖК БСТ, чем полностью парализовал хозяйственную деятельность кооператива, создавая напряжённость и негативные отзывы. Также Евгений активно призывал граждан к написанию заявлений в отношении Романа Василенко с целью возврата пайщиками своих денежных средств, делал он это через каналы Telegram и YouTube. Евгений был единственным, кто имел и имеет доступ к базам данных и ключам. Желая намеренно создать проблемы Роману, он отказывается восстанавливать работу системы Best Way при наличии такой возможности.
Миссия Евгения – это привлечение Романа Василенко к уголовной ответственности, что спровоцирует крах компании и полное прекращение её деятельности, а это в свою очередь 30,000 пайщиков, которые могут стать потерпевшими по уголовному делу. Всё это мне известно из личного общения с Евгением, когда я приезжала к нему в Санкт-Петербург на несколько дней. Мне также достоверно известно, что в настоящее время нет реальных заявлений от недовольных пайщиков, так как в Романа Василенко все верят. Самое главное, что у Евгения есть возможность в любой момент через свой личный ноутбук восстановить работу ЖК Best Way и вернуть доступ к личным кабинетам пайщиков. Однако он этого не делает, преследуя свои личные цели.
Useful info. Lucky me I discovered your web site by chance, and I’m stunned why this twist of fate didn’t happened earlier! I bookmarked it.
reguitti.com.ua/services.html
girlscools.ru/page/39
beritanusantara.co.id/cadir-polimdo-okta-lintong-efendy-rasid-maryke-alelo-masuki-tahap-pertarungan-tingkat-kementerian/
chaussure-lumineuse.fr/chaussure-led-basket-lumineuse-pour-enfant-chaussure-lumineuse/
myanmarfootball.org/gallery/displayimage.php?album=lastup&cat=6&pos=0
Hmm is anyone else experiencing problems with the images on this blog loading? I’m trying to determine if its a problem on my end or if it’s the blog. Any suggestions would be greatly appreciated.
купить диплом в рыбинске
http://windows1.listbb.ru/viewtopic.php?f=3&t=371
http://umorforme.ru/diplom-s-garantiey-kachestva-vyiberite-nas
http://returnrp.listbb.ru/viewtopic.php?f=70&t=615
https://galantclub.od.ua/member.php?u=27724
https://climat-cold.ru/forum/user/3663/
купить диплом россия
What you typed made a bunch of sense. However, think about this, what if you wrote a catchier post title? I am not saying your content is not solid, however suppose you added a title that grabbed people’s attention? I mean %BLOG_TITLE% is kinda boring. You should look at Yahoo’s front page and watch how they write post headlines to grab people interested. You might add a related video or a picture or two to grab readers excited about everything’ve got to say. In my opinion, it could bring your blog a little livelier.
купить диплом воспитателя
http://www.lada-xray.net/member.php?u=2434
http://raceburo.ru/sdelayte-svoi-mechtyi-realnostyu-poluchite-svoy-diplom-zdes
https://goup.hashnode.dev/priobresti-attestaty-o-srednem-obrazovanii
https://wparena.com/wordpress-seo-plugins-dont-help-you-rank-high/#comment-557419
https://goruss.ru/blogs/386/ƒиплом-сегодн€-¬аша-дверь-в-новый-мир
купить диплом в междуреченске
Do you have a spam problem on this site; I also am a blogger, and I was wanting to know your situation; many of us have developed some nice practices and we are looking to exchange strategies with others, why not shoot me an email if interested.
http://fridayad.in/user/profile/2444767
https://saumalkol.com/forum/пїЅпїЅпїЅпїЅпїЅпїЅ-2/2527-пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅ-пїЅ-пїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅ-пїЅпїЅпїЅ-пїЅпїЅпїЅпїЅпїЅпїЅпїЅ.html
http://dp.getbb.ru/viewtopic.php?f=3&t=550
https://na-krychke.ru/com/forum/topic/161/
http://windows1.listbb.ru/viewtopic.php?f=3&t=378
[url=https://rentyachtsincyprus.com/en]hashish with a syringe[/url] 7 grams discounts. 21%
[url=https://rentyachtsincyprus.com/en]powdered heroin – snorting[/url] 50 grams discounts 45%
[url=https://rentyachtsincyprus.com/en]ecstasy under the tongue[/url] – 2 gram discounts 37%
[url=https://rentyachtsincyprus.com/en]hachis con una jeringuilla[/url] 31 grama descuento. 11%
[url=https://rentyachtsincyprus.com/en]heroina en polvo – esnifada[/url] 70 gramas descuento 36%
[url=https://rentyachtsincyprus.com/en]extasis bajo la lengua[/url] – 5 gramas rebajas 45%
[url=https://rentyachtsincyprus.com/en]hasista ruiskulla[/url] 5 grams sale. 38%
[url=https://rentyachtsincyprus.com/en]heroiinijauhe – nuuskaaminen[/url] 55 gram discounts 13%
[url=https://rentyachtsincyprus.com/en]ekstaasi kielen alla[/url] – 5 gram sale 21%
[url=https://rentyachtsincyprus.com/en]haszysz za pomoca strzykawki[/url] 15 gram sleva. 15%
[url=https://rentyachtsincyprus.com/en]sproszkowana heroina – wciaganie[/url] 40 gram sleva 35%
[url=https://rentyachtsincyprus.com/en]ekstaza pod jezykiem[/url] – 5 gramy sale 38%
[url=https://rentyachtsincyprus.com/en]du haschisch avec une seringue [/url]10 grammes sleva. 47%
[url=https://rentyachtsincyprus.com/en]heroine en poudre – sniffer[/url] 30 grammes discounts 32%
[url=https://rentyachtsincyprus.com/en]ecstasy sous la langue[/url] – 20 grammes sleva 17%
A beloved National Park Service ranger died when he tripped, fell and struck his head on a rock during an annual astronomy festival in southwestern Utah, park officials said over the weekend.
[url=https://kpaken17.at]kraken13.at[/url]
Tom Lorig was 78 when he died after the incident at Bryce Canyon National Park late Friday.
kraken14.at
https://kraken13.at-kraken17.at
He was known for his extensive work as a ranger and volunteer at 14 National Park Service sites, including Yosemite National Park, Carlsbad Caverns National Park and Dinosaur National Monument, the park service said in a statement Saturday.
“Tom Lorig served Bryce Canyon, the National Park Service, and the public as an interpretive park ranger, forging connections between the world and these special places that he loved,” Bryce Canyon Superintendent Jim Ireland said in the statement.
hashish with a syringe 5 grams sale. 34%
powdered heroin – snorting 100 grams sleva 31%
ecstasy under the tongue – 5 gram discount 21%
hachis con una jeringuilla 100 grama descuento. 12%
heroina en polvo – esnifada 30 gramas rebajas 23%
extasis bajo la lengua – 5 grama descuento 21%
hasista ruiskulla 3 grams sleva. 23%
heroiinijauhe – nuuskaaminen 55 grams discounts 25%
ekstaasi kielen alla – 1 gram sale 12%
haszysz za pomoca strzykawki 17 gram discounts. 36%
sproszkowana heroina – wciaganie 100 gram sleva 12%
ekstaza pod jezykiem – 11 gram discount 26%
du haschisch avec une seringue 6 gram discounts. 28%
heroine en poudre – sniffer 30 gram sale 15%
ecstasy sous la langue – 15 grammes discounts 32%
купить диплом в твери diplomvash.ru .
купить диплом в томске купить диплом в томске .
Do you have a spam problem on this website; I also am a blogger, and I was wondering your situation; we have created some nice methods and we are looking to exchange solutions with other folks, be sure to shoot me an email if interested.
купить диплом отзывы
https://opaseke.com/users/1243
http://openmotonews.ru/idealnyiy-diplom-dlya-vashego-budushhego-priobretite-pryamo-seychas
http://rabotianadomy.frmbb.ru/viewtopic.php?id=2423#p2858
https://brovary.forum.cool/viewtopic.php?id=5536#p14043
http://sport-faq.ru/diplom-na-zakaz-vasha-lichnaya-doroga-k-uspehu-i-protsvetaniyu
купить свидетельство о браке
Hi! This post could not be written any better! Reading this post reminds me of my previous room mate! He always kept talking about this. I will forward this article to him. Fairly certain he will have a good read. Thanks for sharing!
купить диплом в иркутске
http://prikol100500.ru/vasha-uspeshnaya-karera-nachinaetsya-s-diploma-pokupka-onlayn
https://mamuli.club/forum/topic/23701/
https://films.name/viewforum.php?f=143
http://kisska.net/go.php?url=https://premialnie-diplomans.com/
http://lightsdemons.phorum.pl/viewtopic.php?p=66916#66916
купить диплом вуза
Hello very nice website!! Guy .. Beautiful .. Wonderful .. I will bookmark your blog and take the feeds also? I’m glad to find numerous useful info right here within the post, we want develop more techniques on this regard, thanks for sharing. . . . . .
gadjetforyou.ru/kupit-diplom-10
frugalmechanic.com/tech-blog/tag/sbt
wap-club.ru/guestbook.php?mode=quote&idmsg=210748
eworlddxn.com/ar/product/122
98opt.ru/services/montazh-kholodilnogo-oborudovaniya/
Hi to all, how is all, I think every one is getting more from this site, and your views are fastidious for new users.
buzzchatlive.com/index.php?do=/public/user/blogs/name_Alanpoe/page_7/
connect.nteep.org/blogs/753/Where-can-I-buy-a-diploma-or-certificate-at-an?lang=tr_tr
forum.anomalythegame.com/viewtopic.php?p=609306
cogita.site/news/otchety/andrei-scherbak.-gosudarstvo-i-malyi-biznes-ruka-dayuschaya-i-ruka-karayuschaya
we.riseup.net/wikis/565961
купить свидетельство о рождении diplomvash.ru .
Привет, дорогой читатель!
Предлагаем всем желающим приобрести диплом университетов России по низкой цене с доставкой в руки под ключ
http://obzs.blogspot.com/2014/06/kak-perenesti-ranenogo.html
https://www.unidocs.ru/product/russkoe-menju-ilona-fedotova/reviews/
https://expertmd.me/product/woo-album-3/#comment-252739
http://abk-63.ru/kategorii-voprosov/iz-pisem-fau-fccs-minregion/
https://www.lasergrafics.de/handel/artikel-b/#comment-94804
Добрый день всем!
На нашем сайте вы можете выбрать и приобрести диплом Вуза с гарантией и доставкой в любой регион России по самым низким ценам.
https://briards.mybb.ru/viewtopic.php?id=2001#p49696
http://kochtagebuch.org/doku.php?id=пїЅпїЅпїЅпїЅпїЅпїЅ пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ пїЅпїЅпїЅпїЅпїЅпїЅ пїЅ пїЅпїЅпїЅпїЅпїЅпїЅ пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ
http://maisoncarlos.com/UserProfile/tabid/42/UserID/1938978/Default.aspx
https://school97.ru/vesti/view_profile.php?UID=196193
http://turservisnews.ru/uskorte-svoy-professionalnyiy-rost-kupite-diplom-i-poluchite-neobhodimyie-znaniya
I loved as much as you’ll receive carried out right here. The sketch is attractive, your authored subject matter stylish. nonetheless, you command get bought an nervousness over that you wish be delivering the following. unwell unquestionably come further formerly again as exactly the same nearly very often inside case you shield this hike.
akademiyatepla.ru/catalog/baxi/?goodsCompare=add&id=178
museums.artyx.ru/books/c0003_1.shtml
demo.4admins.ru/viewtopic.php?f=10&t=1549&start=0&view=print
qvmagazine.com/resposible-gambling/
http://www.rolandus.org/forum/profile.php?mode=viewprofile&u=25280
В нашем мире, где диплом становится началом отличной карьеры в любом направлении, многие стараются найти максимально быстрый и простой путь получения образования. Важность наличия официального документа об образовании трудно переоценить. Ведь именно диплом открывает двери перед любым человеком, который собирается вступить в сообщество профессиональных специалистов или учиться в любом ВУЗе.
В данном контексте мы предлагаем быстро получить этот важный документ. Вы имеете возможность приобрести диплом, и это является отличным решением для всех, кто не смог завершить образование или утратил документ. дипломы изготавливаются аккуратно, с особым вниманием ко всем элементам. В итоге вы получите полностью оригинальный документ.
Преимущества данного решения состоят не только в том, что можно оперативно получить диплом. Процесс организован комфортно, с нашей поддержкой. Начиная от выбора нужного образца до консультаций по заполнению персональных данных и доставки по стране — все под абсолютным контролем опытных мастеров.
В итоге, всем, кто хочет найти быстрый способ получения требуемого документа, наша компания предлагает отличное решение. Купить диплом – значит избежать длительного обучения и не теряя времени переходить к достижению личных целей: к поступлению в ВУЗ или к началу успешной карьеры.
http://livetexno.ru
http://kontur-plus.ru
http://unikum-student.ru
http://oskol-sport.ru
http://altaybiotech.ru
Hi there it’s me, I am also visiting this web page on a regular basis, this site is in fact nice and the visitors are truly sharing nice thoughts.
купить диплом швеи
http://transexit.g-talk.ru/viewtopic.php?f=10&t=2000
https://iqtorg.ru/forum/messages/forum2/topic501/message558/?result=new#message558
http://pro-novostroyki.ru/index.php?showtopic=20770
http://kondrateff.5bb.ru/viewtopic.php?id=6291#p11725
https://wiki.gta-zona.ru/index.php/пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅ
где купить диплом о среднем образование
Please let me know if you’re looking for a writer for your weblog. You have some really good articles and I believe I would be a good asset. If you ever want to take some of the load off, I’d absolutely love to write some content for your blog in exchange for a link back to mine. Please send me an email if interested. Cheers!
купить диплом в артеме
https://wiki.mysupp.ru/index.php?title=пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ
http://apso.ru/bitrix/click.php?goto=https://premialnie-diplomans.com/
http://little-witch.ru/forum99.html
http://weekevents.ru/kupit-diplom-v-moskve-professionalnyiy-podhod
https://www.fda.gov.mm/?p=815
купить свидетельство о заключении брака
Здравствуйте!
Приобретите диплом ВУЗа с гарантированной доставкой по России и без предварительной оплаты – надежно и выгодно!
http://little-witch.ru/forum99.html
https://messenger.wepluz.com/read-blog/1618
http://www.volgogradsky.ru/forum/viewtopic.php?p=533840#533840
https://worklife.rolka.me/post.php?fid=4
http://grp.7olimp.ru/viewforum.php?f=1
Hi, everything is going sound here and ofcourse every one is sharing information, that’s in fact excellent, keep up writing.
купить диплом с внесением в реестр
http://drugisaitove.listbb.ru/viewtopic.php?f=2&t=555
https://ya.2bb.ru/viewtopic.php?id=18515#p239450
https://prof-komplekt.com/club/user/4861/blog/10343/
https://heroes.app/blogs/new
http://www.jamsun.net/doku.php?id=пїЅпїЅпїЅпїЅпїЅ_пїЅ_пїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ
купить диплом эколога
Российский завод продает разборные гантели на https://Razbornye-ganteli.ru/ по низким ценам. Для тренировок в квартире – это самый удобный инвентарь с небольшими габаритами и внушительной функциональностью. Продаются в полном наборе с замками и грифами для гантелей.Утяжелители сборные позволяют заниматься с разной нагрузкой. Предлагаем разнообразный набор категорий от ведущих марок в сетевом магазине.
마종 웨이즈
Fang Jifan은 돈에 대해 이야기하는 사람들을 가장 싫어하고 돈에 대해 이야기하는 것은 그를 모욕합니다!Fang Jifan은 어젯밤에 잠을 많이 자지 않았지만 책상에 많은 스케치를 쓰고 그렸습니다.
An outstanding share! I have just forwarded this onto a colleague who was conducting a little research on this. And he in fact ordered me lunch because I stumbled upon it for him… lol. So let me reword this…. Thanks for the meal!! But yeah, thanx for spending some time to talk about this issue here on your website.
купить диплом в оренбурге
https://worklife.rolka.me/viewtopic.php?id=3394#p6625
http://rorp.4admins.ru/viewforum.php?f=33
https://cahaya.my.id/купить-диплом-ваша-дверь-к-новым-возмо
https://www.orderviewdocs.online/doku.php?id=пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅ_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ
https://thehealthbridges.com/read-blog/23880
купить диплом в нефтекамске
At this time it sounds like Drupal is the top blogging platform available right now. (from what I’ve read) Is that what you’re using on your blog?
http://www.phonotope.net/topics/archives/000052.html
onedetailing.pl/mycie-karoserii/23-szampon-samochodowy-meguiar-s-hyper-wash.html
bancaanxudoithuong3d.com/category/game-ban-ca-doi-the?filter_by=popular7
parkdivan.ru/evro-knizhki
roomblock.com/send-rfp.html
Useful info. Fortunate me I discovered your web site unintentionally, and I am shocked why this coincidence didn’t happened in advance! I bookmarked it.
купить диплом в туапсе
https://cds.edu/university-blog-001-2/#comment-401146
http://zdshi-tula.ru/forum/?PAGE_NAME=topic_new&FID=1#postform
http://torica.4adm.ru/viewtopic.php?f=6&t=1972
https://talkitter.com/read-blog/195343
http://newseducation2015.bestbb.ru/viewtopic.php?id=638#p650
купить диплом учителя
This text is invaluable. When can I find out more?
https://ticketsbookmarks.com/story17008571/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%9F%D0%B5%D1%80%D0%BC%D0%B8
http://schoolnumberone.ru/
http://mrkarpiuk.xaa.pl/boards/shopping/member.php?action=profile&uid=33570
https://www.saoluizhotel.com.br/4-springtime-color-schemes-to-try-at-home/
http://venturetraining.net/contact/
Hey excellent blog! Does running a blog such as this take a large amount of work? I’ve absolutely no expertise in programming but I had been hoping to start my own blog soon. Anyhow, if you have any recommendations or techniques for new blog owners please share. I know this is off topic nevertheless I simply needed to ask. Cheers!
http://www.passo.su/forums/index.php?autocom=gallery&req=si&img=2957
http://onlineboxing.net/jforum/user/profile/285562.page
https://keybookmarks.com/story17077024/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B0%D0%B2%D1%82%D0%BE%D0%BC%D0%B5%D1%85%D0%B0%D0%BD%D0%B8%D0%BA%D0%B0
https://aboutalltour.ru/diplomyi-po-vashemu-zaprosu-nadezhno-i-professionalno/
http://dachaweek.ru/page/2
It’s awesome for me to have a site, which is valuable in support of my knowledge. thanks admin
shoelovershub.com/read-blog/475_why-is-the-popularity-of-universities-declining-in-our-time.html?mode=day
programasites.com.br/blog/post/11666/e-mail-marketing-boas-praticas-e-temas-para-abordar
http://www.forum.jrudevels.org/profile.php?mode=viewprofile&u=563048
bossonlighting.com/e_feedback/?page=1178
akademiyatepla.ru/catalog/baxi/?goodsCompare=add&id=178
В нашем мире, где диплом – это начало удачной карьеры в любой сфере, многие пытаются найти максимально простой путь получения образования. Факт наличия официального документа сложно переоценить. Ведь диплом открывает дверь перед людьми, стремящимися начать профессиональную деятельность или продолжить обучение в каком-либо университете.
В данном контексте наша компания предлагает оперативно получить любой необходимый документ. Вы имеете возможность купить диплом, и это будет выгодным решением для всех, кто не смог завершить образование, потерял документ или хочет исправить свои оценки. дипломы изготавливаются аккуратно, с максимальным вниманием ко всем нюансам. На выходе вы получите продукт, 100% соответствующий оригиналу.
Плюсы данного решения заключаются не только в том, что вы сможете оперативно получить диплом. Процесс организовывается просто и легко, с профессиональной поддержкой. От выбора необходимого образца диплома до консультации по заполнению личных данных и доставки по стране — все будет находиться под полным контролем наших мастеров.
В результате, для тех, кто пытается найти оперативный способ получить необходимый документ, наша услуга предлагает выгодное решение. Приобрести диплом – это значит избежать продолжительного процесса обучения и не теряя времени переходить к своим целям, будь то поступление в университет или старт карьеры.
http://cleo-dance.ru
http://kdcmir.ru
http://rada-centr.ru
http://sprchita.ru
http://ipuslu.ru
What a material of un-ambiguity and preserveness of precious knowledge regarding unpredicted emotions.
https://mormoniconoclast.net/
https://exactlybookmarks.com/story17050206/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%83%D0%BD%D0%B8%D0%B2%D0%B5%D1%80%D1%81%D0%B8%D1%82%D0%B5%D1%82%D0%B0
http://trifonov.in/exoplanets/dynamical-analysis-of-multi-planet-systems/hip5364_2_1/
http://plant.geoman.ru/
http://www.asteralaw.com/dhs-provides-relief-to-american-businesses-in-danger-of-suffering-irreparable-harm/workers/
Hello, this weekend is fastidious in favor of me, for the reason that this point in time i am reading this fantastic informative piece of writing here at my house.
http://michiya-cs.com/userinfo.php?uid=30272
https://androidinweb.ru/page/3
http://www.support.ipron.com/communication/forum/index.php?PAGE_NAME=profile_view&UID=17017
https://institutovitae.com/sources-for-digital-marketing/
https://maixepanphat.vn/dich-vu/du-che-su-kien/
https://dr-nona.ru/blog – Dr. Nona
I’m gone to say to my little brother, that he should also pay a visit this website on regular basis to take updated from latest news update.
купить диплом в клинцах
http://pro-novostroyki.ru/index.php?showtopic=20777
http://birsk.mybb.ru/viewtopic.php?id=460#p957
https://sharevita.com/read-blog/22378
https://www.aerocivil.gov.co/atencion/participacion/encuesta/Lists/EncuestaRPAS2019/DispForm.aspx?ID=505
https://wiki.gta-zona.ru/index.php/пїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅ:_пїЅпїЅпїЅпїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅ_пїЅпїЅпїЅпїЅпїЅпїЅпїЅ!
купить диплом в железногорске
Здравствуйте!
