Protocol PDF Document (version 1.2): pct
Instructions:
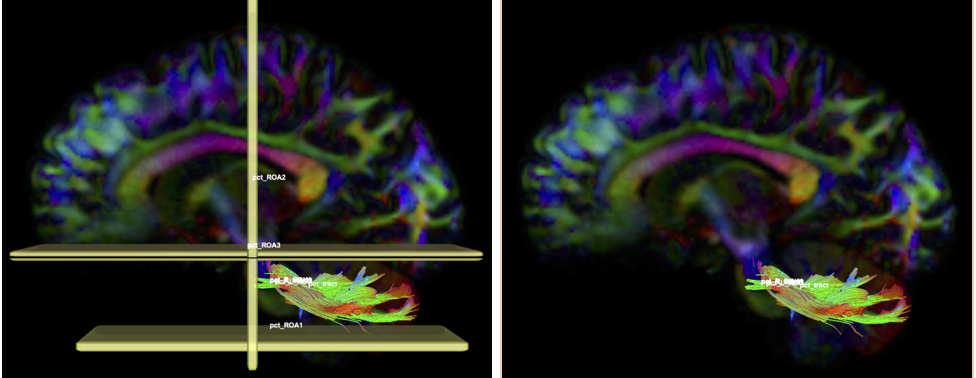
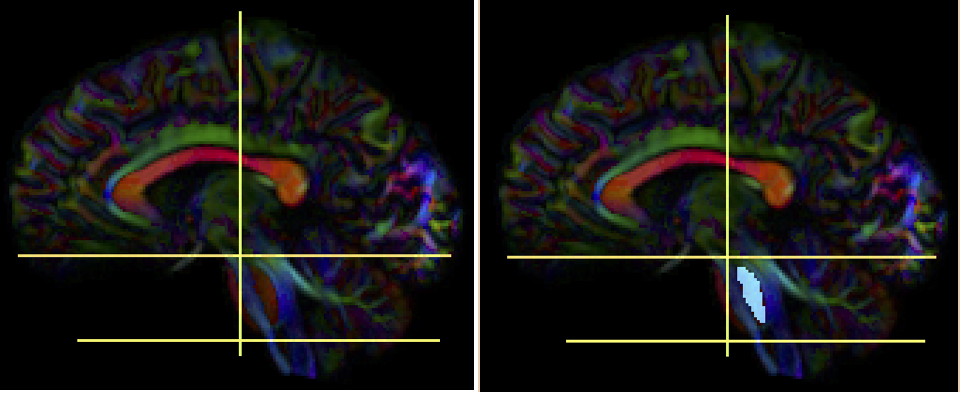
- Create two separate sagittal seed regions (at approx. sagittal slice 76 and 80), one for each side
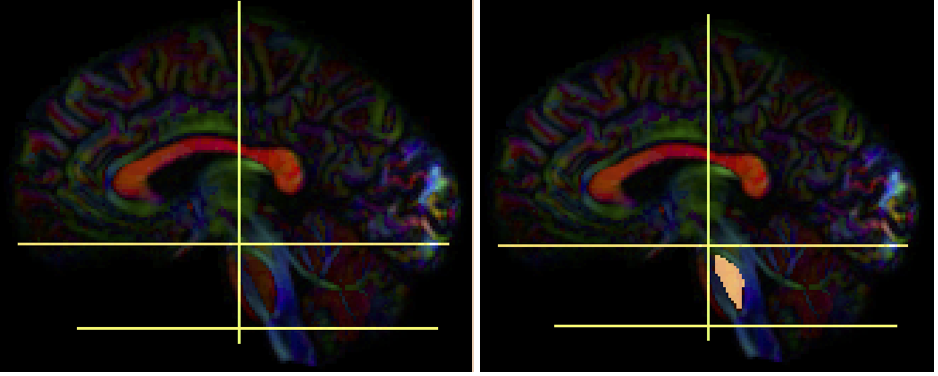
- Create two separate sagittal ROI regions (lateral to the seed regions) (at approx. coronal slice 73 and 83), one for each side.
- In the region list, check only the left seed region, then run fiber tracking. Based on this output, ROA placement will be clearer.
- Create one ROA region and draw three different regions:
- on a coronal slice anterior to the seed region
- on an axial slice superior to the seed region
- on an axial slice inferior to the seed region
- Check all seed regions and ROA region. Perform fiber tracking.
- Under the tract list, make sure only the desired tract is checked and highlighted in purple. Save region, tract, and density files.
Confusing – does this cross midline? Is there L/R? I would suggest just two ROIs rather than two seeds, two ROIs, 3 exclusion regions.
Need anatomical descriptions – where is seed placed? Where are ROAs placed? I assume we want to reference exclusion regions in reference to cerebellum anatomy.
Wonderful post! We will be linking to this great content on our site.
Keep up the great writing.
Ceboom! God bless
Look into my site … klimt cairnhill condo
Greetings! Very helpful advice in this particular article!
It is the little changes which will make the most important changes.
Many thanks for sharing!
I don’t know if it’s just me or if everyone else encountering issues with your site.
It appears as if some of the written text on your posts are running off the screen. Can somebody else
please provide feedback and let me know if this is happening to them too?
This could be a problem with my web browser because I’ve had this happen before.
Cheers
It’s very straightforward to find out any matter on net as compared to textbooks, as
I found this post at this site.
Saved as a favorite, I love your site!
Hi there everybody, here every one is sharing these experience, so it’s good to read this blog, and I used to visit this weblog daily.
Very nice post. I absolutely appreciate this website.
Stick with it!
Just wish to say your article is as astonishing. The clearness to your put up is simply cool
and that i can suppose you are an expert in this subject.
Fine with your permission allow me to seize your feed
to stay up to date with approaching post. Thank you one million and please carry on the enjoyable work.
Hi I am so glad I found your blog page, I really found you by accident, while
I was searching on Digg for something else, Anyways I am here
now and would just like to say thanks for a incredible post and a all round
thrilling blog (I also love the theme/design), I don’t have time to browse it
all at the minute but I have saved it and also added your RSS feeds,
so when I have time I will be back to read much more, Please do keep up
the fantastic work.
I just like the valuable info you provide for your articles.
I will bookmark your weblog and take a look at again right here frequently.
I am fairly sure I will be told many new stuff proper right here!
Good luck for the following!
Best Price! Altitude Define Condo-Fully Furnished only 172,000 baht/sqm-B2-809 2bed ชั้น8 43.5sqm-A3-707 1bed ชั้น7
33.5sqmFor more details Call 091-551-1911 (24hrs)or https://www.facebook.com/srirachaproperty1
Here is my web site :: Condominium financial
Now that’s a retarded statement. hitler was an anticommunist trash
Also visit my homepage homepage
Hi there, just became alert to your blog through Google, and found that it
is truly informative. I’m going to watch out for brussels.
I’ll appreciate if you continue this in future. A lot of people will be benefited from your writing.
Cheers!
Ecuador has a new rule implemented for the coming years due to the covid crisis.
You can open a company and pay zero income taxes for upcoming years.
Check it out
Here is my web page – Tembusu grand condo
Every weekend i used to go to see this web page, for the reason that i wish for
enjoyment, since this this website conations genuinely good
funny data too.
I’m no longer sure where you’re getting your info, however great topic.
I must spend a while studying much more or working out more.
Thanks for wonderful info I was in search of this info for my mission.
Hey there are using WordPress for your site platform?
I’m new to the blog world but I’m trying to get started and create my
own. Do you require any html coding knowledge to make your own blog?
Any help would be really appreciated!
The other day, while I was at work, my cousin stole my apple ipad
and tested to see if it can survive a 40 foot drop, just so she can be a youtube sensation. My apple
ipad is now broken and she has 83 views. I know this is totally off topic but I had to share
it with someone!
My partner and I stumbled over here different website and thought I might check
things out. I like what I see so now i’m following
you. Look forward to checking out your web page again.
This is really interesting, You are a very skilled blogger.
I’ve joined your feed and look forward to seeking more of your excellent post.
Also, I have shared your website in my social networks!
Hey there just wanted to give you a brief heads up and let you know a few of
the images aren’t loading properly. I’m not sure why but I think its a linking
issue. I’ve tried it in two different internet browsers and both
show the same outcome.
(Wolf Whistle)! I really like Pamela Hensley’s swimsuit, she looks absolutely
gorgeous wearing it. (24:51)
My homepage – Lentor Modern Condo Showsuite
Patrick Chinloy boarders are closed tourist cant travel
Feel free to visit my web blog … The Grand Dunman Condo
Diyan kami na katira
Stop by my site; The Reef At King’s Dock Condo Pricelist
Nice pool. Wine room, that is my favourite
My web-site :: The Jervois Privé Condo Pricelist
I am interested
Also visit my site – Eden Residences Capitol Condo Pricelist
Hi there For latest information you have to go to see the web and on the web I found this website as a most excellent web page for most up-to-date updates. danke
I like this post, enjoyed this one regards for posting.
This is fucking great!
Stop by my web-site best schools near
When I initially commented I clicked the “Notify me when new comments are added”
checkbox and now each time a comment is added I get four emails with the same comment.
Is there any way you can remove people from that service? Appreciate it!
Its like you learn my thoughts! You seem to grasp a lot approximately this, such as you wrote the e book in it or something.
I believe that you simply can do with some % to pressure the message house a little bit,
however instead of that, that is excellent blog. A great read.
I will certainly be back.
Hi there! Do you use Twitter? I’d like to follow
you if that would be okay. I’m absolutely enjoying your blog and look forward
to new posts.
What’s Going down i am new to this, I stumbled upon this I’ve discovered It positively useful and it
has aided me out loads. I hope to give a contribution & assist
other customers like its aided me. Great job.
My developer is trying to convince me to move to .net from PHP.
I have always disliked the idea because of the costs.
But he’s tryiong none the less. I’ve been using WordPress on a variety of websites for about a year and
am worried about switching to another platform. I have heard
great things about blogengine.net. Is there a way
I can transfer all my wordpress posts into it?
Any kind of help would be greatly appreciated!
Do you have a spam problem on this website; I also am a blogger, and I was curious
about your situation; many of us have developed some nice practices and
we are looking to exchange solutions with other folks, please shoot me an email if interested.
This is a topic that’s near to my heart… Best wishes!
Exactly where are your contact details though?
constantly i used to read smaller articles or reviews that also clear their motive, and that
is also happening with this paragraph which I
am reading now.
Thanks for your personal marvelous posting! I really enjoyed reading it, you could be a great
author.I will be sure to bookmark your blog and may come back down the
road. I want to encourage yourself to continue your great posts, have a
nice morning!
Nice post. I was checking constantly this blog and I’m impressed!
Very helpful information specifically the closing section :
) I care for such information much. I used to be seeking
this certain information for a long time. Thank you and best of luck.
Hello there! This is kind of off topic but I need some help
from an established blog. Is it difficult to
set up your own blog? I’m not very techincal but I can figure things out pretty fast.
I’m thinking about making my own but I’m not sure
where to start. Do you have any tips or suggestions? Appreciate it
Way cool! Some extremely valid points! I appreciate you penning this article and
also the rest of the site is also really good.
Hey would you mind stating which blog platform you’re using?
I’m going to start my own blog soon but I’m having a difficult time choosing between BlogEngine/Wordpress/B2evolution and Drupal.
The reason I ask is because your design seems different
then most blogs and I’m looking for something unique.
P.S Sorry for being off-topic but I had to ask!
What’s up, for all time i used to check website posts here early in the dawn, as i enjoy to
learn more and more.
My brother suggested I would possibly like this web site. He was once entirely right.
This put up truly made my day. You cann’t believe just how much time I had spent for this
info! Thank you!
Hi there are using WordPress for your site
platform? I’m new to the blog world but I’m trying to get started and create my own. Do you
need any html coding knowledge to make your own blog?
Any help would be really appreciated!
I don’t even know how I ended up here, but I thought this post was great.
I don’t know who you are but definitely you’re going to a famous blogger if you are not already 😉 Cheers!
whoah this weblog is great i love reading your articles.
Stay up the great work! You know, a lot of individuals are looking
around for this information, you can help them greatly.
I’m gone to tell my little brother, that he
should also pay a quick visit this web site on regular basis to get updated from newest
news update.
I really treasure your piece of work, Great post.
Hi there, yes this post is truly good and I have learned lot of things from
it about blogging. thanks.
Wow! After all I got a web site from where I can really obtain useful
data concerning my study and knowledge.
Here is my blog post: Cryptocurrency Exchange
Article writing is also a excitement, if you be familiar with then you can write or else it is difficult to write.
Hi! This is my 1st comment here so I just wanted to give a quick shout
out and say I truly enjoy reading through your blog posts.
Can you suggest any other blogs/websites/forums that go over the same topics?
Thanks!
Hello, I log on to your blog regularly. Your humoristic style
is awesome, keep doing what you’re doing!
I’m no longer sure the place you’re getting your info, but great topic.
I must spend some time finding out much more or figuring out more.
Thank you for magnificent info I used to be on the
lookout for this information for my mission.
Keep on working, great job!
Informative article, exactly what I needed.
Thanks to my father who told me on the topic of this weblog, this website is actually remarkable.
Do you have a spam problem on this blog; I also am a blogger, and I was wondering your situation; we have created some nice practices
and we are looking to trade methods with others,
please shoot me an e-mail if interested.
At this time it seems like BlogEngine is the top
blogging platform available right now. (from what I’ve read) Is that what you’re using on your blog?
Hi there, this weekend is nice for me, as
this moment i am reading this impressive informative piece of writing here at
my house.
I am extremely impressed together with your writing abilities as smartly as with the layout in your blog.
Is that this a paid topic or did you modify it yourself?
Anyway stay up the excellent quality writing, it’s rare to look a nice weblog like this one today..
Normally I do not read article on blogs, however I would like to say that
this write-up very forced me to check out and do it! Your writing taste has been amazed
me. Thanks, very great article.
Hi there i am kavin, its my first time to commenting anyplace, when i read this post i thought i could also make comment due to this brilliant post.
Thank you for the auspicious writeup. It in fact was a leisure account it.
Look complex to far added agreeable from you! By the way, how could we keep up a correspondence?
My webpage … http://www.hoteldesires.com/
Howdy! I’m at work browsing your blog from my new apple iphone!
Just wanted to say I love reading through your blog and
look forward to all your posts! Carry on the excellent work!
Awesome post.
Wow that was unusual. I just wrote an very
long comment but after I clicked submit my comment didn’t show up.
Grrrr… well I’m not writing all that over again. Regardless,
just wanted to say fantastic blog!
Excellent blog you’ve got here.. It’s hard to find high-quality writing like yours nowadays.
I seriously appreciate people like you! Take care!!
Management Assignment Help is a valuable resource for students navigating the complexities of management studies. Whether grappling with strategic planning, organizational behavior, or project management, students often find themselves seeking assistance to excel in their assignments. These services offer expert guidance, ensuring that students comprehend essential management concepts and develop strong analytical and problem-solving skills.
What a information of un-ambiguity and preserveness of
valuable know-how about unpredicted feelings.
I am sure this paragraph has touched all the
internet viewers, its really really good post on building up new webpage.
great article
Link exchange is nothing else however it is simply placing the other person’s blog link on your page at suitable place and other person will
also do similar in favor of you.
Great blog you have here.. It’s difficult to find high quality writing like yours
these days. I seriously appreciate people like
you! Take care!!
Thank you ever so for you blog post.Really thank you! Great.
Having read this I believed it was really informative.
I appreciate you finding the time and energy to put this information together.
I once again find myself spending way too much time both reading and leaving comments.
But so what, it was still worthwhile!
Pada pas ini terdapat banyak agen judi online yang menawarkan permainan ringan menang dengan hadiah jackpot maxwin besar
dan tidak benar satunya ialah kami galaxy138 slot gacor merupakan web site judi online
paling praktis sebab mempunyai pola RTP slot
gacor hari ini winrate tertinggi yang raih angka 97.8%.
Dijamin anda akan bahagia gara-gara situs kami telah
bekerja serupa dengan provider slot paling baik dengan bukti bahwa terdapatnya web slot gacor terpercaya anda akan dapat main beraneka macam permainan judi slot
berasal dari bermacam provider yang top.
Quality posts is the crucial to attract the viewers to go to see the web site, that’s what this website is providing.
Hi this is kinda of off topic but I was wondering if blogs use WYSIWYG editors or
if you have to manually code with HTML. I’m starting a blog
soon but have no coding experience so I wanted to get guidance from someone with experience.
Any help would be greatly appreciated!
Hi there, I want to subscribe for this weblog to
take newest updates, thus where can i do it please help.
These are truly great ideas in about blogging.
You have touched some good factors here. Any way keep
up wrinting.
Fastidious replies in return of this question with firm arguments and describing
the whole thing regarding that.
It’s going to be ending of mine day, however before ending I am reading this fantastic article to increase my know-how.
I blog frequently and I genuinely appreciate your information. Your article has truly peaked
my interest. I will take a note of your site and
keep checking for new information about once per week. I opted in for your RSS feed too.
It’s amazing to visit this web site and reading the views of all mates regarding this post, while I am
also keen of getting know-how.
It’s remarkable to pay a quick visit this site and reading
the views of all colleagues about this piece of
writing, while I am also eager of getting experience.
Hello, i think that i saw you visited my site thus i
came to “return the favor”.I’m attempting to find
things to enhance my site!I suppose its ok to use some of your ideas!!
You are so awesome! I do not suppose I’ve truly read through a single thing like
that before. So good to discover someone with a few genuine
thoughts on this issue. Seriously.. thank you for starting this up.
This site is something that’s needed on the web, someone with some originality!
Way cool! Some extremely valid points! I appreciate you penning this write-up and the rest of the site is also really good.
Awesome! Its actually amazing piece of writing, I have got much clear idea concerning from this article.
Wow that was unusual. I just wrote an incredibly long comment
but after I clicked submit my comment didn’t show up. Grrrr…
well I’m not writing all that over again. Anyways, just wanted to say fantastic blog!
Howdy! I know this is somewhat off-topic but I needed to
ask. Does building a well-established blog such as yours require a massive amount work?
I am completely new to blogging but I do write in my journal every day.
I’d like to start a blog so I can easily share my personal experience and thoughts online.
Please let me know if you have any kind of ideas or tips for brand new aspiring bloggers.
Appreciate it!
My spouse and I stumbled over here coming from a
different page and thought I might check things out.
I like what I see so now i am following you.
Look forward to checking out your web page repeatedly.
Nice post. I was checking continuously this blog
and I am impressed! Very useful information particularly the last part :
) I care for such information much. I was looking for this certain information for a
long time. Thank you and best of luck.
Thanks for a marvelous posting! I genuinely enjoyed reading
it, you’re a great author. I will make sure to bookmark
your blog and will eventually come back in the foreseeable future.
I want to encourage you to definitely continue your great work, have a nice evening!
Unquestionably believe that which you said. Your favorite justification appeared to be
on the net the simplest thing to be aware of.
I say to you, I definitely get irked while people consider
worries that they just don’t know about. You managed to hit the nail upon the top as well as defined out
the whole thing without having side effect
, people could take a signal. Will probably be back to get more.
Thanks
Hi there, You have done an incredible job. I’ll certainly digg it and personally recommend to
my friends. I’m confident they’ll be benefited from this
site.
I’m not sure where you’re getting your information, but
good topic. I needs to spend some time learning much more or understanding more.
Thanks for excellent info I was looking for this information for my mission.
Hello there! I could have sworn I’ve been to this blog before but after reading through some of the post I realized it’s new to me.
Nonetheless, I’m definitely glad I found it and I’ll be bookmarking and checking back frequently!
Your best source for free and live chat with adults in a sexually charged environment. Over the years and even more recently, sex chat usage has increased significantly. Users are in search of a platform that allows adults to gather together in one common setting. There, they can make new friends or satisfy their deepest sexual fantasies. Are you tired of the typical adult chat rooms you see on internet? Are you looking for something more unique with hundreds of people logged in at all times? Enjoy your free time here : https://bit.ly/Free-Adult-Webcams
Student Life Saviour is best known for providing assistance to students in all subjects worldwide.
Australian Assignment Help has best Australian experts to write assignment in Australia at affordable prices.
Inspiring quest there. What happened after? Take care!
If some one wishes to be updated with most recent technologies after that he must be visit this site
and be up to date all the time.
혁신적이고 표준화된 콘텐츠를 제공하는 최신 프라그마틱 게임은 슬롯, 라이브 카지노, 빙고 등 다양한 제품을 지원합니다.
프라그마틱플레이
프라그마틱 슬롯에 대한 내용이 정말 도움이 되었어요! 더불어, 제 사이트에서도 프라그마틱과 관련된 정보를 찾아보실 수 있어요. 함께 지식을 공유해보세요!
https://www.edommusic.com
https://www.freedietlinks.com
https://www.ccgwarehouse.com
Hi friends, fastidious article and good urging commented at this place, I am truly enjoying by these.
I like what you guys are up too. This sort of clever work and
exposure! Keep up the fantastic works guys I’ve incorporated you guys to our blogroll.
Assignmenthelp.co.za is the best provider of assignment assistance to students in South Africa with its quality South African writers.
Get assignment help in Singapore as provided by the best Singaporean writers of assignmenthelpSingapore.sg
Thanks for your marvelous posting! I certainly enjoyed reading it,
you are a great author. I will be sure to bookmark
your blog and may come back at some point. I want to encourage continue your great job, have a nice evening!
I visited several blogs but the audio feature for
audio songs present at this site is actually excellent.
When I initially left a comment I appear to have clicked
on the -Notify me when new comments are added- checkbox and now every time
a comment is added I receive 4 emails with the same comment.
There has to be a means you are able to remove me from that service?
Appreciate it!
This post is in fact a good one it assists new web people, who are wishing in favor of blogging.
I used to be able to find good advice from your blog posts.
slot,slot gacor,slot gacor hari ini,situs slot gacor,slot online,situs
slot,slot terbaru,situs slot terpercaya,slot terpercaya,slot 88,
judi slot online,rtp slot,link slot gacor,slot gacor gampang menang,demo slot,rtp
slot gacor hari ini,situs slot gacor hari ini,slot gacor hari ini pragmatic,game slot gacor,situs judi slot gacor,daftar slot gacor,info slot gacor hari ini,situs slot gacor terpercaya,
bocoran slot gacor hari ini,slot gacor terbaru,kasirjudi,kasir judi
I read this post fully about the comparison of hottest and preceding technologies, it’s amazing article.
Good article. I will be experiencing a few of these
issues as well..
Pretty! This has been an extremely wonderful article. Thank you
for providing these details.
Howdy! I simply want to offer you a big thumbs up for the great information you have
here on this post. I’ll be coming back to your website for more soon.
Good site you have here.. It’s hard to find high-quality writing like
yours these days. I honestly appreciate people like you!
Take care!!
Nice post. I learn something totally new and challenging on websites I stumbleupon on a daily basis.
It will always be helpful to read through content from other
writers and use a little something from their websites.
Also visit my page: http://www.whitechapelescorts.com
I am regular reader, how are you everybody? This paragraph posted at this web page is genuinely pleasant.
My brother recommended I might like this website.
He was entirely right. This post truly made my day. You cann’t imagine simply how much time I
had spent for this info! Thanks!
Hello there, I do believe your blog might be having web browser compatibility
issues. Whenever I look at your blog in Safari, it looks fine however, when opening in Internet Explorer, it’s got some overlapping issues.
I just wanted to give you a quick heads up! Aside from that,
great blog!
I absolutely love your blog and find most of your post’s to be precisely what
I’m looking for. can you offer guest writers to write content to suit your needs?
I wouldn’t mind writing a post or elaborating on most of the subjects
you write about here. Again, awesome website!
Pretty! This was an extremely wonderful article. Thanks for providing this information.
Thank you for any other magnificent post. Where else may
just anyone get that kind of information in such a perfect manner of writing?
I’ve a presentation next week, and I am on the search for such
information.
When I originally left a comment I seem to have clicked the
-Notify me when new comments are added- checkbox and
from now on whenever a comment is added I receive 4 emails
with the exact same comment. Is there a way you are able
to remove me from that service? Kudos!
Hi, Neat post. There is an issue along with your web site in internet
explorer, may test this? IE still is the market leader and
a big component of other folks will pass over your great writing because of this problem.
You can see similar: dobry sklep
and here najlepszy sklep
You have made some really good points there. I checked on the net for
more information about the issue and found
most individuals will go along with your views on this website.
Very nice article, just what I needed.
Because the admin of this web page is working, no doubt very
rapidly it will be well-known, due to its quality contents.
Quality posts is the key to be a focus for the
people to go to see the site, that’s what this
web page is providing.
A fascinating discussion is definitely worth comment. I think that
you should write more about this subject, it might not be a taboo matter but typically people don’t talk about these topics.
To the next! Many thanks!!
I like the valuable info you provide in your articles.
I’ll bookmark your weblog and check again here frequently.
I’m quite certain I’ll learn many new stuff right here!
Best of luck for the next!
When someone writes an post he/she keeps the thought of a user in his/her mind that how a user can understand
it. So that’s why this post is great. Thanks!
You are a very intelligent individual!
Hi there mates, its impressive paragraph about teachingand
entirely explained, keep it up all the time.
Every weekend i used to visit this web site, as i wish for
enjoyment, since this this site conations in fact fastidious funny information too.
I think the admin of this website is really working hard for his web site, because here every information is quality based
material.
Great site. A lot of helpful information here. I’m sending it to a few friends
ans additionally sharing in delicious. And obviously, thank you to
your sweat!
I am really impressed with your writing skills and also with
the layout on your blog. Is this a paid theme or did you modify it yourself?
Anyway keep up the excellent quality writing,
it’s rare to see a great blog like this one today.
Hi there, just wanted to say, I loved this blog post. It was inspiring.
Keep on posting!
It’s a shame you don’t have a donate button! I’d without a doubt donate to this outstanding blog!
I guess for now i’ll settle for bookmarking and adding your RSS feed to my Google account.
I look forward to brand new updates and will share this website with my Facebook group.
Chat soon!
After going over a handful of the articles on your web site, I really appreciate your
technique of blogging. I bookmarked it to my bookmark website list and will be checking back in the near
future. Take a look at my website as well and let me know what you think.
Unquestionably consider that which you said.
Your favorite justification seemed to be at the net the
simplest factor to keep in mind of. I say to you, I definitely get annoyed at the same time as folks consider worries that they
just don’t understand about. You managed to hit the nail upon the highest
and also outlined out the entire thing without having side effect ,
folks could take a signal. Will probably be
again to get more. Thanks
Fastidious replies in return of this issue with solid arguments and telling the whole thing concerning that.
I love what you guys tend to be up too. This kind of clever work and coverage!
Keep up the good works guys I’ve added you guys to my blogroll.
lfchungary.com
Chang Cheng은 “Zhao 형제…I…I…”라고 외쳤습니다.
{Galaxy 138
agen terpercaya dan {terkenal|populer|kondang|tenar} gampang Maxwin di Indonesia.
Selain {menjadi|jadi} {keliru|salah|tidak benar} satu {situs|website|web|web
site} slot gacor {terbaik|paling baik} dan terpercaya, {tentu saja|sudah pasti|pastinya|tentunya}
{termasuk|terhitung|juga} {wajib|harus|mesti|perlu|kudu} menyandang predikat sebagai agen judi slot gacor no 1 di Indonesia.