У нас вы можете заказать диплом Гознака по специальной цене с доставкой в любой регион России без предоплаты – надежно и выгодно!
https://chere.4bb.ru/viewtopic.php?id=6493
http://kugulove.bestbb.ru/viewtopic.php?id=288
http://igpsclub.ru/social/read-blog/5061
https://pantonekid.com/silviontheway-ecuador/#comment-85779
https://narodovmnogo-omsk.ru/forum/user/3495/
Hi there, all is going fine here and ofcourse every one is sharing facts, that’s really good, keep up writing.
https://mediajx.com/story18677519/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%9F%D0%B5%D1%80%D0%BC%D0%B8
https://www.rune-zero.com/index.php?action=profile;u=108583;area=showposts;sa=topics
https://motorcycle-reviews48260.thelateblog.com/27370811/a-secret-weapon-for-marketing
http://toursoul.ru/?p=19056
https://edgarilmlk.look4blog.com/65997995/considerations-to-know-about-marketing
I got this web page from my friend who told me concerning this web page and now this time I am visiting this web site and reading very informative content at this time.
http://www.forum.pankeewa.org.ru/viewtopic.php?p=249664&highlight=
купить аттестат за 11 класс
http://to66minust.ru
купить диплом в верхней пышме
Hi there! This is kind of off topic but I need some help from an established blog. Is it hard to set up your own blog? I’m not very techincal but I can figure things out pretty fast. I’m thinking about creating my own but I’m not sure where to start. Do you have any ideas or suggestions? Appreciate it
ivyfoods.co.uk/product/?category=Non+Basmati+rice&product=Sona+Masoori++Rice
frpworld.com/modules.php?name=Your_Account&op=userinfo&username=titovaong3
http://www.11na11.com/2011/03/pro-vidstan-vid-polya-do-tribun-na-dnipro-areni/
sohapay.com/info/vi/news/trai-nghiem-tien-ich-voi-the-gioi-the-vpbank.html
77lub.ru/products/small-engine-oils/
Hey! This post could not be written any better! Reading this post reminds me of my good old room mate! He always kept talking about this. I will forward this page to him. Pretty sure he will have a good read. Thank you for sharing!
kenhhocsinh.com/tai-sao-ly-thuong-kiet-lai-ket-thuc-cuoc-khang-chien-chong-tong-bang-bien-phap-giang-hoa
http://www.mountbarkertourismwa.com.au/contact_us.php
pmk-polymer.ru/content/kontakty
euroreparmuldersteendam.nl/service-en-onderhoud/
kunigami-minibas.jp/userinfo.php?uid=17375
Hello it’s me, I am also visiting this web page on a regular basis, this site is really good and the visitors are in fact sharing pleasant thoughts.
http://magnitog.ru/katalog-produktsii/magnitnye-igrushki/futbol-magnetiz-mag-2–16071.html
купить диплом гознак
http://uborka-kvartir-v-moskve.ru
купить диплом цена
Hi Dear, are you really visiting this web page daily, if so after that you will absolutely take nice experience.
http://www.tonpentreafc.com/ton_pentre_youth_090409.html
ukrevent.ru/page/12/
online-vuz.ru/viewNews/177/
darknews.ru/page/34
amisite.ru/phpBB2/memberlist.php?mode=joined&order=ASC&start=6850
Greate post. Keep writing such kind of info on your page. Im really impressed by your blog.
Hi there, You’ve performed a fantastic job. I’ll certainly digg it and individually suggest to my friends. I am confident they’ll be benefited from this site.
https://bookmarkbirth.com/story16760785/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BC%D0%B0%D0%B3%D0%B8%D1%81%D1%82%D1%80%D0%B0
https://lapartenza.vn/
http://fundanzaquindio.com/img_8911/
https://www.composites.cz/2010/12/11/sample-post-3/
https://mysocialguides.com/story2412878/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%A0%D0%BE%D1%81%D1%82%D0%BE%D0%B2
I’m gone to inform my little brother, that he should also pay a quick visit this webpage on regular basis to get updated from hottest information.
https://www.scadachem.com/it-admins-in-minor-companies-dissertation-example-6/
https://cuscino.vn/
http://www.geafer.ru/
http://www.sip-vuz.ru/
https://bookmarksparkle.com/story17136231/%D0%B3%D0%B4%D0%B5-%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC
Hi to all, how is everything, I think every one is getting more from this site, and your views are good for new visitors.
https://pankasyno6.com/
https://siambookmark.com/story17133449/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%B2-%D0%A7%D0%B5%D0%BB%D1%8F%D0%B1%D0%B8%D0%BD%D1%81%D0%BA%D0%B5
https://pdtplanilla.com/
http://fabnews.ru/forum/showthread.php?p=76080&mode=threaded
https://ezmarkbookmarks.com/story17184192/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D0%BC%D0%B5%D0%BD%D0%B5%D0%B4%D0%B6%D0%B5%D1%80%D0%B0
Great post but I was wondering if you could write a litte more on this topic? I’d be very thankful if you could elaborate a little bit more. Bless you!
old.kakdelat.ru/company/personal/user/1695/blog/8780/
brisbaneglobaltens.com/tournament/brisbane
shkaranov.ru/Obruchalnie-i-venchalnie-kolca/page7.html
chefrobertsdirect.com/index.php?main_page=page&id=2
circuit-diagrams.com/pr4-PIC16F84A-discolight-effect-with-bass-beat-control.php
May I just say what a relief to uncover somebody that really understands what they are talking about over the internet. You certainly know how to bring an issue to light and make it important. More and more people should check this out and understand this side of your story. I was surprised that you’re not more popular because you most certainly have the gift.
catsandsquirrels.com/tag/squelfie/
ttdinhduong.org/ttdd/tin-tuc/tin-chuyen-mon/679-Thong-bao-chieu-sinh-khoa-dao-tao-ddls.aspx
cbt-v.com.ua/catalog/all
dachkanews.ru/page/10/
cocochampionship.com/en/contact
슬롯 게임 무료
Fang Jifan이 사건을 맡았습니다. “자,이 놈을 잘 때려주세요!”
Great post but I was wondering if you could write a litte more on this subject? I’d be very thankful if you could elaborate a little bit further. Appreciate it!
https://muscle-bg.com/trenirovki/trenirovki-za-masa/397-10-sedmichna-trenirovychna-programa-za-muskulna-masa.html
https://holdenuwvvt.bloggerswise.com/33144707/facts-about-marketing-revealed
https://kbookmarking.com/story17081637/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B0%D1%82%D1%82%D0%B5%D1%81%D1%82%D0%B0%D1%82-%D0%B7%D0%B0-9-%D0%BA%D0%BB%D0%B0%D1%81%D1%81
https://usadba.in.ua/
https://getsocialpr.com/story17673598/%D0%BA%D1%83%D0%BF%D0%B8%D1%82%D1%8C-%D0%B4%D0%B8%D0%BF%D0%BB%D0%BE%D0%BC-%D1%81%D1%82%D0%BE%D0%BC%D0%B0%D1%82%D0%BE%D0%BB%D0%BE%D0%B3%D0%B0
Hello to every body, it’s my first pay a visit of this web site; this website contains awesome and genuinely good stuff in favor of visitors.
turbodom.ru/music/instrumentalnaya/?music/instrumentalnaya&letter=%D0%A4/index.html
supplementgear.com/low-glycemic-fruits/
miupsik.ru/forums/portal.php?page=102
alpha.prime-pc.md/good_info/27721
prachuabwit.ac.th/krusuriya/modules.php?name=Journal&file=display&jid=12564
Hello very nice blog!! Man .. Excellent .. Amazing .. I will bookmark your website and take the feeds also? I am glad to search out so many useful information here in the put up, we want work out extra strategies on this regard, thanks for sharing. . . . . .
https://suamaylanh.com.vn/
https://ann-summers-promo-code05936.blogsvila.com/27006140/rumored-buzz-on-marketing
https://www.bodybuilding.net/members/worksale.html
http://opengadjet.ru/page/3
https://containertraffic.ru/
В современном мире, где диплом – это начало удачной карьеры в любом направлении, многие ищут максимально быстрый и простой путь получения образования. Наличие официального документа переоценить попросту невозможно. Ведь диплом открывает двери перед людьми, стремящимися начать профессиональную деятельность или продолжить обучение в университете.
Мы предлагаем оперативно получить этот важный документ. Вы можете купить диплом, и это становится отличным решением для всех, кто не смог закончить обучение, утратил документ или желает исправить свои оценки. Любой диплом изготавливается аккуратно, с максимальным вниманием ко всем элементам. В результате вы сможете получить 100% оригинальный документ.
Превосходство такого решения состоит не только в том, что можно оперативно получить диплом. Процесс организовывается комфортно, с нашей поддержкой. Начав от выбора необходимого образца до правильного заполнения персональных данных и доставки по стране — все находится под абсолютным контролем квалифицированных специалистов.
Для тех, кто ищет оперативный способ получить необходимый документ, наша услуга предлагает отличное решение. Приобрести диплом – это значит избежать долгого процесса обучения и сразу переходить к достижению личных целей, будь то поступление в ВУЗ или начало профессиональной карьеры.
http://geafer.ru
http://spravka-v-bassein.ru
http://cardioevent.ru
http://extra-art.ru
http://borovskie-izvestiya.ru
I’m not sure why but this web site is loading very slow for me. Is anyone else having this issue or is it a problem on my end? I’ll check back later and see if the problem still exists.
http://www.irc71.ru/index.php/jomsocial/groups/viewbulletin/45-za-kakuyu-tsenu-segodnya-vozmozhno-kupit-khoroshego-kachestva-diplom?groupid=47
купить диплом монтажника
http://diplomvam.ru
купить диплом в рубцовске
Hey! Would you mind if I share your blog with my myspace group? There’s a lot of people that I think would really appreciate your content. Please let me know. Many thanks
http://visayasmed.azurewebsites.net/career-opportunities/hiring-licensed-engineer
купить диплом в дзержинске
http://wmtext.ru
купить диплом в великом новгороде
포춘 슬롯
Fang Jifan은 “왜 이런 말을 나에게 말했고 왜 폐하에게 즉시 말하지 않았습니까? “라고 말했습니다.
Hi i am kavin, its my first time to commenting anywhere, when i read this piece of writing i thought i could also make comment due to this good piece of writing.
http://video-uprazhnenija.ru/category/joga/
куплю диплом о высшем образовании
http://stroiudostoverenie.ru
купить диплом в туапсе
Hello to every body, it’s my first pay a quick visit of this weblog; this web site consists of remarkable and really fine stuff in favor of readers.
http://dmbp.ru/country/russia/country/russia/kaliningrad/ekskursionye-programi/ocharovanie-baltiki
купить свидетельство о рождении
http://fk-arsenal.ru
купить диплом в белогорске
Having read your posts. I believed you have given your readers valuable information. Feel free to visit my website http://allmikki.com/shop/proc/indb.cart.tab.php?action=ok&tab=today&type=delete&returnurl=http://webemail24.com/tips-to-improve-your-email-marketing-strategy and I hope you get additional insights about Crowdfunding as I did upon stumbling across your site.
https://antimafia.se/ – hashish with a syringe 7 gram discount. 31%
https://antimafia.se/ – powdered heroin – snorting 50 grams sleva 13%
https://antimafia.se/ – ecstasy under the tongue – 3 gram sleva 12%
https://antimafia.se/ – hachis con una jeringuilla 100 gramas descuentos. 35%
https://antimafia.se/ – heroina en polvo – esnifada 50 grama descuentos 13%
https://antimafia.se/ – extasis bajo la lengua – 3 gramas descuentos 47%
https://antimafia.se/ – hasista ruiskulla 5 grams sale. 17%
https://antimafia.se/ – heroiinijauhe – nuuskaaminen 100 gram sale 47%
https://antimafia.se/ – ekstaasi kielen alla – 1 gram sale 31%
https://antimafia.se/ – haszysz za pomoca strzykawki 17 gramy sale. 37%
https://antimafia.se/ – sproszkowana heroina – wciaganie 70 gram discount 18%
https://antimafia.se/ – ekstaza pod jezykiem – 11 gramy discount 12%
https://antimafia.se/ – du haschisch avec une seringue 6 gram sale. 15%
https://antimafia.se/ – heroine en poudre – sniffer 30 grammes sale 25%
https://antimafia.se/ – ecstasy sous la langue – 30 grammes discount 35%
Hi, yes this piece of writing is genuinely nice and I have learned lot of things from it on the topic of blogging. thanks.
http://rekshan.ru/realrusrock/
купить диплом о высшем образовании
http://ussursity.ru
купить диплом во владикавказе
Hey there! Do you use Twitter? I’d like to follow you if that would be ok. I’m undoubtedly enjoying your blog and look forward to new updates.
http://skitour.su/member.php?u=1002&tab=visitor_messaging&page=1
куплю диплом
http://seo1st.ru
купить диплом в москве
Right here is the right site for anybody who hopes to find out about this topic. You know so much its almost tough to argue with you (not that I personally will need to…HaHa). You definitely put a new spin on a subject which has been discussed for a long time. Great stuff, just great!
http://www.aescalaproyectos.es/portfolio-view/armarios/
купить диплом в иваново
http://irkutsk-arbitr.ru
купить диплом кандидата наук
“Дело “Лайф-из-Гуд” — “Гермес” — “Бест Вей”: свидетель обвинения объявила себя потерпевшей от следствия
Набойченко Лайф-из-Гуд
6 и 13 июня Приморский районный суд города Санкт-Петербурга, рассматривающий по существу уголовное дело № 1-504/24, связываемое с компаниями “Лайф-из-Гуд”, “Гермес” и кооперативом “Бест Вей”, провел очередные, шестое и седьмое по счету, заседания, посвященные допросу свидетелей обвинения и лиц, признанных следствием потерпевшими в рамках судебного следствия по делу
“К кооперативу претензий не было, следователь предложил подать заявление”
Признанный следствием потерпевший Болян подсудимых не знает. Был клиентом “Гермеса”, а также пайщиком кооператива — но до 2019 года. В 2019-м он вышел из кооператива и из “Гермеса”, ему были возвращены паевые взносы, и никаких претензий к кооперативу у него не было — что он письменно подтвердил, расторгая договоры с этими организациями.
Однако, как Болян отметил на суде, следователь убедил его в том, что он — потерпевший и должен подать заявление на возврат членских взносов. Заявление в МВД писать не хотел, на него вышли сотрудники, сначала претензий к кооперативу не было. Полиция ему объяснила, что можно получить деньги.
Стал клиентом “Гермеса” и пайщиком кооператива через своего консультанта Алексея Виноградова. Виноградов — грамотный маркетолог, он ему верил, тот не работал в кооперативе. Что было предметом договора в “Гермесе”, не помнит. В “Гермес” внес 100 и 700 евро, а в кооператив каждый месяц вносил по 12 тыс. в течение семи месяцев.
Вышел и из кооператива, и из “Гермеса” в 2019 году. Зачем вступал? “Наверное, квартиру купить хотел”. Кооператив вернул ему 70 тыс. паевых взносов, “Гермес” вернул со счета “Виста” 140 тыс. рублей.
В кооперативе деньги вернули почти сразу, удержав вступительный и членские взносы; в “Гермесе” вернули позже через “внутрянку”, но удержали комиссию.
Утверждает, что ему говорили, что можно со счета “Виста” вносить деньги в кооператив. Объясняли, что деньги передаются в доверительное управление трейдерам и брокерам, которые играют на бирже. В кооперативе, как он утверждает, можно было купить место в очереди. По его словам, “Гермес” и кооператив — по сути, одна организация. Требует взыскать с кооператива более 148 тыс. рублей — вступительный и членские взносы, и более 60 тыс. рублей с “Гермеса” — комиссию при выводе средств.
Договор с кооперативом не читал, но ему объяснили, что есть невозвратная часть денег — ее и не вернули, “но хочу попытаться вернуть”. Претензий к кооперативу “как бы и нет, но если вернут взносы, то будет хорошо”.
К Виноградову претензий не предъявлял. “Может, меня и не обманули в кооперативе”, -резюмировал свое выступление в суде Болян.
“Требую выплатить с учетом роста цен на недвижимость”
Признанная следствием потерпевшей Комова была как клиентом “Гермеса”, так и пайщиком кооператива. Подсудимых не знает. Требует более 8800 тыс. с кооператива и более 2700 тыс. с “Гермеса”. При этом из кооператива она не вышла и заявление о выходе не подавала. Сумма требований к кооперативу включает как паевые и членские взносы, так и оценку роста цен на недвижимость, которая не была приобретена.
Утверждает, что можно переводить деньги со счета “Виста” напрямую в кооператив — в подтверждение приводит скрины переписки с консультантами в смартфоне. Суд разъясняет, что доказательство может быть приобщено позднее при надлежащем оформлении.
“Информацию воспринял скептически, но вступил”
Признанный следствием потерпевшим Киреев был только клиентом “Гермеса”. Подсудимых не знает лично — Измайлова видел в видеоролике “Лайф-из-Гуд”. Узнал о “Гермесе” от консультантов Татьяны и Андрея Клейменовых, с которыми знаком с конца 1990-х годов. Вступил в “Гермес” в 2020 году, деньги сразу отобразились на счете. Вносил средства на счет “Виста” небольшими частями, снимал средства со счета для личных нужд. Заявляет требования на более чем 2 млн 300 тыс. рублей.
Когда подписывал договор, в офисе была табличка кооператива, но в офисе находился представитель фирмы “Гермес”, ему сказали, что это два продукта компании “Лайф-из-Гуд”.
В 2022 году начались проблемы с выводом средств. И руководство фирмы пугало блокировкой счета в случае подачи заявления в правоохранительные органы. В хейтерском чате узнал телефон следователя Сапетовой, позвонил и подал заявление.
В чате клиентов компании “Гермес” была информация о том, что счет будет заблокирован в случае подачи заявления в правоохранительные органы. Эта информация была за подписью “администрация”. Он спросил у консультантов: “Кто это — администрация?”, написал вопрос в чате, и его забанили. Других попыток вступать в диалог с “Гермесом” не предпринимал.
Признанный следствием потерпевшим Чернышенко был также только клиентом только компании “Гермес”. Подсудимых не знает. Познакомился с консультантами “Гермеса” во время совместной работы в “Макдоналдсе”. Информацию о “Гермесе” воспринял скептически — “это какой-то обман”, но консультанты его в конце концов уговорили. Они поставили условие, что продадут ему земельный участок, чтобы он вступил в “Гермес”.
“Я хотел их обмануть, — пояснил свидетель, — думал: пусть они продадут, а я в “Гермес” не вступлю. Но они начали уговаривать, и я вступил в апреле 2020 года. Один из консультантов дал расписку на внесение денег на счет в “Гермесе”, я внес через него 230 тыс. И еще раз из жадности 140 тыс. 140 тыс. я вывел, плюс каждый месяц выводил по 5000–7000 руб. С какого-то момента проценты стали падать. В конце я захотел выйти — но не успел. У меня в “Гермесе” осталось 270 тыс., которые я требую вернуть”.
В январе 2023 года Чернышенко подал заявление в полицию, так как прекратился доступ к счету. “Мне предложили 70 тыс. вернуть, чтобы я не ходил в полицию, ссылаясь на то, что помогли мне продать участок за меньшую сумму по документам, чем я его продал на самом деле, но я отказался, сказал, что мне надо 270 тыс.”.
“Я хочу стать потерпевшей. Но Василенко многие благодарны за квартиры”
Свидетель обвинения Галашенкова была только клиентом “Гермеса”. Из подсудимых знает Виктора Ивановича Василенко. Она сидела в зале при допросе двух потерпевших (что запрещено), но заявила, что плохо слышит. Ранее была знакома с Верой Исаевой из компании “Лайф-из-Гуд”.
Вера узнала, что у нее есть офис, и познакомила с Романом Василенко, который снял у нее офис, когда “Лайф-из-Гуд” только начинала раскручиваться. Он снимал офис только за коммунальные платежи. “Я попросила расплатиться за четыре месяца, а он предложил мне вместо денег вступить в “Бест Вей”, но я отказалась, так как квартира не была нужна”. Зато стала клиентом “Гермеса”: “Внесла 15 тыс. евро, но через три месяца на счете оказалось 3400 евро!” Исаева, по словам Галашенковой, оказалась мошенницей.
По словам Галашенковой, она внесла на счет “Виста” более 3 млн рублей. “Когда сказали, что все счета заблокировали, то собрали с нас еще денег, чтобы счета разблокировать, потом пришло сообщение, что открывается новая фирма, но это все оказалось обманом — счета не разблокированы”.
“Я не считала себя потерпевшей, так как не знала, что это можно сделать, — заявила Галашенкова. — Сейчас желаю заявить, что я хочу стать потерпевшей. Считаю, что должна взыскать с “Гермеса” как прямой ущерб, так и упущенную выгоду, так как эта фирма закрылась”.
При этом заявила, что “Василенко молодец, потому что я видела людей, которые купили квартиры, они были ему очень благодарны”.
I like it when folks get together and share thoughts.
Great blog, continue the good work!
https://fakeoff.org/ – hashish with a syringe 7 gram sleva. 34%
https://fakeoff.org/ – powdered heroin – snorting 30 grams discounts 41%
https://fakeoff.org/ – ecstasy under the tongue – 1 gram discounts 33%
https://fakeoff.org/ – hachis con una jeringuilla 31 grama rebajas. 35%
https://fakeoff.org/ – heroina en polvo – esnifada 50 gramas sleva 27%
https://fakeoff.org/ – extasis bajo la lengua – 3 gramas descuento 35%
https://fakeoff.org/ – hasista ruiskulla 10 grams sale. 17%
https://fakeoff.org/ – heroiinijauhe – nuuskaaminen 100 grams discounts 47%
https://fakeoff.org/ – ekstaasi kielen alla – 2 gram sleva 33%
https://fakeoff.org/ – haszysz za pomoca strzykawki 13 gram discount. 38%
https://fakeoff.org/ – sproszkowana heroina – wciaganie 40 gramy discount 13%
https://fakeoff.org/ – ekstaza pod jezykiem – 16 gram discount 42%
https://fakeoff.org/ – du haschisch avec une seringue 15 gram sleva. 23%
https://fakeoff.org/ – heroine en poudre – sniffer 100 grammes sleva 35%
https://fakeoff.org/ – ecstasy sous la langue – 10 grammes sleva 27%
온라인 슬롯 게임 추천
그런데 왜, 마흔 넘은 노인처럼.