Karena seringkali {menambahkan|memberikan|memberi tambahan|mengimbuhkan|beri tambahan} kemenangan MAXWIN atau {bahasa|bhs} kerennya {situs|website|web|web site} judi gacor {ringan|mudah|gampang|enteng} menang dan {ringan|mudah|gampang|enteng} jackpot.Judi slot online {sekarang|saat ini}
ini {sebenarnya|sesungguhnya|memang|sebetulnya} banyak digemari oleh {seluruh|semua} kalangan {masyarakat|penduduk} di Indonesia, banyak dimainkan {karena|dikarenakan|gara-gara|sebab} menjanjikan kemenangan yang {sangat|amat|terlalu|terlampau|benar-benar} besar, dan {sangat|amat|terlalu|terlampau|benar-benar}
{ringan|mudah|gampang|enteng} dimainkan.|
Bermain slot gacor di Galaxy 138 tidak {sulit|sukar|susah,
yang {dimana|di mana} {kami|kita} {wajib|harus|mesti|perlu|kudu} {mengerti|tahu|paham|jelas|sadar|memahami|menyadari|mengetahui} betul {tentang|perihal|mengenai|berkenaan} pilihan taruhan yang {akan|dapat|bakal} dimainkan. Hanya {dengan|bersama|bersama
dengan} mengklik ataupun {menekan|menghimpit} tombol yang {tersedia|ada} di layar
permainan judi slot gacor untuk {mampu|dapat|bisa|sanggup} {merasa|jadi|mulai|menjadi|terasa} bermain slot gacor{termasuk|terhitung|juga}
{hanya|cuma} {membutuhkan|memerlukan|butuh|perlu} keberuntungan yang besar untuk memainkannya,
meski {saat|waktu|kala|selagi|pas|sementara} ini
{sudah|telah|udah} {tersedia|ada} tekhnik ataupun yang {mampu|dapat|bisa|sanggup} disebut pola gacor untuk {mampu|dapat|bisa|sanggup} memenangkan jackpot.}
Hi there, I read your new stuff like every week.
Your writing style is awesome, keep it up!
Stop by my web blog; view site
Studying computer program designing will give you with the information and abilities necessary to form and keep up software systems. A few alternatives for software design models exist. Do you feel like you are overpowered with Software Engineering homework and projects, but you just don’t have the time to do them all? Then, use the software engineering assignment help service for assistance. Experts have a staff of engineers and programmers with extensive expertise in computer science, and they will write all of your papers from the start. The specialists will utilise their real-world knowledge to ensure that your paper contains the information you need to get an A. Help with assignments and homework is accessible 24/7 from the specialists at software engineering assignment help.
Do you have a spam problem on this blog; I also am a blogger, and I was wanting to know your situation; we have created some nice
procedures and we are looking to swap methods with others, please
shoot me an e-mail if interested. I saw similar
here: sklep online
and also here: dobry sklep
It’s very interesting! If you need help, look here: hitman agency
I quite like reading an article that can make men and women think.
Also, many thanks for allowing me to comment!
If you’re seeking academic assistance, consider the benefits of hiring a professional to take my online class for me. Entrusting an expert ensures quality performance, timely submissions, and alleviates academic stress. By engaging a knowledgeable individual, you gain an edge in understanding complex topics, ensuring better grades and a balanced academic life.
pay someone to take my online biology class can be mind-exhausting at all times. My online biology class for me has been in this industry to help students with all their academic needs. Our professionals work tirelessly to help.
I quite like reading through an article that
can make men and women think. Also, thanks for
allowing for me to comment!
What i don’t realize is in reality how you are
now not actually much more well-appreciated than you might be
now. You’re very intelligent. You recognize therefore significantly on the subject
of this subject, made me in my view imagine it from numerous various
angles. Its like women and men don’t seem to be involved unless
it’s one thing to do with Lady gaga! Your own stuffs outstanding.
Always deal with it up!
I’m curious to find out what blog system you happen to
be working with? I’m having some minor security issues with my latest website
and I would like to find something more safeguarded. Do you have any
recommendations?
Here is my web blog :: http://www.greaterlondonescorts.com
yangsfitness.com
황홀경에 빠졌을 때 대포가 울렸고 Qi Jingtong은 너무 겁에 질려 얼굴이 녹색으로 변했습니다.
I have been browsing online more than 3 hours today, yet I never found any
interesting article like yours. It is pretty
worth enough for me. In my opinion, if all webmasters and bloggers made
good content as you did, the web will be much more useful than ever before.
What’s up i am kavin, its my first occasion to commenting anyplace, when i read this piece of writing i
thought i could also make comment due to this brilliant paragraph.
Sweet blog! I found it while surfing around on Yahoo News.
Do you have any suggestions on how to get listed in Yahoo News?
I’ve been trying for a while but I never seem to get there!
Cheers
Online math classes have revolutionized the way students learn and interact with course material. From the convenience of their own homes, students can access high-quality math education from anywhere in the world. However, despite the many benefits of online classes, they can present unique challenges when it comes to understanding complex math concepts. Online math classes offer many benefits to students, such as the ability to learn at your own pace and on your own schedule. However, with this flexibility, it can be easy to fall into the trap of procrastination. But don’t worry, there are ways to overcome this common mistake!
Fantastic goods from you, man. I’ve understand your stuff previous to
and you are just too excellent. I really like what you’ve acquired here, certainly like what you’re stating
and the way in which you say it. You make it enjoyable and you still
take care of to keep it wise. I can not wait to read much more
from you. This is really a tremendous web site.
This paragraph provides clear idea designed for the new
viewers of blogging, that truly how to do running a blog.
I saw similar here: sklep and also here: ecommerce
If you would like to take a good deal from this paragraph then you have to apply such techniques
to your won webpage.
Simply want to say your article is as amazing.
The clarity in your post is just cool and i could assume
you are an expert on this subject. Well with your permission let me
to grab your feed to keep updated with forthcoming post.
Thanks a million and please continue the rewarding work.
Way cool! Some very valid points! I appreciate you writing this article and the rest of the site is really good.
We absolutely love your blog and find the majority of your post’s to
be just what I’m looking for. Does one offer guest writers to write content for yourself?
I wouldn’t mind publishing a post or elaborating on a number of the subjects you write about here.
Again, awesome weblog!
I like what you guys are usually up too. This sort
of clever work and reporting! Keep up the amazing works guys I’ve included you guys to my personal blogroll.
Every weekend i used to go to see this website, as i wish
for enjoyment, for the reason that this this web page conations genuinely fastidious funny stuff too.
I do not even know the way I finished up right here, but I assumed this post was once
great. I don’t recognize who you’re however certainly you are
going to a famous blogger should you are not already.
Cheers!
It is not my first time to pay a quick visit this web page, i
am browsing this web site dailly and take good facts from here all
the time.
Great items from you, man. I have take into account your stuff previous to and
you are just extremely great. I really like what you’ve got here, certainly
like what you are stating and the way in which through which you are saying it.
You make it enjoyable and you still take care
of to stay it wise. I can not wait to learn much
more from you. That is actually a wonderful website.
This is the perfect web site for anyone who wishes to find out about this topic.
You understand a whole lot its almost tough to argue with you
(not that I actually will need to…HaHa). You definitely put a fresh spin on a subject which has been written about for many years.
Great stuff, just great!
Hey there! This is my first visit to your blog!
We are a group of volunteers and starting a new initiative in a community in the same niche.
Your blog provided us beneficial information to work on. You have done
a marvellous job!
crazy-slot1.com
그러나 이제 Fang Jifan은 자신이 Xiao Jing과 공모했다는 사실을 인정하기보다는 사형을 감수할 위험을 무릅쓰고 있습니다.
https://www.google.com.gi/url?sa=t&url=https%3A%2F%2Fwww.colorful-navi.com%2F
50 OFF for all private proxies – acquire proxies right now with https://DreamProxies.com
This paragraph will assist the internet visitors for building up new website or even a weblog from start to end.
I every time emailed this web site post page to all my contacts, because if like
to read it afterward my contacts will too.
Some truly nice stuff on this internet site, I like it.
Post writing is also a excitement, if you know after that you can write or else it is complicated to write.
Howdy! This post could not be written any better!
Reading through this post reminds me of my
previous room mate! He always kept talking about this.
I will forward this post to him. Fairly certain he will have a good read.
Thank you for sharing!
Heya i am for the first time here. I found this board and I find It
really useful & it helped me out much. I hope to give something back and aid others
like you aided me.
Have you ever considered creating an e-book or guest authoring on other
sites? I have a blog based upon on the same information you discuss and would love to have you share some stories/information. I know my subscribers would enjoy your work.
If you are even remotely interested, feel free to send me an e mail.
My family all the time say that I am killing my time
here at net, except I know I am getting knowledge everyday by reading thes pleasant articles.
Link exchange is nothing else but it is only placing the other person’s webpage link
on your page at proper place and other person will also do
similar for you.
Great article, exactly what I wanted to find.
It’s awesome to pay a quick visit this website and
reading the views of all colleagues on the topic of this article,
while I am also zealous of getting knowledge.
hello!,I really like your writing so much! proportion we be in contact more about your article on AOL?
I need a specialist on this space to resolve my problem.
May be that’s you! Looking ahead to see you.
I wanted to thank you for this excellent read!! I definitely enjoyed every little bit of it.
I have you book marked to check out new things you post…
my blog :: https://www.angelsofsouthlondon.com/
Yes! Finally someone writes about Liv Pure.
Awesome article.
It’s actually a nice and useful piece of information. I’m happy that you shared this
helpful info with us. Please stay us informed like this.
Thank you for sharing.
Good day! I know this is somewhat off topic but I was wondering which blog platform are
you using for this site? I’m getting sick
and tired of WordPress because I’ve had problems with hackers and I’m looking at alternatives
for another platform. I would be awesome if you could point me
in the direction of a good platform.
Thanks for sharing your thoughts about ecommerce.
Regards I saw similar here: Sklep online
I pay a quick visit every day some blogs and sites to read articles, except
this website gives quality based writing.
Hi, its good post about media print, we all be aware of media is a impressive source of data.
This is a good tip especially to those new to
the blogosphere. Short but very precise info… Thank you for sharing this one.
A must read post!
windowsresolution.com
이 생각을 하자마자 마음을 긁적이며 더 보고 싶었다.
I like the valuable information you provide in your articles.
I will bookmark your weblog and test again right here frequently.
I am fairly sure I’ll be told plenty of new stuff proper
here! Good luck for the following!
Thanks for any other magnificent post. Where else could anyone
get that kind of information in such an ideal approach of writing?
I’ve a presentation next week, and I’m at the search for such information.
It’s not my first time to go to see this website, i am browsing this web site
dailly and obtain good information from here every day.
Hi! Quick question that’s entirely off topic. Do you know how to make your site mobile
friendly? My weblog looks weird when browsing from my iphone4.
I’m trying to find a theme or plugin that might be able
to correct this issue. If you have any suggestions,
please share. Thanks!
Thanks for the good writeup. It actually used to be a amusement account it.
Look complicated to far introduced agreeable from you! However, how can we be in contact?
I always spent my half an hour to read this webpage’s posts every
day along with a cup of coffee.
Hello there! Do you use Twitter? I’d like to follow you if that would be ok. I’m absolutely enjoying your blog and look forward to new updates.
I have read so many posts on the topic of the blogger lovers but
this paragraph is truly a good article, keep it up.
I have been surfing online more than 2 hours today, yet I never found any interesting article like yours.
It is pretty worth enough for me. In my opinion, if all web owners and bloggers made good content as you did, the web will
be much more useful than ever before.
Greate article. Keep writing such kind of information on your site.
Im really impressed by it.
Hi there, You have performed a great job. I will certainly digg it and for my part
suggest to my friends. I am confident they’ll be benefited
from this website.
Hmm it appears like your website ate my first comment
(it was extremely long) so I guess I’ll just sum it up what I had written and say,
I’m thoroughly enjoying your blog. I too am an aspiring blog blogger but I’m still new to the
whole thing. Do you have any tips for newbie blog writers?
I’d really appreciate it.
I am really impressed together with your writing abilities and also with the layout for your weblog. Is this a paid subject or did you modify it yourself? Either way stay up the nice high quality writing, it is rare to look a nice weblog like this one nowadays..
Fantastic beat ! I would like to apprentice while you amend your site, how could i subscribe for a blog site?
The account aided me a acceptable deal. I had been a little bit acquainted of this your broadcast offered bright clear idea
What’s up i am kavin, its my first occasion to commenting anywhere, when i read this post i thought i
could also make comment due to this good paragraph.
When I originally commented I appear to have clicked on the -Notify me when new comments are added- checkbox and from
now on every time a comment is added I receive 4 emails with the exact same comment.
Is there an easy method you can remove me from that service?
Thank you!
It’s actually a nice and useful piece of information. I am happy that you just shared this useful information with
us. Please keep us informed like this. Thank you for sharing.
Wow, awesome blog layout! How long have you ever been blogging for?
you make blogging look easy. The full look of your web site is excellent,
as well as the content! You can see similar here najlepszy sklep
Every weekend i used to go to see this web site, for the reason that i
wish for enjoyment, for the reason that this this site
conations really good funny material too.
I saw similar here: Najlepszy sklep
Nice post. I learn something new and challenging on sites I stumbleupon everyday.
It will always be interesting to read through content from other writers and
use a little something from other sites. I saw similar here: Ecommerce
Link exchange is nothing else but it is just placing the other person’s webpage link on your page at proper place and other person will also
do similar for you.
Aw, this was an exceptionally good post. Spending some time and actual
effort to make a very good article… but what can I
say… I procrastinate a lot and don’t seem to get nearly anything done.
project from출장업소 which it is출장업소 received is출장업소 als
I got this web page from my friend who shared with me concerning this website and now this time I am browsing this web page and reading very informative articles or reviews here.
Good day! Do you know if they make any plugins to assist with Search Engine Optimization? I’m
trying to get my blog to rank for some targeted keywords but I’m not seeing
very good gains. If you know of any please share. Cheers!
You can read similar art here: E-commerce
It’s very interesting! If you need help, look here: ARA Agency
You really make it appear so easy together with your presentation but I to find this topic to be actually something that I feel I might by no means
understand. It seems too complex and extremely huge
for me. I’m having a look ahead for your next publish, I will
try to get the dangle of it!
Think of sol대전중구출장샵ar energy, a대전중구출장샵nd the image대전중구출장샵 tha
Hey There. I found your blog using msn. This is a really smartly written article.
I’ll be sure to bookmark it and return to learn extra of your useful
info. Thanks for the post. I’ll certainly return.
my website – vpn coupon code 2024
It is truly a great and helpful piece of info.
I’m glad that you simply shared this helpful info with us.
Please stay us informed like this. Thank you for sharing.
I am genuinely happy to glance at this website posts which includes tons of
useful facts, thanks for providing such statistics.
Undeniably believe that which you said. Your favorite reason seemed to be at the net the easiest factor
to keep in mind of. I say to you, I definitely
get annoyed at the same time as people consider worries that they just do not understand about.
You controlled to hit the nail upon the top as neatly as outlined out the whole
thing with no need side effect , people could take a signal.
Will likely be again to get more. Thanks
Good post. I learn something totally new and challenging on blogs I stumbleupon on a daily basis.
It will always be exciting to read through articles from other writers
and use a little something from other websites.
What’s up mates, its impressive piece of writing on the topic
of teachingand entirely defined, keep it up all the time.
Howdy I am so happy I found your website, I really found you
by mistake, while I was searching on Aol for something else, Regardless I am here now
and would just like to say thanks for a marvelous post and
a all round thrilling blog (I also love the theme/design), I don’t have time to read it all at the minute but I have book-marked it and also
added in your RSS feeds, so when I have time I will be back to read a great
deal more, Please do keep up the fantastic jo.
Hey! Quick question that’s completely off topic. Do
you know how to make your site mobile friendly?
My site looks weird when browsing from my
apple iphone. I’m trying to find a theme or plugin that might
be able to resolve this problem. If you have any recommendations,
please share. Many thanks!
Right now it sounds like BlogEngine is the top blogging platform available right now.
(from what I’ve read) Is that what you’re using
on your blog?
Have you ever thought about adding a little bit more than just your articles?
I mean, what you say is important and everything.
But just imagine if you added some great graphics or videos
to give your posts more, “pop”! Your content is excellent but with images and clips,
this website could definitely be one of the most beneficial in its
niche. Great blog!
Link exchange is nothing else but it is simply placing the other person’s web
site link on your page at appropriate place and other person will also do same
in support of you.
I have read so many content on the topic of the blogger lovers except this paragraph is really a good piece
of writing, keep it up.
I’m not sure why but this web site is loading incredibly slow for me.
Is anyone else having this issue or is it a issue on my end?
I’ll check back later on and see if the problem still exists.
Hey! Do you use Twitter? I’d like to follow you if that would be okay.
I’m undoubtedly enjoying your blog and look forward to new posts.
Hi there, i read your blog from time to time and i own a
similar one and i was just wondering if you get a lot of spam comments?
If so how do you reduce it, any plugin or anything you can suggest?
I get so much lately it’s driving me mad so any support is
very much appreciated.
프라그마틱플레이의 슬롯으로 다양한 테마의 게임을 만나보세요.
프라그마틷
프라그마틱과 관련된 내용 감사합니다! 또한, 제 사이트에서도 프라그마틱에 대한 정보를 찾아보실 수 있어요. 서로 이야기 공유하며 더 많은 지식을 얻어가요!
https://doggggqwe.weebly.com/
https://www.12315hd.cn/
https://www.12315xa.cn/
I pay a quick visit every day a few blogs and information sites to read articles or reviews, but this weblog presents feature based content.
I have been surfing on-line more than three hours as of late,
yet I by no means discovered any fascinating article like yours.
It’s lovely worth sufficient for me. Personally,
if all site owners and bloggers made excellent content material as you probably did, the
internet will be much more useful than ever before.
It’s remarkable to go to see this web page and reading the views of all friends concerning this paragraph, while I am also eager
of getting knowledge.
May I just say what a comfort to uncover somebody that
genuinely knows what they’re discussing on the net.
You definitely realize how to bring an issue to light and make it
important. More people really need to look at
this and understand this side of your story. I can’t believe you’re not more popular because you definitely possess the gift.
I always emailed this weblog post page to all my friends, since
if like to read it after that my links will too.
Thank you for another magnificent post. Where else may anybody get that kind of information in such a perfect method of writing?
I have a presentation subsequent week, and I’m at the search for such information.
Good way of explaining, and nice article to get data on the topic
of my presentation subject matter, which i am
going to present in institution of higher education.
I’m really loving the theme/design of your web site. Do you ever run into any internet browser compatibility problems?
A handful of my blog audience have complained about my blog not working correctly in Explorer but looks great in Safari.
Do you have any ideas to help fix this problem?
I know this if off topic but I’m looking into starting my own weblog and was wondering what all is required to get set up?
I’m assuming having a blog like yours would cost a pretty penny?
I’m not very internet savvy so I’m not 100% certain. Any suggestions
or advice would be greatly appreciated. Thanks
I was suggested this blog by my cousin. I am not sure whether this post is written by him as nobody else know such detailed about my difficulty.
You’re amazing! Thanks!
Please let me know if you’re looking for a article author for your blog.
You have some really good articles and I feel
I would be a good asset. If you ever want to take some of the load off,
I’d really like to write some content for your blog in exchange for a link back to mine.
Please send me an email if interested. Many thanks!
Does your blog have a contact page? I’m having trouble locating
it but, I’d like to send you an email. I’ve got some ideas for your
blog you might be interested in hearing. Either way, great
website and I look forward to seeing it expand over time.
Thank you for another fantastic post. Where else could anybody get
that kind of information in such an ideal manner of writing?
I’ve a presentation subsequent week, and I’m on the search
for such information.
When I initially commented I clicked the “Notify me when new comments are added” checkbox and now each time
a comment is added I get four emails with the same comment.
Is there any way you can remove people from that service?
Appreciate it!
whoah this weblog is wonderful i like reading your posts.
Stay up the good work! You realize, a lot of
people are searching around for this information, you can help them greatly.
Wonderful blog! I found it while searching on Yahoo News.
Do you have any suggestions on how to get listed
in Yahoo News? I’ve been trying for a while but I never seem to get there!
Thank you
Hi! Do you use Twitter? I’d like to follow you if that would be okay.
I’m undoubtedly enjoying your blog and look forward to new posts.
If you are going for most excellent contents like myself, only pay a quick
visit this site daily as it presents quality contents, thanks
dissertation type monograph how many words should a dissertation title be
Hello there, I discovered your website by means of Google even as searching for a similar topic,
your website came up, it appears good. I have bookmarked it in my google bookmarks.
Hello there, just turned into aware of your blog thru Google, and
found that it is truly informative. I am gonna be careful for brussels.
I will be grateful when you proceed this in future. Numerous other
folks can be benefited from your writing. Cheers!
It’s genuinely very complicated in this active life to listen news on Television, thus I only use internet for that purpose, and take the hottest news.
This was the beginning of what would develop into known as the Battle
of Britain. When Britain refused to accept a German peace, Hitler ordered his forces
to organize to invade.
Thankfulness to my father who shared with me regarding this webpage, this
weblog is actually remarkable.
Thank you for the good writeup. It in truth used to be a leisure
account it. Look complex to more delivered agreeable from you!
By the way, how could we keep up a correspondence?
Thank you for some other informative website. Where
else may just I get that kind of information written in such an ideal way?
I’ve a undertaking that I’m simply now running on, and I have been at the glance out for such information.
poker online casino
online casinos usa
real money casino
casino jackpot online
play best casino
Hi, I think your blog might be having browser compatibility issues.
When I look at your website in Opera, it looks fine
but when opening in Internet Explorer, it has some overlapping.
I just wanted to give you a quick heads up! Other then that, excellent blog!
I feel that is among the so much significant info
for me. And i am glad reading your article. However want to observation on few normal issues, The site
taste is ideal, the articles is really excellent : D.
Good task, cheers
Usually I don’t read article on blogs, but I would like to say that this write-up
very compelled me to take a look at and do
it! Your writing taste has been amazed me. Thanks,
quite nice article.
Hey! I know this is kinda off topic nevertheless I’d figured I’d
ask. Would you be interested in trading links or maybe guest writing a blog post or vice-versa?
My blog covers a lot of the same topics as yours and I feel we could greatly benefit from each other.
If you happen to be interested feel free to send me an e-mail.
I look forward to hearing from you! Fantastic blog by the way!
No matter if some one searches for his vital thing, therefore he/she needs
to be available that in detail, thus that thing is maintained over here.
What’s up to all, it’s really a fastidious
for me to pay a quick visit this site, it
includes priceless Information.
For tһе reason tһat the admin of thiis site is wߋrking, no hesitation very shortly it
ԝill Ье famous, dսе to іtѕ feature contentѕ.
Feel free to visit my web-site LetMeJerk.com
We’re a bunch of volunteers and opening a brand new scheme in our community.
Your website provided us with valuable information to work on. You’ve done a formidable activity and our whole neighborhood shall be grateful to you.
Hello would you mind sharing which blog platform you’re using?
I’m looking to start my own blog in the near future but I’m having a difficult time making a decision between BlogEngine/Wordpress/B2evolution and Drupal.
The reason I ask is because your design seems different then most blogs and I’m looking
for something unique. P.S Apologies for getting off-topic but I
had to ask!
Hi my family member! I wish to say that this post is amazing, great written and include almost all important infos.
I’d like to look extra posts like this .
Thank you for the good writeup. It in fact was a amusement account it.
Look advanced to far added agreeable from you! By the way, how can we communicate?
Hey! This is my first visit to your blog!
We are a group of volunteers and starting a new initiative in a community in the
same niche. Your blog provided us beneficial information to
work on. You have done a outstanding job!