Общественное здание или его часть зетфликс с оборудованием для публичной демонстрации кинофильмов. Главное помещение кинотеатра – зрительный зал с экраном большого размера и системой воспроизведения звука. В современных кинотеатрах система звуковоспроизведения состоит из нескольких громкоговорителей, обеспечивающих объёмный звук. Первые стационарные кинотеатры назывались в России «электротеатрами»
зетфликс https://zetflixhd.life/
Hi there, all is going sound here and ofcourse every one is sharing data, that’s actually fine, keep up writing.
http://forum.oursson.ru/viewtopic.php?f=117&t=3341&p=10425Г‚
купить диплом логиста
http://energoprofi23.ru
купить диплом в бийске
We absolutely love your blog and find the majority of your post’s to be exactly I’m looking for. Do you offer guest writers to write content in your case? I wouldn’t mind producing a post or elaborating on many of the subjects you write with regards to here. Again, awesome web site!
http://konteiners.ru/izotermicheskii-konteiner-insulated.html
купить диплом в владикавказе
http://281415.ru
купить диплом в усть-илимске
‘Sugary food is a drug for me’: A growing number of children are addicted to ultraprocessed foods
kraken17
Chicago native Jeffrey Odwazny says he has been addicted to ultraprocessed food since he was a child.
“I was driven to eat and eat and eat, and while I would overeat healthy food, what really got me were the candies, the cakes, the pies, the ice cream,” said the 54-year-old former warehouse supervisor
https://kraken-25.at
2krn
“I really gravitated towards the sugary ultraprocessed foods — it was like a physical drive, I had to have it,” he said. “My parents would find hefty bags full of candy wrappers hidden in my closet. I would steal things from stores as a kid and later as an adult.”
Some 12% of the nearly 73 million children and adolescents in the United States today struggle with a similar food addiction, according to research. To be diagnosed, children must meet Yale Food Addiction Scale criteria as stringent as any for alcohol use disorder or other addictions.
“Kids are losing control and eating to the point where they feel physically ill,” said Ashley Gearhardt, a professor of psychology at the University of Michigan in Ann Arbor who conducted the research and developed the Yale addiction scale.
“They have intense cravings and may be sneaking, stealing or hiding ultraprocessed foods,” Gearhardt said. “They may stop going out with friends or doing other activities they used to enjoy in order to stay at home and eat, or they feel too sluggish from overeating to participate in other activities.”
Her research also shows about 14% of adults are clinically addicted to food, predominantly ultraprocessed foods with higher levels of sugar, salt, fat and additives.
For comparison, 10.5% of Americans age 12 or older were diagnosed with alcohol use disorder in 2022, according to the National Survey on Drug Use and Health.
While many people addicted to food will say that their symptoms began to worsen significantly in adolescence, some recall a childhood focused on ultraprocessed food.
“By age 2 or 3, children are likely eating more ultraprocessed foods in any given day than a fruit or vegetable, especially if they’re poor and don’t have enough money in their family to have enough quality food to eat,” Gearhardt said. “Ultraprocessed foods are cheap and literally everywhere, so this is also a social justice issue.”
An addiction to ultraprocessed foods can highjack a young brain’s reward circuitry, putting the primitive “reptilian brain,” or amygdala, in charge — thus bypassing the prefrontal cortex where rational decision-making occurs, said Los Angeles registered dietitian nutritionist David Wiss, who specializes in treating food addiction.
https://dr-Nona.ru/ – Доктор Нона
프라그마틱 무료 슬롯
Xu Pengju의 눈은 여전히 흐릿했고 오랫동안 생각했습니다. “잘 이해가 안 돼요.”
The world’s most liveable cities for 2024
жесткое порно бесплатно
It’s considered among the most beautiful cities in the world to visit, and it seems that Vienna may also be an unbeatable place to live.
The Austrian city has been crowned the most liveable city in the world yet again in the annual list from the Economist Intelligence Unit (EIU), which was released today.
The EIU, a sister organization to The Economist, ranked 173 cities across the globe on a number of significant factors, including health care, culture and environment, stability, infrastructure and education.
Vienna topped the list for the third consecutive year, receiving “perfect” scores in four out of five of the categories — the city was marked lower for culture and environment due to an apparent lack of significant sporting events.
Just behind the Austrian capital, Denmark’s Copenhagen retained its second place position, while Switzerland’s Zurich moved up from sixth place to third on the list.
Australia’s Melbourne fell from third to fourth place, while Canadian city Calgary tied for fifth place with Swiss city Geneva.
Canada’s Vancouver and Australia’s Sydney were in joint seventh place, and Japan’s Osaka and New Zealand’s Auckland rounded out the top 10 in joint ninth place.
«Король» обвинителей «Лайф-из-Гуд» Набойченко – кто он на самом деле?
Свидетель Набойченко
Евгений Набойченко – бывший IT-директор компании «Лайф-из-Гуд», а также сисадмин российского сегмента платежной системы международной инвестиционной компании «Гермес». Он – ключевой свидетель обвинения в так называемом деле «Лайф-из-Гуд» – «Гермес» – кооператива «Бест Вей» и основателя «Лайф-из-Гуд»/«Бест Вей» Романа Василенко, которое рассматривается сейчас судьей Екатериной Богдановой в Приморском районном суде Санкт-Петербурга.
С выполненной им блокировки российского сегмента платежной системы компании «Гермес», а также с его (нетрезвых) сообщений в социальных сетях (сейчас все они им стерты) началась эскалация этого уголовного дела. Но, скорее всего, дело им же самим и его подельниками из питерской полиции и создано.
Вор и дебошир
Наши источники делятся информацией о хищении активов с кошельков клиентов «Гермеса», к которым Евгений Набойченко имел доступ.
Набойченко не гнушался воровать даже у близких. Однажды остановившись у своей любовницы Светланы Н., Евгений избил ее и украл деньги с ее криптовалютного кошелька, пока она пыталась прийти в себя после побоев.
Он бил не только любовницу, но жену и детей, с которыми жил в Краснодарском крае. А после того как его супруга Виктория подала на развод, разгромил, ободрал и ограбил общий дом – унес все, что можно было унести. Надо ли говорить, что Набойченко который год не платит супруге алименты.
Вымогатель
Евгений Набойченко, по словам его бывшей жены, в один прекрасный день решил заработать и с помощью вымогательства. Он шантажировал Романа Василенко – требовал с него деньги: 170 тыс. евро. При этом, по утверждению Виктории Набойченко, угрожал увечьями и самому Роману Василенко, и его супруге, и детям. Похвалялся перед (тогда еще) женой своими матерными сообщениями с угрозами, которые он посылал Василенко и его близким.
Кроме того, вымогал деньги у клиентов «Гермеса»: свидетельства такого рода нам предоставлены.
Алкоголик и наркоман
Деньги нужны Набойченко в том числе для того, чтобы «питать» свою наркотическую зависимость – с некоторых пор он предпочитает шлифовать водку галлюциногенными грибами, об увлечении которыми с упоением рассказывал даже в своих соцсетях.
Он не всегда был алкоголиком и наркоманом – устроившись 10 лет назад в IT-службу, не пил и не употреблял. Но большие деньги, красивая жизнь, которая вдруг стала доступна для простого астраханского парня из низов, развратили его.
Клеветник
Он допустил целый ряд клеветнических высказываний в адрес Романа Василенко – за которые против него заведено уголовное дело. Но расследуется оно ни шатко ни валко, так как расследование тормозит начальник УЭБиПК ГУ МВД России по Санкт-Петербургу и Ленинградской области.
Завербованный органами
Виктория Набойченко сообщает, что за три дня до обыска в их тогда еще совместном доме Евгений уехал и ей сказал, что придут с обыском – но это пустая формальность. Он уже был в сговоре с полицией, так как за три дня был предупрежден об этом обыске.
15 февраля 2022 года Евгений Набойченко был задержан, но отправился не в СИЗО – куда отправили других сотрудников «Лайф-из-Гуд», не был помещен в изолятор или КПЗ, а жил на полицейской квартире.
Он отказался от услуг адвокатов и на этой квартире, по свидетельству одного из полицейских, «бухал с операми». А также каждый день водил оперов по барам и ресторанам – угощал их за свой счет.
В разговоре с женой он очень гордился покровительством начальника УЭБиПК питерского главка МВД.
Наши источники считают, что он украл деньги клиентов «Гермеса», разделив их со своими друзьями из полиции. Вся его дальнейшая жизнь и привычки указывают на то, что он использует ворованные деньги.
Трус и лжец
Набойченко сообщил в социальных сетях, что к нему пришло с адреса fall@mybizon.online письмо, в котором содержится угроза в адрес получателя, призывающая его не являться в суд для дачи показаний. В письме указано, что если он поедет в суд, то не доедет и будет трупом.
Создатель преступного сообщества
Нет сомнений, что Набойченко устроил историю с обрушением платежной системы, чтобы скрыть свое воровство активов клиентов. Также чтобы скрыть свое воровство, он обвинил во всем Василенко и технических специалистов компании «Лайф-из-Гуд», которые находятся сейчас на скамье подсудимых. Оклеветал всех, чтобы отвести подозрения от себя и своих подельников из питерской полиции.
Итак, кризис в «Гермесе» устроил Набойченко. Платежную систему разгромил Набойченко. Активы украл Набойченко, поделившись с питерскими полицейскими.
А теперь сидит и бездельничает, прожигая сворованные деньги. И боится идти в суд – чтобы не превратиться в суде из свидетеля в обвиняемого.
Скорее всего, в суд Набойченко не пускают сами полицейские – опасаясь его неадекватности и перекрестного допроса, на котором вскроется лживость его показаний. То есть силовики заметают следы своего преступления, совершенного с помощью марионетки Набойченко.
«Король» обвинителей «Лайф-из-Гуд» Набойченко – кто он на самом деле?
ПК Бествей
Евгений Набойченко – бывший IT-директор компании «Лайф-из-Гуд», а также сисадмин российского сегмента платежной системы международной инвестиционной компании «Гермес». Он – ключевой свидетель обвинения в так называемом деле «Лайф-из-Гуд» – «Гермес» – кооператива «Бест Вей» и основателя «Лайф-из-Гуд»/«Бест Вей» Романа Василенко, которое рассматривается сейчас судьей Екатериной Богдановой в Приморском районном суде Санкт-Петербурга.
С выполненной им блокировки российского сегмента платежной системы компании «Гермес», а также с его (нетрезвых) сообщений в социальных сетях (сейчас все они им стерты) началась эскалация этого уголовного дела. Но, скорее всего, дело им же самим и его подельниками из питерской полиции и создано.
Вор и дебошир
Наши источники делятся информацией о хищении активов с кошельков клиентов «Гермеса», к которым Евгений Набойченко имел доступ.
Набойченко не гнушался воровать даже у близких. Однажды остановившись у своей любовницы Светланы Н., Евгений избил ее и украл деньги с ее криптовалютного кошелька, пока она пыталась прийти в себя после побоев.
Он бил не только любовницу, но жену и детей, с которыми жил в Краснодарском крае. А после того как его супруга Виктория подала на развод, разгромил, ободрал и ограбил общий дом – унес все, что можно было унести. Надо ли говорить, что Набойченко который год не платит супруге алименты.
Вымогатель
Евгений Набойченко, по словам его бывшей жены, в один прекрасный день решил заработать и с помощью вымогательства. Он шантажировал Романа Василенко – требовал с него деньги: 170 тыс. евро. При этом, по утверждению Виктории Набойченко, угрожал увечьями и самому Роману Василенко, и его супруге, и детям. Похвалялся перед (тогда еще) женой своими матерными сообщениями с угрозами, которые он посылал Василенко и его близким.
Кроме того, вымогал деньги у клиентов «Гермеса»: свидетельства такого рода нам предоставлены.
Алкоголик и наркоман
Деньги нужны Набойченко в том числе для того, чтобы «питать» свою наркотическую зависимость – с некоторых пор он предпочитает шлифовать водку галлюциногенными грибами, об увлечении которыми с упоением рассказывал даже в своих соцсетях.
Он не всегда был алкоголиком и наркоманом – устроившись 10 лет назад в IT-службу, не пил и не употреблял. Но большие деньги, красивая жизнь, которая вдруг стала доступна для простого астраханского парня из низов, развратили его.
Клеветник
Он допустил целый ряд клеветнических высказываний в адрес Романа Василенко – за которые против него заведено уголовное дело. Но расследуется оно ни шатко ни валко, так как расследование тормозит начальник УЭБиПК ГУ МВД России по Санкт-Петербургу и Ленинградской области.
Завербованный органами
Виктория Набойченко сообщает, что за три дня до обыска в их тогда еще совместном доме Евгений уехал и ей сказал, что придут с обыском – но это пустая формальность. Он уже был в сговоре с полицией, так как за три дня был предупрежден об этом обыске.
15 февраля 2022 года Евгений Набойченко был задержан, но отправился не в СИЗО – куда отправили других сотрудников «Лайф-из-Гуд», не был помещен в изолятор или КПЗ, а жил на полицейской квартире.
Он отказался от услуг адвокатов и на этой квартире, по свидетельству одного из полицейских, «бухал с операми». А также каждый день водил оперов по барам и ресторанам – угощал их за свой счет.
В разговоре с женой он очень гордился покровительством начальника УЭБиПК питерского главка МВД.
Наши источники считают, что он украл деньги клиентов «Гермеса», разделив их со своими друзьями из полиции. Вся его дальнейшая жизнь и привычки указывают на то, что он использует ворованные деньги.
Трус и лжец
Набойченко сообщил в социальных сетях, что к нему пришло с адреса fall@mybizon.online письмо, в котором содержится угроза в адрес получателя, призывающая его не являться в суд для дачи показаний. В письме указано, что если он поедет в суд, то не доедет и будет трупом.
Создатель преступного сообщества
Нет сомнений, что Набойченко устроил историю с обрушением платежной системы, чтобы скрыть свое воровство активов клиентов. Также чтобы скрыть свое воровство, он обвинил во всем Василенко и технических специалистов компании «Лайф-из-Гуд», которые находятся сейчас на скамье подсудимых. Оклеветал всех, чтобы отвести подозрения от себя и своих подельников из питерской полиции.
Итак, кризис в «Гермесе» устроил Набойченко. Платежную систему разгромил Набойченко. Активы украл Набойченко, поделившись с питерскими полицейскими.
А теперь сидит и бездельничает, прожигая сворованные деньги. И боится идти в суд – чтобы не превратиться в суде из свидетеля в обвиняемого.
Скорее всего, в суд Набойченко не пускают сами полицейские – опасаясь его неадекватности и перекрестного допроса, на котором вскроется лживость его показаний. То есть силовики заметают следы своего преступления, совершенного с помощью марионетки Набойченко.
The world’s most liveable cities for 2024
гей онлайн
It’s considered among the most beautiful cities in the world to visit, and it seems that Vienna may also be an unbeatable place to live.
The Austrian city has been crowned the most liveable city in the world yet again in the annual list from the Economist Intelligence Unit (EIU), which was released today.
The EIU, a sister organization to The Economist, ranked 173 cities across the globe on a number of significant factors, including health care, culture and environment, stability, infrastructure and education.
Vienna topped the list for the third consecutive year, receiving “perfect” scores in four out of five of the categories — the city was marked lower for culture and environment due to an apparent lack of significant sporting events.
Just behind the Austrian capital, Denmark’s Copenhagen retained its second place position, while Switzerland’s Zurich moved up from sixth place to third on the list.
Australia’s Melbourne fell from third to fourth place, while Canadian city Calgary tied for fifth place with Swiss city Geneva.
Canada’s Vancouver and Australia’s Sydney were in joint seventh place, and Japan’s Osaka and New Zealand’s Auckland rounded out the top 10 in joint ninth place.
Следственная группа по делу «Бест Вей»: следователи или оборотни в погонах?
[url=https://vestnik-jurnal.com/novosti/item/100193-voennyh-unizhayut-tylovye-krysy-opg-kolokolceva]Бест Вей[/url]
Следственная группа по делу кооператива «Бест Вей» во главе со следователями ГСУ ГУ МВД по Санкт-Петербургу и Ленобласти майором юстиции Екатериной Сапетовой и подполковником юстиции Константином Иудичевым играет собственную партию либо для того, чтобы выслужиться перед непосредственным начальством, либо из-за прямой заинтересованности в уничтожении кооператива со стороны конкурентов и/или банков.
Следствие по делу кооператива «Бест Вей» вступило в активную фазу ровно год назад. За это время арестованы четверо технических сотрудников, и уже год они томятся в СИЗО, пятеро граждан объявлены в розыск.
Офис кооператива буквально разгромлен двумя обысками, документация изъята, и следствие запрещает ее копировать, счета и недвижимость арестованы. Под арестом — активы на более чем 15 млрд рублей, хотя ущерб для так называемых потерпевших, пул которых следователи год собирали под разными предлогами, не превышает 115 млн рублей. Это спустя почти полтора года с момента возбуждения уголовного дела, не считая периода длительного проведения оперативно-разыскных мероприятий. Гора родила мышь.
При этом значительная часть этих потерпевших, например, бывшие пайщики, которые внесли невозвратный вступительный взнос, но затем не смогли копить на квартиру и сами вышли из кооператива, а теперь по наущению следствия подали заявление о том, что кооператив им не вернул то, что и не должен был вернуть. Потерпевших 100 с лишним человек, притом что в кооперативе — более 19 тыс. пайщиков, то есть капля в море, да и эти 100 не имеют юридически обоснованных претензий к кооперативу. Среди представленных следствием потерпевших есть и конкуренты — бывшие пайщики, которые еще несколько лет назад вышли из «Бест Вей» и создали «альтернативный» кооператив «Вера».
Следователи обосновывают арест счетов тем, что «деньги могут украсть», хотя они же сами как минимум участвуют во вполне криминальном использовании чужих денег, уже год не давая пайщикам ни приобрести квартиру за счет средств, которые они скопили на счете в кооперативе, ни забрать деньги, при этом предоставляя возможность банкам. Следователи уже год дают возможность банкам пользоваться счетами почти на 4 млрд рублей, притом постоянно пополняющимися, так как большинство пайщиков продолжает вносить паевые и членские взносы.
Парадокс в том, что новая история обиженных дольщиков (в данном случае пайщиков) всероссийского масштаба возникла не из-за того, что у организации не оказалось средств для выполнения обязательств, а из-за того, что следственная группа лишила ее возможности пользования средствами и прямо блокирует выполнение обязательств перед пайщиками.
Адвокатам кооператива несколько раз удавалось добиться снятия арестов с активов кооператива, однако следователи и банки просто игнорируют решения судов. Потом, пользуясь высоким статусом следователей главка, заходят с новым ходатайством об аресте через кабинет судьи и без участия представителей кооператива, и активы снова арестовываются — до следующей успешной апелляции адвокатов, после которой в дело вступают банки, по договоренности со следствием блокирующие счета в рамках процедур комплаенс, игнорируя решения судов.
Должностное преступление
Действия следственной группы не могут квалифицироваться иначе, как должностное преступление, так как в них прослеживается заинтересованность в уничтожении кооператива. Фактически они сами организуют преступление, которое якобы расследуют, целенаправленно разрушают кооператив, а не спасают пайщиков, не нуждающихся в их спасении и 15 февраля проводящих всероссийскую акцию в защиту своего кооператива от следственной группы.
Адвокаты подавали в Следственный комитет заявление о преступлении, но оно было положено под сукно.
К службе не годны
Отдельная тема — вопиющая профнепригодность следственной группы. Иудичев в своих ответах на запросы ничтоже сумняшеся пишет, что Центральный банк признал кооператив финансовой пирамидой, хотя ЦБ, во-первых, не суд, чтобы делать такие «признания», во-вторых, такого признания просто не было: утверждение не соответствует действительности.
ЦБ внес кооператив в так называемый предупредительный список, являющийся инструментом информирования потребителей финансовых услуг об организациях, работа с которыми чревата для граждан рисками, и решение о включении в этот список состоялось после возбуждения уголовного дела, скорее всего, как раз на основе информации о его возбуждении.
Любимая тема следственной группы — якобы незаконность переименования кооператива из жилищного в потребительский: они не могут даже прочитать Жилищный кодекс, где черным по белому написано, что жилищный кооператив — один из видов жилищного. И подобных примеров некомпетентности и выдумок в деятельности следователей масса.
В итоге мы имеем шитое белыми нитками дело, наносящее колоссальный ущерб тысячам пайщиков кооператива, среди которых преобладают малообеспеченные люди.
Возникают вопросы: не пора ли министру Колокольцеву разобраться со своими сотрудниками, не пора ли главе Следкома Бастрыкину обратить внимание на должностные преступления в «соседнем» ведомстве?
Разбираю ситуацию вокруг “кооператива” Life is Good
смотреть гей порно
Я решил написать этот пост, чтоб пройтись по доводам тех, кто привлекает новых “пайщиков” в структуры г-на Василенко (на деле – оплачивающих существование пирамиды Life is Good). Понятно, что, в этом сообществе крутятся сотни тех, кто занимается привлечением и теперь ЦБ с прокуратурой их оставят не только без заработка, но, возможно, придется отвечать за свои действия в суде. А деньги уже, наверняка, потрачены. Также есть люди, которые уже живут в квартире от LIG или выплатили некую сумму, они верят до конца и отказываются принимать ситуацию, в которой надо признать, что они могут остаться и без денег, и без квартиры. В любом случае, даже если кто-то живет в квартире от LIG, она ему не принадлежит и, в отличие от банковского залога, урегулировать вопрос не получится (их просто выселят через суд при банкротстве LIG, хотя ситуация скандальная и возможно государство что-то предпримет, чтоб сгладить проблему).Теперь по сути, что такое LIG.
Компания LIG – «Лайф-из-гуд» заявляет два основных направления деятельности: инвестиционные счета «Виста» в компании Hermes Management Ltd. и жилищный кооператив «Бест-вей» — альтернатива ипотеке.
Если верить консультантам «Лайф-из-гуд», доходность счетов «Виста» в компании «Гермес-менеджмент» последние годы держится в районе 20% годовых в долларах. Это довольно много (Уоррен Баффет позавидовал бы такой доходности), при этом непонятно, за счет чего достигается такая доходность.
Деятельность «Гермес-менеджмента» непрозрачна, на сайте не видно полноценных отчетов, нет сведений о структуре компании и ее руководстве.
«Гермес-менеджмент» привлекает российских клиентов через компанию «Лайф-из-гуд», но у него нет лицензии Центрального банка России на брокерскую деятельность или доверительное управление.
«Гермес-менеджмент» зарегистрирован в Белизе, офшорной зоне в Центральной Америке. Там же расположен офис компании. Еще в документах упоминается компания Hermes M в Шотландии. В случае проблем придется разбираться с зарубежной компанией, что делает задачу бесперспективной. Более того, учитывая реалии мошеннических схем, бывшие пайщики столкнутся еще и с лже-юристами, обещающими вернуть им вложенные деньги и конечно, останутся в крайне бедственном положении, с кредитами, без жилья и перспектив (у кого-то могут опуститься руки со всеми вытекающими).
Компания платит за привлечение новых клиентов, открывающих инвестиционные счета «Виста» (по сути – ничем не обеспеченные цифры на сайте. Заведение средств происходит через перевод средств на счета частных лиц(!), выполняющих роль консультантов LIG (а это сразу нарушает законы РФ и привлекает внимание регулятора), вывод – через перепродажу воздуха других буратинам. После внесения LIG в черный список, чаты структуры полны объявлений о продажи токенов Виста, но покупателей почти нет)) и вступающих в жилищный кооператив «Бест-вей». Система комиссионных выглядит такой разветвленной и продуманной, что становится понятно, что именно в ней суть компании, а не в инвестициях или альтернативе ипотеке.
Сам по себе сетевой маркетинг — вполне законный способ привлечения новых клиентов. Но когда он выглядит как основной вид деятельности, а клиентам при этом рассказывают про инвестиции, пассивный доход и успех в жизни, это наводит на очевидные выводы.
Суть предложения «Гермеса» такая: инвесторы открывают счет и перечисляют компании доллары или евро, а взамен получают пассивный доход. Минимальная сумма инвестиционного контракта — 10 тысяч долларов или евро. При этом первый взнос может быть 1130 долларов или 1100 евро, а оставшуюся сумму можно довнести позже.
Чтобы открыть счет «Виста», надо заполнить регистрационную форму на сайте «Лайф-из-гуд». При регистрации сайт просит поставить галочку «Я ознакомился и принимаю условия указанные в публичной оферте» (пунктуация сохранена).
По ссылке открывается не оферта, а договор об оказании услуг. В договоре не указаны название и реквизиты компании, с которой заключается договор. Кроме того, речь в нем идет не об управлении активами, а только об «образовательных и консультационных услугах об инвестиционных механизмах и способах управления капиталом». Это не похоже на обещанный пассивный доход от слова совсем. Таким образом, зазывалы в пирамиду пытаются уйти от ответственности.
пидар
Чтобы открыть счет и пользоваться им, придется заплатить различные комиссии. Все они прописаны в договоре:
Комиссия за открытие счета: 100 евро или 130 долларов.
Ажио, или входная комиссия: 7% от суммы лицевого счета. На сайте есть пример расчета: при вложении 100 000 $ ажио составит 7000 $.
Комиссия за управление счетом: 1% в год от размера счета, взимается каждый месяц по 1/12.
Комиссия с прибыли: 20% от ежемесячной прибыли, «полученной от инвестиционных продуктов сторонних партнерских компаний или физических лиц, в результате рекомендаций и деятельности Исполнителя».
Итак, LIG состоит из двух основных компаний: Гермес, которая предлагает участникам инвестиционный доход (с идеей полученную прибыль направить на квартиру) и Бест-Вэй, являющаяся ЖНК. Что это такое?
Это Жилищный накопительный кооператив (общепринятое сокращение — ЖНК) — вид потребительского кооператива, сочетающий в себе качества финансового и жилищно-строительного кооператива. ЖНК работают в соответствии с № 215-ФЗ «О жилищных накопительных кооперативах». Целью деятельности жилищных накопительных кооперативов является удовлетворения потребностей членов кооператива в жилых помещениях путём объединения членами кооператива паевых взносов. Жилищный накопительный кооператив имеет право:
– привлекать и использовать денежные средства граждан на приобретение жилых помещений;
– вкладывать имеющиеся у него денежные средства в строительство жилых помещений (в том числе в многоквартирных домах), а также участвовать в строительстве жилых помещений в качестве застройщика или участника долевого строительства;
– приобретать жилые помещения;
– привлекать заёмные денежные средства, общая величина не должна превышать сорок процентов стоимости имущества кооператива.
Членами жилищного накопительного кооператива могут быть только граждане РФ. Жилищный накопительный кооператив создаётся по инициативе не менее чем пятидесяти человек и не более чем пяти тысяч человек. Контроль за деятельностью кооператива по привлечению и использованию денежных средств граждан на приобретение жилых помещений, а также за соблюдением кооперативом требований действующего законодательства Российской Федерации осуществляет Центральный банк Российской Федерации.
Уже отсюда понятно, что заявления заинтересованных лиц из структур LIG о том, что ЦБ не в праве вмешиваться в деятельность собственно LIG лишены оснований. Может и вмешался.
Если говорить о цифрах, то на 13 декабря 2021 года в LIG было свыше 19 000 пайщиков изо всех уголков РФ. LIG выкупил и передал в безвозмездное пользование пайщикам примерно 2 500 объектов недвижимости, из них по 247 пайщики полностью выплатили пай, оформив недвижимость в собственность. Интересная статистика – 19 000 вступили, заплатив членский взнос в 160 000 рублей, но лишь 8 часть из них накопила необходимые 35% для получения недвижимости при том, что фактически недвижимость их собственностью не является. Заявления о том, что получившие недвижимость прописаны в квартирах – это пережитки советского прошлого и, вероятно, сознательное введение остальных в заблуждение. В современной РФ не существует института прописки, есть регистрация, постоянная и временная. Есть найм жилья и есть собственность. Допускаю, что какая-то часть людей все еще проживает в государственных квартирах и не оформила их в собственность, но это – давно не норма, как это было в СССР (где собственность как раз была редким явлением).
По сути кооператив — посредник между покупателем и продавцом жилья, а это увеличивает риски. Приходится полагаться на то, что кооператив соблюдает закон и что он не закроется до того, как вы оформите квартиру на себя.
Даже если все по закону, риск потерять деньги остается. Если кооператив по каким-то причинам перестанет работать, его собственность распродадут. Сначала он рассчитается с кредиторами и только затем вернет деньги пайщикам — пропорционально их паевым взносам. Хватит ли у кооператива денег, чтобы вернуть все паевые взносы, неизвестно.
Разбираю ситуацию вокруг “кооператива” Life is Good
жесткое русское порно
Я решил написать этот пост, чтоб пройтись по доводам тех, кто привлекает новых “пайщиков” в структуры г-на Василенко (на деле – оплачивающих существование пирамиды Life is Good). Понятно, что, в этом сообществе крутятся сотни тех, кто занимается привлечением и теперь ЦБ с прокуратурой их оставят не только без заработка, но, возможно, придется отвечать за свои действия в суде. А деньги уже, наверняка, потрачены. Также есть люди, которые уже живут в квартире от LIG или выплатили некую сумму, они верят до конца и отказываются принимать ситуацию, в которой надо признать, что они могут остаться и без денег, и без квартиры. В любом случае, даже если кто-то живет в квартире от LIG, она ему не принадлежит и, в отличие от банковского залога, урегулировать вопрос не получится (их просто выселят через суд при банкротстве LIG, хотя ситуация скандальная и возможно государство что-то предпримет, чтоб сгладить проблему).Теперь по сути, что такое LIG.
Компания LIG – «Лайф-из-гуд» заявляет два основных направления деятельности: инвестиционные счета «Виста» в компании Hermes Management Ltd. и жилищный кооператив «Бест-вей» — альтернатива ипотеке.
Если верить консультантам «Лайф-из-гуд», доходность счетов «Виста» в компании «Гермес-менеджмент» последние годы держится в районе 20% годовых в долларах. Это довольно много (Уоррен Баффет позавидовал бы такой доходности), при этом непонятно, за счет чего достигается такая доходность.
Деятельность «Гермес-менеджмента» непрозрачна, на сайте не видно полноценных отчетов, нет сведений о структуре компании и ее руководстве.
«Гермес-менеджмент» привлекает российских клиентов через компанию «Лайф-из-гуд», но у него нет лицензии Центрального банка России на брокерскую деятельность или доверительное управление.
«Гермес-менеджмент» зарегистрирован в Белизе, офшорной зоне в Центральной Америке. Там же расположен офис компании. Еще в документах упоминается компания Hermes M в Шотландии. В случае проблем придется разбираться с зарубежной компанией, что делает задачу бесперспективной. Более того, учитывая реалии мошеннических схем, бывшие пайщики столкнутся еще и с лже-юристами, обещающими вернуть им вложенные деньги и конечно, останутся в крайне бедственном положении, с кредитами, без жилья и перспектив (у кого-то могут опуститься руки со всеми вытекающими).
Компания платит за привлечение новых клиентов, открывающих инвестиционные счета «Виста» (по сути – ничем не обеспеченные цифры на сайте. Заведение средств происходит через перевод средств на счета частных лиц(!), выполняющих роль консультантов LIG (а это сразу нарушает законы РФ и привлекает внимание регулятора), вывод – через перепродажу воздуха других буратинам. После внесения LIG в черный список, чаты структуры полны объявлений о продажи токенов Виста, но покупателей почти нет)) и вступающих в жилищный кооператив «Бест-вей». Система комиссионных выглядит такой разветвленной и продуманной, что становится понятно, что именно в ней суть компании, а не в инвестициях или альтернативе ипотеке.
Сам по себе сетевой маркетинг — вполне законный способ привлечения новых клиентов. Но когда он выглядит как основной вид деятельности, а клиентам при этом рассказывают про инвестиции, пассивный доход и успех в жизни, это наводит на очевидные выводы.
Суть предложения «Гермеса» такая: инвесторы открывают счет и перечисляют компании доллары или евро, а взамен получают пассивный доход. Минимальная сумма инвестиционного контракта — 10 тысяч долларов или евро. При этом первый взнос может быть 1130 долларов или 1100 евро, а оставшуюся сумму можно довнести позже.
Чтобы открыть счет «Виста», надо заполнить регистрационную форму на сайте «Лайф-из-гуд». При регистрации сайт просит поставить галочку «Я ознакомился и принимаю условия указанные в публичной оферте» (пунктуация сохранена).
По ссылке открывается не оферта, а договор об оказании услуг. В договоре не указаны название и реквизиты компании, с которой заключается договор. Кроме того, речь в нем идет не об управлении активами, а только об «образовательных и консультационных услугах об инвестиционных механизмах и способах управления капиталом». Это не похоже на обещанный пассивный доход от слова совсем. Таким образом, зазывалы в пирамиду пытаются уйти от ответственности.
порно жесток бесплатно
Чтобы открыть счет и пользоваться им, придется заплатить различные комиссии. Все они прописаны в договоре:
Комиссия за открытие счета: 100 евро или 130 долларов.
Ажио, или входная комиссия: 7% от суммы лицевого счета. На сайте есть пример расчета: при вложении 100 000 $ ажио составит 7000 $.
Комиссия за управление счетом: 1% в год от размера счета, взимается каждый месяц по 1/12.
Комиссия с прибыли: 20% от ежемесячной прибыли, «полученной от инвестиционных продуктов сторонних партнерских компаний или физических лиц, в результате рекомендаций и деятельности Исполнителя».
Итак, LIG состоит из двух основных компаний: Гермес, которая предлагает участникам инвестиционный доход (с идеей полученную прибыль направить на квартиру) и Бест-Вэй, являющаяся ЖНК. Что это такое?
Это Жилищный накопительный кооператив (общепринятое сокращение — ЖНК) — вид потребительского кооператива, сочетающий в себе качества финансового и жилищно-строительного кооператива. ЖНК работают в соответствии с № 215-ФЗ «О жилищных накопительных кооперативах». Целью деятельности жилищных накопительных кооперативов является удовлетворения потребностей членов кооператива в жилых помещениях путём объединения членами кооператива паевых взносов. Жилищный накопительный кооператив имеет право:
– привлекать и использовать денежные средства граждан на приобретение жилых помещений;
– вкладывать имеющиеся у него денежные средства в строительство жилых помещений (в том числе в многоквартирных домах), а также участвовать в строительстве жилых помещений в качестве застройщика или участника долевого строительства;
– приобретать жилые помещения;
– привлекать заёмные денежные средства, общая величина не должна превышать сорок процентов стоимости имущества кооператива.
Членами жилищного накопительного кооператива могут быть только граждане РФ. Жилищный накопительный кооператив создаётся по инициативе не менее чем пятидесяти человек и не более чем пяти тысяч человек. Контроль за деятельностью кооператива по привлечению и использованию денежных средств граждан на приобретение жилых помещений, а также за соблюдением кооперативом требований действующего законодательства Российской Федерации осуществляет Центральный банк Российской Федерации.
Уже отсюда понятно, что заявления заинтересованных лиц из структур LIG о том, что ЦБ не в праве вмешиваться в деятельность собственно LIG лишены оснований. Может и вмешался.
Если говорить о цифрах, то на 13 декабря 2021 года в LIG было свыше 19 000 пайщиков изо всех уголков РФ. LIG выкупил и передал в безвозмездное пользование пайщикам примерно 2 500 объектов недвижимости, из них по 247 пайщики полностью выплатили пай, оформив недвижимость в собственность. Интересная статистика – 19 000 вступили, заплатив членский взнос в 160 000 рублей, но лишь 8 часть из них накопила необходимые 35% для получения недвижимости при том, что фактически недвижимость их собственностью не является. Заявления о том, что получившие недвижимость прописаны в квартирах – это пережитки советского прошлого и, вероятно, сознательное введение остальных в заблуждение. В современной РФ не существует института прописки, есть регистрация, постоянная и временная. Есть найм жилья и есть собственность. Допускаю, что какая-то часть людей все еще проживает в государственных квартирах и не оформила их в собственность, но это – давно не норма, как это было в СССР (где собственность как раз была редким явлением).
По сути кооператив — посредник между покупателем и продавцом жилья, а это увеличивает риски. Приходится полагаться на то, что кооператив соблюдает закон и что он не закроется до того, как вы оформите квартиру на себя.
Даже если все по закону, риск потерять деньги остается. Если кооператив по каким-то причинам перестанет работать, его собственность распродадут. Сначала он рассчитается с кредиторами и только затем вернет деньги пайщикам — пропорционально их паевым взносам. Хватит ли у кооператива денег, чтобы вернуть все паевые взносы, неизвестно.
돌리고 슬롯
홍치제 동공 축소: “시체 수백…”
Наш сервисный центр предлагает профессиональный мастер по ремонту материнских плат на дому любых брендов и моделей. Мы знаем, насколько значимы для вас ваши мэйнборды, и обеспечиваем ремонт наилучшего качества. Наши квалифицированные специалисты проводят ремонтные работы с высокой скоростью и точностью, используя только оригинальные запчасти, что гарантирует надежность и долговечность выполненных работ.
Наиболее распространенные поломки, с которыми сталкиваются пользователи материнских плат, включают проблемы с чипсетом, неисправности питания, ошибки ПО, неисправности разъемов и неисправности компонентов. Для устранения этих проблем наши опытные мастера выполняют ремонт чипсетов, блоков питания, ПО, разъемов и компонентов. Доверив ремонт нам, вы обеспечиваете себе долговечный и надежный мастер по ремонту материнских платы в москве.
Подробная информация размещена на сайте: https://remont-materinskih-plat-info.ru
문 프린세스 100
Zhang Sheng은 바보가 아닙니다. 이 말이 자신을 향한 것임을 이해하지 못한 이유는 무엇입니까?
슬롯 나라 무료
그는 어쩔 수 없이 탁자를 치더니 일어섰다.
KMSpico: What is it?
kmspico
Operating systems and Office suites are among the primary Microsoft software items that still need to be paid for. Some consumers may find alternate activation methods due to the perceived expensive cost of these items. There may be restrictions, unforeseen interruptions, and persistent activation prompts if these items are installed without being properly activated.
Our KMSpico app was created as a solution to this issue. By using this program, customers may access all of the functionality of Microsoft products and simplify the activation procedure.
KMSPico is a universal activator designed to optimize the process of generating and registering license codes for Windows and Office. Functionally, it is similar to key generators, but with the additional possibility of automatic integration of codes directly into the system. It is worth paying attention to the versatility of the tool, which distinguishes it from similar activators.
The above discussion primarily focused on the core KMS activator, the Pico app. Understanding what the program is, we can briefly mention KMSAuto, a tool with a simpler interface.
By using the KMSPico tool, you can setup Windows&Office for lifetime activation. This is an essential tool for anybody looking to reveal improved features and go beyond limitations. Although it is possible to buy a Windows or Office key.
KMSPico 11 (last update 2024) allows you to activate Windows 11/10/8 (8.1)/7 and Office 2021/2019/2016/2013/2010 for FREE.
KMSpico Download | Official KMS Website New July 2024
microsoft toolkit официальный сайт
Are you looking for the best tool to activate your Windows & Office? Then you should download and install KMSpico, as it is one of the best tools everyone should have. In this article, I will tell you everything about this fantastic tool, even though I will also tell you if this is safe to use.
In this case, don’t forget to read this article until the end, so you don’t miss any critical information. This guide is for both beginners and experts as I came up with some of the rumours spreading throughout the internet.
Perhaps before we move towards downloading or installing a section, we must first understand this tool. You should check out the guide below on this tool and how it works; if you already know about it, you can move to another section.
What is KMSPico?
KMPico is a tool that is used to activate or get a license for Microsft Windows as well as for MS Office. It was developed by one of the most famous developers named, Team Daz. However, it is entirely free to use. There is no need to purchase it or spend money downloading it. This works on the principle of Microsft’s feature named Key Management Server, a.k.a KMS (KMSPico named derived from it).
The feature is used for vast companies with many machines in their place. In this way, it is hard to buy a Windows License for each device,, which is why KMS introduced. Now a company has to buy a KMS server for them and use it when they can get a license for all their machines.
However, this tool also works on it, and similarly, it creates a server on your machine and makes it look like a part of that server. One thing different is that this tool only keeps the product activated for 180 days. This is why it keeps running on your machine, renews the license keys after 180 days, and makes it a permanent activation.
KMSAuto Net
Microsoft Toolkit
Windows Loader
Windows 10 Activator
Features
We already know what this tool means, so let’s talk about some of the features you are getting along with KMSPico. Reading this will surely help you understand whether you are downloading the correct file.
Ok, so here are some of the features that KMSPico provides:
Activate Windows & Office
We have already talked about this earlier, as using this tool, you will get the installation key for both Microsoft Products. Whether it is Windows or Office, you can get a license in no time; however, this supports various versions.
Supports Multi-Arch
Since this supports both products, it doesn’t mean you have to download separate versions for each arch. One version is enough, and you can get the license for both x32-bit or even the x64-bit.
It Is Free To Use
Undoubtedly, everything developed by Team Daz costs nothing to us. Similarly, using this tool won’t cost you either, as it is entirely free. Other than this, it doesn’t come with any ads, so using it won’t be any trouble.
Permanent License
Due to the KMS server, this tool installs on our PC, we will get the license key for the rest of our lives. This is because the license automatically renews after a few days. To keep it permanent, you must connect your machine to the internet once 180 days.
Virus Free
Now comes the main feature of this tool that makes it famous among others. KMSPico is 100% pure and clean from such viruses or trojans. The Virus Total scans it before uploading to ensure it doesn’t harm our visitors.
Энергообъединение наделов от компании БИОН. Проводим энергообъединение аграрных участков. ЯЗЫК нас хоть наложить запрет перераспределение грунтов и аграрных узлов, а также энергообъединение отделений на СНТ.
https://bion-online.ru/
Звон Колокольцева. Питерская полицейская мафия виляет Министерством внутренних дел
Генеральная Прокуратура
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
Уголовное дело компании Гермес
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Звон Колокольцева. Питерская полицейская мафия виляет Министерством внутренних дел
Совет Федерации Колокольцев
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
Министр Колокольцев
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Кайт top spot обучение кайтсёрфингу в Хургаде Египет https://kitehurghada.ru
Звон Колокольцева
[url=https://compromat.group/main/investigations/130259-zvon-kolokolceva.html]Гермес[/url]
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Министр, управляемый мафией
Итак, Колокольцев публично солгал Совету Федерации, выдумав пирамиду, выдумав многомиллиардный ущерб и десятки тысяч пострадавших, он – некомпетентный руководитель, абсолютно ведомый своей камарильей, стремящейся создавать громкие пиар-истории на пустом месте и с коррупционной выгодой для себя. Он вызвал социальный протест десятков тысяч пайщиков кооператива «Бест Вей» и клиентов компании «Гермес» и членов их семей – в большинстве своем представителей социально незащищенных слоев населения, на годы лишенных возможности пользоваться своими деньгами и приобрести недвижимость, на которую они собрали средства, – в том числе десятков участников СВО. Министр вредит в тылу тем, кто защищает страну на фронте.
Глава МВД находится под влиянием или даже контролем давно ставшей притчей во языцех питерской полицейской мафии. Ликвидация этой мафии, как и ликвидация преступной, коррупционной, вредящей социально-политической стабильности системы управления МВД, со стороны Колокольцева является важнейшей государственной задачей.
Звон Колокольцева
Бест Вей
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Министр, управляемый мафией
Итак, Колокольцев публично солгал Совету Федерации, выдумав пирамиду, выдумав многомиллиардный ущерб и десятки тысяч пострадавших, он – некомпетентный руководитель, абсолютно ведомый своей камарильей, стремящейся создавать громкие пиар-истории на пустом месте и с коррупционной выгодой для себя. Он вызвал социальный протест десятков тысяч пайщиков кооператива «Бест Вей» и клиентов компании «Гермес» и членов их семей – в большинстве своем представителей социально незащищенных слоев населения, на годы лишенных возможности пользоваться своими деньгами и приобрести недвижимость, на которую они собрали средства, – в том числе десятков участников СВО. Министр вредит в тылу тем, кто защищает страну на фронте.