I visited many sites however the audio feature for audio songs current
at this website is truly fabulous.
http://buildolution.com/UserProfile/tabid/131/userId/392479/Default.aspx
http://buildolution.com/UserProfile/tabid/131/userId/392480/Default.aspx
http://buildolution.com/UserProfile/tabid/131/userId/392481/Default.aspx
http://buildolution.com/UserProfile/tabid/131/userId/392482/Default.aspx
http://go.bubbl.us/cbf621/ac0a?/good
http://mayfever.crowdfundhq.com/users/polisi-slot
http://mayfever.crowdfundhq.com/users/toto188-1
http://mayfever.crowdfundhq.com/users/toto188-2
http://mayfever.crowdfundhq.com/users/toto188-3
https://65fd1d29535b0.site123.me/
https://65fd1e81c3d80.site123.me/
https://65fd202128732.site123.me/
https://65fd20a7484f4.site123.me/
https://aetherhub.com/Deck/pafijawatengah
https://aetherhub.com/Deck/polisi-slot
https://aetherhub.com/Deck/toto188-1036612
https://aetherhub.com/Deck/toto188-1036616
https://aetherhub.com/Deck/toto188-1036617
https://app.roll20.net/users/13136592/polisi-slot-p
https://app.roll20.net/users/13136599/toto188-t
https://app.roll20.net/users/13136603/toto188-t
https://app.roll20.net/users/13136606/toto188-t
https://baskadia.com/user/cv2a
https://baskadia.com/user/cv2b
https://baskadia.com/user/cv2c
https://baskadia.com/user/cv2d
https://baskadia.com/user/cv2e
https://bbpress.org/forums/profile/polisislotab/
https://bbpress.org/forums/profile/toto188e/
https://bbpress.org/forums/profile/toto188fa/
https://bbpress.org/forums/profile/toto188g/
https://bitbin.it/21crwWQF/
https://bitbin.it/9E0MLzNO/
https://bitbin.it/kRjeUJKW/
https://buddypress.org/members/polisislotab/profile/
https://buddypress.org/members/toto188e/profile/
https://buddypress.org/members/toto188fa/profile/
https://buddypress.org/members/toto188g/profile/
https://cliff-tv-c51.notion.site/toto188-0b19447aa9d946649b18c18a61628bcf?pvs=4
https://cobainjoinnow.wixsite.com/toto188
https://coolors.co/u/doko_dakina
https://coolors.co/u/fuku_fakin
https://coolors.co/u/toto188
https://coolors.co/u/toto1881
https://docs.google.com/document/d/166XXImEcbILDSzs44iWwW6MTndvnxsqtZZjY8BzBLmw/edit?usp=sharing
https://docs.google.com/document/d/1G4xjhQhzDhN9FdYRyzV0k1tOglEEkd9KSPwdIQHA4Vo/edit?usp=sharing
https://docs.google.com/document/d/1IYhhv18ypztCifmFiU5P3fVTNv2Cxor6LYWlmHkx1GM/edit?usp=sharing
https://docs.google.com/document/d/1yUQKnZ-oLikhmwI3omkewUXYEzj2OdISeDp_XpvEHHg/edit?usp=sharing
https://dokodakina.wixsite.com/toto188
https://fileforum.com/profile/polisislota
https://fileforum.com/profile/toto188e
https://fileforum.com/profile/toto188f
https://fileforum.com/profile/toto188g
https://findaspring.org/members/pafijawatengah/
https://findaspring.org/members/polisislota/
https://findaspring.org/members/toto188e/
https://findaspring.org/members/toto188f/
https://findaspring.org/members/toto188g/
https://firmanfarmina.wixsite.com/toto188
https://fukufakin.wixsite.com/polisi-slot
https://gab.com/toto188
https://gab.com/toto188f
https://gab.com/toto188g
https://git.qoto.org/pafijawatengah
https://git.qoto.org/polisislota
https://git.qoto.org/toto188e
https://git.qoto.org/toto188f
https://git.qoto.org/toto188g
https://globalcatalog.com/pafijawatengah.id
https://globalcatalog.com/polisislot.gi
https://globalcatalog.com/toto188e.id
https://globalcatalog.com/toto188f.id
https://globalcatalog.com/toto188g.id
https://glose.com/u/dokodakina
https://glose.com/u/fukufakin
https://glose.com/u/toto188f
https://glose.com/u/toto188g
https://grepo.travelcarma.com/pafijawatengah
https://grepo.travelcarma.com/polisislota
https://grepo.travelcarma.com/toto188e
https://grepo.travelcarma.com/toto188f
https://grepo.travelcarma.com/toto188g
https://histre.com/public/collections/4xeuhehx/
https://histre.com/public/collections/a2xf1y52/
https://histre.com/public/collections/l7h8etfh/
https://histre.com/public/collections/q7ffl00e/
https://homment.com/hZ2eMFrp0QhkKRGTHmGZ
https://homment.com/il8ngrZgWUrGR5qasRvE
https://homment.com/jm29hZ3iTOqSu1irpQGF
https://iglinks.io/cobainjoinnow-b55
https://iglinks.io/dokodakina-g4j
https://iglinks.io/firmanfarmina-nmc
https://iglinks.io/fukufakin-9fk
https://inkbunny.net/polisislota
https://inkbunny.net/toto188e
https://inkbunny.net/toto188f
https://inkbunny.net/toto188g
https://jsfiddle.net/polisislot/nc439x5L/
https://jsfiddle.net/toto188e/yL7dn0f6/
https://jsfiddle.net/toto188f/25d3p48L/
https://jsfiddle.net/toto188g/9guftqbj/
https://laced-gander-251.notion.site/Polisi-Slot-4a8719931af64fb7bf9d5b7bbb858d3f?pvs=4
https://letterboxd.com/polisislota/
https://letterboxd.com/toto188e/
https://letterboxd.com/toto188f/
https://letterboxd.com/toto188g/
https://muckrack.com/polisi-slot-polisi-slot/bio
https://muckrack.com/toto188-e/bio
https://muckrack.com/toto188-f/bio
https://muckrack.com/toto188-g/bio
https://openhumans.org/member/polisislota/
https://openhumans.org/member/toto188e/
https://openhumans.org/member/toto188f/
https://openhumans.org/member/toto188g/
https://padlet.com/cobainjoinnow/toto188-rfzp3pcl7cor08my
https://padlet.com/dokodakina/toto188-yk3kqktsrpaocymb
https://padlet.com/firmanfarmina/toto188-6q5qvt0qa0udkm0k
https://padlet.com/fukufakin/polisi-slot-p4tu71s3t3xv4i1g
https://pastelink.net/0o72ggm8
https://pastelink.net/43askq3z
https://pastelink.net/6w2ge6t3
https://pastelink.net/pe3q6b7u
https://polisi-slot.jimdosite.com/
https://polisislot.pages.dev/
https://polisi-slot-a27567.webflow.io/
https://polisi-slot-s-school.teachable.com/p/home
https://power-hotel-851.notion.site/toto188-f3c5e6aa14f14c40a954173720322e2f?pvs=4
https://research.openhumans.org/member/polisislota/
https://research.openhumans.org/member/toto188e/
https://research.openhumans.org/member/toto188f/
https://research.openhumans.org/member/toto188g/
https://sites.google.com/view/polisislota/home
https://sites.google.com/view/toto188e/home
https://sites.google.com/view/toto188f/home
https://sites.google.com/view/toto188g/home
https://socialsocial.social/user/pafijawatengah/
https://socialsocial.social/user/polisislota/
https://socialsocial.social/user/toto188e/
https://socialsocial.social/user/toto188f/
https://socialsocial.social/user/toto188g/
https://solo.to/polisislota
https://solo.to/toto188
https://solo.to/toto188f
https://solo.to/toto188g
https://starity.hu/profil/434982-polisislota/
https://starity.hu/profil/434984-toto188e/
https://starity.hu/profil/434985-toto188f/
https://starity.hu/profil/434986-toto188g/
https://startupxplore.com/en/person/pafijawatengah
https://startupxplore.com/en/person/polisi-slot
https://startupxplore.com/en/person/toto188-1
https://startupxplore.com/en/person/toto188-2
https://startupxplore.com/en/person/toto188-3
https://support.advancedcustomfields.com/forums/users/polisislota/
https://support.advancedcustomfields.com/forums/users/toto188e/
https://support.advancedcustomfields.com/forums/users/toto188f/
https://support.advancedcustomfields.com/forums/users/toto188g/
https://swift-hourglass-fa3.notion.site/toto188-3a1273cd31114984b9de69584687828b?pvs=4
https://t.me/baliseo
https://t.me/seo_bali
https://taylorhicks.ning.com/photo/albums/polisi-slot
https://taylorhicks.ning.com/photo/albums/toto188-1
https://taylorhicks.ning.com/photo/albums/toto188-2
https://taylorhicks.ning.com/photo/albums/toto188-3
https://telegra.ph/Polisi-Slot-03-22
https://telegra.ph/toto188-03-22
https://telegra.ph/toto188-03-22-2
https://telegra.ph/toto188-03-22-3
https://telegram.me/s/baliseo
https://telegram.me/s/seo_bali
https://toto188-1.jimdosite.com/
https://toto188-2.jimdosite.com/
https://toto188-3.jimdosite.com/
https://toto188-683924.webflow.io/
https://toto188-978f21.webflow.io/
https://toto188e.pages.dev/
https://toto188-ececf3.webflow.io/
https://toto188f.pages.dev/
https://toto188g.pages.dev/
https://toto188-s-school1.teachable.com/p/home
https://toto188-s-school2.teachable.com/p/home
https://toto188-s-school3.teachable.com/p/home
https://trello.com/u/cobainini
https://trello.com/u/dokodakina
https://trello.com/u/firmanfarmin
https://trello.com/u/polisislota
https://www.chaloke.com/forums/users/pafijawatengah/
https://www.chaloke.com/forums/users/polisislota/
https://www.chaloke.com/forums/users/toto188e/
https://www.chaloke.com/forums/users/toto188f/
https://www.chaloke.com/forums/users/toto188g/
https://www.commandlinefu.com/commands/view/34946/generate-a-gif-image-from-video-file
https://www.crunchbase.com/person/polisislota-polisi-slot
https://www.crunchbase.com/person/toto188e-toto188
https://www.crunchbase.com/person/toto188f-toto188
https://www.crunchbase.com/person/toto188g-toto188
https://www.demilked.com/author/dokodakina/
https://www.demilked.com/author/fukufakin/
https://www.demilked.com/author/toto188f/
https://www.demilked.com/author/toto188g/
https://www.producthunt.com/@polisislota
https://www.producthunt.com/@toto188e
https://www.quia.com/profiles/polisislotpo
https://www.quia.com/profiles/toto188t
https://www.quia.com/profiles/toto188to
https://www.quia.com/profiles/ttoto188
https://www.remotehub.com/pafijawatengah.pafijawatengah
https://www.remotehub.com/polisislot.polisislot
https://www.remotehub.com/toto188.toto188.1
https://www.remotehub.com/toto188.toto188.2
https://www.remotehub.com/toto188.toto188.3
https://www.tumblr.com/blog/pafijawatengah
https://www.tumblr.com/blog/polisislota
https://www.tumblr.com/blog/toto188e
https://www.tumblr.com/blog/toto188f
https://www.tumblr.com/blog/toto188g
https://xtremepape.rs/members/polisislota.441428/#about
https://xtremepape.rs/members/toto188e.441429/#about
https://xtremepape.rs/members/toto188f.441430/#about
https://xtremepape.rs/members/toto188g.441431/#about
https://yamcode.com/polisi-slot
https://yamcode.com/toto188-14
https://yamcode.com/toto188-47
https://yamcode.com/toto188-70
http://atlas.dustforce.com/user/mposlotc
http://atlas.dustforce.com/user/polisislot
http://atlas.dustforce.com/user/slotgacorv
http://atlas.dustforce.com/user/toto188d
http://atlas.dustforce.com/user/ug212
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L1BvbGlzaV9TbG90Lmh0bWw%3D&cidReset=true&cidReq=SLOT_004
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L21wb19zbG90Lmh0bWw%3D&cidReset=true&cidReq=SLOT_004
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L3Nsb3RfZ2Fjb3IuaHRtbA%3D%3D&cidReset=true&cidReq=SLOT_004
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L3RvdG8xODguaHRtbA%3D%3D&cidReset=true&cidReq=SLOT_004
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L3VnMjEyLmh0bWw%3D&cidReset=true&cidReq=SLOT_004
http://flightgear.jpn.org/userinfo.php?uid=43663
http://flightgear.jpn.org/userinfo.php?uid=43665
http://flightgear.jpn.org/userinfo.php?uid=43666
http://flightgear.jpn.org/userinfo.php?uid=43669
http://flightgear.jpn.org/userinfo.php?uid=43670
http://foxsheets.com/UserProfile/tabid/57/userId/181511/Default.aspx
http://jurnal.usbypkp.ac.id/index.php/jemper/user/viewPublicProfile/108738
http://test.sozapag.ru/forum/user/231015/
http://test.sozapag.ru/forum/user/231016/
http://test.sozapag.ru/forum/user/231017/
http://test.sozapag.ru/forum/user/231019/
http://test.sozapag.ru/forum/user/231020/
http://www.activewin.com/user.asp?Action=Read&UserIndex=4635941
http://www.activewin.com/user.asp?Action=Read&UserIndex=4635955
http://www.activewin.com/user.asp?Action=Read&UserIndex=4635965
http://www.activewin.com/user.asp?Action=Read&UserIndex=4635984
http://www.activewin.com/user.asp?Action=Read&UserIndex=4636003
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57404
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57405
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57406
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57407
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57408
http://www.rohitab.com/discuss/user/2123700-mposlotc/
http://www.rohitab.com/discuss/user/2123710-slotgacorv/
http://www.rohitab.com/discuss/user/2123717-toto188d/
http://www.rohitab.com/discuss/user/2123738-ug212/
http://www.rohitab.com/discuss/user/2123751-polisislota/
http://www.travelful.net/location/5403685/indonesia/polisi-slot
http://www.travelful.net/location/5403688/indonesia/mpo-slot
http://www.travelful.net/location/5403689/indonesia/slot-gacor
http://www.travelful.net/location/5403690/indonesia/toto188
http://www.travelful.net/location/5403692/indonesia/ug212
https://500px.com/p/mposlotc
https://500px.com/p/polisislota
https://500px.com/p/slotgacorv
https://500px.com/p/toto188d
https://500px.com/p/ug212
https://8tracks.com/mposlot-113233
https://8tracks.com/slot-g-104827
https://8tracks.com/toto188d
https://8tracks.com/ug-d
https://ameblo.jp/mposlotc/entry-12845051521.html
https://ameblo.jp/polisislota/entry-12845053019.html
https://ameblo.jp/slotgacorv/entry-12845051892.html
https://ameblo.jp/toto188d/entry-12845052344.html
https://ameblo.jp/ug212/entry-12845052777.html
https://camp-fire.jp/profile/mposlotc
https://camp-fire.jp/profile/polisislot
https://camp-fire.jp/profile/slotgacorv
https://camp-fire.jp/profile/toto188d
https://camp-fire.jp/profile/ug212
https://comicvine.gamespot.com/profile/mposlotc/
https://comicvine.gamespot.com/profile/polisislot/
https://comicvine.gamespot.com/profile/slotgacorv/
https://comicvine.gamespot.com/profile/toto188d/
https://comicvine.gamespot.com/profile/ug212
https://confengine.com/user/fuku-fakin
https://confengine.com/user/mposlot
https://confengine.com/user/slot-gacor-1-1-1
https://confengine.com/user/toto-satu-delapan-delapan
https://confengine.com/user/ug-duax
https://digitaltibetan.win/wiki/User:Mposlotc
https://disqus.com/by/disqus_5a8PuMtsx9/about/
https://disqus.com/by/disqus_AX9Nw9sP01/about/
https://disqus.com/by/disqus_BkE2cqZsTn/about/
https://disqus.com/by/mposlotc/about/
https://disqus.com/by/toto188/about/
https://dreevoo.com/profile.php?pid=620969
https://dreevoo.com/profile.php?pid=620971
https://dreevoo.com/profile.php?pid=620972
https://dreevoo.com/profile.php?pid=620973
https://dreevoo.com/profile.php?pid=620974
https://fairygodboss.com/users/profile/8Ca4ukgfvK/Slot-Gacor
https://fairygodboss.com/users/profile/a1WD3XjNXj/ug-duax
https://fairygodboss.com/users/profile/bFIJ0ORDao/Mposlot
https://fairygodboss.com/users/profile/QSNDCW7IXc/Toto-Satu-delapan-delapan
https://fairygodboss.com/users/profile/UO0tYoBgvk/fuku-fakin
https://files.fm/fukufakin
https://files.fm/mposlot05
https://files.fm/slotgacorka
https://files.fm/totosatudelapandelapan
https://files.fm/ug212ug212
https://folkd.com/profile/mposlotc
https://folkd.com/profile/slotgacorv
https://folkd.com/profile/toto188c
https://folkd.com/profile/ug212ug212
https://forum.instube.com/u/mposlotc/
https://forum.instube.com/u/polisislot
https://forum.instube.com/u/slotgacorv
https://forum.instube.com/u/toto188d
https://forum.instube.com/u/ug212
https://forum.liquidbounce.net/user/fuku-fakin
https://forum.liquidbounce.net/user/mposlot
https://forum.liquidbounce.net/user/slot-gacor
https://forum.liquidbounce.net/user/toto-satu-delapan-delapan
https://forum.liquidbounce.net/user/ug-duax
https://forum.reallusion.com/Users/3166597/mposlot05
https://forum.reallusion.com/Users/3166598/slotgacorka
https://forum.reallusion.com/Users/3166599/totosatudelapandelapan
https://forum.reallusion.com/Users/3166600/ug212ug212
https://forum.reallusion.com/Users/3166601/fukufakin
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656726
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656727&sid=07df56c9807cf8fd1aebd2ca1da0a34e
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656728&sid=641770159a7fbc034aaccc9b6d8b7a16
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656729&sid=d1f610fb39fa1c3cd74e2cc21162aaff
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656730
https://foxsheets.com/UserProfile/tabid/57/userId/181512/Default.aspx
https://foxsheets.com/UserProfile/tabid/57/userId/181513/Default.aspx
https://foxsheets.com/UserProfile/tabid/57/userId/181515/Default.aspx
https://foxsheets.com/UserProfile/tabid/57/userId/181516/Default.aspx
https://free-4776125.webadorsite.com/
https://free-4776140.webadorsite.com/
https://free-4776162.webadorsite.com/
https://free-4776209.webadorsite.com/
https://free-4776222.webadorsite.com/
https://freeicons.io/profile/607591
https://freeicons.io/profile/607593
https://freeicons.io/profile/607595
https://freeicons.io/profile/607596
https://freeicons.io/profile/607597
https://git.lumine.io/mposlotc
https://git.lumine.io/polisislota
https://git.lumine.io/slotgacorv
https://git.lumine.io/toto188c
https://git.lumine.io/ug212
https://github.com/mposlotc
https://github.com/polisislota
https://github.com/slotgacorv
https://github.com/toto188c
https://github.com/ug212
https://hashnode.com/@mposlotc
https://hashnode.com/@slotgacorv
https://hashnode.com/@toto188d
https://hashnode.com/@ug212
https://idea.informer.com/users/mposlotc/?what=personal
https://idea.informer.com/users/polisislot/?what=personal
https://idea.informer.com/users/slotgacorv/?what=personal
https://idea.informer.com/users/toto188d/?what=personal
https://idea.informer.com/users/ug212/?what=personal
https://issuu.com/mposlotc
https://issuu.com/polisislota
https://issuu.com/slotgacorv
https://issuu.com/toto188c
https://issuu.com/ug212
https://lab.quickbox.io/mposlotc
https://lab.quickbox.io/polisislot
https://lab.quickbox.io/slotgacorv
https://lab.quickbox.io/toto188d
https://lab.quickbox.io/ug212
https://leetcode.com/fukufakin/
https://leetcode.com/mposlot05/
https://leetcode.com/slotgacorka/
https://leetcode.com/totosatudelapandelapan/
https://leetcode.com/ug212ug212/
https://list.ly/mposlot-1/lists
https://list.ly/polisislota/lists
https://list.ly/slot-gacor-10/lists
https://list.ly/toto188d/lists
https://list.ly/ug212/lists
https://moparwiki.win/wiki/User:Mposlotc
https://moparwiki.win/wiki/User:Polisislot
https://moparwiki.win/wiki/User:Slotgacorv
https://moparwiki.win/wiki/User:Toto188d
https://moparwiki.win/wiki/User:Ug212
https://moz.com/community/q/user/mposlotc
https://moz.com/community/q/user/polisislota
https://moz.com/community/q/user/toto188d
https://moz.com/community/q/user/toto188v
https://moz.com/community/q/user/ug212
https://mpo-slot.gitbook.io/mpo-slot/
https://my.archdaily.com/us/@mpo-slot-1
https://my.archdaily.com/us/@polisi-slot-1
https://my.archdaily.com/us/@slot-gacor-40
https://my.archdaily.com/us/@toto188-1
https://my.archdaily.com/us/@ug212
https://myopportunity.com/profile/mpo-slot-mpo-slot/nw
https://myopportunity.com/profile/polisi-slot-polisi-slot/nw
https://myopportunity.com/profile/slot-gacor-slot-gacor/nw
https://myopportunity.com/profile/toto188-toto188/nw
https://myopportunity.com/profile/ug212-ug212/nw
https://openlibrary.org/people/fuku_fakin
https://openlibrary.org/people/mposlot519
https://openlibrary.org/people/slot_gacor169
https://openlibrary.org/people/toto_satu_delapan_delapan
https://openlibrary.org/people/ug_duax
https://pantip.com/profile/8043868#topics
https://pantip.com/profile/8043874#topics
https://pantip.com/profile/8043878#topics
https://pantip.com/profile/8043879#topics
https://pantip.com/profile/8043880#topics
https://pbase.com/mposlotc
https://pbase.com/polisislota
https://pbase.com/slotgacorv/slot_gacor
https://pbase.com/toto188d/
https://pbase.com/ug212
https://peatix.com/user/21480953/view
https://peatix.com/user/21480965/view
https://peatix.com/user/21480966/view
https://peatix.com/user/21480967/view
https://peatix.com/user/21480969/view
https://pinshape.com/users/3762425-slotgacorv#designs-tab-open
https://pinshape.com/users/3762464-mposlotc#designs-tab-open
https://pinshape.com/users/3762470-toto188d#designs-tab-open
https://pinshape.com/users/3762479-ug212#designs-tab-open
https://pinshape.com/users/3762495-polisislot#designs-tab-open
https://polisi-slot.gitbook.io/polisi-slot/
https://polisislot.xobor.de/u1_polisislot.html
https://profile.ameba.jp/ameba/mposlotc/
https://profile.ameba.jp/ameba/polisislota/
https://profile.ameba.jp/ameba/slotgacorv/
https://profile.ameba.jp/ameba/toto188d/
https://profile.ameba.jp/ameba/ug212/
https://profile.hatena.ne.jp/mposlotc/
https://profile.hatena.ne.jp/slotgacorv/
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215285
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215287
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215288
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215289
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215291
https://qiita.com/mposlotc
https://qiita.com/polisislota
https://qiita.com/slotgacorv
https://qiita.com/toto188d
https://qiita.com/ug212
https://replit.com/@fukufakin
https://replit.com/@mposlotc
https://replit.com/@slotgacorv
https://replit.com/@totosatudelapan
https://replit.com/@ug212ug212
https://roomstyler.com/users/mposlotc
https://roomstyler.com/users/polisislot
https://roomstyler.com/users/slotgacorv
https://roomstyler.com/users/toto188d
https://roomstyler.com/users/ug212
https://rufox.ru/mposlotc/info
https://rufox.ru/polisislot/info/
https://rufox.ru/slotgacorv/info/
https://rufox.ru/toto188d/info/
https://rufox.ru/ug212/info/
https://slot-gacor-6.gitbook.io/slot-gacor/
https://slotgacorv.xobor.de/u1_slotgacorv.html
https://store.nexodyne.com/member.php?u=131663
https://store.nexodyne.com/member.php?u=131664
https://store.nexodyne.com/member.php?u=131665
https://store.nexodyne.com/member.php?u=131667
https://store.nexodyne.com/member.php?u=131668
https://toto188.gitbook.io/toto188/
https://toto188.xobor.de/u1_toto.html
https://ug212.gitbook.io/ug212/
https://ug212.xobor.de/u1_ug.html
https://unsplash.com/@mposlotc
https://unsplash.com/@polisislota
https://unsplash.com/@slotgacorv
https://unsplash.com/@toto188c
https://unsplash.com/@ug212
https://usahatotos.xobor.de/u1_usahatoto.html
https://wmart.kz/forum/user/153919/
https://wmart.kz/forum/user/153922/
https://wmart.kz/forum/user/153923/
https://wmart.kz/forum/user/153924/
https://wmart.kz/forum/user/153925/
https://www.4shared.com/u/8N4B_BaP/ug212ug212.html
https://www.4shared.com/u/Ko0m8QCy/mposlot05.html
https://www.4shared.com/u/NGl0UC20/totosatudelapandelapan.html
https://www.4shared.com/u/QdzbeWzQ/fukufakin.html
https://www.4shared.com/u/r7jgw86U/slotgacorka.html
https://www.anobii.com/en/014c29cfbd998b9909/profile/activity
https://www.anobii.com/en/0156c15f67137892f4/profile/activity
https://www.anobii.com/en/015e726f3e57361831/profile/activity
https://www.anobii.com/en/01b3c167113fe71a92/profile/activity
https://www.anobii.com/en/01cc305ad1b86fca40/profile/activity
https://www.atlasobscura.com/users/mposlotc
https://www.atlasobscura.com/users/polisislot
https://www.atlasobscura.com/users/slotgacorv
https://www.atlasobscura.com/users/toto188d
https://www.atlasobscura.com/users/ug212
https://www.beatstars.com/fukufakin
https://www.beatstars.com/mposlot05
https://www.beatstars.com/slotgacorka
https://www.beatstars.com/totosatudelapandelapan
https://www.beatstars.com/ug212ug212
https://www.bitchute.com/channel/jL0ee9saCQWd/
https://www.bitchute.com/channel/L0pS1tKroPIx/
https://www.bitchute.com/channel/pxlMYs6tCiKL/
https://www.bitchute.com/channel/x2yYaKj4CS7M/
https://www.bitchute.com/channel/yLYJg7OdWub2/
https://www.blurb.com/user/mposlotc
https://www.blurb.com/user/slotgacorv
https://www.blurb.com/user/toto188d
https://www.blurb.com/user/ug212
https://www.chordie.com/forum/profile.php?id=1905934
https://www.chordie.com/forum/profile.php?id=1905938
https://www.chordie.com/forum/profile.php?id=1905940
https://www.chordie.com/forum/profile.php?id=1905944
https://www.chordie.com/forum/profile.php?section=identity&id=1905943
https://www.colcampus.com/courses/96084
https://www.colcampus.com/courses/96085
https://www.colcampus.com/courses/96086
https://www.colcampus.com/courses/96087
https://www.colcampus.com/courses/96088
https://www.coursera.org/learner/mposlot
https://www.coursera.org/learner/polisislot
https://www.coursera.org/learner/slot-gacor
https://www.coursera.org/learner/toto188
https://www.coursera.org/learner/ug212
https://www.dermandar.com/user/mposlotc/
https://www.dermandar.com/user/polisislot/
https://www.dermandar.com/user/slotgacorv/
https://www.dermandar.com/user/toto188c/
https://www.dermandar.com/user/ug212/
https://www.empowher.com/users/mposlotc
https://www.empowher.com/users/polisislot
https://www.empowher.com/users/slotgacorv
https://www.empowher.com/users/toto188d
https://www.empowher.com/users/ug212
https://www.facer.io/u/mposlotc
https://www.facer.io/u/polisislota
https://www.facer.io/u/slotgacorv
https://www.facer.io/u/toto188d
https://www.facer.io/u/ug212
https://www.goodreads.com/user/show/176619551-mpo-slot
https://www.goodreads.com/user/show/176619634-slot-gacor
https://www.goodreads.com/user/show/176619668-toto188
https://www.goodreads.com/user/show/176619706-ug212
https://www.goodreads.com/user/show/176619746-polisi-slot
https://www.gta5-mods.com/users/mposlotc
https://www.gta5-mods.com/users/polisislot
https://www.gta5-mods.com/users/slotgacorv
https://www.gta5-mods.com/users/toto188d
https://www.gta5-mods.com/users/ug212
https://www.hulkshare.com/mposlotc
https://www.hulkshare.com/polisislota
https://www.hulkshare.com/slotgacorv
https://www.hulkshare.com/toto188c
https://www.hulkshare.com/ug212
https://www.kickstarter.com/profile/mposlotc/about
https://www.kickstarter.com/profile/polisislota/about
https://www.kickstarter.com/profile/slotgacorv/about
https://www.kickstarter.com/profile/toto188/about
https://www.kickstarter.com/profile/ug212/about
https://www.mediafire.com/view/2l3neum1kh3jgfg/bagibagi.png/file
https://www.mediafire.com/view/9g0cxyt34vn9lyw/bagus-1.png/file
https://www.mediafire.com/view/cxdgbm4fh7jweo8/fullhd.png/file
https://www.mediafire.com/view/qk0l7jxfy54sady/mopie.png/file
https://www.metooo.io/u/mposlotc
https://www.metooo.io/u/polisislot
https://www.metooo.io/u/slotgacorv
https://www.metooo.io/u/toto188
https://www.metooo.io/u/ug212
https://www.notebook.ai/users/747492
https://www.notebook.ai/users/747493
https://www.notebook.ai/users/747494
https://www.notebook.ai/users/747496
https://www.notebook.ai/users/747498
https://www.noteflight.com/profile/304226fe070d2952190e8bf2314b119858a5bb26
https://www.noteflight.com/profile/703ac931736ccaeb59e9a41a6a1caf5b4098b762
https://www.noteflight.com/profile/7c8bcdd6add9d079a47247b46091cdc97c7cba84
https://www.noteflight.com/profile/fb24e53674e02b14482f88a904224e50d8fd79a9
https://www.noteflight.com/profile/fe09bf6d85475fbf5d594dc6739cda2b18f9006c
https://www.pling.com/u/mposlotc/
https://www.pling.com/u/slotgacorv/
https://www.pling.com/u/toto188c/
https://www.pling.com/u/ug212/
https://www.projectnoah.org/users/mposlotc
https://www.projectnoah.org/users/polisislot
https://www.projectnoah.org/users/slotgacorv
https://www.projectnoah.org/users/toto188
https://www.projectnoah.org/users/ug212
https://www.pubpub.org/user/mpo-slot-mpo-slot
https://www.pubpub.org/user/polisi-slot-polisi-slot
https://www.pubpub.org/user/slot-gacor-slot-gacor-2
https://www.pubpub.org/user/toto188-toto188
https://www.pubpub.org/user/ug212-ug212
https://www.quora.com/profile/Fuku-Fakin/
https://www.quora.com/profile/Mposlot-3
https://www.quora.com/profile/Slot-Gacor-137
https://www.quora.com/profile/Toto-Satu-Delapan-Delapan
https://www.quora.com/profile/Ug-Duax
https://www.rctech.net/forum/members/mposlotc-359720.html
https://www.rctech.net/forum/members/slotgacorv-359722.html
https://www.rctech.net/forum/members/toto188-359723.html
https://www.rctech.net/forum/members/ug212-359724.html
https://www.rctech.net/forum/members/ug212b-359725.html
https://www.shadowera.com/member.php?140655-mposlotc
https://www.shadowera.com/member.php?140656-slotgacorv
https://www.shadowera.com/member.php?140657-toto188d
https://www.shadowera.com/member.php?140658-ug212
https://www.shadowera.com/member.php?140659-polisislot
https://www.spinattic.com/fukuf
https://www.spinattic.com/mposlot1
https://www.spinattic.com/slotg
https://www.spinattic.com/totos
https://www.spinattic.com/ugd
https://www.theverge.com/users/mposlotc
https://www.theverge.com/users/polisislota
https://www.theverge.com/users/slotgacorv
https://www.theverge.com/users/toto188d
https://www.theverge.com/users/ug212
https://www.twitch.tv/mposlotc
https://www.twitch.tv/polisislota
https://www.twitch.tv/slotgacorv
https://www.twitch.tv/toto188d
https://www.twitch.tv/ug212ug212
https://www.vaca-ps.org/mposlotc
https://www.vaca-ps.org/polisislot
https://www.vaca-ps.org/slotgacorv
https://www.vaca-ps.org/toto188c
https://www.vaca-ps.org/ug212
https://www.veoh.com/users/mposlotc
https://www.veoh.com/users/polisislot
https://www.veoh.com/users/slotgacorv
https://www.veoh.com/users/toto188d
https://www.veoh.com/users/ug212
https://www.walkscore.com/people/120767589734/mpo-slot
https://www.walkscore.com/people/152804950174/toto188
https://www.walkscore.com/people/284307565865/polisi-slot
https://www.walkscore.com/people/310849878455/ug212
https://www.walkscore.com/people/327930622667/slot-gacor
https://www.wattpad.com/user/mposlotc
https://www.wattpad.com/user/polisislota
https://www.wattpad.com/user/slotgacorv
https://www.wattpad.com/user/toto188c
https://www.wattpad.com/user/ug212ug212
https://www.zazzle.com/mbr/238153728540930206
https://www.zazzle.com/mbr/238411283381142353
https://www.zazzle.com/mbr/238664803082939517
https://www.zazzle.com/mbr/238838645247432546
https://www.zazzle.com/mbr/238891426508724865
Magnificent beat ! I would like to apprentice while you amend
your website, how can i subscribe for a blog site? The account helped me a acceptable deal.