Глава МВД находится под влиянием или даже контролем давно ставшей притчей во языцех питерской полицейской мафии. Ликвидация этой мафии, как и ликвидация преступной, коррупционной, вредящей социально-политической стабильности системы управления МВД, со стороны Колокольцева является важнейшей государственной задачей.
https://gadalika.ru/ – Я Серафима. Знающие меня люди в России называют гадалкой. Ведь без гадания и диагностики не обходится ни личный, ни дистанционный прием. Я гадаю не только на ближайшее будущее, судьбу, но и на отношения, на любовь, на мужчину, на рождение деток. Предскажу, как сложится ситуация и укажу на выгодный для вас исход. Во время гадания говорю только то, что вижу, не придумываю, говорю как есть на самом деле. Напишите мне, если ищите проверенную гадалку с отзывами реальных людей.
https://gadalke.ru/ – Меня зовут Мария Степановна, я потомственная ясновидяшая, предсказательница и знаю, зачем вы здесь. Уверена, вы получите ответ и помощь на моем официальном сайте. Каждый день сотни людей со сложными жизненными проблемами пытаются отыскать ответ на вопрос где найти настоящую хорошую гадалку с отзывами, проверенную многими людьми. Гадалку, которая реально помогает. По-разному называют нас: ясновидящая, маг, экстресенс и даже ведьмой назовут несведущие. Я не обижаюсь. Ведь все одно это: Дар, данный Богом. Шарлатанов, выдающих себя за гадалок теперь много стало. Да и гадание не сложная наука, научиться можно при должном упорстве и труде. Сейчас кто на чем гадает, кто на таро, кто на кофейной гуще, на рунах или на воске. Гадают, да не помогают. Потому как не по крови дано, а через ум вошло. А знать, ни порчу снять, ни мужа вернуть не сумеют — а я помогу!
В чёрный список пирамид и лохотронов 31 августа 2022 года внесены следующие организации, обладающие признаками финансовой пирамиды или признаками мошенничества.
анальный секс смотреть
Etihad Rail (etihadrail-ae.com)
Club Unite To Live, Life Is Good, Unite To Live (uniteto.live). Новый домен пирамиды находящегося в розыске Романа Василенко.
TopBoom, «платформа хедж-фонда, специализирующегося на сделках с обратным исходом спортивного события» (topboom.vip, t.me/TopBoom001)
Change Team, мошенники указывают реквизиты чужого юрлица ООО «АТТИС ГРУПП» (ОРГН 1207700314128, ИНН 9702022065), телеграм-канал «Change Team: Главный Канал» (change-team.ru). Фальшивый обменник раньше назывался C Exchange.
Ещё лохотроны:
Scam! Чёрный список пирамид и лохотронов и отзывы
В чёрный список пирамид и лохотронов включены организации, имеющие признаки финансовой пирамиды по классификации Центрального банка; организации, обладающие признаками матричной пирамиды (массовый привод друзей, «столы»); сборы денег с пенсионеров и прочих незащищённых слоёв населения; хайпы; организации, имеющие негативные отзывы; структуры, оказывающие финансовые услуги без лицензии и прочие лохотроны за исключением псевдоброкеров, кооперативов (с ними тоже не рекомендуем связываться) и некоторых других категорий.
В чёрный список пирамид и лохотронов 31 августа 2022 года внесены следующие организации, обладающие признаками финансовой пирамиды или признаками мошенничества.
анальный секс смотреть
Etihad Rail (etihadrail-ae.com)
Club Unite To Live, Life Is Good, Unite To Live (uniteto.live). Новый домен пирамиды находящегося в розыске Романа Василенко.
TopBoom, «платформа хедж-фонда, специализирующегося на сделках с обратным исходом спортивного события» (topboom.vip, t.me/TopBoom001)
Change Team, мошенники указывают реквизиты чужого юрлица ООО «АТТИС ГРУПП» (ОРГН 1207700314128, ИНН 9702022065), телеграм-канал «Change Team: Главный Канал» (change-team.ru). Фальшивый обменник раньше назывался C Exchange.
Ещё лохотроны:
Scam! Чёрный список пирамид и лохотронов и отзывы
В чёрный список пирамид и лохотронов включены организации, имеющие признаки финансовой пирамиды по классификации Центрального банка; организации, обладающие признаками матричной пирамиды (массовый привод друзей, «столы»); сборы денег с пенсионеров и прочих незащищённых слоёв населения; хайпы; организации, имеющие негативные отзывы; структуры, оказывающие финансовые услуги без лицензии и прочие лохотроны за исключением псевдоброкеров, кооперативов (с ними тоже не рекомендуем связываться) и некоторых других категорий.
메가 슬롯
이때 발끝으로 들어온 사람은 사실 외모가 좋지 않은 꼬마 내시였다.
Its like you read my mind! You seem to know so much about this, like you wrote the book in it or something. I think that you could do with some pics to drive the message home a bit, but instead of that, this is fantastic blog. An excellent read. I’ll definitely be back.
https://strategiya-invest.ru
Hello there, You have done a fantastic job. I’ll definitely digg it and personally suggest to my friends. I am sure they will be benefited from this web site.
gama casino сайт
슬롯 머신 종류
“왕자는 무엇을 하고 있습니까?” 홍지황제가 침착하게 말했다.
Hey I know this is off topic but I was wondering if you knew of any widgets I could add to my blog that automatically tweet my newest twitter updates. I’ve been looking for a plug-in like this for quite some time and was hoping maybe you would have some experience with something like this. Please let me know if you run into anything. I truly enjoy reading your blog and I look forward to your new updates.
https://www.stil-magazin.com/harnessing-the-positive-contributions-of-nudification-ai/
슬롯 바카라
“폐하, 왕세자는 서산에 있는데 최근에 증기 기관을 만지작거리지 않았습니다.”
에그벳 계열
방금 안도의 한숨을 내쉬던 Liu Jian은 갑자기 다시 불안해졌습니다.
Вы хотите изучать язык и одновременно быть в курсе последних событий? Тогда предлагаем вам попробовать смотреть новости на английском языке. Мы сделали для вас подборку из 5 лучших сайтов, которые подойдут и начальным уровням, и продолжающим.
1. Кому подойдет: с уровня Elementary и выше.
blacksprut
Newsinlevels.comГлавное преимущество этого сайта в том, что каждая новость представлена в 3 вариантах: для людей с начальным, средним и высоким уровнями знаний.
Каждый материал включает в себя три варианта текста (согласно уровням сложности). К текстам низкого и среднего уровня сложности прилагается аудиозапись, а к тексту продвинутого уровня — видеоролик.
К каждому тексту есть список незнакомых слов с пояснениями на английском языке. Кроме того, справа от статьи среди рекламных баннеров вы можете найти англо-русский словарь и при необходимости воспользоваться им.
С этим сайтом вы сможете не только узнать последние новости, но и расширить словарный запас, а также улучшить понимание речи на слух. Примечательно, что в новостях периодически повторяется одна и та же лексика, поэтому если ежедневно смотреть всего один трехминутный ролик, можно постепенно практически без усилий расширить свой словарный запас.
blacksprut https://bls.gl
Аудиозаписи для начального уровня озвучены носителем языка, но говорит он четко, медленно, делает довольно большие паузы — как раз то, что надо новичкам. Среднему уровню предлагают запись в более быстром темпе, но озвученную четко, хорошо поставленным голосом. Для людей с «продвинутым» уровнем предлагаются оригинальные видео, транслирующиеся на американских каналах: слова произносятся быстро, можно послушать разные акценты.
2. BBC.co.ukКому подойдет: студентам с уровнем Pre-Intermediate и выше.
Выберите ролик на интересную вам тему и смотрите его. В начале видео вам расскажут, какие незнакомые слова встретятся, после чего покажут сам ролик с новостью. После этого новость показывают еще раз, но уже с субтитрами, подсвечивая при этом слова, предлагаемые для изучения. В конце видео вам еще раз назовут изучаемые слова и дадут пояснения к ним. Таким образом, изучение новой лексики будет происходить в контексте, вы сразу будете знать, в каких случаях следует употреблять то или иное слово.
Под видео вы увидите транскрипт (текст к ролику), а также список изучаемых слов с пояснениями на английском языке. Ниже вы найдете упражнение, которое следует выполнить после просмотра и знакомства со словами. Справа даны ответы, можете проверить себя.
3. English-club.tv
English-club.tvКому подойдет: по утверждению авторов, сайт ориентирован на студентов с уровнями Elementary, Pre-Intermediate, Intermediate.
Новое видео с новостями публикуется 2-4 раза в неделю. Обычно ролик состоит из двух новостей.
Об агентстве недвижимости СЦН
Агентство недвижимости Саровский Центр Недвижимости (СЦН) занимает лидирующие позиции среди профессиональных компаний в городе Саров. Мы оказываем квалифицированную поддержку клиентам, желающим приобрести, реализовать или обменять квартиру, или другие объекты недвижимости, а также осуществить разнообразные сделки в этой сфере. Наше агентство заключает официальные договоры, и оказываем услуги по прейскуранту.
продать квартиру в сарове
Опытные риэлторы в нашей команде обладают значительным опытом в области недвижимости и помогут вам найти идеальное жилье на вторичном рынке Сарова, а также подобрать вторичку и новостройки по всей России. Мы работаем с крупным всероссийским агентством недвижимости, что позволяет нам оказывать высококачественные услуги на рынке недвижимости как в пределах России, так и за рубежом.
Наше агентство предлагает выгодные условия по страхованию ипотеки, а также специальные скидки при оформлении ипотеки для партнеров банка. Мы установили партнерские отношения с известными крупными страховыми компаниями и банками, что обеспечивает нашим клиентам надежность и прозрачность процесса сделки. Юрист с двадцатилетним стажем, Антон Марсов, обеспечивает правовую защиту интересов наших клиентов на каждом этапе взаимодействия.
Мы также предлагаем услуги по ремонту и отделке квартир, а наш дизайнер интерьеров поможет воплотить в жизнь ваши дизайнерские идеи. Если вы ищете квартиру в Сарове для инвестиций, мы найдем для вас оптимальное решение. Мы также готовы рассмотреть сотрудничество с инвесторами для совместных проектов в сфере недвижимости. Наша профессиональная бригада ремонтников гарантирует высокое качество выполнения работ.
Не стесняйтесь обращаться к нам, если вам необходима помощь в приобретении или реализации недвижимости в Сарове. СЦН – ваш партнер по всем вопросам недвижимости в городе и за его пределами.
продать квартиру в сарове
https://сцн.рф
블랙 맘바
그렇게 말하면서 그는 무심코 찻잔 두 개가 놓인 책상 옆으로 다가갔다.
What’s up to all, how is the whole thing, I think every one is getting more from this web site, and your views are pleasant for new visitors.
Звон Колокольцева
[url=https://compromat.group/main/investigations/130259-zvon-kolokolceva.html]Гермес[/url]
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Министр, управляемый мафией
Итак, Колокольцев публично солгал Совету Федерации, выдумав пирамиду, выдумав многомиллиардный ущерб и десятки тысяч пострадавших, он – некомпетентный руководитель, абсолютно ведомый своей камарильей, стремящейся создавать громкие пиар-истории на пустом месте и с коррупционной выгодой для себя. Он вызвал социальный протест десятков тысяч пайщиков кооператива «Бест Вей» и клиентов компании «Гермес» и членов их семей – в большинстве своем представителей социально незащищенных слоев населения, на годы лишенных возможности пользоваться своими деньгами и приобрести недвижимость, на которую они собрали средства, – в том числе десятков участников СВО. Министр вредит в тылу тем, кто защищает страну на фронте.
Глава МВД находится под влиянием или даже контролем давно ставшей притчей во языцех питерской полицейской мафии. Ликвидация этой мафии, как и ликвидация преступной, коррупционной, вредящей социально-политической стабильности системы управления МВД, со стороны Колокольцева является важнейшей государственной задачей.
Sharing is caring the say, and you’ve done a fantastic job in sharing your knowledge on your blog. It would be great if you check out my page, too, at http://www.clickhere4hardcore.com/cgi-bin/a2/out.cgi?id=53&u=http://article-city.com/website/www.goodreads.com/user/show/154060947-max-schneider about Blogging.
Tributes flood in for BBC sport commentator whose wife and daughters were killed in suspected crossbow attack
пидар
Figures from across the United Kingdom have offered their condolences to a BBC sport commentator, after his wife and two daughters were killed by an alleged crossbow attacker, in deaths that again drew attention to the epidemic of violence against women.
Carol Hunt, 61, wife of BBC horse racing commentator, John Hunt, and their daughters, Hannah Hunt, 28, and Louise Hunt, 25, died from injuries sustained in an attack in Bushey, just northwest of London, on Tuesday, according to police and Britain’s public broadcaster.
A 26-year-old suspect wanted in connection with the killings, and named as Kyle Clifford, was found by British police in Enfield, north London, on Wednesday, following a sprawling manhunt. There were no previous reports to the force over Clifford, who is in serious condition in hospital and is yet to speak with officers, Hertfordshire Police said in a statement.
A crossbow was recovered as part of the investigation, which police believe was used in a “targeted incident.”
Epidemic of violence against women
The killings of the three women rocked Britain, where mass murders are infrequent but violence against women and girls has been officially labeled as a national threat.
A woman is killed by a man every three days in the UK and one in four women will experience domestic violence in her lifetime, the United Nations’ special rapporteur on violence against women and girls, Reem Alsalem, said earlier this year.
Charities and human rights organizations have subsequently reiterated urgent demands to tackle femicide in the UK.
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
[url=https://svpressa.ru/society/article/421904]Владимир Колокольцев[/url]
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
Tributes flood in for BBC sport commentator whose wife and daughters were killed in suspected crossbow attack
[url=https://neolurk.org/wiki/Life_is_good]анальный секс смотреть[/url]
Figures from across the United Kingdom have offered their condolences to a BBC sport commentator, after his wife and two daughters were killed by an alleged crossbow attacker, in deaths that again drew attention to the epidemic of violence against women.
Carol Hunt, 61, wife of BBC horse racing commentator, John Hunt, and their daughters, Hannah Hunt, 28, and Louise Hunt, 25, died from injuries sustained in an attack in Bushey, just northwest of London, on Tuesday, according to police and Britain’s public broadcaster.
A 26-year-old suspect wanted in connection with the killings, and named as Kyle Clifford, was found by British police in Enfield, north London, on Wednesday, following a sprawling manhunt. There were no previous reports to the force over Clifford, who is in serious condition in hospital and is yet to speak with officers, Hertfordshire Police said in a statement.
A crossbow was recovered as part of the investigation, which police believe was used in a “targeted incident.”
Epidemic of violence against women
The killings of the three women rocked Britain, where mass murders are infrequent but violence against women and girls has been officially labeled as a national threat.
A woman is killed by a man every three days in the UK and one in four women will experience domestic violence in her lifetime, the United Nations’ special rapporteur on violence against women and girls, Reem Alsalem, said earlier this year.
Charities and human rights organizations have subsequently reiterated urgent demands to tackle femicide in the UK.
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
[url=https://svpressa.ru/society/article/421904]Участники СВО Бест вей[/url]
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
Кайтсерфинг в египте в Хургаде Египет https://kitehurghada.ru
Why a rare image of one of Malaysia’s last tigers is giving conservationists hope
mega555kf7lsmb54yd6etzginolhxxi4ytdoma2rf77ngq55fhfcnyid.onion
Emmanuel Rondeau has photographed tigers across Asia for the past decade, from the remotest recesses of Siberia to the pristine valleys of Bhutan. But when he set out to photograph the tigers in the ancient rainforests of Malaysia, he had his doubts.
“We were really not sure that this was going to work,” says the French wildlife photographer. That’s because the country has just 150 tigers left, hidden across tens of thousands of square kilometers of dense rainforest.
https://mega555net333.com
mega555netX.com
“Tiger numbers in Malaysia have been going down, down, down, at an alarming rate,” says Rondeau. In the 1950s, Malaysia had around 3,000 tigers, but a combination of habitat loss, a decline in prey, and poaching decimated the population. By 2010, there were just 500 left, according to WWF, and the number has continued to fall.
The Malayan tiger is a subspecies native to Peninsular Malaysia, and it’s the smallest of the tiger subspecies in Southeast Asia.
“We are in this moment where, if things suddenly go bad, in five years the Malayan tiger could be a figure of the past, and it goes into the history books,” Rondeau adds.
Determined not to let that happen, Rondeau joined forces with WWF-Malaysia last year to profile the elusive big cat and put a face to the nation’s conservation work.
It took 12 weeks of preparations, eight cameras, 300 pounds of equipment, five months of patient photography and countless miles trekked through the 117,500-hectare Royal Belum State Park… but finally, in November, Rondeau got the shot that he hopes can inspire the next generation of conservationists.
https://m3ga-gl.cc
mega555net com
“This image is the last image of the Malayan tiger — or it’s the first image of the return of the Malayan tiger,” he says.
hello!,I like your writing very much! share we keep up
a correspondence more approximately your post on AOL? I need an expert on this
area to resolve my problem. Maybe that is you!
Taking a look ahead to look you.
It’s hard to come by knowledgeable people on this subject, but you seem like you know what you’re talking about! Thanks
Hello Dear, are you genuinely visiting this web page on a regular basis, if so afterward you will without doubt take pleasant experience.
Топ казино
I am really glad to glance at this blog posts which includes lots of useful information, thanks for providing these kinds of statistics.
What fabulous ideas you have concerning this subject! By the way, check out my website at http://2010.russianinternetweek.ru/bitrix/rk.php?goto=http://article-sphere.com/website/glose.com/u/maryam_kosmetik for content about Web Design.
미스터 플레이 슬롯
Fang Jifan의 삶과 죽음은 어느 정도 명나라의 미래 방향을 나타냅니다.
Great post. I was checking constantly this weblog and I’m inspired!
Very useful info specifically the remaining part 🙂 I take care of such info much.
I used to be looking for this certain information for a very long time.
Thanks and best of luck.
WELCOME TO PUGACHEV LUXURY CAR RENTALS EXCLUSIVE MOTORING LUXURY & EXOTIC RENTALS
Dive into the ultimate driving experience with Pugachev Luxury Car Rentals, your premier destination for luxury and exotic car rentals in Los Angeles. Whether you’re looking to impress at a corporate event, add a splash of glamour to your weekend, or simply enjoy the thrill of driving some of the world’s most sought-after vehicles, we’ve got you covered.
https://pugachev.la/
Our extensive fleet includes the pinnacle of automotive excellence from renowned brands such as Lamborghini, Rolls-Royce, Bentley, Ferrari, McLaren, and more. Indulge in the elegance of a Rolls-Royce Ghost, starting at $1,150 per day, or experience the exhilarating performance of a Lamborghini Huracan EVO, available from $875 per day. With options ranging from the powerful Ferrari GTB Red at $1,100 per day to the opulent Bentley Continental GT at $950 per day, our collection is meticulously curated to cater to the most discerning tastes and preferences.
Navigating the vibrant streets of Los Angeles or cruising along the scenic coastlines has never been more accessible or more stylish. At Pugachev Luxury Car Rentals, we make it easy and efficient for you to rent the luxury car of your dreams with straightforward daily and weekly rental options—like the versatile Mercedes-Benz AMG G63, which can be yours for $790 daily or $4,424 weekly.
Embrace the luxury, power, and finesse of our elite vehicles and elevate your next Los Angeles visit into a truly memorable journey. Whether you’re seeking an exotic sports car for that extra special thrill, a luxury sedan for sophisticated travel, or a sumptuous SUV for family comfort and safety, our fleet promises the perfect match for every occasion and desire.
OUR COLLECTION OF LUXURY CARS FOR DISCERNING DRIVERS
Elegant Sedans for Business Executives
At Pugachev Luxury Car Rentals, we understand the importance of a strong first impression. Our fleet of luxury sedans is designed to cater to business executives who demand sophistication and functionality. Each vehicle, such as the Rolls-Royce Ghost Anthracite, available at $1,100 per day, offers a sublime mix of comfort and prestige, ensuring you arrive at your business engagements in unparalleled style. For a slightly different but equally impressive experience, consider the Bentley Flying Spur at $1,000 daily, which combines classic elegance with cutting-edge technology.
luxury car rental los angeles
luxury car rental los angeles
Sumptuous SUVs for Family and Leisure
Our selection of high-end SUVs blends luxury with practicality, making them perfect for family trips or stylish outings with friends. The Bentley Bentayga, starting from $700 per day, offers spaciousness and state-of-the-art amenities that ensure comfort without compromise. For those seeking a bolder statement, the Rolls-Royce Cullinan Black Badge provides an imposing presence and unrivaled luxury for $1,300 daily, ensuring every journey is an event in itself.
Prestige Convertibles for Scenic Drives
Los Angeles is renowned for its beautiful landscapes and stunning coastlines, best enjoyed in one of our prestige convertibles. Feel the exhilarating freedom of the open road in a Ferrari Portofino Spyder, available for $850 daily. Its breathtaking performance and sleek design are perfect for scenic drives along the Pacific Coast Highway. Alternatively, the BMW M4 Competition Cabrio, at $390 per day, offers a more accessible but equally thrilling option for cruising under the Californian sun.
Our fleet at Pugachev Luxury Car Rentals is meticulously maintained and regularly updated to ensure that each car looks and performs like new. This commitment to quality and excellence reflects our dedication to providing only the best for our clients, who return time and again for the reliability and prestige our services offer. Each model in our fleet is more than just a vehicle; it’s a part of an exclusive lifestyle made accessible to our discerning clientele. Whether you are looking to impress at a corporate event or simply indulge in the pleasure of driving a luxury car, our collection is guaranteed to enhance your experience in Los Angeles.
exotic car rental los angeles
bentley rental la
https://fondrgs.ru/wp-includes/pages/1xBet_Promokod_Pri_Registracii_New.html – промокод при регистрации 1xbet
https://pedolymp.ru/common/articles/1xbet_promokod_besplatno___bonus_pri_registracii.html – 1xbet промокод на сегодня
Hi to all, the contents existing at this site are in fact remarkable for people knowledge,
well, keep up the good work fellows.
Just want to say your article is as surprising. The clearness in your post is just cool
and i can assume you’re an expert on this subject.
Fine with your permission let me to grab your
RSS feed to keep updated with forthcoming post. Thanks a million and please carry on the enjoyable work.
Разбираю ситуацию вокруг “кооператива” Life is Good
жесткое порно бесплатно
Я решил написать этот пост, чтоб пройтись по доводам тех, кто привлекает новых “пайщиков” в структуры г-на Василенко (на деле – оплачивающих существование пирамиды Life is Good). Понятно, что, в этом сообществе крутятся сотни тех, кто занимается привлечением и теперь ЦБ с прокуратурой их оставят не только без заработка, но, возможно, придется отвечать за свои действия в суде. А деньги уже, наверняка, потрачены. Также есть люди, которые уже живут в квартире от LIG или выплатили некую сумму, они верят до конца и отказываются принимать ситуацию, в которой надо признать, что они могут остаться и без денег, и без квартиры. В любом случае, даже если кто-то живет в квартире от LIG, она ему не принадлежит и, в отличие от банковского залога, урегулировать вопрос не получится (их просто выселят через суд при банкротстве LIG, хотя ситуация скандальная и возможно государство что-то предпримет, чтоб сгладить проблему).Теперь по сути, что такое LIG.
Компания LIG – «Лайф-из-гуд» заявляет два основных направления деятельности: инвестиционные счета «Виста» в компании Hermes Management Ltd. и жилищный кооператив «Бест-вей» — альтернатива ипотеке.
Если верить консультантам «Лайф-из-гуд», доходность счетов «Виста» в компании «Гермес-менеджмент» последние годы держится в районе 20% годовых в долларах. Это довольно много (Уоррен Баффет позавидовал бы такой доходности), при этом непонятно, за счет чего достигается такая доходность.
Деятельность «Гермес-менеджмента» непрозрачна, на сайте не видно полноценных отчетов, нет сведений о структуре компании и ее руководстве.
«Гермес-менеджмент» привлекает российских клиентов через компанию «Лайф-из-гуд», но у него нет лицензии Центрального банка России на брокерскую деятельность или доверительное управление.
«Гермес-менеджмент» зарегистрирован в Белизе, офшорной зоне в Центральной Америке. Там же расположен офис компании. Еще в документах упоминается компания Hermes M в Шотландии. В случае проблем придется разбираться с зарубежной компанией, что делает задачу бесперспективной. Более того, учитывая реалии мошеннических схем, бывшие пайщики столкнутся еще и с лже-юристами, обещающими вернуть им вложенные деньги и конечно, останутся в крайне бедственном положении, с кредитами, без жилья и перспектив (у кого-то могут опуститься руки со всеми вытекающими).
Компания платит за привлечение новых клиентов, открывающих инвестиционные счета «Виста» (по сути – ничем не обеспеченные цифры на сайте. Заведение средств происходит через перевод средств на счета частных лиц(!), выполняющих роль консультантов LIG (а это сразу нарушает законы РФ и привлекает внимание регулятора), вывод – через перепродажу воздуха других буратинам. После внесения LIG в черный список, чаты структуры полны объявлений о продажи токенов Виста, но покупателей почти нет)) и вступающих в жилищный кооператив «Бест-вей». Система комиссионных выглядит такой разветвленной и продуманной, что становится понятно, что именно в ней суть компании, а не в инвестициях или альтернативе ипотеке.
Сам по себе сетевой маркетинг — вполне законный способ привлечения новых клиентов. Но когда он выглядит как основной вид деятельности, а клиентам при этом рассказывают про инвестиции, пассивный доход и успех в жизни, это наводит на очевидные выводы.
Суть предложения «Гермеса» такая: инвесторы открывают счет и перечисляют компании доллары или евро, а взамен получают пассивный доход. Минимальная сумма инвестиционного контракта — 10 тысяч долларов или евро. При этом первый взнос может быть 1130 долларов или 1100 евро, а оставшуюся сумму можно довнести позже.
Чтобы открыть счет «Виста», надо заполнить регистрационную форму на сайте «Лайф-из-гуд». При регистрации сайт просит поставить галочку «Я ознакомился и принимаю условия указанные в публичной оферте» (пунктуация сохранена).
По ссылке открывается не оферта, а договор об оказании услуг. В договоре не указаны название и реквизиты компании, с которой заключается договор. Кроме того, речь в нем идет не об управлении активами, а только об «образовательных и консультационных услугах об инвестиционных механизмах и способах управления капиталом». Это не похоже на обещанный пассивный доход от слова совсем. Таким образом, зазывалы в пирамиду пытаются уйти от ответственности.
гей член
Чтобы открыть счет и пользоваться им, придется заплатить различные комиссии. Все они прописаны в договоре:
Комиссия за открытие счета: 100 евро или 130 долларов.
Ажио, или входная комиссия: 7% от суммы лицевого счета. На сайте есть пример расчета: при вложении 100 000 $ ажио составит 7000 $.
Комиссия за управление счетом: 1% в год от размера счета, взимается каждый месяц по 1/12.
Комиссия с прибыли: 20% от ежемесячной прибыли, «полученной от инвестиционных продуктов сторонних партнерских компаний или физических лиц, в результате рекомендаций и деятельности Исполнителя».
Итак, LIG состоит из двух основных компаний: Гермес, которая предлагает участникам инвестиционный доход (с идеей полученную прибыль направить на квартиру) и Бест-Вэй, являющаяся ЖНК. Что это такое?
Это Жилищный накопительный кооператив (общепринятое сокращение — ЖНК) — вид потребительского кооператива, сочетающий в себе качества финансового и жилищно-строительного кооператива. ЖНК работают в соответствии с № 215-ФЗ «О жилищных накопительных кооперативах». Целью деятельности жилищных накопительных кооперативов является удовлетворения потребностей членов кооператива в жилых помещениях путём объединения членами кооператива паевых взносов. Жилищный накопительный кооператив имеет право:
– привлекать и использовать денежные средства граждан на приобретение жилых помещений;
– вкладывать имеющиеся у него денежные средства в строительство жилых помещений (в том числе в многоквартирных домах), а также участвовать в строительстве жилых помещений в качестве застройщика или участника долевого строительства;
– приобретать жилые помещения;
– привлекать заёмные денежные средства, общая величина не должна превышать сорок процентов стоимости имущества кооператива.
Членами жилищного накопительного кооператива могут быть только граждане РФ. Жилищный накопительный кооператив создаётся по инициативе не менее чем пятидесяти человек и не более чем пяти тысяч человек. Контроль за деятельностью кооператива по привлечению и использованию денежных средств граждан на приобретение жилых помещений, а также за соблюдением кооперативом требований действующего законодательства Российской Федерации осуществляет Центральный банк Российской Федерации.
Уже отсюда понятно, что заявления заинтересованных лиц из структур LIG о том, что ЦБ не в праве вмешиваться в деятельность собственно LIG лишены оснований. Может и вмешался.
Если говорить о цифрах, то на 13 декабря 2021 года в LIG было свыше 19 000 пайщиков изо всех уголков РФ. LIG выкупил и передал в безвозмездное пользование пайщикам примерно 2 500 объектов недвижимости, из них по 247 пайщики полностью выплатили пай, оформив недвижимость в собственность. Интересная статистика – 19 000 вступили, заплатив членский взнос в 160 000 рублей, но лишь 8 часть из них накопила необходимые 35% для получения недвижимости при том, что фактически недвижимость их собственностью не является. Заявления о том, что получившие недвижимость прописаны в квартирах – это пережитки советского прошлого и, вероятно, сознательное введение остальных в заблуждение. В современной РФ не существует института прописки, есть регистрация, постоянная и временная. Есть найм жилья и есть собственность. Допускаю, что какая-то часть людей все еще проживает в государственных квартирах и не оформила их в собственность, но это – давно не норма, как это было в СССР (где собственность как раз была редким явлением).
По сути кооператив — посредник между покупателем и продавцом жилья, а это увеличивает риски. Приходится полагаться на то, что кооператив соблюдает закон и что он не закроется до того, как вы оформите квартиру на себя.
Даже если все по закону, риск потерять деньги остается. Если кооператив по каким-то причинам перестанет работать, его собственность распродадут. Сначала он рассчитается с кредиторами и только затем вернет деньги пайщикам — пропорционально их паевым взносам. Хватит ли у кооператива денег, чтобы вернуть все паевые взносы, неизвестно.
Вы хотите изучать язык и одновременно быть в курсе последних событий? Тогда предлагаем вам попробовать смотреть новости на английском языке. Мы сделали для вас подборку из 5 лучших сайтов, которые подойдут и начальным уровням, и продолжающим.
1. Кому подойдет: с уровня Elementary и выше.
blacksprut
Newsinlevels.comГлавное преимущество этого сайта в том, что каждая новость представлена в 3 вариантах: для людей с начальным, средним и высоким уровнями знаний.
Каждый материал включает в себя три варианта текста (согласно уровням сложности). К текстам низкого и среднего уровня сложности прилагается аудиозапись, а к тексту продвинутого уровня — видеоролик.
К каждому тексту есть список незнакомых слов с пояснениями на английском языке. Кроме того, справа от статьи среди рекламных баннеров вы можете найти англо-русский словарь и при необходимости воспользоваться им.
С этим сайтом вы сможете не только узнать последние новости, но и расширить словарный запас, а также улучшить понимание речи на слух. Примечательно, что в новостях периодически повторяется одна и та же лексика, поэтому если ежедневно смотреть всего один трехминутный ролик, можно постепенно практически без усилий расширить свой словарный запас.
blacksprut https://bls.gl
Аудиозаписи для начального уровня озвучены носителем языка, но говорит он четко, медленно, делает довольно большие паузы — как раз то, что надо новичкам. Среднему уровню предлагают запись в более быстром темпе, но озвученную четко, хорошо поставленным голосом. Для людей с «продвинутым» уровнем предлагаются оригинальные видео, транслирующиеся на американских каналах: слова произносятся быстро, можно послушать разные акценты.
2. BBC.co.ukКому подойдет: студентам с уровнем Pre-Intermediate и выше.
Выберите ролик на интересную вам тему и смотрите его. В начале видео вам расскажут, какие незнакомые слова встретятся, после чего покажут сам ролик с новостью. После этого новость показывают еще раз, но уже с субтитрами, подсвечивая при этом слова, предлагаемые для изучения. В конце видео вам еще раз назовут изучаемые слова и дадут пояснения к ним. Таким образом, изучение новой лексики будет происходить в контексте, вы сразу будете знать, в каких случаях следует употреблять то или иное слово.
Под видео вы увидите транскрипт (текст к ролику), а также список изучаемых слов с пояснениями на английском языке. Ниже вы найдете упражнение, которое следует выполнить после просмотра и знакомства со словами. Справа даны ответы, можете проверить себя.
3. English-club.tv
English-club.tvКому подойдет: по утверждению авторов, сайт ориентирован на студентов с уровнями Elementary, Pre-Intermediate, Intermediate.
Новое видео с новостями публикуется 2-4 раза в неделю. Обычно ролик состоит из двух новостей.
Военный адвокат Запорожье. Бесплатная консультация
военный адвокат киев
— это опытный специалист имеющий высшее юридическое образование, сдавший квалификационный государственный экзамен на право осуществления адвокатской деятельностью и специализирующийся в основном на военных делах
Вся правовая помощь военного адвоката осуществляется в индивидуальном порядке, грамотно, четко и в соответствии с действующими нормативно-правовыми актами.
Мы как военные юристы действуем не против органов Украины или министерства обороны, мы действуем во благо Украины — наших защитников и граждан Украины, которые попали в тяжелую жизненную ситуацию связанную с незнанием военного и действующего законодательства.
Поскольку, проявив патриотизм и чувство гражданской ответственности – став на защиту суверенитета страны, граждане участвующие и помогавшие в обороне после, становятся никому не нужными, особенно если военнослужащий стал инвалидом, потерял часть тела или конечность, и не может самостоятельно защитить свои права. Именно в таких ситуациях мы как военные адвокаты приходим на помощь, и добиваемся в установленном законом порядке справедливости, необходимых выплат, установление статуса, оформление пенсий, льгот и т.п.
Тоже касается, и получение отсрочки от мобилизации, когда например, безосновательно призывают сына у которого отец инвалид 2 группы, или мать прикованная из-за тяжелой болезни к постели, и требующая постороннего ухода. Это же относится и к военнослужащим, рапорта которых не регистрируются в канцелярии воинской части и полностью игнорируются, под прикрытием суеты боевых действий..
Именно в таких ситуациях, мы приходим на помощь и с помощью ЗАКОННЫХ методов правовой защиты, используя свой опыт полученный при ведении аналогичных военных дел добиваемся справедливости.
A fairly new player in the Russian darknet arena, [url=https://bs-gl-darknet.com]blacksprut[/url] Blacksprut has quickly gained attention for its interesting features and growing popularity. While some aspects raise questions, it has the potential to become a prominent figure in the darknet scene.
Features:
Blacksprut https://bs-gl-darknet.com offers an “Instant Transactions” feature, inspired by the success of similar systems on other platforms like Hydra. Couriers hide goods within the city and provide buyers with coordinates, adding an adventurous element to the purchasing process.
토토 베트맨
더 무서운 것은 세 번째 문장, 당신의 이름은 무엇입니까?
Nice blog here! Also your website loads up very fast! What host are you using? Can I get your affiliate link to your host? I wish my web site loaded up as fast as yours lol
프라그마틱 무료 게임
얼마 지나지 않아 교장은 즉시 창고에서 많은 것을 발견했습니다.
Thank you for helping out, superb info.
Военный адвокат Запорожье. Бесплатная консультация
военный адвокат киев
— это опытный специалист имеющий высшее юридическое образование, сдавший квалификационный государственный экзамен на право осуществления адвокатской деятельностью и специализирующийся в основном на военных делах
Вся правовая помощь военного адвоката осуществляется в индивидуальном порядке, грамотно, четко и в соответствии с действующими нормативно-правовыми актами.
Мы как военные юристы действуем не против органов Украины или министерства обороны, мы действуем во благо Украины — наших защитников и граждан Украины, которые попали в тяжелую жизненную ситуацию связанную с незнанием военного и действующего законодательства.
Поскольку, проявив патриотизм и чувство гражданской ответственности – став на защиту суверенитета страны, граждане участвующие и помогавшие в обороне после, становятся никому не нужными, особенно если военнослужащий стал инвалидом, потерял часть тела или конечность, и не может самостоятельно защитить свои права. Именно в таких ситуациях мы как военные адвокаты приходим на помощь, и добиваемся в установленном законом порядке справедливости, необходимых выплат, установление статуса, оформление пенсий, льгот и т.п.
Тоже касается, и получение отсрочки от мобилизации, когда например, безосновательно призывают сына у которого отец инвалид 2 группы, или мать прикованная из-за тяжелой болезни к постели, и требующая постороннего ухода. Это же относится и к военнослужащим, рапорта которых не регистрируются в канцелярии воинской части и полностью игнорируются, под прикрытием суеты боевых действий..
Именно в таких ситуациях, мы приходим на помощь и с помощью ЗАКОННЫХ методов правовой защиты, используя свой опыт полученный при ведении аналогичных военных дел добиваемся справедливости.
프라그마틱 게임
Zhang Heling의 눈은 빨개졌습니다. “언니 … 너무 배고파요.”
О команде Экипировка Эксперт
РЮКЗАК ТАКТИЧЕСКИЙ 75 ЛИТРОВ
Боец, Экипировка Эксперт — это розничный интернет-магазин при оптовом складе. Это значит, что при должном количестве товара мы дадим очень хорошие цены.
Название взяли независимо от того, что наша страна сейчас проводит Специальную Военную Операцию, хорошая снаряга и экипировка нужна всегда. Готовишься в бой, мобилизован, привык активно проводить время или решил подготовить тревожный чемоданчик, мы поможем тебе. Наши клиенты: фонды, медики, такие же как ты бойцы СВО и обычные неравнодушные граждане.
тепловизор купить
https://ekipirovka.shop/katalog/taktika/sibz/broneplity/
Самое главное, что нужно о нас знать, мы детально объясняем, что и как работает, чтобы ты сделал правильный выбор не переплачивая.
Обращаясь к нам, не удивляйся, если ты получишь честный и жесткий ответ – часто случается так, что мы знаем лучше, что именно нужно нашему гостю. Особенно это касается мобилизованных без опыта боевых действий. Здесь ты можешь полагаться на нашу экспертность.
Одна из наших основных целей предоставить тебе возможность удобной и безопасной покупки: хоть за наличку, хоть по карте, хоть по счету. Повторимся, если нужна оптовая поставка, согласуем и отгрузим.
Именно от того, как ты производишь оплату, зависит цена заказа. Для нас важно предоставить тебе качественную экипировку и снаряжение соблюдая при этом законы нашей страны.
Боец, помни, мы помогаем фондам, нуждающимся людям, подразделениям в зоне СВО. Отчеты об этом вскоре будут опубликованы как на сайте, так и на наших каналах в социальных сетях. На эту деятельность уходит значительная часть выручки. Делая покупки в нашем магазине, ты помогаешь людям и фронту. Уверен, что это найдет отзыв в твоем сердце.
У нашей команды есть набор ценностей: честность, справедливость, сопереживание, взаимопомощь, мужество, патриотичность. Уверены, ты их разделяешь, и мы легко найдем общий язык.
Ну а если что-то пойдет не так, не руби с плеча, объясни, где мы ошиблись и поверь, мы разберемся и исправим.
Наш девиз “In hostem omnia licita” – по отношению к врагу дозволено все. Возьми этот девиз, он поможет тебе принять правильное решение в трудной ситуации, с честью выполнить боевую задачу и вернуться домой живым и здоровым!
Изготовление металлоконструкций также продуктов в Иркутске. https://smt38.ru/
Мы производим Уши, Котлеты,Тефтели сверху 100% БЕЗ Химии, СВЕРХ ГМО (а) также прочего. Доставка числом Иркутску
https://olkhon138.ru/
온라인 슬롯 사이트
주된 이유는 그들이 이 거대한 괴물에 겁을 먹고 항상 이곳이 안전하지 않다고 느끼기 때문입니다.
Кайт top spot обучение кайтсёрфингу в Хургаде Египет https://kitehurghada.ru
Меня просто выворачивает от этих новостей. «Бест Вей» – это спасение для многих семей военнослужащих. Люди доверяли кооперативу свои деньги, чтобы получить квартиры, а теперь их средства заморожены, и они не могут даже забрать свои деньги. Это несправедливо!Эти обвинения против «Бест Вей» просто смешны. Как можно обвинять кооператив, который помогает людям, в создании финансовой пирамиды? Это полный абсурд. Почему государство не защищает тех, кто защищает нашу страну? Вместо этого их топят и делают из них преступников. Пайщики уже выходят на протесты, пишут обращения, требуют справедливости. И я с ними. Давайте поддержим «Бест Вей» и покажем, что мы не позволим так обращаться с теми, кто помогает нашим военным и их семьям. Требуем немедленного разблокирования счетов и прекращения этого позорного дела!
Бест Вей
Воины-пайщики оказались без защиты в тылу
Пайщики «Бест Вей» — участники СВО готовы защищать свой кооператив
Многие пайщики кооператива «Бест Вей» и те, в чьих интересах приобретаются квартиры в кооперативе, — участники СВО. И они сами, и их родственники возмущены событиями, происходящими вокруг кооператива. Ведь защищая интересы страны на фронте, в тылу они не защищены от того, что не могут приобрести недвижимость из-за того, что счета кооператива с почти 4 млрд рублей уже более двух лет находятся под арестом — притом, что сумма ущерба по уголовному делу, рассматриваемому Приморским районным судом Санкт-Петербурга, согласно обвинительному заключению, составляет 282 млн рублей.
«Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию»
Наталья Пригаро, мать пайщика кооператива — участника СВО из Нефтеюганска Даниила Пригаро:
— Сын расплатился за однокомнатную квартиру, приобретенную с помощью кооператива, еще два года назад, подал пакет документов на оформление квартиры в собственность в Росреестр в начале сентября 2022 года — и все это время не может зарегистрировать собственность на нее из-за того, что в Росреестре она значится как арестованная. В конце сентября 2022 года он был мобилизован. Сын брал кредит, чтобы расплатиться за квартиру с кооперативом. Планировал продать эту квартиру и взять квартиру побольше в ипотеку. Однако неоднократно накладывался арест на недвижимость кооператива, несмотря на то что сейчас ареста нет — последний арест, накладывавшийся осенью 2023 года, 19 февраля истек, ходатайства о новом аресте дважды отклонены судом, по Росреестру арест с квартиры до сих пор не снят. В результате кредит приходится платить, квартира в собственность до сих пор не оформлена.
У сына нет никакой физической и финансовой возможности выяснять отношения с Росреестром — в административном или судебном порядке, он приезжает в отпуска на неделю-полторы, ему не до сутяжничества. Я нахожусь в Анапе, приобрела квартиру с помощью кооператива — сын не имеет возможности сделать на меня генеральную доверенность, а я не могу из Анапы приехать в Нефтеюганск и решать вопросы в Росреестре — кроме поезда, сейчас, сами понимаете, ничего не ходит (конец цитаты).
«На безобразие, которое происходит с кооперативом, мы пишем жалобы во все инстанции, — говорит она. — Надеемся, новое руководство Министерства обороны поможет урегулировать ситуацию».
«Надежда, что все разрешится, остается — не может же все быть коррумпировано»
Елена Бойко — пайщик кооператива из Новосибирска и мать солдата, воющего в СВО, для которого она собиралась приобрести одну из двух квартир в кооперативе. «Я стала пайщиком кооператива в 2021 году. В январе 2021 года подала заявление на покупку двух квартир с помощью кооператива по накопительной программе. У меня двое детей — сын и дочь, им нужно жилье. В 2020 году взяли в ипотеку загородный дом — причем оформили на сына как единственного, кому ее могли одобрить, а в 2021 году он ушел в армию как призывник, а потом подписал контракт. Его относительно недавно отправили „за ленточку“. Думали, что поживем в ипотечном доме, и тут как раз подойдет очередь приобретать квартиры с помощью кооператива. Мы останемся жить в частном доме, а дети въедут в кооперативные квартиры».
«Ипотека — это разорительно, — подчеркивает Елена, — мы взяли 2,5 млн, а за 30 лет отдадим 6 млн. Когда появился кооператив, для нас это была возможность предоставить каждому из детей жилье: за 10 лет или раньше они расплатятся с кооперативом, причем с минимальной по сравнению ипотекой переплатой, и у них будет собственное жилье. Из-за репрессий в отношении кооператива мы можем быть этой возможности лишены».