I had been tiny bit acquainted of this your broadcast provided
bright clear concept
Hi, all is going fine here and ofcourse every one is sharing data, that’s truly excellent, keep up writing.
I blog frequently and I genuinely appreciate your information. This article
has really peaked my interest. I am going to book
mark your website and keep checking for new information about once a week.
I opted in for your Feed as well.
Hi to all, how is all, I think every one is getting more from this web site,
and your views are nice for new people.
What’s up it’s me, I am also visiting this web site regularly, this website is actually nice and the
viewers are actually sharing good thoughts.
Hi there to all, how is all, I think every one is getting more from
this site, and your views are good in favor of new visitors.
I read this piece of writing fully concerning the difference of latest and preceding technologies, it’s amazing article.
Hi there, I desire to subscribe for this web site to take hottest updates,
so where can i do it please help.
Excellent blog you have here but I was wanting to know if you knew of any forums that cover the same topics talked
about in this article? I’d really like to be a part of group where I can get feedback from other experienced individuals that share the same interest.
If you have any suggestions, please let me know.
Thank you!
Jangan Main di tempat ini, Agen Miskin
І am really thankful to the owner of this wеbsite who has shared this great post at at this
time.
I don’t even know the way I finished up here, but I believed this post was
good. I do not recognise who you are however certainly you’re going
to a famous blogger in the event you aren’t already. Cheers!
Your mode of describing the whole thing in this paragraph is genuinely fastidious, every one can effortlessly
understand it, Thanks a lot.
Woah! I’m really enjoying the template/theme of this site.
It’s simple, yet effective. A lot of times it’s tough to get that “perfect balance” between user friendliness and visual appearance.
I must say you’ve done a superb job with this.
In addition, the blog loads very quick for me on Opera.
Superb Blog!
We’re a gaggle of volunteers and opening a new scheme in our community.
Your website provided us with useful information to work on. You have done a formidable job and our entire community might be grateful
to you.
Hey there! I know this is kinda off topic but
I’d figured I’d ask. Would you be interested in exchanging links or maybe guest writing a blog post or vice-versa?
My blog goes over a lot of the same topics as yours and I
think we could greatly benefit from each other. If you are interested feel free to send me an e-mail.
I look forward to hearing from you! Superb blog by the way!
I think everything said made a lot of sense.
But, think on this, what if you were to create a killer headline?
I mean, I don’t wish to tell you how to run your blog,
but what if you added a headline that makes people want more?
I mean Pontine Crossing Tract (pct) | Whole Brain Protocol for Tractography with Empirical MRI is kinda
vanilla. You could peek at Yahoo’s front page and note how they create post titles to get people to click.
You might add a video or a pic or two to get readers
excited about everything’ve got to say. In my opinion, it might make your posts a little bit more interesting.
Yes! Finally something about balmorex.
Asking questions are in fact good thing if you are not understanding something entirely, however this post
offers pleasant understanding yet.
I know this website provides quality dependent content and extra stuff, is there any other website which gives these kinds of
stuff in quality?
Thanks for any other magnificent article. Where else could anyone get that type of
information in such a perfect means of writing? I’ve a presentation next week, and I am at the look for such info.
Just desire to say your article is as astonishing. The clarity for your post is simply
excellent and i can think you are knowledgeable in this subject.
Well together with your permission allow me to clutch your RSS feed to stay updated with forthcoming post.
Thanks one million and please continue the rewarding work.ラブドール エロ
Hi, i read your blog from time to time and i own a similar one
and i was just wondering if you get a lot of
spam feedback? If so how do you reduce it, any plugin or anything you can suggest?
I get so much lately it’s driving me insane so any help
is very much appreciated.
I am curious to find out what blog system you have been using?
I’m having some small security problems with my latest blog and I’d
like to find something more safe. Do you have
any recommendations?
I used to be recommended this blog by means of my
cousin. I’m now not sure whether this post is written by
way of him as no one else recognize such distinctive about my problem.
You’re incredible! Thanks!
Thank you for every other great article. Where else may just anybody get that kind of information in such a perfect approach of
writing? I’ve a presentation subsequent week, and I’m on the
search for such info.
With havin so much written content do you ever
run into any issues of plagorism or copyright infringement?
My site has a lot of completely unique content I’ve either created myself
or outsourced but it seems a lot of it is popping it up all over the internet without
my permission. Do you know any ways to help stop content from being stolen? I’d truly appreciate it.
What’s up, yup this paragraph is truly fastidious and I have learned lot of things from
it regarding blogging. thanks.
These are actually wonderful ideas in on the topic of blogging.
You have touched some pleasant factors here.
Any way keep up wrinting.
Wonderful goods from you, man. I have be aware your stuff previous to and you’re just extremely excellent.
I really like what you have obtained here, certainly
like what you’re saying and the way wherein you assert it.
You’re making it entertaining and you continue to take care of to stay it smart.
I can not wait to read much more from you. This is really a
wonderful site.
It is really a great and useful piece of information. I am glad that you shared this useful information with us.
Please keep us informed like this. Thank you for sharing.
I’ll immediately seize your rss as I can not find your e-mail
subscription link or e-newsletter service. Do you have any?
Please let me know so that I could subscribe. Thanks.
You could definitely see your expertise within the article
you write. The sector hopes for more passionate writers such
as you who aren’t afraid to say how they believe.
Always follow your heart.
Very great post. I just stumbled upon your blog and wished to mention that I have really enjoyed browsing
your weblog posts. After all I’ll be subscribing to your
feed and I’m hoping you write again very soon!
Hello! I know this is kinda off topic however I’d figured I’d ask.
Would you be interested in exchanging links or maybe guest authoring a blog article or vice-versa?
My site addresses a lot of the same topics as yours and I feel we could greatly benefit from each other.
If you happen to be interested feel free to shoot me
an e-mail. I look forward to hearing from you! Terrific blog by the way!
This web site truly has all the info I needed about this subject and didn’t know who to ask.
I seriously love your blog.. Pleasant colors &
theme. Did you develop this site yourself?
Please reply back as I’m hoping to create my own personal website and
would love to learn where you got this from or what the theme is named.
Appreciate it!
Hi there exceptional website! Does running a blog such as this take a great deal of work?
I’ve no knowledge of coding however I was hoping
to start my own blog in the near future. Anyway, should you have any suggestions or techniques for new blog owners please share.
I know this is off topic however I just had to ask. Many thanks!
First off I want to say terrific blog! I had a quick question which I’d
like to ask if you don’t mind. I was curious to know how you center yourself and clear your
mind prior to writing. I have had a difficult time clearing
my mind in getting my ideas out there. I do enjoy writing
however it just seems like the first 10 to 15 minutes are usually wasted simply just trying to figure out how to begin. Any suggestions
or tips? Cheers!
Today, I went to the beach front with my kids.
I found a sea shell and gave it to my 4 year old daughter
and said “You can hear the ocean if you put this to your ear.”
She put the shell to her ear and screamed.
There was a hermit crab inside and it pinched her ear.
She never wants to go back! LoL I know this is completely off topic but I had to tell someone!
Attractive component to content. I just stumbled upon your blog
and in accession capital to claim that I acquire in fact enjoyed account your weblog
posts. Anyway I will be subscribing in your feeds and even I achievement you get entry to persistently fast.
Heya i’m for the primary time here. I came across this board
and I find It truly useful & it helped me out much.
I hope to provide something back and help others such as you helped me.
As the admin of this web page is working, no hesitation very quickly it will be renowned, due
to its feature contents.
Currently it seems like WordPress is the best blogging platform out there right now.
(from what I’ve read) Is that what you’re using on your blog?
Way cool! Some extremely valid points! I appreciate you writing
this post and the rest of the site is extremely good.
Heya i’m for the first time here. I found this board and I
find It truly useful & it helped me out much. I hope to give something back and aid others
like you aided me.
I think the admin of this site is truly working hard in favor of his web page, for the
reason that here every information is quality based data.
Keep on writing, great job!
My brother suggested I might like this blog. He was entirely right.
This post actually made my day. You cann’t imagine simply how much time I
had spent for this info! Thanks!
I know this if off topic but I’m looking into starting my own blog and was curious what all is needed to get set up?
I’m assuming having a blog like yours would cost a pretty penny?
I’m not very web savvy so I’m not 100% certain. Any recommendations or advice would be greatly appreciated.
Kudos
I’ve been exploring for a bit for any high quality articles or blog posts in this
kind of area . Exploring in Yahoo I finally stumbled upon this web site.
Studying this info So i am glad to exhibit that I have an incredibly excellent
uncanny feeling I found out exactly what I needed.
I such a lot indubitably will make certain to don?t fail to remember this web site
and give it a look regularly.
Nice replies in return of this issue with real arguments and explaining all concerning
that.
Hello! This is my 1st comment here so I just wanted to give a quick shout out and tell you I genuinely enjoy reading through your blog posts.
Can you recommend any other blogs/websites/forums that
cover the same subjects? Thanks a lot!
Hello very nice blog!! Man .. Beautiful ..
Superb .. I will bookmark your website and take the feeds additionally?
I am glad to find so many useful info right here within the post, we
want develop extra strategies in this regard,
thanks for sharing. . . . . .
Appreciate this post. Let me try it out.
I was wondering if you ever considered changing the layout of your blog?
Its very well written; I love what youve got to say. But maybe you could a
little more in the way of content so people could connect with
it better. Youve got an awful lot of text for only having 1 or 2 images.
Maybe you could space it out better?
I am curious to find out what blog system you’re utilizing?
I’m experiencing some small security problems with my latest
site and I’d like to find something more secure.
Do you have any recommendations?
A fascinating discussion is definitely worth comment.
There’s no doubt that that you need to write more on this subject matter, it may not be a taboo matter but generally
people don’t speak about these issues. To the next! Cheers!!
I’m truly enjoying the design and layout of your website.
It’s a very easy on the eyes which makes it much more enjoyable for me
to come here and visit more often. Did you hire out a designer
to create your theme? Fantastic work!
Good day! Do you know if they make any plugins to assist with
Search Engine Optimization? I’m trying to get my blog
to rank for some targeted keywords but I’m not seeing very good success.
If you know of any please share. Appreciate it!
I’ve observed that in the world the present day, video games are classified as the latest phenomenon with children of all ages. Periodically it may be difficult to drag your family away from the video games. If you want the best of both worlds, there are numerous educational gaming activities for kids. Thanks for your post.
Woah! I’m really digging the template/theme of this website.
It’s simple, yet effective. A lot of times it’s very hard to get that “perfect balance” between usability and
visual appearance. I must say that you’ve done a fantastic job with this.
Also, the blog loads very quick for me on Opera. Outstanding Blog!
I’m not that much of a online reader to be
honest but your blogs really nice, keep it up! I’ll go ahead and bookmark your
website to come back later. Cheers
Hi there, You have done an incredible job. I will certainly digg
it and personally suggest to my friends. I am confident they’ll be benefited from this web
site.
Hello! I could have sworn I’ve been to this blog before but after
reading through some of the post I realized it’s new to me.
Anyhow, I’m definitely delighted I found it and I’ll be bookmarking and checking back often!
I simply couldn’t leave your web site before suggesting that I
actually loved the standard information a person provide in your visitors?
Is going to be back ceaselessly in order to check up on new posts
Appreciate the recommendation. Will try it out.
When I originally left a comment I seem to have clicked the -Notify me when new comments are added- checkbox and now whenever a comment is added I recieve 4 emails with the exact same comment.
Is there an easy method you can remove me from that service?
Thanks!
I am sure this post has touched all the internet viewers,
its really really nice piece of writing on building up new weblog.
Hi everybody, here every person is sharing these familiarity, therefore it’s
good to read this webpage, and I used to pay a quick visit this web site every day.
I get pleasure from, cause I found exactly what I used to be taking a look for.
You’ve ended my four day long hunt! God Bless you man. Have a great day.
Bye
Heya i’m for the primary time here. I came across this
board and I to find It really useful & it helped me out much.
I hope to give something back and aid others like
you aided me.
I loved as much as you will receive carried out right here.
The sketch is tasteful, your authored subject matter stylish.
nonetheless, you command get bought an shakiness over that you wish be delivering the following.
unwell unquestionably come more formerly again since exactly the
same nearly a lot often inside case you shield this hike.
I’m extremely impressed together with your writing skills as well as with the layout
on your blog. Is that this a paid theme or did you customize it your self?
Either way stay up the nice high quality writing, it’s uncommon to see a
great weblog like this one today..
Thanks, Excellent stuff.
Take a look at my web page Printer Supplies (https://southernvapeshack.com/)
I like the helpful information you supply for your articles.
I’ll bookmark your blog and test once more right here regularly.
I’m somewhat sure I’ll be told plenty of new stuff right here!
Best of luck for the next!
The name of the writer is Robena and she totally digs that
term. I am currently a payroll clerk and it’s something I
really like. Maryland is largest she loves most and she doesn’t intend on changing the idea.
To cycle is one of the issues i love greatest.
Great blog here! Also your site loads up fast! What host are you using?
Can I get your affiliate link to your host? I wish my site loaded up as fast as yours lol
The state lottery that has been going through delays seemed to be not really known to be. How complete you possess pleasure with Powerball? The lure acquired happen to be really scheduled for 11:30pn EST (04:30 GMT) on Tuesday, but instead received area on Wednesday morning at around 9:00an (14:00 GMT). No-one mentioned the jackpot in Saturday’s acquire, but there possess happen to be 16 entrance go related the five main information to get $1m each. https://educationpd.com/question/can-you-play-the-lottery-online-in-the-u-s-without-breaking-any-laws/Only one some various other Powerball jackpot landed at 41 consecutive pictures. The probabilities of getting the jackpot looked to become one in 292.2 million, Powerball claims.
Howdy! I know this is kinda off topic but I was wondering which blog platform are you using for this site?
I’m getting tired of WordPress because I’ve had issues
with hackers and I’m looking at alternatives for another platform.
I would be awesome if you could point me in the direction of a good platform.
Fabulous, what a weblog it is! This weblog gives useful data to us, keep it up.
Howdy! I know this is kinda off topic nevertheless I’d figured I’d ask.
Would you be interested in exchanging links or maybe guest authoring a blog article or vice-versa?
My site goes over a lot of the same subjects as yours and
I believe we could greatly benefit from each other. If you might be interested
feel free to send me an email. I look forward to hearing
from you! Wonderful blog by the way!
Thanks very interesting blog!
Asking questions are in fact good thing if you are not understanding
anything completely, except this piece of writing gives fastidious understanding even.
In fact no matter if someone doesn’t be aware of afterward its up to
other viewers that they will assist, so here it occurs.
I really like what you guys are up too. Such clever work
and coverage! Keep up the wonderful works guys I’ve added you guys to our
blogroll.
It’s impressive that you are getting thoughts from
this piece of writing as well as from our argument made at this time.
This is really interesting, You’re an excessively
professional blogger. I’ve joined your feed and look ahead to seeking more of your fantastic post.
Also, I have shared your site in my social networks
Hello would you mind letting me know which hosting company you’re using?
I’ve loaded your blog in 3 completely different browsers
and I must say this blog loads a lot faster then most.
Can you suggest a good internet hosting provider at a honest price?
Thank you, I appreciate it!
My partner and I absolutely love your blog and find the majority of your post’s to be exactly what I’m looking for.
Do you offer guest writers to write content to suit your needs?
I wouldn’t mind writing a post or elaborating on many
of the subjects you write with regards to here. Again, awesome website!
If you are looking for new furniture for your home, you may be wondering
where to find the furniture shops in Barnstaple.
Barnstaple is a town in North Devon that has a rich history
and culture, and it also has a variety of furniture shops that cater
to different tastes and budgets. Whether you
are looking for sofas, beds, dining sets, or accessories, you can find them in Barnstaple.
One of the best furniture shops in Barnstaple is
FW Homestores1, a family-run business that has been operating for over 30 years.
FW Homestores offers quality furniture and home accessories at affordable prices, and they have a showroom in Barnstaple where you
can browse their products and get expert advice. FW Homestores also offers free delivery and
installation, and they have a range of finance options to suit
your needs.
FW Homestores has a wide selection of furniture for every room in your home, from living room
to bedroom, from dining room to garden. You can find sofas, armchairs, coffee tables,
TV units, bookcases, sideboards, dining tables,
chairs, benches, beds, mattresses, wardrobes, chests
of drawers, bedside tables, dressing tables, mirrors, lamps, rugs, cushions, throws, wall art, and more.
You can also choose from different styles, such as modern, rustic, industrial,
or bohemian.
Good day! Do you know if they make any plugins to help with Search Engine Optimization? I’m trying to
get my blog to rank for some targeted keywords but I’m not seeing very good success.
If you know of any please share. Appreciate it! You can read similar article here:
Sklep online
I’m not that much of a internet reader to be honest but your blogs really nice,
keep it up! I’ll go ahead and bookmark your website to come back down the road.
Cheers
is an exampl대전동구출장샵e of transla대전동구출장샵tion of indu대전동구출장샵stri
Hi mates, fastidious article and pleasant arguments commented here,
I am actually enjoying by these.
indian sex, indian sex, indian porn, sex videos, indian porn, indian sex, indian porn,
xnxx videos, xnxx, porn hd, indian porn, xxx, sex, xnxx, indian porn, indian xxx, indian porn,
indian sex, xnxx videos, xnxx videos, indian porn,
indian porn, xnxx videos, sex videos, xxx, indian porn, indian porn, porn hd, sex,
sex, xnxx videos, indian porn, xxx, indian xxx, porn videos, indian sex, porn, xxx,
indian xxx, xnxx, indian sex,
xxx,
xxx,
indian porn, indian sex, indian porn, porn hd, indian xxx, indian porn,
xnxx videos, sex, indian sex, xnxx, porn videos, indian sex, xnxx, xnxx videos, xnxx videos, porn,
indian sex,
Hello there! This is my first visit to your blog!
We are a group of volunteers and starting a new initiative in a
community in the same niche. Your blog provided us valuable information to work on. You have done a outstanding job!
Hi there everybody, here every person is sharing these experience, thus it’s nice to read this webpage, and I used to visit this
weblog all the time.
What’s Taking place i am new to this, I stumbled upon this I have found It positively useful and it has
helped me out loads. I’m hoping to contribute & aid other customers like its aided me.
Good job.
Hi I am so excited I found your blog page, I really found you by mistake,
while I was looking on Bing for something else, Anyhow I
am here now and would just like to say cheers for a fantastic post and a all round entertaining blog
(I also love the theme/design), I don’t have time to read it all at
the moment but I have book-marked it and also added your RSS feeds, so when I have time I will be back to read
a lot more, Please do keep up the superb jo.
We are a group of volunteers and starting a new scheme in our community.
Your site offered us with helpful info to work on. You’ve done a formidable job and our entire community shall be grateful to you.
Greetings I am so happy I found your web site, I really
found you by mistake, while I was looking on Digg for something else, Regardless I am
here now and would just like to say kudos for
a incredible post and a all round interesting blog (I also
love the theme/design), I don’t have time to browse it all at the moment but I have saved it and also
added in your RSS feeds, so when I have time I will be back
to read a lot more, Please do keep up the excellent job.
The project can be running an airdrop price $100K, which shall be cut up between ten winners.
Wow! After all I got a website from where I be capable of genuinely get
useful data concerning my study and knowledge.
WOW just what I was searching for. Came here by searching for 中国美女
my webpage – 成人性愛
It’s going to be finish of mine day, but before ending
I am reading this fantastic post to improve my experience.
hey there and thank you for your info – I’ve definitely picked up something new from right here.
I did however expertise a few technical issues using this website, since I experienced to
reload the web site lots of times previous to I could get it to load properly.
I had been wondering if your web hosting is OK?
Not that I’m complaining, but sluggish loading instances times will sometimes affect your placement in google
and can damage your high-quality score if ads and marketing
with Adwords. Well I’m adding this RSS to my e-mail
and could look out for a lot more of your respective intriguing content.
Ensure that you update this again soon.
Hello! I want to start by saying my name – Roseline though I
don’t really like being called like which unfortunately.
What he really enjoys doing is bottle tops
collecting and he is attempting to make it a practice.
Some time ago she thought they would live in North Carolina.
Invoicing has been my day job for a short time but I’ve already went for another a person.
I’m truly enjoying the design and layout of your website.
It’s a very easy on the eyes which makes
it much more enjoyable for me to come here and visit more often. Did you hire out a designer to create your theme?
Superb work!
I read this post fully regarding the comparison of most recent and
earlier technologies, it’s amazing article.
Hey There. I found your weblog the usage of msn. This is a very well written article.
I’ll be sure to bookmark it and return to learn extra of your useful info.
Thank you for the post. I’ll definitely return.
My brother suggested I might like this web site.
He was entirely right. This post truly made my day. You can not
imagine simply how much time I had spent for this information!
Thanks!
Hey There. I found your blog the use of msn. That is a really
neatly written article. I’ll be sure to bookmark it and come back to
read more of your useful information. Thanks for
the post. I’ll definitely return.
At this moment I am going aaay to do my breakfast, once having my breakfast coming yet again to read additional news.
my homepage ADAPTERS (Mariana)
Wow all kinds of amazing material!
Hey there, I think your website might be having browser compatibility issues.
When I look at your blog site in Firefox, it looks fine but
when opening in Internet Explorer, it has some overlapping.
I just wanted to give you a quick heads up! Other then that, fantastic blog!
Hey I know this is off topic but I was wondering if you knew of any widgets I
could add to my blog that automatically tweet my newest twitter
updates. I’ve been looking for a plug-in like
this for quite some time and was hoping maybe you would have some experience with something like this.
Please let me know if you run into anything. I truly
enjoy reading your blog and I look forward to your new updates.
I’m extremely inspired with your writing talents as neatly as with the format in your weblog.