«Я всем сердцем переживаю за ситуацию с кооперативом, заявление о выходе из кооператива пока не пишу — надеюсь, что его деятельность „разморозится“, — говорит Елена. — Обращаемся во все инстанции, в прокуратуру — приходят отписки. Смысл происходящего я понимаю хорошо — когда-то занималась кредитным потребительским кооперативом: нас тогда закрыл Центральный банк. Нашел формальный повод. Но настоящая причина была в том, что мы мешали банкам обирать клиентов. Но надежда на то, что все разрешится, остается — не может же быть все коррумпировано».
«Больше всего в нашей ситуации возмущает то, что наши воины стоят на стороне государства на фронте, а здесь, в тылу, их интересы нарушаются государственными структурами. Такого быть не должно!»
«Для многих кооператив — единственная возможность приобрести квартиру»
Лариса Зискинд — пайщица «Бест Вей», переехавшая с помощью кооператива с семьей — с дочерью, зятем и маленьким внуком - с Дальнего Востока в Белгородскую область. Ее зять, живущий в кооперативной квартире, находится на фронте СВО. «С помощью кооператива я приобрела квартиру в двухэтажном доме в селе Пушкарное Белгородской области — мы с дочерью переехали сюда. Мы уже два года расплачиваемся за него с кооперативом. Не до конца сделали ремонт, кухни фактически нет: моем посуду в ванной. Финансовых средств не хватает, так как зять уже год на СВО, пока ни разу не был в отпуске, и перевести деньги с его карты в ВТБ сейчас непросто, потому что карта — в расположении, до ближайшего банка ехать далеко, а он сам — на линии фронта. Мы фактически живем с выплат на ребенка, на мне кредиты — у меня полпенсии уходит на их оплату. Квартира по Росреестру находится под арестом, хотя арест по квартирам кооператива арест истек — а я боюсь ехать в Белгород, чтобы решать по ней вопрос. Я с ребенком на улицу гулять не выхожу в нынешней военной обстановке — сирены воздушной тревоги постоянные. Побыстрее бы наши войска создали санитарную зону в Харьковской области!»
По поводу покупки квартиры Лариса сообщает, что «ипотека точно никак не светила. Кооператив — это 14 тыс. в месяц и всего лишь 10 лет. В кооперативе даже студентка может приобрести квартиру, почти не переплачивая. При покупке через банк придется заплатить еще как минимум за одну квартиру».
«Среди пайщиков кооператива очень много пенсионеров, социально незащищенных людей, это была их единственная возможность приобрести квартиру, — говорит Лариса. — Из-за репрессий против кооператива мы можем лишиться единственного жилья. Мы куда только не писали в защиту кооператива. Но, видимо, слишком большой куш. Кооперативу не дают даже платить налоги. Надеемся, что отношение к кооперативу в результате справедливого разбирательства изменится в лучшую сторону».
«Мы очень устали. Но боремся»
Людмила Гарина — пайщица кооператива из Саратова, зять которой, проживающий вместе с ней в кооперативной квартире, участвует в СВО. «Я проживаю в кооперативной квартире почти три года вместе с дочерью и внуком, вносим ежемесячные платежи через госуслуги».
«Возможности взять ипотеку не было, — говорит Людмила. — Фактически у нас один работающий человек — дочь, зять мобилизован, внук — студент, внучке — четыре года. Мы с мужем пенсионеры».
«У нас с кооперативом проблем не было никаких, — говорит она, — но сейчас уверенности в завтрашнем дне нет. Пишем во все возможные инстанции, в том числе в Администрацию президента, ходили на прием к Володину. Пока ничего не помогает. Вся надежда на то, что суд разберется».
«Хотелось, чтобы вопрос о возобновлении работы кооператива быстрее решился, — говорит она, — тогда стало бы проще. Когда кооператив заработает, будет совершенно иная ситуация. Мы очень устали. Но боремся».
«20 тыс. пайщиков кооператива не так просто остановить»
Алла Яровая — пайщик кооператива из Пятигорска и представитель пайщицы Людмилы Дворцовой, сын которой, Игорь Дворцов, находился на СВО, а теперь служит внутри страны. «Людмила приобрела с помощью кооператива в Пятигорске квартиру для своего сына. Вступила в кооператив она в 2017 году, сначала находилась в накопительной программе — накапливала на первоначальный паевый взнос, накопила, потом стояла в очереди на приобретение жилье, приобрела с помощью кооператива однокомнатную квартиру за 1450 тыс. рублей. Потом приобрела квартиру и постепенно полностью за нее расплатилась. Работала для этого на двух работах, в том числе санитаркой в инфекционном отделении».
Но оформить квартиру в собственность не удалось, так как она оказалась под арестом, поясняет Алла. И хотя арест 19 февраля истек и больше не накладывался, суд первой инстанции в Пятигорске этот арест подтвердил. «По вопросу получения права собственности и снятия ареста мы обратились в Пятигорский городской суд, где судья очень предвзято отнесся к решению вопроса и мы получили отказ, — говорит Алла, — Хотя аналогичный вопрос рассматривался в Минераловодском суде и там судья вынесла положительное решение и в настоящий момент пайщик получил право собственности на свою квартиру. Предстоит рассмотрение в апелляционной инстанции — несмотря на то, что рассматривать нечего: ареста на недвижимость кооператива сейчас нет. А на квартиру Людмилы Дворцовой — почему-то есть».
«Кооператив — единственная возможность для людей со средним достатком получить жилье, — говорит Алла, — Нерешенный вопрос — налоги. Мы вынуждены платить налоги с юридических лиц, но мы же физические лица. К депутатам — с нас берут налог как с юрлица. Почему мы должны оплачивать — мы же физлица? Во многих регионах это сделано. Надеемся добиться того, чтобы это было сделано по всей стране».
«20 тыс. пайщиков кооператива не так просто остановить, — подчеркивает пайщица. — Это наши деньги, и мы за них боремся. Обязательно отстоим наш кооператив!»
«Активно участвую в защите кооператива»
У пайщицы кооператива, жительницы из Ленинградской области Татьяны Власенко старший сын находится на СВО с сентября 2022 года. «Три месяца проходил обучение, а потом оказался „за ленточкой“, — говорит Татьяна. — Мы должны были в конце лета 2022 года купить квартиру с помощью кооператива — квартиру планировали большую. Но работа кооператива оказалась заблокирована, и кооператив не может ничего купить уже более двух лет, так как и его счета, и его деятельность заблокированы. Деньги пайщиков, собранные для приобретения квартир, при этом обесцениваются».
«Я активно участвую в защите нашего кооператива, — говорит Татьяна. — Езжу на все суды в Санкт-Петербурге, хотя живу в деревне — ехать два с половиной, а то и три часа, мне 64 года, а путь неблизкий. И сын в этом поддерживает меня».
«Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются»
У пайщицы из Хабаровска Натальи Ромашко на СВО было два племянника — один, к сожалению, погиб. Планировалась покупка большой квартиры в Хабаровске, Наталья состояла в накопительной программе. «Вот уже более двух лет работа кооператива заблокирована, деньги обесцениваются, — говорит Наталья. — Я участвую в защите кооператива, пишу письма в прокуратуру, в высшие государственные инстанции, потому что нарушаются наши права, нас лишают возможности приобрести квартиру вне ипотечной системы. Пока приходят одни отписки, но борьбу мы продолжаем!»
«Ситуация — как на СВО»
Пайщица Светлана Иванова из Новосибирска, волонтер СВО, планировала приобрести с помощью кооператива две квартиры — для себя и для сына, который воюет на СВО добровольцем. «До этого я продала квартиру и сейчас вообще без собственного жилья, — говорит она. — Вступила в кооператив по накопительной программе, рассматривали для приобретения квартиры Крым и Новосибирск. В СВО участвую как волонтер — не остаюсь в стороне, когда Родине нужна помощь».
«От ситуации с кооперативом очень многие люди пострадали, — говорит Светлана. — Ситуация — как на СВО. И еще больше пострадают, если реализуется плохой сценарий. Хочется надеяться на то, что все образуется, правда восторжествует. Заявление о выходе из кооператива и возврате паевых средств я не подавала. Готова продолжить участие в нем, готова приобрести недвижимость с его помощью: это самый лучший вариант покупки квартиры».
«Сдаваться в планах нет»
Пайщица Марина Янковская родом из Хабаровска, она переехала в Новороссийск, ее супруг погиб на СВО. «У нас уже подходила очередь на покупку квартиры в Новороссийске для нашей семьи, мы должны были подбирать объект, — рассказывает она. — Но два года назад все остановилось. Хотели приобрести двухкомнатную квартиру в Новороссийске. Деньги лежат на арестованных счетах кооператива — а недвижимость за это время подорожала. Надеемся хоть что-то приобрести после того, как счета разблокируют».
«Мы всячески поддерживаем кооператив, — говорит Марина. — Записывали видео, что мы не обманутые, а вполне довольные пайщики, объясняли, что это никакая не пирамида, а организация, созданная в помощь людям. Ждем, когда снимут эти аресты. Сдаваться в планах нет».
Колокольцев – предатель Родины
Армия восстанет против ОПГ Колокольцева и Плугина
Целенаправленно разрушая кооператив для военных «Бест Вей», захватывая его активы в интересах своей камарильи, глава МВД и его команда ударили по тылу армии, наказав тысячи пайщиков кооператива – военных и членов их семей.
Приморский районный суд Санкт-Петербурга (судья Екатерина Богданова) сейчас рассматривает так называемое уголовное дело «Лайф-из-Гуд» – «Гермес» – «Бест Вей». В рамках этого дела два года почти непрерывно арестованы счета кооператива «Бест Вей», пайщиками которого являются тысячи военнослужащих, в том числе участники СВО, на которых около 4 млрд рублей. В уголовном деле речь идет прежде всего об обязательствах иностранной инвесткомпании «Гермес» перед более чем 200 клиентами этой компании.
Следствие ГУ МВД по Санкт-Петербургу и прокуратура города на Неве объявили кооператив, «Гермес» и «Лайф-из-Гуд» частью некоего единого холдинга, которого в природе никогда не существовало. Кооператив объявлен гражданским ответчиком по уголовному делу.
Общая сумма претензий потерпевших, согласно обвинительному заключению, 282 млн рублей – даже их (незаконное) изъятие для погашения долгов перед клиентами «Гермеса» никак бы не сказалось ни на операционной деятельности, ни на ликвидности кооператива. Но из раза в раз арестовываются абсолютно все средства на счетах, что полностью блокирует как покупку квартир кооперативом, так и возврат денег пайщикам, которые изъявили желание забрать свои паевые взносы.
Недвижимость, на которую претендовали пайщики, обесценивается, деньги обесцениваются – и этот беспредел продолжается уже более двух лет по ходатайствам ГСУ питерского главка МВД и Прокуратуры Санкт-Петербурга.
Кто-то влиятельный пытается наложить лапу именно на кооперативные 4 млрд – хотя это деньги пайщиков: физических лиц, рядовых граждан России, и как минимум соучастниками грабежа являются сотрудники правоохранительных органов.
Армейский кооператив
Кооператив изначально создавался военнослужащими – одной из его задач в 2014 году было решение жилищной проблемы военнослужащих, увольняемых в запас, эта задача сохранилась и в последующие годы. Из почти 20 тыс. его пайщиков – тысячи военнослужащих со всей России. Значительная часть из них – участники СВО.
В результате действий ОПГ Колокольцева сотни семей военнослужащих, участников СВО стали потерпевшими от действий правоохранительных органов. Налицо преступные действия против участников СВО.
Камарилья, которая устроила репрессии против кооператива – руководители следственной группы Сапетова и Винокуров, начальник ГСУ питерского главка МВД Негрозов и начальник этого главка Плугин, замминистра внутренних дел по следствию Лебедев, пресс-секретарь министра Волк, транслировавшая на всю страну ложь про десятки тысяч потерпевших от работы кооператива, сам министр внутренних дел Колокольцев, – ведет вредительскую деятельность, все эти люди – предатели Родины.
Разоряет экономику и подрывает тыл
Колокольцев и Ко разрушают экономику страны, фабрикуют дела. При этом его камарилья наживается на честном бизнесе.
Колокольцев – миллиардер! Его боевая подруга генеральша Волк – миллиардерша! Зам. начальника питерского ГСУ МВД полковник Винокуров, который управляет Колокольцевым, заставляя его подписываться под липовым уголовным делом «Лайф-из-Гуд» – «Гермес» – «Бест Вей», – миллиардер! Колокольцев превратил МВД в коммерческое антинародное образование и зарабатывает деньги на простых людях.
Колокольцев и Ко прицепили звезды на погоны и возомнили себя офицерами – на деле они тыловые вороватые крысы. Настоящие офицеры против бутафорских чинуш со звездами на погонах.
Министр внутренних дел дестабилизирует ситуацию в народе и армии, разоряет армию в тылу. Колокольцев со своими тыловыми крысами подрывают доверие народа к власти и отбивают желание людей идти на военную службу, потому что благодаря таким, как он, государство не может обеспечить безопасность военных в тылу.
Армия против Колокольцева
Армия восстанет против ОПГ Колокольцева. Военные требуют за совершенные преступления против народа, за воровство и коррупцию арестовать Колокольцева, Негрозова, Винокурова, Сапетову, оправдать и отпустить невинных людей – рядовых сотрудников «Лайф-из-Гуд», которые более двух лет томятся в тюрьме.
Новый министр обороны Белоусов наверняка поддержит своих бойцов, поможет им защитить собственные и своих семей имущественные интересы в тылу: по-другому и быть не может.
Одного любителя запускать руку в казну и любителя генеральш – Шойгу – уже отправили на пенсионную должность, Колокольцев должен последовать за ним: это – в интересах страны, в интересах народа!
Title: Como Contatar o Suporte ao Cliente do Spin Casino: Seu Guia Completo
spin jogos
Description: Descubra como contatar o suporte do Spin Casino por chat, e-mail, telefone e redes sociais. Resolva suas duvidas de forma rapida e eficaz!
H1: Como posso entrar em contato com o suporte ao cliente do Spin Casino?
Bem-vindo ao guia completo de como entrar em contato com o suporte ao cliente do Spin Casino! Neste artigo, exploraremos todas as opcoes disponiveis para que voce possa resolver qualquer questao de maneira rapida e eficiente. Entenderemos por que um suporte ao cliente acessivel e crucial para uma experiencia de jogo online satisfatoria e como o Spin Casino se destaca neste aspecto.
H2: Metodos de Contato Disponiveis
A acessibilidade e eficiencia no suporte ao cliente sao cruciais para uma experiencia satisfatoria em qualquer cassino online. No Spin Casino, os jogadores tem a disposicao diversos metodos para entrar em contato com o suporte. Exploraremos cada um deles detalhadamente.
H3: Chat ao Vivo
A opcao de chat ao vivo e ideal para solucionar problemas de forma rapida e direta. Disponivel diretamente atraves do site do Spin Casino, esse servico esta ativo 24 horas por dia, sete dias por semana, garantindo assistencia imediata a qualquer momento. Para acessa-lo, basta clicar no icone de chat no canto inferior direito da pagina inicial e iniciar uma conversa com um dos atendentes. Usuarios costumam receber respostas em poucos minutos, o que torna este metodo eficiente para questoes urgentes.
H3: E-mail
Para aqueles que preferem documentar suas interacoes ou tem questoes que requerem detalhamento, o e-mail e uma excelente opcao. O Spin Casino oferece um endereco de e-mail dedicado ao suporte: support@spincasino.com. Ao enviar um e-mail, e aconselhavel incluir detalhes como o nome de usuario e a descricao precisa do problema. O tempo de resposta tipico pode variar de algumas horas a um dia util, dependendo do volume de consultas.
H3: Telefone
Embora menos comum na era digital, o suporte por telefone ainda e oferecido pelo Spin Casino para quem prefere uma abordagem mais pessoal. Os jogadores podem encontrar os numeros de telefone aplicaveis na secao de contato do site, disponiveis para diversas regioes. Este metodo e particularmente util quando o problema requer uma explicacao mais detalhada ou quando a solucao e complexa e as instrucoes passo a passo sao necessarias. E importante verificar os horarios de atendimento para garantir que a ligacao seja feita durante os periodos em que a equipe esta disponivel para ajudar.
Cada um desses metodos de contato foi projetado para proporcionar uma experiencia suave e eficaz, garantindo que todas as questoes e preocupacoes dos jogadores sejam atendidas com a atencao que merecem.
H2: Suporte atraves das Redes Sociais
O Spin Casino tambem oferece suporte aos seus usuarios atraves de diversas plataformas de redes sociais, reconhecendo a importancia e a conveniencia desses canais no dia a dia dos jogadores. Os clientes podem entrar em contato com a equipe de suporte por meio de perfis oficiais no Facebook, Twitter e Instagram. Esses canais sao particularmente uteis para consultas rapidas ou para obter atualizacoes sobre promocoes e eventos. Para enviar uma mensagem privada, basta acessar o perfil desejado e clicar em “mensagem” ou “contato”. E importante notar que as respostas podem nao ser tao imediatas quanto no chat ao vivo, mas a equipe se esforca para responder todas as interacoes o mais rapido possivel.
H2: Dicas para um Contato Eficaz com o Suporte
Para garantir que sua experiencia de contato com o suporte do Spin Casino seja o mais eficiente possivel, considere as seguintes dicas:
Prepare-se Antecipadamente: Antes de entrar em contato, tenha em maos todas as informacoes relevantes, como detalhes da conta, descricoes de problemas e qualquer correspondencia anterior.
Seja Claro e Conciso: Descreva seu problema de forma clara e direta, fornecendo todos os detalhes necessarios para que a equipe de suporte possa entender e resolver a questao rapidamente.
Escolha o Metodo de Contato Adequado: Dependendo da urgencia e complexidade do seu problema, escolha o metodo de contato mais adequado. Use o chat ao vivo para questoes urgentes e e-mail para problemas que requerem explicacoes detalhadas.
H2: Contato Adicional
Alem dos canais de suporte convencionais, o Spin Casino incentiva os usuarios a fornecerem feedback sobre suas experiencias. Se, por algum motivo, uma questao nao for resolvida satisfatoriamente atraves dos canais regulares de suporte, os jogadores sao encorajados a enviar suas reclamacoes ou sugestoes atraves de um formulario especial disponivel no site. Esse compromisso com a melhoria continua do servico mostra o empenho do casino em manter uma relacao transparente e positiva com seus clientes. Alem disso, essa comunicacao adicional ajuda a garantir que todas as preocupacoes sejam devidamente registradas e tratadas de maneira adequada.
Esses elementos sao cruciais para manter um relacionamento saudavel e responsivo entre o casino e seus jogadores, assegurando que cada interacao contribua para uma experiencia de jogo positiva e segura.
Ao final deste guia, voce esta agora equipado com todas as informacoes necessarias para contatar o suporte ao cliente do Spin Casino de maneira eficiente. Lembre-se de escolher o metodo de contato que melhor se adapta a sua necessidade, seja ela uma urgencia ou uma questao que requer detalhamento. O compromisso do Spin Casino com um suporte de alta qualidade garante que sua experiencia de jogo sera sempre segura e agradavel. Nao hesite em utilizar esses recursos sempre que necessario!
FAQ:
Qual e o metodo mais rapido de contato com o suporte do Spin Casino?
O chat ao vivo e o metodo mais rapido, operando 24/7 diretamente pelo site.
Como posso enviar um e-mail para o suporte do Spin Casino?
Envie um e-mail para support@spincasino.com com seu nome de usuario e a descricao do problema.
O suporte ao cliente do Spin Casino esta disponivel em quais redes sociais?
O suporte esta ativo no Facebook, Twitter e Instagram. Envie uma mensagem privada atraves do perfil desejado.
Como posso dar feedback sobre o suporte recebido?
Utilize o formulario especial no site para enviar reclamacoes ou sugestoes, ajudando a melhorar continuamente o servico.
This is the right site for everyone who would like to find out about this topic. You know so much its almost hard to argue with you (not that I actually will need to…HaHa). You definitely put a new spin on a subject that’s been written about for decades. Wonderful stuff, just excellent!
cratosroyalbet casino
Кайт школа обучение кайтсёрфингу в Хургаде Египет https://kitehurghada.ru
Кайт это обучение кайтсёрфингу в Хургаде Египет https://kitehurghada.ru
i-guru – Ремонт Iphone и другой техники Apple в Минске
замена экрана iphone 15 Plus в Минске
Профессиональный ремонт любых устройств из линейки Apple, без посредников
80% ремонтов iPhone занимает около 20 минут
Несложные ремонты делаются быстро, модульный ремонт в большинстве случаев занимает не более 20 минут.
Почему выбирают i-guru
Ремонт в тот же день
Даже большинство сложных ремонтов выполняем день в день.
Гарантировано низкие цены
Мы беремся за сложные задачи, не передавая их сторонним исполнителям, и зарабатываем не как посредники.
Честная гарантия
Предлагаем честную гарантию на выполненные работы до 12 месцев в зависимости от вида работ.
Компания Септик-Нара-купить септик в наро-фоминске
занимается продажей, установкой и обслуживанием септиков в Наро-Фоминске Наро-Фоминском районе. Основное направление деятельности нашей компании именно установка под ключ септиков любых видов и размеров. С момента основания нашей компании, мы произвели монтаж более 1000 септиков по Московской и Калужской области. Благодаря этому у нас огромный опыт работы с любыми станциями, представленными в нашем регионе!
Если Вы решили купить септик для дома или дачи, мы с радостью поможем Вам с выбором модели, доставим и установим септик на вашем участке в кратчайшие сроки.
Мы занимаемся продажей септиков таких марок: Топас Юнилос Астра Евролос Тверь Аквалос Дочиста Фекалов Волгарь Удача. Мы работаем напрямую с производителями септиков, поэтому Вы можете быть уверены, что не переплачиваете ни копейки. Вся продукция в нашей компании имеет соответствующие сертификаты и лицензии. Время выезда на осмотр, установки или привоза оборудования согласовывается с клиентами и выполняется в срок. Мы заботимся о своей репутации, поэтому выполняем работу надежно и быстро.
Если Вы не знаете, какой септик больше всего Вам подходит, мы предоставим консультацию и выезд специалиста на Ваш участок абсолютно бесплатно!