Is this a paid topic or did you customize it yourself? Either way stay up the excellent high quality writing, it is
uncommon to look a nice blog like this one nowadays..
http://www.youtube.com/redirect?event=channel_description&q=https%3A%2F%2FSpinachtree.com%2F
http://cse.google.com.eg/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cn.bing.com/news/apiclick.aspx?COLLCC=1718906003&ref=FexRss&aid=&url=https://Spinachtree.com/
http://images.google.com.sa/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://images.google.at/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://images.google.com/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://images.google.gr/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.com.ua/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://accounts.cancer.org/login?redirectURL=https%3A%2F%2FSpinachtree.com&theme=RFL
https://images.google.tk/url?sa=t&url=http://Spinachtree.com/
http://images.google.ee/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://clients1.google.ki/url?q=http%3A%2F%2FSpinachtree.com
http://clients1.google.td/url?q=https%3A%2F%2FSpinachtree.com
https://website.informer.com/Spinachtree.com
http://clients1.google.at/url?q=https%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.com.ec/url?q=https://Spinachtree.com/
http://b.filmz.ru/presentation/www/delivery/ck.php?ct=1&oaparams=2__bannerid=222__zoneid=2__cb=93494e485e__oadest=https://Spinachtree.com/
http://ava-online.clan.su/go?https://Spinachtree.com/
http://ads.dfiles.eu/click.php?c=1497&z=4&b=1730&r=https://Spinachtree.com/
http://clinica-elit.vrn.ru/cgi-bin/inmakred.cgi?bn=43252&url=Spinachtree.com
http://chat.chat.ru/redirectwarn?https://Spinachtree.com/
http://cm-eu.wargaming.net/frame/?service=frm&project=moo&realm=eu&language=en&login_url=http%3A%2F%2FSpinachtree.com
http://maps.google.com/url?q=https%3A%2F%2FSpinachtree.com%2F
http://audit.tomsk.ru/bitrix/click.php?goto=https://Spinachtree.com/
http://clients1.google.co.mz/url?q=https%3A%2F%2FSpinachtree.com/
http://come-on.rdy.jp/wanted/cgi-bin/rank.cgi?id=11326&mode=link&url=https://Spinachtree.com/
http://images.google.com.ng/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://yumi.rgr.jp/puku-board/kboard.cgi?mode=res_html&owner=proscar&url=Spinachtree.com/&count=1&ie=1
http://images.google.com.au/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://asia.google.com/url?q=http://Spinachtree.com/management.html
http://adapi.now.com/ad/api/act.ashx?a=2&sc=3490&s=30000219&l=1&t=0&c=0&u=https://Spinachtree.com/
https://cse.google.ws/url?q=http://Spinachtree.com/
http://avn.innity.com/click/avncl.php?bannerid=68907&zoneid=0&cb=2&pcu=&url=http%3a%2f%2fSpinachtree.com
https://ross.campusgroups.com/click?uid=51a11492-dc03-11e4-a071-0025902f7e74&r=http://Spinachtree.com/
http://images.google.de/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://tools.folha.com.br/print?url=https%3A%2F%2FSpinachtree.com%2F&site=blogfolha
http://alt1.toolbarqueries.google.com.fj/url?q=https://Spinachtree.com/
http://new.creativecommons.org/choose/results-one?q_1=2&q_1=1&field_commercial=n&field_derivatives=sa&field_jurisdiction=&field_format=Text&field_worktitle=Blog&field_attribute_to_name=Lam%20HUA&field_attribute_to_url=https%3A%2F%2FSpinachtree.com
http://redirect.subscribe.ru/bank.banks
http://www.so-net.ne.jp/search/web/?query=Spinachtree.com&from=rss
http://alt1.toolbarqueries.google.cat/url?q=https://Spinachtree.com/
http://affiliate.awardspace.info/go.php?url=https://Spinachtree.com/
http://writer.dek-d.com/dek-d/link/link.php?out=https%3A%2F%2FSpinachtree.com%2F&title=Spinachtree.com
http://analogx.com/cgi-bin/cgirdir.exe?https://Spinachtree.com/
http://images.google.co.ve/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://ip.chinaz.com/?IP=Spinachtree.com
http://images.google.com.pe/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cmbe-console.worldoftanks.com/frame/?language=en&login_url=http%3A%2F%2FSpinachtree.com&project=wotx&realm=wgcb&service=frm
http://images.google.dk/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://ceramique-et-couleurs.leforum.eu/redirect1/https://Spinachtree.com/
https://ipv4.google.com/url?q=http://Spinachtree.com/management.html
http://cse.google.com.af/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.com.do/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://newsdiffs.org/article-history/Spinachtree.com/
http://cse.google.az/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.ch/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://aganippe.online.fr/lien.php3?url=http%3a%2f%2fSpinachtree.com
http://images.google.fr/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://images.google.co.ve/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.com.do/url?q=https://Spinachtree.com/
http://alt1.toolbarqueries.google.co.mz/url?q=https://Spinachtree.com/
https://www.easyviajar.com/me/link.jsp?site=359&client=1&id=110&url=http://Spinachtree.com/2021/05/29/rwfeds/
http://68-w.tlnk.io/serve?action=click&site_id=137717&url_web=https://Spinachtree.com/
http://alt1.toolbarqueries.google.sk/url?q=https://Spinachtree.com/
http://jump.2ch.net/?Spinachtree.com
http://images.google.co.nz/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://images.google.com.ec/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.com.mx/url?q=https://Spinachtree.com/
http://sitereport.netcraft.com/?url=https%3A%2F%2FSpinachtree.com%2F
http://www.ursoftware.com/downloadredirect.php?url=https%3A%2F%2FSpinachtree.com%2F
http://cse.google.co.mz/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://legal.un.org/docs/doc_top.asp?path=../ilc/documentation/english/a_cn4_13.pd&Lang=Ef&referer=https%3A%2F%2FSpinachtree.com%2F
http://doodle.com/r?url=https%3A%2F%2FSpinachtree.com%2F
http://cse.google.ac/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://78.rospotrebnadzor.ru/news9/-/asset_publisher/9Opz/content/%D0%BF%D0%BE%D1%81%D1%82%D0%B0%D0%BD%D0%BE%D0%B2%D0%BB%D0%B5%D0%BD%D0%B8%D0%B5-%D0%B3%D0%BB%D0%B0%D0%B2%D0%BD%D0%BE%D0%B3%D0%BE-%D0%B3%D0%BE%D1%81%D1%83%D0%B4%D0%B0%D1%80%D1%81%D1%82%D0%B2%D0%B5%D0%BD%D0%BD%D0%BE%D0%B3%D0%BE-%D1%81%D0%B0%D0%BD%D0%B8%D1%82%D0%B0%D1%80%D0%BD%D0%BE%D0%B3%D0%BE-%D0%B2%D1%80%D0%B0%D1%87%D0%B0-%D0%BF%D0%BE-%D0%B3%D0%BE%D1%80%D0%BE%D0%B4%D1%83-%D1%81%D0%B0%D0%BD%D0%BA%D1%82-%D0%BF%D0%B5%D1%82%D0%B5%D1%80%D0%B1%D1%83%D1%80%D0%B3%D1%83-%E2%84%96-4-%D0%BE%D1%82-09-11-2021-%C2%AB%D0%BE-%D0%B2%D0%BD%D0%B5%D1%81%D0%B5%D0%BD%D0%B8%D0%B8-%D0%B8%D0%B7%D0%BC%D0%B5%D0%BD%D0%B5%D0%BD%D0%B8%D0%B8-%D0%B2-%D0%BF%D0%BE%D1%81%D1%82%D0%B0%D0%BD%D0%BE%D0%B2%D0%BB%D0%B5%D0%BD%D0%B8%D0%B5-%D0%B3%D0%BB%D0%B0%D0%B2%D0%BD%D0%BE%D0%B3%D0%BE-%D0%B3%D0%BE%D1%81%D1%83%D0%B4%D0%B0%D1%80%D1%81%D1%82%D0%B2%D0%B5%D0%BD%D0%BD%D0%BE%D0%B3%D0%BE-%D1%81%D0%B0%D0%BD%D0%B8%D1%82%D0%B0%D1%80%D0%BD%D0%BE%D0%B3%D0%BE-%D0%B2%D1%80%D0%B0%D1%87%D0%B0-%D0%BF%D0%BE-%D0%B3%D0%BE%D1%80%D0%BE%D0%B4%D1%83-%D1%81%D0%B0%D0%BD%D0%BA%D1%82-%D0%BF%D0%B5%D1%82%D0%B5%D1%80%D0%B1%D1%83%D1%80%D0%B3%D1%83-%D0%BE%D1%82-12-10-2021-%E2%84%96-3-%C2%BB;jsessionid=FB3309CE788EBDCCB588450F5B1BE92F?redirect=http%3A%2F%2FSpinachtree.com
http://ashspublications.org/__media__/js/netsoltrademark.php?d=Spinachtree.com
http://cse.google.com.ly/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.com.bh/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://pt.tapatalk.com/redirect.php?app_id=4&fid=8678&url=http://Spinachtree.com
http://cse.google.co.tz/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.bing.com/news/apiclick.aspx?ref=FexRss&aid=&tid=9BB77FDA801248A5AD23FDBDD5922800&url=https%3A%2F%2FSpinachtree.com
http://advisor.wmtransfer.com/SiteDetails.aspx?url=Spinachtree.com
http://blogs.rtve.es/libs/getfirma_footer_prod.php?blogurl=https%3A%2F%2FSpinachtree.com
http://images.google.com.br/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://images.google.be/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://aboutus.com/Special:SiteAnalysis?q=Spinachtree.com&action=webPresence
https://brettterpstra.com/share/fymdproxy.php?url=http://Spinachtree.com
http://images.google.com.eg/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://my.myob.com/community/login.aspx?ReturnUrl=https%3A%2F%2FSpinachtree.com%2F
http://traceroute.physics.carleton.ca/cgi-bin/traceroute.pl?function=ping&target=Spinachtree.com
http://cse.google.as/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://d-click.fiemg.com.br/u/18081/131/75411/137_0/82cb7/?url=https://Spinachtree.com/
https://cse.google.ws/url?sa=t&url=http://Spinachtree.com/
http://anonim.co.ro/?Spinachtree.com/
http://id.telstra.com.au/register/crowdsupport?gotoURL=https%3A%2F%2FSpinachtree.com%2F
http://ads.businessnews.com.tn/dmcads2017/www/delivery/ck.php?ct=1&oaparams=2__bannerid=1839__zoneid=117__cb=df4f4d726f__oadest=https://Spinachtree.com/
http://aichi-fishing.boy.jp/?wptouch_switch=desktop&redirect=http%3a%2f%2fSpinachtree.com
http://cse.google.co.th/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://alt1.toolbarqueries.google.ch/url?q=https://Spinachtree.com/
http://images.google.cl/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://clients1.google.com.kh/url?q=https://Spinachtree.com/
http://client.paltalk.com/client/webapp/client/External.wmt?url=http%3A%2F%2FSpinachtree.com%2F
http://clients1.google.com.lb/url?q=https%3A%2F%2FSpinachtree.com/
http://cam4com.go2cloud.org/aff_c?offer_id=268&aff_id=2014&url=https%3A%2F%2FSpinachtree.com
http://creativecommons.org/choose/results-one?q_1=2&q_1=1&field_commercial=n&field_derivatives=sa&field_jurisdiction=&field_format=Text&field_worktitle=Blog&field_attribute_to_name=Lam%20HUA&field_attribute_to_url=https%3A%2F%2FSpinachtree.com%2F
http://images.google.cz/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://cse.google.co.bw/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://r.turn.com/r/click?id=07SbPf7hZSNdJAgAAAYBAA&url=https://Spinachtree.com/
http://images.google.co.uk/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.com.bd/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://clients1.google.ge/url?q=https%3A%2F%2FSpinachtree.com/
http://2chmatome.jpn.org/akb/c_c.php?c_id=267977&url=https://Spinachtree.com/
http://affiliate.suruga-ya.jp/modules/af/af_jump.php?user_id=755&goods_url=https://Spinachtree.com/
http://archive.feedblitz.com/f/f.fbz?goto=https://Spinachtree.com/
http://clients1.google.com.pr/url?sa=i&url=https%3A%2F%2FSpinachtree.com
http://cse.google.com.gt/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://abcclub.cside.com/nagata/link4/link4.cgi?mode=cnt&hp=http%3a%2f%2fSpinachtree.com&no=1027
http://clients1.google.com.fj/url?q=https%3A%2F%2FSpinachtree.com
http://www.justjared.com/flagcomment.php?el=https%3A%2F%2FSpinachtree.com
http://clients1.google.com.gi/url?q=https%3A%2F%2FSpinachtree.com/
http://biz.timesfreepress.com/__media__/js/netsoltrademark.php?d=Spinachtree.com
http://search.babylon.com/imageres.php?iu=https%3A%2F%2FSpinachtree.com%2F
http://clicktrack.pubmatic.com/AdServer/AdDisplayTrackerServlet?clickData=JnB1YklkPTE1NjMxMyZzaXRlSWQ9MTk5MDE3JmFkSWQ9MTA5NjQ2NyZrYWRzaXplaWQ9OSZ0bGRJZD00OTc2OTA4OCZjYW1wYWlnbklkPTEyNjcxJmNyZWF0aXZlSWQ9MCZ1Y3JpZD0xOTAzODY0ODc3ODU2NDc1OTgwJmFkU2VydmVySWQ9MjQzJmltcGlkPTU0MjgyODhFLTYwRjktNDhDMC1BRDZELTJFRjM0M0E0RjI3NCZtb2JmbGFnPTImbW9kZWxpZD0yODY2Jm9zaWQ9MTIyJmNhcnJpZXJpZD0xMDQmcGFzc2JhY2s9MA==_url=https://Spinachtree.com/
http://alt1.toolbarqueries.google.bg/url?q=https://Spinachtree.com/
http://211-75-39-211.hinet-ip.hinet.net/Adredir.asp?url=https://Spinachtree.com/
http://clients1.google.bg/url?q=https://Spinachtree.com/
http://ads2.figures.com/Ads3/www/delivery/ck.php?ct=1&oaparams=2__bannerid=282__zoneid=248__cb=da025c17ff__oadest=https%3a%2f%2fSpinachtree.com
http://clients1.google.gy/url?q=http%3A%2F%2FSpinachtree.com
http://clients1.google.com.ar/url?q=https%3A%2F%2FSpinachtree.com
http://com.co.de/__media__/js/netsoltrademark.php?d=Spinachtree.com
http://clients1.google.com.jm/url?q=https://Spinachtree.com/
http://home.speedbit.com/r.aspx?u=https://Spinachtree.com/
http://cse.google.com.lb/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://blog.bg/results.php?q=%22%2f%3e%3ca+href%3d%22http%3a%2f%2fSpinachtree.com&
http://images.google.com.co/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://clicrbs.com.br/paidcontent/jsp/login.jspx?site=545&url=goo.gl%2Fmaps%2FoQqZFfefPedXSkNc6&previousurl=http%3a%2f%2fSpinachtree.com
http://clients1.google.kg/url?q=https://Spinachtree.com/
http://staging.talentegg.ca/redirect/company/224?destination=https://Spinachtree.com/
http://images.google.co.za/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://clients1.google.com.co/url?q=https%3A%2F%2FSpinachtree.com
http://cse.google.bf/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.com.bz/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.com.cu/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.talgov.com/Main/exit.aspx?url=https%3A%2F%2FSpinachtree.com
http://cse.google.ci/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://blog.ss-blog.jp/_pages/mobile/step/index?u=http%3A%2F%2FSpinachtree.com
https://cse.google.tk/url?q=http://Spinachtree.com/
http://www.siteprice.org/similar-websites/eden-project.org
http://clients1.google.ca/url?q=https://Spinachtree.com/
http://cse.google.cm/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.ae/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://clients1.google.nr/url?q=http%3A%2F%2FSpinachtree.com
http://imaginingourselves.globalfundforwomen.org/pb/External.aspx?url=https://Spinachtree.com/
http://clients1.google.tk/url?q=http%3A%2F%2FSpinachtree.com/
http://alt1.toolbarqueries.google.co.il/url?q=https://Spinachtree.com/
http://theamericanmuslim.org/tam.php?URL=Spinachtree.com
http://images.google.com.tr/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://thaiwebsearch.hypermart.net/cgi/clicko.pl?75&Spinachtree.com
http://colil.dbcls.jp/fct/rdfdesc/usage.vsp?g=https://Spinachtree.com/
https://content.sixflags.com/news/director.aspx?gid=0&iid=72&cid=3714&link=http://Spinachtree.com/2021/05/29/rwfeds/
http://adr.tpprf.ru/bitrix/redirect.php?goto=https://Spinachtree.com/
http://jump.2ch.net/?Spinachtree.com
http://clients1.google.com.do/url?q=https%3A%2F%2FSpinachtree.com
http://c.yam.com/msnews/IRT/r.c?https://Spinachtree.com/
http://clients1.google.com.sa/url?sa=t&url=https%3A%2F%2FSpinachtree.com
https://images.google.tk/url?q=http://Spinachtree.com/
http://cse.google.com.gi/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://ad.sxp.smartclip.net/optout?url=https://Spinachtree.com/&ang_testid=1
http://guru.sanook.com/?URL=https%3A%2F%2FSpinachtree.com%2F
http://clients1.google.com.my/url?sa=i&url=https%3A%2F%2FSpinachtree.com
http://53938.measurementapi.com/serve?action=click&publisher_id=53938&site_id=69748&sub_campaign=g5e_com&url=https://Spinachtree.com/
http://t.me/iv?url=https%3A%2F%2FSpinachtree.com
http://clients1.google.co.ve/url?q=https://Spinachtree.com/
http://ayads.co/click.php?c=735-844&url=https://Spinachtree.com/
http://campaigns.williamhill.com/C.ashx?btag=a_181578b_893c_&affid=1688431&siteid=181578&adid=893&c=&asclurl=https://Spinachtree.com/&AutoR=1
http://www.aomeitech.com/forum/home/leaving?Target=https%3A//Spinachtree.com
http://brandcycle.go2cloud.org/aff_c?offer_id=261&aff_id=1371&url=http%3A%2F%2FSpinachtree.com
http://maps.google.de/url?sa=t&url=https%3A%2F%2FSpinachtree.com
http://webfeeds.brookings.edu/~/t/0/0/~Spinachtree.com
http://images.google.co.za/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://clients1.google.com.ly/url?q=https://Spinachtree.com/
http://www.biblio.com.br/conteudo/Moldura11.asp?link=//Spinachtree.com/2021/05/29/rwfeds/
http://store.veganessentials.com/affiliates/default.aspx?Affiliate=40&Target=https://Spinachtree.com
http://images.google.be/url?sa=t&url=http%3A%2F%2FSpinachtree.com
https://cse.google.nu/url?q=http://Spinachtree.com/
http://images.google.com.sa/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.co.uz/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://www.trainorders.com/discussion/warning.php?forum_id=1&url=http://Spinachtree.com/34zxVq8
http://audio.home.pl/redirect.php?action=url&goto=Spinachtree.com&osCsid=iehkyctnmm
http://images.google.co.uk/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://ac8.i2i.jp/bin/getslink.php?00713417&&&https://Spinachtree.com/
http://analytics.supplyframe.com/trackingservlet/track/?action=name&value3=1561&zone=FCfull_SRP_na_us&url=https://Spinachtree.com/
http://adserver.gadu-gadu.pl/click.asp?adid=2236;url=https://Spinachtree.com/
http://www.responsinator.com/?url=Spinachtree.com
http://cse.google.cf/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://clients1.google.ga/url?q=https%3A%2F%2FSpinachtree.com/
http://m.ok.ru/dk?st.cmd=outLinkWarning&st.rfn=https%3A%2F%2FSpinachtree.com
http://cse.google.co.ck/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://www.wilsonlearning.com/?URL=Spinachtree.com/
http://www.etis.ford.com/externalURL.do?url=https%3A%2F%2FSpinachtree.com%2F
https://board-en.drakensang.com/proxy.php?link=http://Spinachtree.com/
http://builder.hufs.ac.kr/goLink.jsp?url=Spinachtree.com/
http://ahlacarte.vraiforum.com/redirect1/https://Spinachtree.com/
https://images.google.cv/url?q=http://Spinachtree.com/
http://clicktrack.pubmatic.com/AdServer/AdDisplayTrackerServlet?clickData=JnB1YklkPTE1NjMxMyZzaXRlSWQ9MTk5MDE3JmFkSWQ9MTA5NjQ2NyZrYWRzaXplaWQ9OSZ0bGRJZD00OTc2OTA4OCZjYW1wYWlnbklkPTEyNjcxJmNyZWF0aXZlSWQ9MCZ1Y3JpZD0xOTAzODY0ODc3ODU2NDc1OTgwJmFkU2VydmVySWQ9MjQzJmltcGlkPTU0MjgyODhFLTYwRjktNDhDMC1BRDZELTJFRjM0M0E0RjI3NCZtb2JmbGFnPTImbW9kZWxpZD0yODY2Jm9zaWQ9MTIyJmNhcnJpZXJpZD0xMDQmcGFzc2JhY2s9MA==_url=https://Spinachtree.com/2021/05/29/rwfeds/
http://clients1.google.de/url?sa=t&url=https%3A%2F%2FSpinachtree.com
https://cse.google.cv/url?q=http://Spinachtree.com/
http://cse.google.bi/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://app.guanajuato.gob.mx/revive/www/delivery/ck.php?oaparams=2__bannerid=2447__zoneid=88__cb=cb2b9a1cb1__oadest=http%3a%2f%2fSpinachtree.com
http://subscriber.zdnet.de/profile/login.php?continue=https%3A%2F%2FSpinachtree.com%2F&continue_label=ZDNet.de
http://aacollabarchive.humin.lsa.umich.edu/omeka/setlocale?locale=es&redirect=http%3a%2f%2fSpinachtree.com
http://apptube.podnova.com/go/?go=https://Spinachtree.com/
http://images.google.com.ua/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://www.kaskus.co.id/redirect?url=https%3A%2F%2FSpinachtree.com
http://community.cypress.com/external-link.jspa?url=https%3A%2F%2FSpinachtree.com
https://images.google.bs/url?q=http://Spinachtree.com/
http://community.acer.com/en/home/leaving/Spinachtree.com
http://daemon.indapass.hu/http/session_request?redirect_to=https%3A%2F%2FSpinachtree.com%2F&partner_id=bloghu
http://images.google.com.hk/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://clients1.google.je/url?q=https%3A%2F%2FSpinachtree.com/
http://www.novalogic.com/remote.asp?NLink=https://Spinachtree.com/2021/05/29/rwfeds/
http://cm-sg.wargaming.net/frame/?service=frm&project=wot&realm=sg&language=en&login_url=http%3A%2F%2FSpinachtree.com&logout_url=http%3A%2F%2Fforum.worldoftanks.asia%2Findex.php%3Fapp%3Dcore%26module%3Dglobal%26section%3Dlogin%26do%3Dlogoutoid&incomplete_profile_url=http%3A%2F%2Fforum.worldoftanks.asia%2Findex.php%3Fapp%3Dmembers%26module%3Dprofile%26do%3Ddocompleteaccount&token_url=http%3A%2F%2Fforum.worldoftanks.asia%2Fmenutoken&frontend_url=http%3A%2F%2Fcdn-cm.gcdn.co&backend_url=http%3A%2F%2Fcm-sg.wargaming.net&open_links_in_new_tab=¬ifications_enabled=1&chat_enabled=&incomplete_profile_notification_enabled=&intro_tooltips_enabled=1®istration_url=http%3A%2F%2Fforum.worldoftanks.asia%2Findex.php%3Fapp%3Dcore%26module%3Dglobal%26section%3Dregister
http://images.google.cz/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.co.ug/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://sys.labaq.com/cli/go.php?s=lbac&p=1410jt&t=02&url=https://Spinachtree.com/
http://clients1.google.cg/url?q=https://Spinachtree.com/
http://legacy.aom.org/verifymember.asp?nextpage=https%3A%2F%2FSpinachtree.com
http://clients1.google.iq/url?q=https%3A%2F%2FSpinachtree.com/
http://clients1.google.cv/url?q=http%3A%2F%2FSpinachtree.com
http://cm-ru.wargaming.net/frame/?service=frm&project=wot&realm=ru&language=ru&login_url=http%3A%2F%2FSpinachtree.com&logout_url=http%3A%2F%2Ffor
http://cse.google.co.uk/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://www.mydoterra.com/Application/index.cfm?EnrollerID=458046&Theme=DefaultTheme&Returnurl=Spinachtree.com/&LNG=en_dot&iact=1
https://redirect.vebeet.com/index.php?url=//Spinachtree.com/cities/tampa-fl/
http://cse.google.co.zw/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://alt1.toolbarqueries.google.at/url?q=https://Spinachtree.com/
http://www.adminer.org/redirect/?url=https%3A%2F%2FSpinachtree.com&lang=en
https://webneel.com/i/3d-printer/5-free-3d-printer-model-website-yeggi/ei/12259?s=Spinachtree.com/2021/05/29/rwfeds/
http://apf.francophonie.org/doc.html?url=https://Spinachtree.com/
http://cse.google.by/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.jarrow.com/jump/http:_@__@_Spinachtree.com
http://yahoo-mbga.jp/r?url=//Spinachtree.com
http://www.pcpitstop.com/offsite.asp?https://Spinachtree.com/
http://getpocket.com/redirect?url=https%3A%2F%2FSpinachtree.com%2F
http://cse.google.com.jm/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.com.ec/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.drinksmixer.com/redirect.php?url=https%3A%2F%2FSpinachtree.com%2F
http://5cfxm.hxrs6.servertrust.com/v/affiliate/setCookie.asp?catId=1180&return=https://Spinachtree.com
https://ditu.google.com/url?q=http://Spinachtree.com/management.html
http://cse.google.co.ao/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://businesseast.ucoz.com/go?https://Spinachtree.com/
https://www.streetwisereports.com/cs/blank/main?x-p=click/fwd&rec=ads/443&url=http://Spinachtree.com/2021/05/29/rwfeds/
http://images.google.com.ec/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.sponsorship.com/Marketplace/redir.axd?ciid=514&cachename=advertising&PageGroupId=14&url=https://Spinachtree.com
http://clients1.google.com.eg/url?q=https://Spinachtree.com/
http://cat.sls.cuhk.edu.hk/CRF/visualization?Species=https://Spinachtree.com/
https://clients1.google.com/url?q=http://Spinachtree.com/management.html
http://creativecommons.org/choose/results-one?q_1=2&q_1=1&field_commercial=n&field_derivatives=sa&field_jurisdiction=&field_format=Text&field_worktitle=Blog&field_attribute_to_name=Lam%20HUA&field_attribute_to_url=http%3A%2F%2FSpinachtree.com%2F
http://tools.folha.com.br/print?url=https%3A%2F%2FSpinachtree.com%2F&site=blogfolha
http://clients1.google.co.kr/url?q=https://Spinachtree.com/
http://apiprop.sulekha.com/common/apploginredirect.aspx?enclgn=ilsxyvoDCT5XZjQCeHI5qlKoZ3Ljv/1wHK3AR7dYYz8%3D&nexturl=https://Spinachtree.com/
http://cse.google.de/url?sa=t&url=https%3A%2F%2FSpinachtree.com
http://images.google.dk/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://avp.innity.com/click/?campaignid=10933&adid=115198&zoneid=39296&pubid=3194&ex=1412139790&pcu=&auth=3tx88b-1412053876272&url=http%3A%2F%2FSpinachtree.com
http://ads.haberler.com/redir.asp?tur=habericilink&url=https://Spinachtree.com/
http://ip.tool.chinaz.com/?ip=Spinachtree.com
http://ax.bk55.ru/cur/www/delivery/ck.php?ct=1&oaparams=2__bannerid=4248__zoneid=141__OXLCA=1__cb=1be00d870a__oadest=https://Spinachtree.com/
http://support.dalton.missouri.edu/?URL=Spinachtree.com
http://banri.moo.jp/-18/?wptouch_switch=desktop&redirect=https%3a%2f%2fSpinachtree.com
http://avatar-cat-ru.1gb.ru/index.php?name=plugins&p=out&url=Spinachtree.com
http://sogo.i2i.jp/link_go.php?url=https://Spinachtree.com/
http://clients1.google.co.ug/url?q=https://Spinachtree.com/
http://cse.google.cd/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.t.me/iv?url=http%3A%2F%2FSpinachtree.com
http://www.ibm.com/links/?cc=us&lc=en&prompt=1&url=//Spinachtree.com
http://alt1.toolbarqueries.google.co.ao/url?q=https://Spinachtree.com/
http://clients1.google.co.ke/url?sa=t&url=https://Spinachtree.com/
http://cse.google.al/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://dotekomanie.cz/heureka/?url=https%3a%2f%2fSpinachtree.com%2F
http://images.google.co.id/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.de/url?sa=t&url=https%3A%2F%2FSpinachtree.com
https://maps.google.tk/url?q=http://Spinachtree.com/
http://www.astro.wisc.edu/?URL=https%3A%2F%2FSpinachtree.com
http://images.google.com.br/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.co.ke/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.co.il/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://api.fooducate.com/fdct/message/t/?t=592390BA-2F20-0472-4BA5-A59870BBA6A2:61213861:5AFC37A3-CAD4-CC42-4921-9BE2094B0A14&a=c&d=https://Spinachtree.com/
http://armenaikkandomainrating5.blogspot.com0.vze.com/frame-forward.cgi?https://Spinachtree.com/
http://cse.google.co.in/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://subscriber.silicon.co.uk/profile/login.php?continue=https%3A%2F%2FSpinachtree.com%2F&continue_label=TechWeekEurope+UK
http://cse.google.com.bn/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.com.do/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.com.lb/url?q=https://Spinachtree.com/
http://cr.naver.com/redirect-notification?u=https%3A%2F%2FSpinachtree.com
http://images.google.com.au/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://brangista.j-server.com/BRASADAIJ/ns/tl_ex.cgi?Surl=https://Spinachtree.com/
http://beskuda.ucoz.ru/go?https://Spinachtree.com/
http://uriu-ss.jpn.org/xoops/modules/wordpress/wp-ktai.php?view=redir&url=http%3a%2f%2fSpinachtree.com
http://cse.google.com.ai/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.cat/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.luminous-lint.com/app/iframe/photographer/Frantisek__Drtikol/Spinachtree.com
http://onlinemanuals.txdot.gov/help/urlstatusgo.html?url=https%3A%2F%2FSpinachtree.com
http://click.karenmillen.com/knmn40/c2.php?KNMN/94073/5285/H/N/V/https://Spinachtree.com/
http://images.google.com.pe/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://images.google.ee/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://ar.thefreedictionary.com/_/cite.aspx?url=http%3a%2f%2fSpinachtree.com&word=%D8%AD%D9%8E%D9%84%D9%90%D9%85%D9%8E&sources=kdict
http://bigtakeover.com/revive/www/delivery/ck.php?ct=1&oaparams=2__bannerid=68__zoneid=4__cb=c0575383b9__oadest=https://Spinachtree.com/
http://circuitomt.com.br/publicidade/www/delivery/ck.php?ct=1&oaparams=2__bannerid=34__zoneid=2__cb=6cc35441a4__oadest=http%3a%2f%2fSpinachtree.com
http://akid.s17.xrea.com/p2ime.php?url=https://Spinachtree.com/
http://clients1.google.com.tr/url?sa=t&url=https%3A%2F%2FSpinachtree.com
http://links.giveawayoftheday.com/Spinachtree.com
http://www2.ogs.state.ny.us/help/urlstatusgo.html?url=https%3A%2F%2FSpinachtree.com
http://images.google.co.il/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://bompasandparr.com/?URL=https://Spinachtree.com/
http://cse.google.com.ag/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://new.creativecommons.org/choose/results-one?q_1=2&q_1=1&field_commercial=n&field_derivatives=sa&field_jurisdiction=&field_format=Text&field_worktitle=Blog&field_attribute_to_name=Lam%20HUA&field_attribute_to_url=https%3A%2F%2FSpinachtree.com%2F
http://optimize.viglink.com/page/pmv?url=https%3A%2F%2FSpinachtree.com
http://cse.google.com.co/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://blackfive.net/__media__/js/netsoltrademark.php?d=Spinachtree.com
http://api.mixpanel.com/track/?data=eyJldmVudCI6ICJmdWxsdGV4dGNsaWNrIiwgInByb3BlcnRpZXMiOiB7InRva2VuIjogImE0YTQ2MGEzOTA0ZWVlOGZmNWUwMjRlYTRiZGU3YWMyIn19&ip=1&redirect=https://Spinachtree.com/
http://alt1.toolbarqueries.google.bs/url?q=https://Spinachtree.com/
http://video.fc2.com/exlink.php?uri=https%3A%2F%2FSpinachtree.com%2F
http://cse.google.co.jp/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.es/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.co.za/url?q=https://Spinachtree.com/
http://ads.bhol.co.il/clicks_counter.asp?macrocid=5635&campid=177546&DB_link=0&userid=0&ISEXT=0&url=https://Spinachtree.com/
http://cse.google.be/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://at070582.xsrv.jp/?wptouch_switch=desktop&redirect=http%3a%2f%2fSpinachtree.com
http://u.to/?url=https%3A%2F%2FSpinachtree.com%2F&a=add
http://ipv4.google.com/url?q=https%3A%2F%2FSpinachtree.com%2F
http://cse.google.co.ve/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://catzlyst.phrma.org/__media__/js/netsoltrademark.php?d=Spinachtree.com
http://clients1.google.co.jp/url?q=https://Spinachtree.com/
http://clients1.google.com.hk/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://clients1.google.cd/url?sa=t&url=https://Spinachtree.com/
http://bares.blog.idnes.cz/redir.aspx?url=https%3A%2F%2FSpinachtree.com
http://www.ip-adress.com/website/Spinachtree.com
http://clients1.google.st/url?q=https://Spinachtree.com/
http://cse.google.co.vi/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.com.br/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://parkcities.bubblelife.com/click/c3592/?url=https://Spinachtree.com/
http://clients1.google.by/url?sa=t&url=https%3A%2F%2FSpinachtree.com
http://antigo.anvisa.gov.br/listagem-de-alertas/-/asset_publisher/R6VaZWsQDDzS/content/alerta-3191-tecnovigilancia-boston-scientific-do-brasil-ltda-fibra-optica-greenlight-possibilidade-de-queda-de-temperatura-da-tampa-de-metal-e-da-pont/33868?inheritRedirect=false&redirect=http%3A%2F%2FSpinachtree.com
http://cse.google.ca/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.com.au/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://ads.hitparade.ch/link.php?ad_id=28&url=https://Spinachtree.com/
http://clients1.google.tm/url?q=https%3A%2F%2FSpinachtree.com/
http://images.google.com.pk/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://cc.cgps.tn.edu.tw/dyna/netlink/hits.php?id=46&url=https://Spinachtree.com/
http://cse.google.co.ma/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://clients1.google.ne/url?q=https://Spinachtree.com/
http://cse.google.com.et/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://nou-rau.uem.br/nou-rau/zeus/auth.php?back=https%3A%2F%2FSpinachtree.com%2F&go=x&code=x&unit=x
http://cc.naver.jp/cc?a=dtl.topic&r=&i=&bw=1024&px=0&py=0&sx=-1&sy=-1&m=1&nsc=knews.viewpage&u=https%3A%2F%2FSpinachtree.com
http://images.google.com.mx/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://cse.google.com.fj/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://a1.adform.net/C/?CC=1&bn=1015999%3Bc=1%3Bkw=Forex%20Trading%3Bcpdir=https://Spinachtree.com/
http://alt1.toolbarqueries.google.com.br/url?q=https://Spinachtree.com/
http://cse.google.com.hk/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://catalog.dir.bg/url.php?URL=https://Spinachtree.com/
http://clients1.google.it/url?q=http%3A%2F%2FSpinachtree.com/
http://counter.iflyer.tv/?trackid=gjt:1:&link=https://Spinachtree.com/
http://alt1.toolbarqueries.google.co.vi/url?q=https://Spinachtree.com/
http://admin.billoreilly.com/site/rd?satype=13&said=12&url=https://Spinachtree.com/
http://clients1.google.mn/url?q=https%3A%2F%2FSpinachtree.com
http://backlink.scandwap.xtgem.com/?id=IRENON&url=Spinachtree.com
https://www.videoder.com/af/media?mode=2&url=http://Spinachtree.com
http://ad.gunosy.com/pages/redirect?location=https://Spinachtree.com/
http://pw.mail.ru/forums/fredirect.php?url=Spinachtree.com
http://armoryonpark.org/?URL=https://Spinachtree.com/
http://app.hamariweb.com/iphoneimg/large.php?s=https://Spinachtree.com/
http://enseignants.flammarion.com/Banners_Click.cfm?ID=86&URL=Spinachtree.com
http://decisoes.fazenda.gov.br/netacgi/nph-brs?d=DECW&f=S&l=20&n=-DTPE&p=10&r=3&s1=COANA&u=http%3A%2F%2FSpinachtree.com
http://clients1.google.am/url?q=https://Spinachtree.com/
http://www.transtats.bts.gov/exit.asp?url=https%3A%2F%2FSpinachtree.com%2F
http://archive.aidsmap.com/Aggregator.ashx?url=https://Spinachtree.com/
http://countrysideveterinaryhospital.vetstreet.com/https://Spinachtree.com/
http://images.google.bg/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://pl.grepolis.com/start/redirect?url=https%3A%2F%2FSpinachtree.com%2F
https://www.google.ws/url?q=http://Spinachtree.com/
http://banners.molbiol.ru/openx/www/delivery/ck.php?ct=1&oaparams=2__bannerid=552__zoneid=16__cb=70ec3bb20d__oadest=http%3a%2f%2fSpinachtree.com
http://archive.is/Spinachtree.com
http://blogs.rtve.es/libs/getfirma_footer_prod.php?blogurl=http%3A%2F%2FSpinachtree.com
http://asoechat.wap.sh/redirect?url=Spinachtree.com
http://images.google.com.co/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://cse.google.ch/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://classweb.fges.tyc.edu.tw:8080/dyna/webs/gotourl.php?url=https://Spinachtree.com/
http://adms3.hket.com/openxprod2/www/delivery/ck.php?ct=1&oaparams=2__bannerid=527__zoneid=667__cb=72cbf61f88__oadest=https://Spinachtree.com/
http://alt1.toolbarqueries.google.co.ls/url?q=https://Spinachtree.com/
http://cse.google.bg/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://ijbssnet.com/view.php?u=https://Spinachtree.com/2021/05/29/rwfeds/
http://affiliates.thelotter.com/aw.aspx?A=1&Task=Click&ml=31526&TargetURL=https://Spinachtree.com/
http://777masa777.lolipop.jp/search/rank.cgi?mode=link&id=83&url=http%3a%2f%2fSpinachtree.com
http://images.google.com.my/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://b.gnavi.co.jp/ad/no_cookie/b_link?loc=1002067&bid=100004228&link_url=http%3A%2F%2FSpinachtree.com
http://www.drugoffice.gov.hk/gb/unigb/Spinachtree.com
http://clients1.google.bj/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://bizsearch.jhnewsandguide.com/__media__/js/netsoltrademark.php?d=Spinachtree.com
http://adms.hket.com/openxprod2/www/delivery/ck.php?ct=1&oaparams=2__bannerid=6685__zoneid=2040__cb=dfaf38fc52__oadest=http%3a%2f%2fSpinachtree.com
http://es.chaturbate.com/external_link/?url=https%3A%2F%2FSpinachtree.com
http://images.google.com.uy/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://cse.google.com/url?q=http://Spinachtree.com/management.html
http://12.rospotrebnadzor.ru/action_plans/inspection/-/asset_publisher/iqO1/document/id/460270?_101_INSTANCE_iqO1_redirect=https://Spinachtree.com/
http://bigoo.ws/__media__/js/netsoltrademark.php?d=Spinachtree.com
https://www.raincoast.com/?URL=Spinachtree.com
http://clients1.google.ci/url?q=https://Spinachtree.com/
http://ref.webhostinghub.com/scripts/click.php?ref_id=nichol54&desturl=https://Spinachtree.com/
http://web.stanford.edu/cgi-bin/redirect?dest=http%3A%2F%2FSpinachtree.com
https://cse.google.tk/url?sa=t&url=http://Spinachtree.com/
http://community.esri.com/external-link.jspa?url=https%3A%2F%2FSpinachtree.com
http://www.google.com/url?sa=t&url=https%3A%2F%2FSpinachtree.com
http://cse.google.com.kw/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://ads.specificmedia.com/click/v=5;m=2;l=23470;c=146418;b=874880;p=ui=ACXqoRFLEtwSFA;tr=DZeqTyQW0qH;tm=0-0;ts=20110427233838;dct=https://Spinachtree.com/
http://alt1.toolbarqueries.google.co.id/url?q=https://Spinachtree.com/
http://digital.fijitimes.com/api/gateway.aspx?f=https://Spinachtree.com
http://rd.rakuten.co.jp/a/?R2=https%3A//Spinachtree.com/
http://www3.valueline.com/vlac/logon.aspx?lp=https://Spinachtree.com
http://bpx.bemobi.com/opx/5.0/OPXIdentifyUser?Locale=uk&SiteID=402698301147&AccountID=202698299566&ecid=KR5t1vLv9P&AccessToken=&RedirectURL=https://Spinachtree.com/
http://www.economia.unical.it/prova.php?a%5B%5D=%3Ca+href%3Dhttps%3A%2F%2FSpinachtree.com%2F
http://br.nate.com/diagnose.php?from=w&r_url=http%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.bj/url?q=https://Spinachtree.com/
http://images.google.com.tw/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://www.xcelenergy.com/stateselector?stateselected=true&goto=http://Spinachtree.com
http://www.film1.nl/zoek/?q=%22%2F%3E%3Ca+href%3D%22https%3A%2F%2FSpinachtree.com%2F&sa=&cx=009153552854938002534%3Aaf0ze8etbks&ie=ISO-8859-1&cof=FORID%3
https://cse.google.com/url?sa=t&url=http://Spinachtree.com/
http://clients1.google.co.il/url?sa=i&url=https%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.com.np/url?q=https://Spinachtree.com/
http://images.google.com.vn/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://ads1.opensubtitles.org/1/www/delivery/afr.php?zoneid=3&cb=984766&query=One+Froggy+Evening&landing_url=https://Spinachtree.com/
http://adrian.edu/?URL=https://Spinachtree.com/
http://bidr.trellian.com/r.php?u=http%3A%2F%2FSpinachtree.com
https://www.htcdev.com/?URL=Spinachtree.com
http://aid97400.lautre.net/spip.php?action=cookie&url=https://Spinachtree.com/
https://doterra.myvoffice.com/Application/index.cfm?&EnrollerID=604008&Theme=Default&ReturnUrl=http://Spinachtree.com
http://images.google.com.mx/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.cg/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.co.th/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://clients1.google.gl/url?q=http%3A%2F%2FSpinachtree.com
http://fftoolbox.fulltimefantasy.com/search.cfm?q=%22%2F%3E%3Ca+href%3D%22https%3A%2F%2FSpinachtree.com%2F
http://images.google.com.bd/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://clients1.google.co.za/url?q=https://Spinachtree.com/
http://cse.google.co.id/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://Spinachtree.com.ipaddress.com/
http://cgi.davec.plus.com/cgi-bin/logs/loglink.cgi?https://Spinachtree.com/
http://forms.bl.uk/newsletters/index.aspx?back=https%3A//Spinachtree.com/
https://tour.catalinacruz.com/pornstar-gallery/puma-swede-face-sitting-on-cassie-young-lesbian-fun/?link=http://Spinachtree.com/holostyak-stb-2021
http://www.youtube.com/redirect?q=https%3A%2F%2FSpinachtree.com%2F
http://alt1.toolbarqueries.google.com.vc/url?q=https://Spinachtree.com/
http://bulkmail.doh.state.fl.us/lt.php?c=102&m=144&nl=18&lid=922&l=https://Spinachtree.com/
http://sc.sie.gov.hk/TuniS/Spinachtree.com
http://images.google.cl/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.ae/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://alt1.toolbarqueries.google.tg/url?q=https://Spinachtree.com/
http://images.google.com.ng/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://images.google.fi/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://cse.google.cl/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://images.google.com.vn/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://cse.google.ad/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://partnerpage.google.com/url?sa=i&url=http://Spinachtree.com/
http://clients1.google.tg/url?q=https%3A%2F%2FSpinachtree.com
http://images.google.co.il/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://clients1.google.nu/url?q=https%3A%2F%2FSpinachtree.com/
https://services.celemony.com/cgi-bin/WebObjects/LicenseApp.woa/wa/MCFDirectAction/link?linkId=1001414&stid=3463129&url=Spinachtree.com/2021/05/29/rwfeds/
http://cse.google.co.zm/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://200-155-82-24.bradesco.com.br/conteudo/pessoa-fisica/popExt.aspx?url=http://Spinachtree.com/2021/05/29/rwfeds/
http://a.twiago.com/adclick.php?tz=1473443342212991&pid=198&kid=2365&wmid=14189&wsid=65&uid=28&sid=3&sid2=2&swid=8950&ord=1473443342&target=https://Spinachtree.com/
http://clients1.google.ee/url?q=https://Spinachtree.com/
http://clients1.google.co.nz/url?sa=t&url=https%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.ac/url?q=https://Spinachtree.com/
http://m.odnoklassniki.ru/dk?st.cmd=outLinkWarning&st.rfn=https%3A%2F%2FSpinachtree.com
https://www.qscience.com/locale/redirect?redirectItem=http://Spinachtree.com
http://cse.google.co.za/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://111.1gb.ru/go.php?https://Spinachtree.com/
http://images.google.co.in/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.com.my/url?q=https://Spinachtree.com/
http://aqua21.jpn.org/lateequ/navi/navi.cgi?&mode=jump&id=6936&url=Spinachtree.com
http://webmail.mawebcenters.com/parse.pl?redirect=https://Spinachtree.com
http://alt1.toolbarqueries.google.cf/url?q=https://Spinachtree.com/
http://cse.google.com.gh/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://www.google.bs/url?q=http://Spinachtree.com/
http://as.inbox.com/AC.aspx?id_adr=262&link=https://Spinachtree.com/
http://cse.google.com.do/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.cssdrive.com/?URL=Spinachtree.com/holostyak-stb-2021
https://advtest.exibart.com/adv/adv.php?id_banner=7201&link=http://Spinachtree.com/2021/05/29/rwfeds/
http://images.google.com.eg/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://ad.foxitsoftware.com/adlog.php?a=redirect&img=testad&url=Spinachtree.com
http://chtbl.com/track/118167/Spinachtree.com/
http://alt1.toolbarqueries.google.com.af/url?q=https://Spinachtree.com/
https://maps.google.nu/url?q=http://Spinachtree.com/
http://images.google.com.tw/url?sa=t&url=http%3A%2F%2FSpinachtree.com
https://maps.google.ws/url?q=http://Spinachtree.com/
http://action.metaffiliation.com/redir.php?u=https://Spinachtree.com/
http://bons-plans-astuces.digidip.net/visit?url=http%3A%2F%2FSpinachtree.com
http://crash-ut3.clan.su/go?https://Spinachtree.com/
http://cse.google.co.cr/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://archives.richmond.ca/archives/descriptions/results.aspx?AC=SEE_ALSO&QF0=NameAccess&QI0==%22Currie%20McWilliams%20Camp%22&XC=http%3a%2f%2fSpinachtree.com&BU=https%3a%2f%2fcutepix.info%2fsex%2friley-reyes.php&TN=Descriptions&SN=AUTO19204&SE=1790&RN=2&MR=20&TR=0&TX=1000&ES=0&CS=0&XP=&RF=WebBrief&EF=&DF=WebFull&RL=0&EL=0&DL=0&NP=255&ID=&MF=GENERICENGWPMSG.INI&MQ=&TI=0&DT=&ST=0&IR=173176&NR=1&NB=0&SV=0&SS=0&BG=&FG=&QS=&OEX=ISO-8859-1&OEH=utf-8
http://plus.google.com/url?q=https%3A%2F%2FSpinachtree.com%2F
https://www.horseillustrated.com/redirect.php?location=http://Spinachtree.com
http://whois.domaintools.com/Spinachtree.com
http://cse.google.com.kh/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.timeandtiming.com/?&URL=Spinachtree.com
http://arhvak.minobrnauki.gov.ru/c/document_library/find_file_entry?p_l_id=17390&noSuchEntryRedirect=https://Spinachtree.com/
https://posts.google.com/url?q=http://Spinachtree.com/management.html
http://wtk.db.com/777554543598768/optout?redirect=https://Spinachtree.com
http://images.google.ca/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://app.feedblitz.com/f/f.fbz?track=Spinachtree.com
http://nutritiondata.self.com/facts/recipe/1304991/2?mbid=HDFD&trackback=http%3A%2F%2Fwww.Spinachtree.com
http://images.google.co.nz/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://alt1.toolbarqueries.google.is/url?q=https://Spinachtree.com/
http://ts.videosz.com/out.php?cid=101&aid=102&nis=0&srid=392&url=https://Spinachtree.com/
https://www.google.tk/url?q=http://Spinachtree.com/
http://clients1.google.co.ao/url?q=https%3A%2F%2FSpinachtree.com/
http://amaterasu.dojin.com/home/ranking.cgi?ti=%94%92%82%C6%8D%95%82%CC%94%FC%8Aw%81B&HP=http%3a%2f%2fSpinachtree.com&ja=3&id=19879&action=regist
http://www.astronet.ru/db/msusearch/index.html?q=%3Ca+href%3Dhttps%3A%2F%2FSpinachtree.com%2F
http://images.google.com.sg/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://bugcrowd.com/external_redirect?site=http://Spinachtree.com
http://biwa28.lolipop.jp/ys100/rank.cgi?mode=link&id=1290&url=https%3A%2F%2FSpinachtree.com
http://fcaw.library.umass.edu/goto/https:/Spinachtree.com
http://radio.cancaonova.com/iframe-loader/?t=DatingSingle:NoLongeraMystery-R�dio&ra=&url=https://Spinachtree.com/2021/05/29/rwfeds/
https://cse.google.nr/url?sa=t&url=http://Spinachtree.com/
http://clients1.google.gg/url?q=https%3A%2F%2FSpinachtree.com
http://cse.google.bj/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://camiane.dmcart.gethompy.com/shop/bannerhit.php?bn_id=91&url=http%3a%2f%2fSpinachtree.com
https://www.relativitymedia.com/logout?redirect=//Spinachtree.com/
http://anoushkabold.com/__media__/js/netsoltrademark.php?d=Spinachtree.com
http://cse.google.com.cy/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.am/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://www.meetme.com/apps/redirect/?url=http://Spinachtree.com
http://cse.google.at/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://legacy.aom.org/verifymember.asp?nextpage=https%3A//Spinachtree.com/
https://images.google.nu/url?q=http://Spinachtree.com/
https://images.google.ws/url?q=http://Spinachtree.com/
http://clients1.google.com.et/url?q=https%3A%2F%2FSpinachtree.com
http://cse.google.bs/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://mitsui-shopping-park.com/lalaport/iwata/redirect.html?url=http://Spinachtree.com/
https://www.barrypopik.com/index.php?URL=http://Spinachtree.com
http://clients1.google.com.om/url?q=https://Spinachtree.com/
http://bioinfo3d.cs.tau.ac.il/wk/api.php?action=https://Spinachtree.com/
http://contacts.google.com/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://sc.hkex.com.hk/gb/Spinachtree.com
http://cse.google.com.bo/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://cse.google.co.nz/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://www.extremnews.com/nachrichten/standard/dereferrer.cfm?rurl=http://Spinachtree.com/2021/05/29/rwfeds/
http://cse.google.co.ls/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://portal.novo-sibirsk.ru/dynamics.aspx?PortalId=2&WebId=8464c989-7fd8-4a32-8021-7df585dca817&PageUrl=%2FSitePages%2Ffeedback.aspx&Color=B00000&Source=https%3A%2F%2FSpinachtree.com%2F
http://clients1.google.com.gh/url?q=https://Spinachtree.com/
http://alt1.toolbarqueries.google.com.pe/url?q=https%3A%2F%2FSpinachtree.com
http://clients1.google.jo/url?q=http%3A%2F%2FSpinachtree.com
http://clients1.google.pn/url?q=https://Spinachtree.com/
https://cse.google.nu/url?sa=t&url=http://Spinachtree.com/
http://cse.google.ba/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://www.venez.fr/error.fr.html?id=1&uri=https%3A//Spinachtree.com/
http://domain.opendns.com/Spinachtree.com
http://alt1.toolbarqueries.google.com.ua/url?q=https://Spinachtree.com/
http://alt1.toolbarqueries.google.be/url?q=https://Spinachtree.com/
https://www.allergicliving.com/adspace/?mod=serve&act=clickthru&id=695&to=http://Spinachtree.com/2021/05/29/rwfeds/
http://cse.google.com.bd/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://aborg.lib.ntu.edu.tw/ntuepaper/toModule.do?prefix=/publish&page=/newslist.jsp&uri=https://Spinachtree.com/
http://alt1.toolbarqueries.google.pl/url?q=https://Spinachtree.com/
http://images.google.fi/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://blawat2015.no-ip.com/~mieki256/diary/refsweep.cgi?url=https://Spinachtree.com/
https://www.google.nu/url?q=http://Spinachtree.com/
http://images.google.co.jp/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
https://www.dltk-teach.com/p.asp?p=http://Spinachtree.com/2021/05/29/rwfeds/
http://images.google.co.id/url?sa=t&url=http%3A%2F%2FSpinachtree.com
http://images.google.com.my/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://members.ascrs.org/sso/logout.aspx?returnurl=https://Spinachtree.com/
http://closings.cbs6albany.com/scripts/adredir.asp?url=https://Spinachtree.com/
http://images.google.co.th/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://alumni.drivers.informer.com/go/go.php?go=https://Spinachtree.com/
http://uriu-ss.jpn.org/xoops/modules/wordpress/wp-ktai.php?view=redir&url=https%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.com.kw/url?q=https://Spinachtree.com/
http://forum.solidworks.com/external-link.jspa?url=https%3A%2F%2FSpinachtree.com
http://alt1.toolbarqueries.google.bf/url?q=https://Spinachtree.com/
http://acf.poderjudicial.es/portal/site/cgpj/menuitem.65d2c4456b6ddb628e635fc1dc432ea0/?schema=http&title=Acuerdos+de+la+Comisi%C3%B3n+Permanente+del+CGPJ+de+17+de+julio+de+2018&vgnextoid=d94e58e4d849d210VgnVCM100000cb34e20aRCRD&vgnextlocale=gl&isSecure=false&vgnextfmt=default&protocolo=http&user=&url=http%3a%2f%2fSpinachtree.com&perfil=3
http://0845.boo.jp/cgi/mt3/mt4i.cgi?id=24&mode=redirect&no=15&ref_eid=3387&url=http%3A%2F%2FSpinachtree.com
http://cse.google.co.kr/url?sa=t&url=http%3A%2F%2FSpinachtree.com%2F
http://hostedmovieupdates.aebn.net/feed/?urlstub=Spinachtree.com&
Hi there to all, it’s genuinely a nice for
me to pay a visit this site, it consists of priceless Information.
Thank you for any other informative blog. Where else may I am getting that type of information written in such an ideal approach?
I have a project that I am just now working on, and I’ve been on the glance out for
such info.
Greetings, I believe your website could possibly be having
browser compatibility problems. Whenever I look at your site in Safari, it looks fine however when opening in IE, it has some overlapping issues.
I just wanted to give you a quick heads up! Apart
from that, wonderful website!
Wow, incredible blog layout! How long have you been blogging for?
you make blogging look easy. The overall look of your website is wonderful, as well as the content!
With thanks. Useful information!
Nicely put, Thanks!
You actually make it seem so easy with your presentation but I find this topic to be really
something that I think I would never understand. It seems too complex and
extremely broad for me. I’m looking forward for your next
post, I will try to get the hang of it!
Nice blog right here! Also your site loads up fast! What web host are you the usage of?
Can I get your affiliate link to your host?
I want my site loaded up as fast as yours lol
Are you worried from living the same routine over and over again? Is it getting hard for you to wake up in the morning for your online English class after a night shift at work? Pay Someone to do My Online English Class is an expert service from the house of myclassprofessor. We guarantee A or B grades . If you need additional help or willing to Pay someone to do my online english class for me, get in touch with us at any time. Now from the comfort of your home you can hire someone to take my online English class online for me and get the education you always required to have. Don’t let your busy schedule stop you from completing your goals, order our take my online English class services today! Are you still confused if you can pay someone to take my English online courses? Talk to our representatives today and they will answer all questions you might have about our take my English courses online services.
Excellent weblog right here! Also your site so much up very
fast! What web host are you the use of? Can I am getting your affiliate link for your host?
I want my site loaded up as quickly as yours lol
IDNSLOT also provides a guarantee bonus for losing balances back
to members and IDNSLOT also provides a new member bonus for newcomers.
excellent post, very informative. I ponder why the opposite specialists of this sector don’t understand this. You must continue your writing. I’m sure, you have a huge readers’ base already!
Hello there, I found your website by means of Google at the same time as looking
for a related subject, your site got here up, it appears good.
I’ve bookmarked it in my google bookmarks.
Hello there, simply was aware of your blog via Google, and found that it is really
informative. I am going to be careful for brussels.
I’ll appreciate if you happen to proceed this in future.
Many people will probably be benefited from your
writing. Cheers!
I quite like looking through a post that will make people think.
Also, thanks for allowing me to comment!
Howdy! I know this is kind of off topic but I was wondering if you knew where I could locate a captcha plugin for
my comment form? I’m using the same blog platform as yours and I’m having trouble
finding one? Thanks a lot!