Благодаря приобретению качественных септиков в нашей компании, каждый клиент получает большое количество преимуществ:
» компактные размеры и небольшой вес устройства;
» доступная стоимость очистной установки;
» быстрый и простой монтаж;
» невысокая стоимость эксплуатации;
» отсутствие неприятных запахов;
» высокая производительность;
» длительный срок эксплуатации;
» высокая степень очистки;
» практически полностью автономный режим работы.
Кайт обучение кайтсёрфингу в Хургаде Египет https://kitehurghada.ru
https://tezfiles.cc/
Tezfiles Premium account owners are specified infinite download speeds to let go b exonerate them access their files in their cloud storage entirely quickly.
Tributes flood in for BBC sport commentator whose wife and daughters were killed in suspected crossbow attack
порно анальный секс
Figures from across the United Kingdom have offered their condolences to a BBC sport commentator, after his wife and two daughters were killed by an alleged crossbow attacker, in deaths that again drew attention to the epidemic of violence against women.
Carol Hunt, 61, wife of BBC horse racing commentator, John Hunt, and their daughters, Hannah Hunt, 28, and Louise Hunt, 25, died from injuries sustained in an attack in Bushey, just northwest of London, on Tuesday, according to police and Britain’s public broadcaster.
A 26-year-old suspect wanted in connection with the killings, and named as Kyle Clifford, was found by British police in Enfield, north London, on Wednesday, following a sprawling manhunt. There were no previous reports to the force over Clifford, who is in serious condition in hospital and is yet to speak with officers, Hertfordshire Police said in a statement.
A crossbow was recovered as part of the investigation, which police believe was used in a “targeted incident.”
Epidemic of violence against women
The killings of the three women rocked Britain, where mass murders are infrequent but violence against women and girls has been officially labeled as a national threat.
A woman is killed by a man every three days in the UK and one in four women will experience domestic violence in her lifetime, the United Nations’ special rapporteur on violence against women and girls, Reem Alsalem, said earlier this year.
Charities and human rights organizations have subsequently reiterated urgent demands to tackle femicide in the UK.
mvp 토토 사이트
이 수도에서의 생활은 힘들지만 이전보다 몇 배나 낫습니다.
A fairly new player in the Russian darknet arena, blacksprut Blacksprut has quickly gained attention for its interesting features and growing popularity. While some aspects raise questions, it has the potential to become a prominent figure in the darknet scene.
Features:
Blacksprut https://bs-gl-darknet.com offers an “Instant Transactions” feature, inspired by the success of similar systems on other platforms like Hydra. Couriers hide goods within the city and provide buyers with coordinates, adding an adventurous element to the purchasing process.
Ирина Волк: расследование уголовного дела пирамиды Life is Good завершено
жесткое групповое порно
Официальный представитель МВД России Ирина Волк сообщила об окончании предварительного расследования уголовного дела Life is Good. Материалы с утвержденным обвинительным заключением в отношении десяти человек направлены в Приморский районный суд Санкт-Петербурга для рассмотрения по существу. Они обвиняются в организации деятельности по привлечению денежных средств и мошенничестве, совершенных в особо крупном размере в составе преступного сообщества. Всего, по версии следствия, в состав криминальной организации входили 35 соучастников. Потерпевшими по уголовному делу признаны 221 человек – люди заключили договоры с целью получения пассивного дохода. Общая сумма материального ущерба составила свыше 280 млн рублей.
«Кроме того, в деятельность финансовой пирамиды были вовлечены более 18 тысяч россиян. Они вложили в жилищный кооператив более 15 миллиардов рублей, два миллиарда из которых организаторы криминальной схемы присвоили. Десять предполагаемых участников преступного сообщества были задержаны в феврале 2022 года оперативниками ГУЭБиПК МВД России совместно с коллегами из Санкт-Петербурга и сотрудниками ФСБ России. Остальные обвиняемые в противоправной деятельности объявлены в розыск. Возбуждено уголовное дело по признакам преступления, предусмотренного частью 3 статьи 210, частью 4 статьи 159, частью 2 статьи 172.2 УК РФ. Расследование было взято на контроль руководством Следственного департамента МВД России», – сообщила Ирина Волк.
По информации МВД, в общей сложности полицейскими и сотрудниками ФСБ проведено более 100 обысков по местам проживания фигурантов и в офисах подконтрольных им организаций. Изъята компьютерная техника, средства связи, документы и другие предметы, имеющие доказательственное значение.
«В качестве обеспечительной меры по возмещению причиненного материального ущерба наложен арест на более чем 2300 находящихся в собственности кооператива квартир, а также на денежные средства на счетах компании в сумме свыше 3,7 млрд рублей. Также арестованы активы фигурантов на общую сумму порядка 1 млрд рублей», – рассказала генерал-майор полиции.
По версии следствия, противоправная деятельность велась с 2014 года на территории девяти регионов России. Злоумышленниками были созданы подконтрольные организации с сетью региональных филиалов, которые работали под единым брендом «Лайф из Гуд». Гражданам предлагали стать пайщиками кооператива, обещая либо новое жилье, либо получение дохода в размере до 30% годовых. При этом реальным инвестированием вкладов фигуранты не занимались. Приобретение недвижимости и обещанные выплаты осуществлялись за счет привлечения средств новых клиентов. Кроме того, граждан вводили в заблуждение об уровне доходности вложений, указывая в договорах параметры гораздо ниже обещанных. Полученные от вкладчиков деньги похищались.
Ранее стало известно, что Приморский районный суд Санкт-Петербурга заочно арестовал топ-лидера инвестиционного проекта Life is Good в Набережных Челнах Сергея Санникова и его жену Юлию. Также суд избрал меру пресечения в виде заключения под стражу и в отношении директора холдинга Игоря Маланчука. В июле этого года они были объявлены в федеральный розыск – соответствующая информация появилась в базе данных министерства.
플레이 슬롯
Zhu Houzhao는 “코가 아버지만큼 크지 않다는 것뿐입니다. “라고 말했습니다.
Thank you a bunch for sharing this with all of us you really realize what you are speaking approximately! Bookmarked. Kindly additionally consult with my web site =). We could have a link alternate contract among us
https://thetvdb.plex.tv/movies/real-gangsters
What’s up it’s me, I am also visiting this website daily, this site is actually pleasant and the people are really sharing nice thoughts.
https://thetvdb.plex.tv/movies/real-gangsters
Кооператив для военных
Как пайщики — участники СВО относятся к событиям вокруг «Бест Вей»
[url=https://svpressa.ru/society/article/423652]Социальная программа для участников СВО[/url]
Потребительский кооператив «Бест Вей» оказался затронут уголовным делом, касающимся в основном иностранной инвесткомпании «Гермес», которое сейчас рассматривается Приморским районным судом Санкт-Петербурга. Более двух лет более 3,5 млрд рублей на счетах кооператива почти непрерывно арестованы по ходатайству сначала ГСУ ГУ МВД по Санкт-Петербургу, а затем Прокуратуры Санкт-Петербурга: пайщики не имеют возможности ни приобрести недвижимость, ни вернуть средства.
По данным совета потребительского кооператива «Бест Вей», в числе его пайщиков, страдающих от блокировки средств, тысячи военнослужащих, в том числе сотни участников СВО, часть из которых успела приобрести квартиру, часть собирает или собрала первоначальный взнос, а часть — планировала вступить в кооператив. «СП» пообщалась с некоторыми из них и их родственниками, чтобы узнать отношение к «Бест Вей» и событиям вокруг кооператива.
«К кооперативу отношение очень хорошее»
Александр Голдман на СВО с мая 2022 года как доброволец, три ранения. Пришел на СВО рядовым, сейчас — начальник штаба батальона. Был пайщиком кооператива, сейчас пайщик — его мама.
«С помощью кооператива в 2019 году приобретена двухкомнатная квартира во Владивостоке, в которой проживает мама, — рассказывает он. — Расплачиваемся за нее, в ближайшие месяцы намерены погасить задолженность перед кооперативом и оформить квартиру в собственность. К кооперативу отношение очень хорошее, полностью его поддерживаю — он дает возможность без больших переплат приобрести недвижимость. К действиям в отношении кооператива отношусь отрицательно, так как „Бест Вей“ — единственная возможность приобрести жилье в рассрочку».
«Происходящее вокруг кооператива вызывает шок»
Гвардии рядовой Глушков Иван Васильевич — пайщик кооператива из Челябинской области. Танкист, мобилизованный, проходил службу в 15-й отдельной гвардейской мотострелковой Александрийской бригаде 2-й гвардейской общевойсковой армии ЦВО. Погиб 11 октября 2023 года.
Рассказывает его вдова Татьяна Неручева:
«Мы внесли первоначальный паевый взнос и во второй половине 2021 года встали в очередь на приобретение квартиры в Челябинске, когда начались события вокруг кооператива — была заблокирована его возможность приобретать недвижимость и заблокированы его счета. Наша очередь должна была подойти примерно через год. Мы заявили для приобретения небольшую квартиру, но планировали при покупке увеличить ее стоимость до 3 млн и купить двухкомнатную квартиру — с увеличением первоначального паевого взноса: уставом кооператива это позволяется, а затем переехать из Коркино в Челябинск. Сейчас 3 млн хватит только на небольшую квартиру-студию метров 25, то есть мы понесли материальный ущерб из-за блокирования деятельности кооператива, так как лишены были возможности приобрести квартиру, на которую собрали первоначальный паевый взнос. Работа кооператива заблокирована, счета арестованы уже более двух лет. В наследование пая я только вступаю, так как муж очень долго считался пропавшим без вести — долго шла экспертиза, и свидетельство о смерти мы получили только 18 июня этого года».
Татьяна — юрист: «Как у юриста у меня происходящее вокруг кооператива вызывает шок, и любой непредвзятый юрист вам скажет то же самое». «Кооператив, — подчеркивает она, — абсолютно прозрачен, он полностью соответствует законодательству о кооперации — что и подтверждалось многократно государственными органами и судами. Если бы мы с мужем не были уверены, что все прозрачно и законно, мы бы не вкладывали в него деньги. Мы не подавали заявление о выходе из кооператива. Я, несмотря ни на что, жду счастливого завершения рукотворного кризиса вокруг кооператива. Для меня это еще и память о муже — он продал долю в квартире, которая ему принадлежала, чтобы вложиться в кооператив. Кроме того, мне хочется понять — до какого маразма может дойти ситуация у нас в государстве в плане незаконных действий в отношении организации, которая по тем или иным причинам не понравилась каким-то чиновникам?».
«Рассчитываю, что ситуация закончится благополучно»
Сергей Логинов, рядовой, мобилизованный, представлен к награде «Честь и доблесть».
«Пять лет назад я стал пайщиком кооператива, участвовал в накопительной программе, планировал прибрести однокомнатную квартиру в Самарской области. Уже нужно было подбирать объект недвижимости и вставать в очередь на покупку, как работа кооператива была заблокирована по инициативе правоохранительных органов, и это продолжается уже более двух лет. По-прежнему надеюсь получить квартиру и рассчитываю, что ситуация закончится благополучно. Поддерживаю кооператив».
«В кооперативе минимум переплат — несопоставимо с ипотекой»
Егор Ивков, офицер флота, дирижер военного оркестра, пайщик кооператива с 2019 года.
«Я нахожусь в накопительной программе — планировал покупку квартиры в Санкт-Петербурге. До постановки в очередь на покупку дело не дошло, но сумма внесена серьезная — и на два года все зависло. Есть друзья-военнослужащие, которые также являются пайщиками и тоже накапливали деньги на первоначальный паевый взнос — они, как и я, не могут ни продолжать накапливать, ни получить деньги обратно, потому что счета арестованы. Отношение наше к ситуации, создавшейся вокруг кооператива, крайне негативное».
«Кооператив поддерживаю, — говорит пайщик. — Моя сестра живет в квартире, приобретенной с помощью „Бест Вей“ — у нее многодетная семья, сейчас кооперативная квартира переходит в ее собственность. Минимум переплат — несопоставимо с ипотекой. Надеюсь, что ситуация с арестом счетов разрешится в ближайшее время».
Звон Колокольцева. Питерская полицейская мафия виляет Министерством внутренних дел
Потерпевшие Бествей
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
ГСУ Санкт-Петербурга беспредел
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
Звон Колокольцева. Питерская полицейская мафия виляет Министерством внутренних дел
Прокуратура Санкт-Петербурга
Поздравляем вас, гражданин министр, соврамши!
Выступая прошлой осенью в Совете Федерации, министр внутренних дел Владимир Колокольцев рассказывал, о так называемом уголовном деле «Лайф-из-Гуд» – «Гермес» – «Бест Вей», обещал миллиарды рублей ущерба и десятки тысяч потерпевших. Пресс-служба МВД под руководством его боевой подруги Ирины Волк заявила о том, что вскрыта деятельность крупнейшей в истории России финансовой пирамиды.
Однако в уголовном деле, расследованном или, вернее сказать, изготовленном ГСУ питерского главка МВД, которое в феврале начал рассматривать Приморский районный суд Санкт-Петербурга, 282 млн рублей ущерба и 221 лицо, признанное следствием потерпевшим: никаких миллиардов и десятков тысяч потерпевших.
Министр и его Волк публично солгали – на основе информации, переданной замначальника ГСУ руководителем следственной группы полковником юстиции А.Н. Винокуровым, фактически даже руководившей СГ его заместительницей майором, а затем подполковником юстиции Е.А. Сапетовой по материалам, состряпанным опером УЭБиПК питерского главка МВД майором полиции А.Ю. Машевским, при попустительстве (или соучастии?) начальника ГСУ Негрозова и замначальника Следственного департамента федерального министерства Вохмянина. Полковник и подполковник с примерно пятого-шестого уровня иерархии МВД виляет генералом полиции Российской Федерации, постоянным членом Совета безопасности России – это позор для государства.
Так называемые потерпевшие и реально пострадавшие
Большинство «потерпевших» на суде заявляют суммы около 1–2 млн рублей, при этом в ходе судебного следствия выясняется, что они, как правило, получали немалый доход, причем, они еще и налоговые/валютные преступники, так как этот доход не декларировали. Среди «потерпевших» есть граждане, заявляющие смехотворные суммы в 50–70 тыс., то есть количество потерпевших специально накручивалось следствием. И даже с этим накручиванием удалось набрать так мало – учитывая, что у «Гермеса», по данным самого же следствия, более 200 тыс. клиентов в России, а в кооперативе «Бест Вей» – около 20 тыс. пайщиков.
То есть большинство и клиентов «Гермеса», и пайщиков кооператива не считают себя потерпевшими от деятельности этих организаций.
Судя по тысячам обращений во все инстанции, нескольким волнам митингов, прокатившихся по России, они считают себя потерпевшими от деятельности органов внутренних дел.
Ведь именно завербованный питерским УЭБиПК сисадмин российской платежной системы «Гермеса» Набойченко заблокирован и разгромил в феврале 2022 года эту платежную систему, повесив на сайте дисклеймер: «Обращайтесь в правоохранительные органы», что на месяцы прекратило вывод средств. Именно действия правоохранительных органов в отношении компании до и после затруднили вывод средств. Дело в том, что для вывода средств многими использовался механизм p2p, позволяющий не платить комиссию, то есть для вывода средств нужно, чтобы кто-то вносил средства (что, понятно, резко сократилось из-за уголовного дела) и происходил обмен. Однако этот способ не единственный, вывод средств так или иначе осуществляется.
Тысячи пайщиков кооператива тем более не считают себя потерпевшими от его деятельности – потому что именно правоохранительные органы воспрепятствовали приобретению недвижимости с помощью кооператива, а она из-за более чем двухлетнего ареста его счетов, на которых около 4 млрд рублей, не может быть приобретена по прежней цене.
Именно правоохранительные органы прямо запрещают выплаты пайщикам кооператива, решившим забрать свой пай, – даже по исполнительным листам судов. И клиентам «Гермеса», и пайщикам кооператива правоохранительными органами нанесен колоссальный ущерб – и материальный, и моральный, который они намерены взыскать с государства.
«Следователи»-преступники должны сидеть в тюрьме
Весьма скромный результат следствия МВД был достигнут откровенно преступным путем.
Прокуратура Санкт-Петербург
1. Некоторые из преступлений следствия были фактически признаны судами. 1 декабря прошлого года Приморский районный суд города Санкт-Петербурга признал незаконным, нарушающим УПК фактический отказ кооперативу в ознакомлении с материалами уголовного дела.При рассмотрении дела в суде выяснилось, что следственная группа ГСУ питерского главка МВД, формально руководимая замначальника ГСУ полковником юстиции А.Н. Винокуровым, а фактически – подполковником юстиции Е.А. Сапетовой, подделала документы. Автор подделки – Сапетова – еще в феврале была уволена из ГСУ «по собственному желанию».
Уличенная адвокатами кооператива в нарушении УПК, следственная группа составила письмо об удовлетворении ходатайства задним числом и попыталась представить дело так, что кооператив не получил письмо по своей вине. Ложь была выявлена в том числе и с помощью системы электронного документооборота питерского главка МВД.
2. Подделка документов была вынужденным преступлением для сокрытия более серьезного: незаконного содержания под стражей. Следственная группа грубо нарушила права гражданских истцов и ответчиков, потому что без этого нарушения она не успела за 30 суток до истечения предельного срока содержания четверых обвиняемых под стражей начать ознакомление обвиняемых с материалами дела –а это было единственное основание продления им срока содержания под стражей свыше предельного.
Следственная группа из-за спешки даже толком не смогла завершить следственные действия, незаконно вела параллельное расследование по «резервному» делу, но позднее, отбросив стыд, из-за отсутствия материала для составления «нужного» обвинительного заключения, незаконно продолжила расследование «основного» дела, в том числе проводила следственные действия, которые, согласно УПК, невозможны после начала ознакомления обвиняемых с материалами дела. Все эти ухищрения были необходимы для того, чтобы ни в коем случае не выпускать обвиняемых и продолжать держать их в заложниках.
3. Де-факто происходит уголовное наказание неосужденных людей – четверо подсудимых уже второй год сидят в тюрьме. При этом наказываются явно ни в чем неповинные люди – технические сотрудники «Лайф-из-Гуд»: даже если предположить, что действительно работала пирамида – что опровергается показаниями свидетелей самого обвинения, которые сообщают суду, что компания «Гермес» хорошо работала, они были довольны получаемым доходом, и проблемы начались после того, как российская платежная система компании была обрушена завербованным полицией петербургским сисадмином компании Набойченко. Все подсудимые – технические сотрудники компании «Лайф-из-Гуд» и ни к каким управленческим решениям отношения никогда не имели. Их взяли в заложники для того, чтобы они дали показания на руководство компании.
4. Еще одно преступление – заведомо подложное постановление руководителя следственной группы А.Н. Винокурова о привлечении кооператива «Бест Вей» в качестве гражданского ответчика на 16 млрд рублей, тогда как сумма ущерба в уголовном деле –282 млн, и в деле нет ни одного искового заявления – даже на 100 рублей.
5. Следствие стимулировало двух особенно активных так называемых потерпевших написать заявления о моральном ущербе на миллиард (!) рублей каждое – исключительно для ложного обоснования ареста активов кооператива, но понятно, что это ничтожные документы, так как моральный ущерб во всех случаях, не связанных с причинением смерти, присуждается российскими судами в размере не более десятков тысяч рублей.
6. Ни один из «потерпевших» не доказал обоснованность своих претензий в гражданском суде. При этом арестованы активы кооператива почти на 4 млрд рублей и активы частных лиц на такую же сумму. Это не что иное, как попытка захвата активов при участии органов внутренних дел некой заинтересованной группой клиентов «Гермеса» – необязательно из числа «потерпевших», то есть коррупционное преступление, которое упорно игнорирует ГУСБ МВД.
Механизм для такого захвата есть – это передача средств под управление Федерального общественно-государственного фонда по защите прав вкладчиков и акционеров. Понятно, почему не ограничиваются активами подсудимых и обвиняемых: этого недостаточно для удовлетворения аппетитов тех, кто стоит за заказным уголовным делом. И понятно, почему одно юридическое лицо – кооператив «Бест Вей» – незаконно пытаются привлечь к ответственности за другое – компанию «Гермес»: активы «Гермеса» – за рубежом.
7. Следствие, а теперь и прокуратура совершают еще одно преступление: незаконно удерживает средства пайщиков кооператива, отказываясь их вернуть, то есть совершают хищение.
하이퍼노바 인피니티 릴스
이 비명은 거의 비참한 지경에 이르렀습니다.
Только лучшее сайт вавада подробности на сайте https://amabel.freeforums.net/thread/1496/vavada-casino
МОСКВА, 7 мая — РИА Новости. Правительство России, возглавляемое Михаилом Мишустиным, уйдет в отставку во вторник.
Сразу после инаугурации президента Владимира Путина нынешний кабинет министров сложит полномочия перед вступившим в должность главой государства.
Прогон по комментария
Кайт top spot обучение кайтсёрфингу в Хургаде Египет https://t.me/detivetrachat
슬롯 카지노
“즉시… 선생님을 위해 책을 편집하고, 서둘러!” Jiang Chen의 흥분된 목소리가 떨리고 있었습니다.
Как можно верить тем, кто обещал миллиарды ущерба, а на деле показал всего 282 миллиона? “Гермес” и “Бествей” работали честно, предоставляя своим клиентам возможность зарабатывать. Но теперь их обвиняют в том, чего они не совершали. Полиция подделала документы, нарушила законы и держит невинных людей в тюрьме. На суде выяснилось, что большинство “пострадавших” сами получили доход и даже не декларировали его. Мы, клиенты этих компаний, считаем, что настоящими преступниками являются те, кто ведёт это фальшивое расследование. Мы будем добиваться справедливости и наказания для тех, кто нам вредит.
Что за правовой абсурд происходит в нашей стране? Следственная группа утверждает, что мы должны 16 миллиардов рублей, но никаких исковых требований к кооперативу ‘Бест Вей’ в деле нет. Суд это подтвердил, но счета и имущество нашего кооператива продолжают быть арестованными. Это что за издевательство над людьми? Мы не позволим, чтобы нас просто так грабили! Мы будем бороться за свои права, за свои деньги и за свою собственность. Этот беспредел со стороны органов внутренних дел должен прекратиться, и виновные должны понести наказание. Мы требуем справедливости и немедленного разблокирования наших счетов!
Кайт лагерь обучение кайтсёрфингу в Хургаде Египет https://t.me/detivetrachat