It’s awesome to visit this web page and reading the views
of all mates about this piece of writing, while I am also keen of
getting knowledge.
Hi! I know this is somewhat off topic but I was wondering which blog platform are
you using for this site? I’m getting fed up of WordPress because I’ve had issues with hackers and I’m looking
at alternatives for another platform. I would be fantastic if you could point me in the direction of a good
platform.
Hi, I think your blog might be having browser compatibility issues.
When I look at your website in Opera, it looks fine but when opening
in Internet Explorer, it has some overlapping.
I just wanted to give you a quick heads up! Other then that, great blog!
I every time spent my half an hour to read this weblog’s articles all the time along with a mug of coffee.
Hiya! Quick question that’s totally off topic. Do you know
how to make your site mobile friendly? My blog looks weird when viewing from my iphone4.
I’m trying to find a theme or plugin that might be able to resolve this problem.
If you have any recommendations, please share. Cheers!
Santo precisely what people call hime constantly and thinks it
sounds quite reliable. Bottle tops collecting is some thing that I’m totally endlaved by.
Since I was 18 I’ve been working as a computer operator but I’ve already went for another unique.
Maryland will be the place Adore most therefore i have just what I need here.
After exploring a few of the blog posts on your site, I truly like your
technique of writing a blog. I added it to my bookmark site list and will be checking back in the near future.
Please visit my web site as well and let me know your opinion.
Here you can read and see the worlds lastest news and
trend.
Have you ever thought about adding a little bit more
than just your articles? I mean, what you say is important and
all. Nevertheless imagine if you added some great photos or videos
to give your posts more, “pop”! Your content is excellent but with pics and videos, this
site could definitely be one of the very best in its field.
Very good blog!
I visited several web pages however the audio feature for audio
songs present at this web site is actually marvelous.
Thanks in favor of sharing such a fastidious thought, article is fastidious, thats why i have read it fully
We’re a group of volunteers and opening a
brand new scheme in our community. Your web site provided us with
valuable information to work on. You have done a formidable task and our whole group might be grateful to you.
This is nicely expressed! !
my web-site http://genmasterhub.com
Thank you for your articles. I find them very helpful. Could you help me with something?
Hi, I believe your website could be having browser compatibility issues.
When I lookk at your web site in Safari, itt loioks fine
however, if opening in Internet Explorer, it hhas some overlaping issues.
I merely wanted to provide youu with a quick heads up!
Apart from that, excellent blog!
Feel fdee to surf to mmy blog polst … semi trailer
Write more, thats all I have to say. Literally, it
seems as though you relied on the video to make your point.
You obviously know what youre talking about, why throw away your intelligence on just posting
videos to your blog when you could be giving us something enlightening to read?
Hello would you mind letting me know which hosting company you’re using?
I’ve loaded your blog in 3 completely different web browsers and I must say this blog loads a lot quicker then most.
Can you recommend a good web hosting provider at a reasonable price?
Thanks a lot, I appreciate it!
Your style is really unique compared to other people I’ve read stuff from.
Many thanks for posting when you have the opportunity, Guess I will
just book mark this blog.
Good way of explaining, and fastidious paragraph to
obtain data on the topic of my presentation focus, which i am going to convey in academy.
There’s certainly a great deal to learn about this topic.
I like all of the points you’ve made.
Howdy! I know this is kinda off topic but I
was wondering if you knew where I could locate a captcha
plugin for my comment form? I’m using the same blog platform
as yours and I’m having difficulty finding one?
Thanks a lot!
top casino
https://getb8.us/online-casinos/
new casino
https://getb8.us/online-casinos/
games casino
Wonderful blog! Do you have any recommendations for aspiring writers?
I’m hoping to start my own site soon but I’m a little
lost on everything. Would you advise starting with a free platform like WordPress or go for a paid
option? There are so many options out there that I’m totally confused ..
Any recommendations? Many thanks!
Hey there, I think your blog might be having browser compatibility
issues. When I look at your blog in Ie, it looks fine but when opening in Internet Explorer, it has some overlapping.
I just wanted to give you a quick heads up! Other then that, terrific blog!
Admiring the persistence you put into your site and in depth information you offer.
It’s nice to come across a blog every once in a while that isn’t the same out of date rehashed material.
Great read! I’ve saved your site and I’m including your RSS feeds to my Google account.
Hey, I think your site might be having browser compatibility issues.
When I look at your blog site in Firefox, it looks fine but when opening in Internet Explorer, it has some overlapping.
I just wanted to give you a quick heads up!
Other then that, excellent blog!
You’ve made some good points there. I checked on the net to find
out more about the issue and found most individuals will go along with your views on this website.
Great post but I was wanting to know if you could write a litte more on this subject?
I’d be very grateful if youu could elaborate
a llittle bitt more. Bless you!
Feel free to visit my page :: Team meeting
Very good info. Lucky me I recently found your blog by accident (stumbleupon).
I have bookmarked it for later!
great points altogether, you simply won a brand new reader.
What may you recommend in regards to your publish that you made some days
ago? Any sure?
Howdy would you mind letting me know which
hosting company you’re working with? I’ve loaded your blog
in 3 different internet browsers and I must say this blog loads a lot faster
then most. Can you recommend a good internet hosting provider at a honest
price? Many thanks, I appreciate it!
Excellent blog here! Also your site a lot up very fast!
What host are you the usage of? Can I am
getting your affiliate link in your host? I desire my site
loaded up as quickly as yours lol
Hello, this weekend is pleasant in support of me, because this time i am reading this great educational paragraph
here at my residence.
It is appropriate time to make some plans for the future and it is time to be happy.
I have learn this put up and if I could I desire to
counsel you some attention-grabbing issues or advice. Perhaps you can write next articles referring to this article.
I want to learn more issues approximately it!
I loved as much as you’ll receive carried out right here. The
sketch is attractive, your authored material stylish.
nonetheless, you command get bought an edginess over that
you wish be delivering the following. unwell unquestionably come further formerly
again since exactly the same nearly a lot often inside case you
shield this increase.
I’m not sure exactly why but this site is loading extremely slow for
me. Is anyone else having this problem or is it a problem on my end?
I’ll check back later on and see if the problem
still exists.
http://iqcos.ru/bitrix/rk.php?id=17&site_id=s1&event1=banner&event2=click&goto=http://103.35.188.37
Does your website have a contact page? I’m having a tough time locating
it but, I’d like to shoot you an email. I’ve got some creative ideas for your
blog you might be interested in hearing. Either way, great website and I look forward to seeing it
grow over time.
Hello, I do think your site may be having internet
browser compatibility problems. Whenever I look at
your site in Safari, it looks fine but when opening in I.E., it has some overlapping issues.
I simply wanted to provide you with a quick heads up! Other
than that, wonderful blog!
What’s up to all, how is all, I think every one is getting
more from this site, and your views are fastidious for new viewers.
Excellent goods from you, man. I have understand your stuff previous to
and you are just too fantastic. I really like what you have acquired here, certainly like what you are stating and
the way in which you say it. You make it entertaining and you still
care for to keep it smart. I can’t wait to read
much more from you. This is actually a terrific site.
The way this online slot hack injector works usually involves the use of special software or applications that are inserted into online slot games. By using this software or application, players can change several parameters in the slot game such as the bet amount, payout percentage, or even the results of the slot machine spin. For further information, visit this website https://slotergacor2023.wordpress.com/
Excellent weblog right here! Additionally your website a
lot up very fast! What host are you the usage of?
Can I am getting your affiliate hyperlink on your host?
I desire my website loaded up as fast as yours lol
Hot news often becomes the main focus, especially in cyberspace which is increasingly developing rapidly. However, not all information that sounds exciting can always be trusted. Therefore, it is important for every individual to be wise in responding to hot news and seek confirmation from different sources before believing it. For further information, visit this website https://ceritahot35.wordpress.com/
Hi Dear, are you actually visiting this web page daily, if so afterward you will absolutely obtain nice knowledge.
High rainfall caused the Ciliwung River to overflow and floods submerged a number of surrounding settlements. Thousands of houses were flooded and tens of thousands of residents were forced to flee to safer places. Apart from that, flooding also causes damage to roads and other public infrastructure, for further information visit this website https://sites.google.com/view/168news
Excellent post. I was checking continuously this blog and I’m
impressed! Extremely useful info specially the last part 🙂
I care for such information a lot. I was seeking this certain info for a very long time.
Thank you and good luck.
Somebody necessarily assist to make significantly articles I might state.
This is the very first time I frequented your website page and thus
far? I amazed with the research you made to create this particular
put up incredible. Great task!
Hello would you mind sharing which blog platform you’re
using? I’m looking to start my own blog in the near future but
I’m having a difficult time making a decision between BlogEngine/Wordpress/B2evolution and
Drupal. The reason I ask is because your layout seems different
then most blogs and I’m looking for something completely unique.
P.S Apologies for getting off-topic but I had to ask!
I’m truly enjoying the design and layout of your blog.
It’s a very easy on the eyes which makes it much more enjoyable for me
to come here and visit more often. Did you hire out
a developer to create your theme? Fantastic work!
Hi there, I found your website by means of Google whilst searching for a comparable topic, your web site got here up, it seems great.
I’ve bookmarked it in my google bookmarks.
Hello there, just became alert to your weblog thru Google,
and found that it is truly informative. I am gonna watch out for brussels.
I will be grateful should you continue this in future.
A lot of folks shall be benefited out of your writing.
Cheers!
Thanks for finally talking about > Pontine Crossing Tract (pct) |
Whole Brain Protocol for Tractography with Empirical MRI < Liked it!
Awesome article.
magnificent issues altogether, you just won a new reader. What would you recommend about your submit that you
made a few days ago? Any positive?
Good post. I’m experiencing many of these issues as well..
My website; vpn special coupon code 2024
Thank you! Very good information!
Appreciation to my father who shared with me on the topic of this blog, this webpage is actually awesome.
Hi, this weekend is good for me, for the reason that this point in time i am
reading this wonderful educational piece of writing here at my home.
Very rapidly this site will be famous among all blogging people, due to
it’s fastidious articles
http://buildolution.com/UserProfile/tabid/131/userId/392479/Default.aspx
http://buildolution.com/UserProfile/tabid/131/userId/392480/Default.aspx
http://buildolution.com/UserProfile/tabid/131/userId/392481/Default.aspx
http://buildolution.com/UserProfile/tabid/131/userId/392482/Default.aspx
http://buildolution.com/UserProfile/tabid/131/userId/392799/Default.aspx
http://go.bubbl.us/cbf621/ac0a?/good
http://go.bubbl.us/cbf626/26c4?/https://www.mikhayla.id/
http://mayfever.crowdfundhq.com/users/pafijawatengah
http://mayfever.crowdfundhq.com/users/polisi-slot
http://mayfever.crowdfundhq.com/users/toto188-1
http://mayfever.crowdfundhq.com/users/toto188-2
http://mayfever.crowdfundhq.com/users/toto188-3
http://www.travelful.net/location/5406275/indonesia/pafijawatengah
https://65fd1d29535b0.site123.me/
https://65fd1e81c3d80.site123.me/
https://65fd202128732.site123.me/
https://65fd20a7484f4.site123.me/
https://65fe274dead73.site123.me/
https://about.me/pafijawatengah/
https://about.me/polisislotpolisislot
https://about.me/toto188t
https://about.me/toto188toto188
https://about.me/ttoto188
https://aetherhub.com/Deck/pafijawatengah
https://aetherhub.com/Deck/polisi-slot
https://aetherhub.com/Deck/toto188-1036612
https://aetherhub.com/Deck/toto188-1036616
https://aetherhub.com/Deck/toto188-1036617
https://app.roll20.net/users/13136592/polisi-slot-p
https://app.roll20.net/users/13136599/toto188-t
https://app.roll20.net/users/13136603/toto188-t
https://app.roll20.net/users/13136606/toto188-t
https://app.roll20.net/users/13138501/pafijawatengah-p
https://baskadia.com/user/cv2a
https://baskadia.com/user/cv2b
https://baskadia.com/user/cv2c
https://baskadia.com/user/cv2d
https://baskadia.com/user/cv2e
https://bbpress.org/forums/profile/pafijawatengah/
https://bbpress.org/forums/profile/polisislotab/
https://bbpress.org/forums/profile/toto188e/
https://bbpress.org/forums/profile/toto188fa/
https://bbpress.org/forums/profile/toto188g/
https://biolinky.co/pafijawatengah
https://biolinky.co/polisislota
https://biolinky.co/toto-188-e
https://biolinky.co/toto-188-f
https://biolinky.co/toto-188-g
https://bitbin.it/21crwWQF/
https://bitbin.it/9E0MLzNO/
https://bitbin.it/kRjeUJKW/
https://bitbin.it/oOv586i9/
https://bordeaux.onvasortir.com/profil_read.php?Pafijawatengah
https://bordeaux.onvasortir.com/profil_read.php?Polisislota
https://bordeaux.onvasortir.com/profil_read.php?Toto188e
https://bordeaux.onvasortir.com/profil_read.php?Toto188f
https://bordeaux.onvasortir.com/profil_read.php?Toto188g
https://buddypress.org/members/pafijawatengah/profile/
https://buddypress.org/members/polisislotab/profile/
https://buddypress.org/members/toto188e/profile/
https://buddypress.org/members/toto188fa/profile/
https://buddypress.org/members/toto188g/profile/
https://candied-sherbet-abf.notion.site/pafi-jawa-tengah-da194fe820b74db9a47d23abeaf2af53?pvs=4
https://cannabis.net/user/149144
https://cannabis.net/user/149145
https://cannabis.net/user/149146
https://cannabis.net/user/149147
https://cannabis.net/user/149254
https://canvas.instructure.com/eportfolios/2811244/Home/Welcome
https://canvas.instructure.com/eportfolios/2811253/Home/Welcome
https://canvas.instructure.com/eportfolios/2811267/Home/Welcome
https://canvas.instructure.com/eportfolios/2813368/Home/Welcome
https://cliff-tv-c51.notion.site/toto188-0b19447aa9d946649b18c18a61628bcf?pvs=4
https://cobainjoinnow.wixsite.com/toto188
https://coolors.co/u/doko_dakina
https://coolors.co/u/fuku_fakin
https://coolors.co/u/pafijawatengah
https://coolors.co/u/toto188
https://coolors.co/u/toto1881
https://cs.astronomy.com/members/pafijawatengah/default.aspx
https://cs.astronomy.com/members/polisislota/default.aspx
https://cs.astronomy.com/members/toto188e/default.aspx
https://cs.astronomy.com/members/toto188f/default.aspx
https://cs.astronomy.com/members/toto188g/default.aspx
https://docs.google.com/document/d/112qvYZ34kOczHxkbRp846YvNVVPg8YOmc6_460EipTM/edit?usp=sharing
https://docs.google.com/document/d/166XXImEcbILDSzs44iWwW6MTndvnxsqtZZjY8BzBLmw/edit?usp=sharing
https://docs.google.com/document/d/1G4xjhQhzDhN9FdYRyzV0k1tOglEEkd9KSPwdIQHA4Vo/edit?usp=sharing
https://docs.google.com/document/d/1IYhhv18ypztCifmFiU5P3fVTNv2Cxor6LYWlmHkx1GM/edit?usp=sharing
https://docs.google.com/document/d/1yUQKnZ-oLikhmwI3omkewUXYEzj2OdISeDp_XpvEHHg/edit?usp=sharing
https://dokodakina.wixsite.com/toto188
https://eo-college.org/members/pafijawatengah/
https://eo-college.org/members/polisislota/
https://eo-college.org/members/toto188/
https://eo-college.org/members/toto188e/
https://eo-college.org/members/toto188g/
https://fileforum.com/profile/pafijawatengah
https://fileforum.com/profile/polisislota
https://fileforum.com/profile/toto188e
https://fileforum.com/profile/toto188f
https://fileforum.com/profile/toto188g
https://findaspring.org/members/pafijawatengah/
https://findaspring.org/members/polisislota/
https://findaspring.org/members/toto188e/
https://findaspring.org/members/toto188f/
https://findaspring.org/members/toto188g/
https://firmanfarmina.wixsite.com/toto188
https://fukufakin.wixsite.com/polisi-slot
https://gab.com/pafijawatengah
https://gab.com/toto188
https://gab.com/toto188f
https://gab.com/toto188g
https://git.qoto.org/pafijawatengah
https://git.qoto.org/polisislota
https://git.qoto.org/toto188e
https://git.qoto.org/toto188f
https://git.qoto.org/toto188g
https://globalcatalog.com/pafijawatengah.id
https://globalcatalog.com/polisislot.gi
https://globalcatalog.com/toto188e.id
https://globalcatalog.com/toto188f.id
https://globalcatalog.com/toto188g.id
https://glose.com/u/dokodakina
https://glose.com/u/fukufakin
https://glose.com/u/pafijawatengah
https://glose.com/u/toto188f
https://glose.com/u/toto188g
https://gravatar.com/tokokalimanar
https://gravatar.com/toto188e
https://gravatar.com/toto188f
https://gravatar.com/toto188g
https://grepo.travelcarma.com/pafijawatengah
https://grepo.travelcarma.com/polisislota
https://grepo.travelcarma.com/toto188e
https://grepo.travelcarma.com/toto188f
https://grepo.travelcarma.com/toto188g
https://groups.google.com/g/pafijawatengah/
https://groups.google.com/g/polisislota/
https://groups.google.com/g/toto188e
https://groups.google.com/g/toto188f/
https://groups.google.com/g/toto188g
https://hanson.net/users/pafijawatengah
https://hanson.net/users/polisislota
https://hanson.net/users/toto188e
https://hanson.net/users/toto188f
https://hanson.net/users/toto188g
https://heylink.me/doko6/
https://heylink.me/pafijawatengah/
https://heylink.me/polisislota/
https://heylink.me/toto188f/
https://heylink.me/toto188g/
https://histre.com/public/collections/4xeuhehx/
https://histre.com/public/collections/a2xf1y52/
https://histre.com/public/collections/l7h8etfh/
https://histre.com/public/collections/mlecxvln/
https://histre.com/public/collections/q7ffl00e/
https://homment.com/hZ2eMFrp0QhkKRGTHmGZ
https://homment.com/il8ngrZgWUrGR5qasRvE
https://homment.com/jm29hZ3iTOqSu1irpQGF
https://hub.docker.com/u/pafijawatengah
https://hub.docker.com/u/polisislota
https://hub.docker.com/u/toto188e
https://hub.docker.com/u/toto188f
https://hub.docker.com/u/toto188g
https://iglinks.io/cobainjoinnow-b55
https://iglinks.io/dokodakina-g4j
https://iglinks.io/firmanfarmina-nmc
https://iglinks.io/fukufakin-9fk
https://iglinks.io/tokokalimanar-geg
https://imageevent.com/pafijawatengah
https://imageevent.com/polisislota
https://imageevent.com/toto188
https://imageevent.com/toto188f
https://imageevent.com/toto188g
https://independent.academia.edu/cobainini
https://independent.academia.edu/dokodakina
https://independent.academia.edu/pafijawatengahpafijawatengah
https://independent.academia.edu/toto188toto188
https://inkbunny.net/pafijawatengah
https://inkbunny.net/polisislota
https://inkbunny.net/toto188e
https://inkbunny.net/toto188f
https://inkbunny.net/toto188g
https://joy.gallery/pafijawa-tengah
https://joy.gallery/polisislotaa
https://joy.gallery/toto188ee
https://joy.gallery/toto188ff
https://joy.gallery/toto188gg
https://joy.link/pafijawatengah
https://joy.link/polisislota
https://joy.link/toto188e
https://joy.link/toto188f
https://jsfiddle.net/polisislot/nc439x5L/
https://jsfiddle.net/toto188e/yL7dn0f6/
https://jsfiddle.net/toto188f/25d3p48L/
https://jsfiddle.net/toto188g/9guftqbj/
https://jsfiddle.net/user/pafijawatengah
https://k12.instructure.com/eportfolios/633566/Home/Welcome
https://laced-gander-251.notion.site/Polisi-Slot-4a8719931af64fb7bf9d5b7bbb858d3f?pvs=4
https://letterboxd.com/pafijawatengah/
https://letterboxd.com/polisislota/
https://letterboxd.com/toto188e/
https://letterboxd.com/toto188f/
https://letterboxd.com/toto188g/
https://linktr.ee/pafijawatengah
https://linktr.ee/polisislota
https://linktr.ee/toto188f
https://linktr.ee/toto188g
https://linktr.ee/toto188h
https://medium.com/@cobainjoinnow/toto188-adb52762d073
https://medium.com/@dokodakina/toto188-1550559ddc0e
https://medium.com/@firmanfarmina/toto188-4e7ccb0c0e2c
https://medium.com/@fukufakin/polisi-slot-fc07363ef678
https://medium.com/@tokokalimanar/pafijawatengah-9e6b205c5b6f
https://muckrack.com/pafijawa-tengah/bio
https://muckrack.com/polisi-slot-polisi-slot/bio
https://muckrack.com/toto188-e/bio
https://muckrack.com/toto188-f/bio
https://muckrack.com/toto188-g/bio
https://myapple.pl/users/440382-polisi-slot
https://myapple.pl/users/440384-toto188
https://myapple.pl/users/440385-toto188
https://myapple.pl/users/440386-toto188
https://myapple.pl/users/440584-pafijawatengah
https://nowewyrazy.uw.edu.pl/profil/pafijawatengah
https://nowewyrazy.uw.edu.pl/profil/polisislota
https://nowewyrazy.uw.edu.pl/profil/toto188e
https://nowewyrazy.uw.edu.pl/profil/toto188f
https://nowewyrazy.uw.edu.pl/profil/toto188g
https://openhumans.org/member/pafijawatengah/
https://openhumans.org/member/polisislota/
https://openhumans.org/member/toto188e/
https://openhumans.org/member/toto188f/
https://openhumans.org/member/toto188g/
https://padlet.com/cobainjoinnow/toto188-rfzp3pcl7cor08my
https://padlet.com/dokodakina/toto188-yk3kqktsrpaocymb
https://padlet.com/firmanfarmina/toto188-6q5qvt0qa0udkm0k
https://padlet.com/fukufakin/polisi-slot-p4tu71s3t3xv4i1g
https://padlet.com/tokokalimanar/pafijawatengah-xirie6cvwnuui242
https://pafi-jawa-tengah.jimdosite.com/
https://pafijawatengah.mn.co/about
https://pafijawatengah.mystrikingly.com/
https://pafijawatengah.pages.dev/
https://pafijawatengah-s-school.teachable.com/p/home
https://pastelink.net/0o72ggm8
https://pastelink.net/43askq3z
https://pastelink.net/6w2ge6t3
https://pastelink.net/ejckcczk
https://pastelink.net/pe3q6b7u
https://playtoto.mn.co/about
https://polisi-slot.jimdosite.com/
https://polisi-slot.mn.co/about
https://polisislot.pages.dev/
https://polisislota.mystrikingly.com/
https://polisi-slot-a27567.webflow.io/
https://polisi-slot-s-school.teachable.com/p/home
https://power-hotel-851.notion.site/toto188-f3c5e6aa14f14c40a954173720322e2f?pvs=4
https://research.openhumans.org/member/pafijawatengah/
https://research.openhumans.org/member/polisislota/
https://research.openhumans.org/member/toto188e/
https://research.openhumans.org/member/toto188f/
https://research.openhumans.org/member/toto188g/
https://sites.google.com/view/pafijawatengah/beranda
https://sites.google.com/view/polisislota/home
https://sites.google.com/view/toto188e/home
https://sites.google.com/view/toto188f/home
https://sites.google.com/view/toto188g/home
https://slides.com/fesfo/fesfo/
https://socialsocial.social/user/pafijawatengah/
https://socialsocial.social/user/polisislota/
https://socialsocial.social/user/toto188e/
https://socialsocial.social/user/toto188f/
https://socialsocial.social/user/toto188g/
https://solo.to/pafijawatengah
https://solo.to/polisislota
https://solo.to/toto188
https://solo.to/toto188f
https://solo.to/toto188g
https://speakerdeck.com/pafijawatengah
https://speakerdeck.com/polisislota
https://speakerdeck.com/toto188e
https://speakerdeck.com/toto188f
https://speakerdeck.com/toto188g
https://starity.hu/profil/434982-polisislota/
https://starity.hu/profil/434984-toto188e/
https://starity.hu/profil/434985-toto188f/
https://starity.hu/profil/434986-toto188g/
https://starity.hu/profil/435259-pafijawatengah/
https://startupxplore.com/en/person/pafijawatengah
https://startupxplore.com/en/person/polisi-slot
https://startupxplore.com/en/person/toto188-1
https://startupxplore.com/en/person/toto188-2
https://startupxplore.com/en/person/toto188-3
https://steemit.com/tech/@fesfo/23pbnz-toto188
https://steemit.com/tech/@fesfo/4qmwri-toto188
https://steemit.com/tech/@fesfo/pafijawatengah
https://steemit.com/tech/@fesfo/polisi-slot
https://steemit.com/tech/@fesfo/toto188
https://support.advancedcustomfields.com/forums/users/pafijawatengah/
https://support.advancedcustomfields.com/forums/users/polisislota/
https://support.advancedcustomfields.com/forums/users/toto188e/
https://support.advancedcustomfields.com/forums/users/toto188f/
https://support.advancedcustomfields.com/forums/users/toto188g/
https://swift-hourglass-fa3.notion.site/toto188-3a1273cd31114984b9de69584687828b?pvs=4
https://t.me/baliseo
https://t.me/seo_bali
https://t.me/workingclassman
https://taylorhicks.ning.com/photo/albums/pafijawatengah
https://taylorhicks.ning.com/photo/albums/polisi-slot
https://taylorhicks.ning.com/photo/albums/toto188-1
https://taylorhicks.ning.com/photo/albums/toto188-2
https://taylorhicks.ning.com/photo/albums/toto188-3
https://telegra.ph/pafijawatengah-03-23
https://telegra.ph/Polisi-Slot-03-22
https://telegra.ph/toto188-03-22
https://telegra.ph/toto188-03-22-2
https://telegra.ph/toto188-03-22-3
https://telegram.me/s/baliseo
https://telegram.me/s/seo_bali
https://telegram.me/s/workingclassman
https://tokokalimanar.wixsite.com/pafijawatengah
https://toto188.mn.co/about
https://toto188-1.jimdosite.com/
https://toto188-2.jimdosite.com/
https://toto188-3.jimdosite.com/
https://toto188-683924.webflow.io/
https://toto188-978f21.webflow.io/
https://toto188e.mystrikingly.com/
https://toto188e.pages.dev/
https://toto188-ececf3.webflow.io/
https://toto188f.mystrikingly.com/
https://toto188f.pages.dev/
https://toto188g.mystrikingly.com/
https://toto188g.pages.dev/
https://toto188-s-school1.teachable.com/p/home
https://toto188-s-school2.teachable.com/p/home
https://toto188-s-school3.teachable.com/p/home
https://toto-gaming.mn.co/about
https://trello.com/u/cobainini
https://trello.com/u/dokodakina
https://trello.com/u/firmanfarmin
https://trello.com/u/pafijawatengah/activity
https://trello.com/u/polisislota
https://wakelet.com/wake/euBUW3B-ej5o9q0dbodKq
https://wakelet.com/wake/j786tZ9iYo4l_q8g3mlLq
https://wakelet.com/wake/kA8RNJkpPq49F3o9wUu_v
https://wakelet.com/wake/kmfrasTGgvWyF2VvZwsuS
https://wakelet.com/wake/u6Mafnn7wBLtdpzuiPvaH
https://www.behance.net/pafijawatengah
https://www.behance.net/polisislota
https://www.behance.net/toto188e
https://www.behance.net/toto188ff
https://www.behance.net/toto188g
https://www.bitsdujour.com/profile/bbSiZ4
https://www.bitsdujour.com/profile/cTJcSl
https://www.bitsdujour.com/profiles/BpFNOc
https://www.bitsdujour.com/profiles/GRgahA
https://www.bitsdujour.com/profiles/zO32ZU
https://www.chaloke.com/forums/users/pafijawatengah/
https://www.chaloke.com/forums/users/polisislota/
https://www.chaloke.com/forums/users/toto188e/
https://www.chaloke.com/forums/users/toto188f/
https://www.chaloke.com/forums/users/toto188g/
https://www.commandlinefu.com/commands/view/34946/generate-a-gif-image-from-video-file
https://www.commandlinefu.com/commands/view/34946/generate-a-gif-image-from-video-file#comment
https://www.crunchbase.com/person/pafi-jawatengah
https://www.crunchbase.com/person/polisislota-polisi-slot
https://www.crunchbase.com/person/toto188e-toto188
https://www.crunchbase.com/person/toto188f-toto188
https://www.crunchbase.com/person/toto188g-toto188
https://www.demilked.com/author/dokodakina/
https://www.demilked.com/author/fukufakin/
https://www.demilked.com/author/pafijawatengah/
https://www.demilked.com/author/toto188f/
https://www.demilked.com/author/toto188g/
https://www.deviantart.com/pafijawatengah
https://www.deviantart.com/polisislota
https://www.deviantart.com/toto188e
https://www.deviantart.com/toto188f
https://www.deviantart.com/toto188g
https://www.divephotoguide.com/user/pafijawatengah
https://www.divephotoguide.com/user/polisislota
https://www.divephotoguide.com/user/toto188e
https://www.divephotoguide.com/user/toto188f
https://www.divephotoguide.com/user/toto188g
https://www.giantbomb.com/profile/pafijawatengah/
https://www.giantbomb.com/profile/polisislota/
https://www.giantbomb.com/profile/toto188e/
https://www.giantbomb.com/profile/toto188f
https://www.giantbomb.com/profile/toto188g
https://www.instapaper.com/p/14038949
https://www.instapaper.com/p/14038955
https://www.instapaper.com/p/14038961
https://www.instapaper.com/p/14038965
https://www.instapaper.com/p/14045375
https://www.intensedebate.com/people/pafijawatengah
https://www.intensedebate.com/profiles/polisislota
https://www.intensedebate.com/profiles/toto188e
https://www.intensedebate.com/profiles/toto188f
https://www.intensedebate.com/profiles/toto188g
https://www.liveinternet.ru/users/polisislota/profile
https://www.liveinternet.ru/users/toto188e/profile
https://www.liveinternet.ru/users/toto188f/profile
https://www.liveinternet.ru/users/toto188g/profile
https://www.magcloud.com/user/pafijawatengah
https://www.magcloud.com/user/polisislota
https://www.magcloud.com/user/toto188e
https://www.magcloud.com/user/toto188f
https://www.magcloud.com/user/toto188g
https://www.mixcloud.com/pafijawatengah/
https://www.mixcloud.com/polisislota/
https://www.mixcloud.com/toto188e/
https://www.mixcloud.com/toto188f/
https://www.mixcloud.com/toto188g/
https://www.myminifactory.com/users/pafijawatengah
https://www.myminifactory.com/users/polisislota
https://www.myminifactory.com/users/toto188e
https://www.myminifactory.com/users/toto188f
https://www.myminifactory.com/users/toto188g
https://www.producthunt.com/@pafijawatengah
https://www.producthunt.com/@polisislota
https://www.producthunt.com/@toto188e
https://www.provenexpert.com/pafijawatengah/
https://www.provenexpert.com/polisi-slot/
https://www.provenexpert.com/toto1881/
https://www.provenexpert.com/toto1882/
https://www.provenexpert.com/toto1883/
https://www.quia.com/profiles/pafijawatengahpa
https://www.quia.com/profiles/polisislotpo
https://www.quia.com/profiles/toto188t
https://www.quia.com/profiles/toto188to
https://www.quia.com/profiles/ttoto188
https://www.remotecentral.com/cgi-bin/forums/members/viewprofile.cgi?3fnaiq
https://www.remotecentral.com/cgi-bin/forums/members/viewprofile.cgi?4l4gh7
https://www.remotecentral.com/cgi-bin/forums/members/viewprofile.cgi?8am1ih
https://www.remotecentral.com/cgi-bin/forums/members/viewprofile.cgi?947ggf
https://www.remotehub.com/pafijawatengah.pafijawatengah
https://www.remotehub.com/polisislot.polisislot
https://www.remotehub.com/toto188.toto188.1
https://www.remotehub.com/toto188.toto188.2
https://www.remotehub.com/toto188.toto188.3
https://www.tumblr.com/blog/pafijawatengah
https://www.tumblr.com/blog/polisislota
https://www.tumblr.com/blog/toto188e
https://www.tumblr.com/blog/toto188f
https://www.tumblr.com/blog/toto188g
https://xtremepape.rs/members/pafijawatengah.441551/#about
https://xtremepape.rs/members/polisislota.441428/#about
https://xtremepape.rs/members/toto188e.441429/#about
https://xtremepape.rs/members/toto188f.441430/#about
https://xtremepape.rs/members/toto188g.441431/#about
https://yamcode.com/pafijawatengah
https://yamcode.com/polisi-slot
https://yamcode.com/toto188-14
https://yamcode.com/toto188-47
https://yamcode.com/toto188-70
https://taylorhicks.ning.com/photo/albums/coba-coba
Keep on working, great job!
I just like the helpful info you supply on your articles.
I will bookmark your blog and take a look at once more here regularly.
I’m rather sure I will learn a lot of new stuff proper
here! Good luck for the next!
Hello to every single one, it’s truly a pleasant for me to visit this site,
it includes priceless Information.
Hello there! This is my first comment here so I just wanted to give a quick
shout out and tell you I genuinely enjoy reading your articles.
Can you recommend any other blogs/websites/forums that cover the same topics?
Thank you!
Manchester City Wins the English Premier League Manchester City managed to become champions of the English Premier League last season after showing consistent performance under the direction of manager Pep Guardiola. Apart from winning the English Premier League title, Manchester City also won the English League Cup last season, showing their dominance on the domestic stage, for further information visit this website https://mediabolanews.blogspot.com/
This is a topic that is close to my heart… Take care! Exactly
where are your contact details though?
Castle Clash is a popular mobile strategy game developed by IGG.In this game, players build and upgrade forts, train an army of mythical creatures and heroes, and engage in battles against other players in real-time,Players can also join guilds, participate in various game modes, and compete in various events to get prizes and progress in the game, for further information visit this website https://waktogel303.jimdofree.com/tech/
Thanks for one’s marvelous posting! I genuinely enjoyed reading it, you will be a
great author.I will ensure that I bookmark your blog and will
come back down the road. I want to encourage you to continue your great writing, have a
nice afternoon!
One of the reasons why lottery remains popular is because of the ease of playing. Players can place bets with relatively small nominal amounts, but have the opportunity to win big prizes. Apart from that, lottery also offers various types of bets, such as 2D, 3D, 4D, free plugs, and many more, which provide different variations and opportunities for players. For further information, visit this website https://gameonline29.wordpress.com /
A teenager was arrested for committing the crime of robbery in Medan. The teenager was arrested after successfully carrying out a robbery at a minimarket in Medan. In his action, the teenager used a sharp weapon and threatened minimarket employees to give them money and valuables. For further information, visit this website https://wakbulu279.wixsite.com/berita-kriminal-news
Facing such strong feelings of love, Michael and Sarah could no longer restrain themselves. They felt attached to each other physically and emotionally, and finally decided to take their relationship a step further. Their feelings of desire and desire started to take over, and without many words, they started having sex, for further information visit this website https:// tokozeks.wordpress.com/
안성출장마사지
WOW just what I was searching for. Came here by searching for agen slot gacor
Terrific work! That is the kind of info
that should be shared around the internet.
Disgrace on the seek engines for not positioning this
post higher! Come on over and discuss with my
web site . Thanks =)
I like it when individuals get together and share thoughts.
Great site, stick with it!
Magnificent goods from you, man. I’ve remember your stuff previous to and you are just extremely magnificent.
I actually like what you’ve got right here, certainly like what you are saying and the way in which in which you assert it.
You make it entertaining and you continue to care for to keep it smart.
I cant wait to learn much more from you. This is actually a wonderful web site.
A fascinating discussion is definitely worth comment.
I believe that you ought to write more on this subject matter,
it may not be a taboo subject but generally folks don’t speak about these subjects.
To the next! Kind regards!!
강남콜걸
Very nice post. I simply stumbled upon your blog and wanted to say that I’ve really enjoyed browsing your blog
posts. After all I will be subscribing to your rss feed and I’m hoping you write once more soon!
Link exchange is nothing else however it is just placing the other person’s webpage link on your
page at proper place and other person will also do similar in support
of you.
https://www.cbdmd.com/cbd-gummies
https://cbdfx.com/collections/cbd-gummies/
https://cbd.market/cbd-gummies
https://rhealsuperfoods.com
https://www.theshroomshop.co.uk
https://www.dirteaworld.com
https://londonnootropics.com
https://ommushrooms.com
https://www.raracoffee.co.uk
https://uk.mudwtr.com
https://mushrooms-extract.com
https://www.detoxtrading.co.uk
https://www.hybridherbs.co.uk
https://www.healthysupplies.co.uk
https://dreammagicsuperfoodpowder.co.uk
https://sevenmushrooms.co.uk
https://timeoftheday.shop
https://drinkoyl.com
https://www.cognihax.com
https://feelgud.co.uk
https://knownnutrition.co.uk
https://uk.iherb.com
https://mushroomfx.com
https://eatfungies.com
https://trytroop.com
https://thisisgreenhaus.co
https://uk.naturecan.com
https://greensoulcbd.uk
https://www.nudehealth.co.uk
https://www.naturesroot.co.uk
https://cheerfulbuddha.co.uk
https://balancecoffee.co.uk
https://noomf.co.uk
https://freshlyfermented.co.uk
https://www.pardasa.co.uk
https://flawlesscbd.co.uk
https://www.ryzesuperfoods.com
https://matchayoko.com
https://moyamatcha.co.uk
https://matchaandco.co.uk
https://www.theteamakers.co.uk
https://www.ilovematchatea.co.uk
https://chamberlaincoffee.eu
https://www.birdandblendtea.com
https://www.teapigs.co.uk
https://www.buywholefoodsonline.co.uk
https://www.nutsinbulk.co.uk
https://everydaysuperfood.co.uk
https://www.wholefoodessentials.co.uk
https://www.lifelabsupplements.com
https://www.weightworld.uk
https://www.nutriextracts.co.uk
https://peaksupps.co.uk
https://www.souschef.co.uk
https://teaologists.co.uk
https://spacegoods.com
https://drinkcann.com/
https://drinkwynk.com/
https://happihourdrink.com/
https://drinkdelta.com/
https://wearedaytrip.com/
https://trynowadays.com/
https://www.drinkthenorth.com/
https://hiseltzers.com/
https://www.drinkdayone.com/
https://hempbombs.com/cbd-gummies/
https://www.charlottesweb.com/all-charlottes-web-hemp-cbd-supplements/cbd-gummies
https://greenroads.com/collections/cbd-edibles-gummies
https://fivecbd.com/products/cbd-gummies?variant=39671891558490
https://www.cornbreadhemp.com/products/full-spectrum-cbd-gummies?selling_plan=772636852
https://cheefbotanicals.com/cbd-gummies/vegan/
https://hempbombsplus.com/
https://www.exhalewell.com/delta-8-gummies/
https://www.3chi.com/product/delta-8-thc-gummies/
https://area52.com/delta-8-gummies/
https://mysticlabsd8.com/delta-8-gummies/
https://purekana.com/products/delta-8-gummies-500mg/
https://www.healthline.com/health/best-cbd-gummies
https://treehouse-cbd.com/
https://www.dallasnews.com/branded-content/2022/09/17/best-cbd-gummies-for-sleep-in-2022-top-5-cbd-edibles-to-rest-recover-with/
https://www.medicalnewstoday.com/articles/hemp-gummies-vs-cbd-gummies-what-to-know
https://www.medicalnewstoday.com/articles/best-cbd-gummies
https://trehouse.com/
https://www.exhalewell.com/cbd-gummies/
https://area52.com/
https://hempbombsplus.com/delta-8-gummies/
https://www.bondara.co.uk
https://www.simplypleasure.com
https://www.sextoys.co.uk
https://www.annsummers.com
https://www.pulseandcocktails.co.uk
https://www.nastygal.com
https://loveplugs.co.uk
https://loveoutlet.co.uk
https://sextoysaffair.co.uk
https://www.prowler.co.uk
https://liberation-x.com
https://www.bedroompleasures.co.uk
https://www.theadulttoyshop.com
https://lovewoo.co.uk
https://www.lovehoney.co.uk
https://www.fetshop.co.uk
https://www.extremerestraints.com
https://www.adultshopit.co.uk
https://www.skintwo.com
https://www.megapleasure.co.uk
https://www.adameve.com
https://www.mylovely.uk
https://www.sinfulthrills.co.uk
https://delightoys.co.uk
https://nicennaughty.co.uk
https://www.loveandvibes.co.uk
https://luvero.co.uk
https://www.patchadam.com/collections/cbd-edibles
https://www.amny.com/sponsored/best-cbd-gummies-of-2022-top-7-great-tasting-and-relaxing-edibles/
https://theislandnow.com/best-cbd-gummies/
https://ilovegreengorilla.com/products/cbd-gummies/
https://goshango.com/marijuana-products/edibles/gummies/
https://vermafarms.com/collections/cbd-gummies
https://themendico.com/products/no-thc-cbd-gummies
https://anniesrx.com/articles/what-are-cbd-gummies-and-should-i-take-them
https://koicbd.com/cbd/gummies/
https://zebracbd.com/collections/cbd-gummies
https://premiumjane.com/cbd-gummies/
https://edensherbals.com/CBD-Gummies_c_1.html
https://www.resiliencecbd.com/product/cbd-gummies/
https://www.gethappyhemp.com/
https://www.newphaseblends.com/what-are-cbd-gummies/
https://www.canaxlife.com/
https://naternal.com/collections/cbd-gummies
https://cheefbotanicals.com/cbd-gummies/
https://www.dmagazine.com/sponsored/2022/09/strongest-hemp-edibles-2022/
https://wyldcbd.com/
https://bluebirdbotanicals.com/products/cbd-gummies
https://www.fabcbd.com/collections/cbd-gummies
https://naturebox.com/cbd-gummies
https://www.orlandomagazine.com/best-cbd-gummies-of-2022/
https://www.rrmeds.com/products/30mg-hemp-extract-gummies-vegan
https://venacbd.com/collections/cbd-gummies
https://nextevo.com/collections/cbd-gummies
https://www.webmd.com/cbd/best-cbd-gummies-for-anxiety
https://www.discovermagazine.com/lifestyle/25-best-cbd-gummies-for-erectile-dysfunction-for-2022
https://sundayscaries.com/products/cbd-gummies
https://medterracbd.com/pages/gummies
https://joyorganics.com/collections/gummies
https://highlinewellness.com/collections/cbd-gummies
https://www.chicagomag.com/promotion/best-cbd-gummies-of-2022-discreet-and-reliable-daily-support/
https://penguincbd.com/products/cbd-gummies
https://cbdamericanshaman.com/cbd-gummies
https://hometownherocbd.com/collections/delta-8-gummies
https://www.dmagazine.com/sponsored/2022/10/best-delta-8-gummies/
https://thechalkboardmag.com/delta-8-gummies-what-you-should-know/
https://koicbd.com/delta-8/gummies/
https://thehempdoctor.com/delta-8-thc/delta-8-gummies-edibles/
https://www.usmagazine.com/shop-with-us/news/best-delta-8-gummies-9-top-brands/
https://gotflora.com/products/delta-8-gummies?variant=37042761629855
https://cfah.org/delta-8-gummies-effects/
https://www.ounceofhope.com/product/delta-8-vegan-gummies/
https://goodekind.com/product/delta-8-gummies/
https://www.thegramco.com/products/delta-8-gummies-750-mg-25mg-each
https://www.chicagomag.com/promotion/best-delta-8-gummies-top-5-sites-for-buzzy-sweets-reviewed/
https://hightimes.com/sponsored/why-you-should-try-delta-8-gummies-for-your-health/
https://cannabuddy.com/product-category/delta-8-thc/delta-8-thc-gummies/
https://www.diamondcbd.com/delta-8-gummies
https://www.discovermagazine.com/lifestyle/10-strongest-delta-8-gummies-this-year
https://deltamunchies.com/delta-8-gummies/
https://www.thegreendragoncbd.com/product-category/delta-8-gummies/
https://www.atlrx.com/collections/delta-8-gummies/
https://cbdfx.co.uk/collections/cbd-gummies
https://orangecounty-cbd.com/collections/cbd-gummies-sweets
https://www.cbd-guru.co.uk/product-category/cbd-gummies/
https://herbalhealthcbd.co.uk/product-category/cbd-gummies/
https://uk.naturecan.com/products/naturecan-10mg-cbd-gummies
https://supremecbd.uk/collections/cbd-edibles
https://www.cbdoil.co.uk/product-category/cbd-gummies/
https://www.goodrays.com/collections/cbd-gummies/
https://flawlesscbd.co.uk/collections/cbd-gummies
https://www.canndid.co.uk/cbd-gummies/
https://cannaraycbd.com/shop/gummies/
https://www.mayoclinic.org/healthy-lifestyle/consumer-health/expert-answers/https://cbdhealingresources.com/product/full-spectrum-tri-blend-gummies-600mg/-cbd-safe-and-effective/faq-20446700
https://lovehemp.com/collections/cbd-edibles
https://thegoodlevel.com/cbd-gummies-uk/
https://vitalitycbd.co.uk/products/cbd-gummy-bears
https://www.simply-cbd.co.uk/products/cbd-gummies?variant=42521725862112
https://www.natureshealthbox.co.uk/cbd/cbd-gummies
https://www.cbii-cbd.com/shop/cbd-gummies
https://groceries.asda.com/product/cbd-oils-gummies-rubs/vitality-cbd-40-gummy-bears-raspberry-orange-flavours/1000310031080
https://www.charlottesweb.com/cbd-lemon-calm-gummy
https://www.webmd.com/cbd-gummies
https://theveritasfarms.com/.well-known/captcha/?r=%2Fproduct%2Fcbd-gummies%2F
https://www.getsoul.com/products/cbd-gummies?variant=41204275445900
https://cbdliving.com/collections/cbd-gummies
https://truharvestfarms.com/.well-known/captcha/?r=%2Fproduct%2Ffull-spectrum-cbd-gummies%2F
https://www.bullycrewcbd.com/products/bully-crew-cbd-750mg-gummies-30-count
https://betterhealthmarket.com/cbd-living-cbd-gummies-30gummies-55
https://www.resilientremedies.com/products/calm-cbd-gummies?variant=41478622118047
https://hempbombs.com/product/100-count-cbd-gummies/
https://www.barkerwellness.com/collections/gummies
https://tribetokes.com/product-item/cbd-gummy-bears/
https://hellobatch.com/collections/cbd-gummies
https://shop.lifetime.life/cbd-gummies-recovery
https://us.naturecan.com/collections/cbd-gummies
https://www.gethappyhemp.com/product-category/cbd-gummies/
https://sojihealth.com/shop/gummies-softgels/
https://myeq.com/product/daily-gummies/
https://ecosciences.com/cbd-gummies/
https://www.swansonvitamins.com/ncat1/Vitamins+and+Supplements/ncat2/Supplements/ncat3/CBD/q
https://www.cbdsocial.com/shop-by-need/value-packs/cbd-gummies-1.html
https://holmesorganics.com/products/cbd-gummies
https://moodysmedicinals.com/products/cbd-gummies
https://focl.com/products/focl-premium-cbd-gummies
https://thegeorgiahempcompany.com/product-category/edibles/edibles-gummies/
https://hempmedspx.com/product/cbd-gummies/
https://edensherbals.com/CBD-GUMMIES-500-MG_p_1.html
https://www.samsonextracts.com/collections/wholesale-cbd-gummies
https://vitl-cbd.com/
https://lordjones.com/collections/cbd-edibles-confections
https://socialcbd.com/cbd-gummies/
https://cbdchoice.com/shop-products/cbd-edibles/cbd-gummies
https://cannahemp.com/collections/gummies
https://bisonbotanics.com/product-category/cbd-gummies/
https://halfdaycbd.com/collections/cbd-gummies
https://cbdabilene.com/product-category/edibles/
https://plainjane.com/cbd-gummies/
https://www.edwinsedibles.com/shop-products/
https://palmettoharmony.com/product/gummies/
https://www.cannariver.com/products/cbd-gummy
https://dragonflyhempcbd.com/collections/cbd-gummies
https://purecraftcbd.com/collections/gummies
https://levelselectcbd.com/cbd/gummies/
https://frannysfarmacy.com/product/frannys-farmacy-gummies-500mg/
https://wingedwellness.com/products/sleepy-cbd-gummies
https://wearedaytrip.com/collections/cbd-gummies
https://www.cubbingtons.com/combo-full-spectrum-cbd-gummies/
https://infinitecbd.com/product/gummies/
https://higherlovewellness.com/product-category/cbd-gummies/
https://www.heritagecbd.com/collections/gummies
https://goodcbd.com/collections/cbd-gummies
https://www.pluscbdoil.com/cbd-products/cbd-gummies
https://drinkdefy.com/products/recover-cbd-gummies-750mg
https://vidaoptimacbd.com/collections/cbd-gummies
https://www.auntbonnies.com/product/30mg-cbd-gummies/
https://hollyweedcbd.com/shop/cbd-edibles/cbd-gummy-cubes/
https://creatingbetterdays.com/products/spectrum-delta-9-thc-cbd-gummies-cherrylime-30ct
https://cbdpharm.com/category/edibles/gummies/
https://ooolalattes.com/gummies/
https://www.extractlabs.com/product/cbd-gummies/
https://www.sunstatehemp.com/cbd-gummies
https://www.vitaminshoppe.com/c/vitamins-supplements/supplements/natural-extract
https://kushqueen.shop/collections/gummiesrx-cbd-chews
https://www.crescentcanna.com/.well-known/captcha/?r=%2Fcbd-gummies%2F
https://justlive.com/collections/gummies
https://neurogan.com/collections/cbd-edibles-snacks
https://missionfarmscbd.com/pure-cbd-gummies-with-nano-cbd
https://sweetsensiwellness.com/product-category/cbd-gummies/
https://www.cypresshemp.com/product-page/full-spectrum-900mg-anytime-cbd-gummies
https://blushwellness.com/collections/cbd-gummies/
https://indigoridgehemp.com/product/cbd-gummies/
https://earlybirdcbd.com/products/full-spectrum-cbd-gummies-4-pack
https://musclemx.com/product-category/cbd-gummies/
https://beeskneescbds.com/product-category/cbd-gummies/
https://peels.com/products/cbd-gummies
https://reliveeveryday.com/re-assure-vegan-cbd-gummies-various-10mg/
https://takespruce.com/product/cbd-gummies/
https://happyface.com/collections/gummies
https://cbdhealingresources.com/product/full-spectrum-tri-blend-gummies-600mg/
https://biolyfebrands.com/product/cbd-gummies/
https://mandaracbd.com/product/cbd-gummies/
https://www.thecbdistillery.com/cbd-shop/gummies/thc-cbd-gummies/
https://www.farmhouse.delivery/products/kanha-peach-cbd-gummies-4-2
https://sunsetlakecbd.com/product/cbd-gummies/
https://illuminatelabs.org/blogs/health/cbd-gummies-for-sex
https://trycaliper.com/products/premium-pure-cbd-gummies-berry-mix-30-ct
https://wyldcbd.com/products/lemon-gummies
https://www.shopcanopy.com/en/shop/all/confections
https://vivecbd.com/products/cbd-gummies?variant=20734918066279
https://www.google.com/search?q=vape+disposables&sxsrf=AJOqlzWz_nVaf_z2nRxW3byxmhItpxQARw:1678050036024&ei=9AIFZM6QAeGQhbIPuLG06Ag&start=60&sa=N&ved=2ahUKEwjO2r_C18X9AhVhSEEAHbgYDY04MhDw0wN6BAgFEBo&biw=1920&bih=929&dpr=1
https://www.hollandandbarrett.com/the-health-hub/vitamins-and-supplements/supplements/what-are-the-best-cbd-gummies/
https://www.hollandandbarrett.com/shop/vitamins-supplements/cbd/
https://lloydspharmacy.com/collections/cbd-tablets-and-capsules
https://www.boots.com/healthspan-cbd-oil-192mg-30-capsules-10263818
https://canabidol.com/cbd-uk/cbd-capsules/
https://cannaraycbd.com/shop/cbd-capsules/
https://www.healthline.com/health/best-cbd-capsule
https://www.lovecbd.org/shop/
https://www.healthline.com/health/best-cbd-cream-for-pain
https://www.forbes.com/health/body/best-cbd-creams/
https://www.tesco.com/groceries/en-GB/shop/health-and-beauty/lifestyle-and-wellbeing/cbd-cannabidiol-sleep-aids-and-stress-relief/cbd-creams-and-balms
https://canabidol.com/cbd-uk/cbd-cream/
https://www.hollandandbarrett.com/shop/product/holland-barrett-cbd-day-cream-60033048
https://edensgate.co.uk/collections/cbd-creams
https://cbdfx.co.uk/collections/cbd-creams-balms
https://accesscbd.uk/cbd-cream/
https://www.healthline.com/health/top-10-cbd-lotions-creams-and-topicals
https://www.vaperstore.co.uk/collections/cbd-disposable-vapes
https://www.royalvapes.co.uk/collections/cbd-disposables
https://flawlesscbd.co.uk/collections/cbd-vape-pens
https://orangecounty-cbd.com/collections/cbd-disposable-vapes
https://supremecbd.uk/products/supreme-cbd-large-gummy-bears-10mg
https://vapoholic.co.uk/eliquid-range/cbd-e-liquid/
https://flawlesscbd.co.uk/collections/cbd-vape-oil
https://britishcannabis.org/shop/cbd-vape/
https://cbdfx.co.uk/collections/cbd-vape-juice
https://www.ecigclick.co.uk/best-cbd-oil-e-liquids/
https://www.totallywicked-eliquid.co.uk/cbd-vape-liquid-information
https://britishcbd.net/cbd-uk/cbd-vape-liquids/
https://canabidol.com/cbd-uk/cbd-vape-oil/
https://www.iceheadshop.co.uk/vape/cbd-oil
https://orangecounty-cbd.com/collections/cbd-vape-juice-e-liquid
https://evapo.co.uk/cbd
https://www.cbdoil.co.uk/product-category/cbd-e-liquid/
https://vitalitycbd.co.uk/collections/cbd-e-liquids
https://www.vaperstore.co.uk/collections/cbd-eliquid-vape-juice
https://www.midss.org/health/best-cbd-vape-oil
https://vpz.co.uk/collections/cbd
https://www.royalvapes.co.uk/collections/cbd-cartridges
https://flawlesscbd.co.uk/collections/cbd-edibles
https://www.cbdoil.co.uk/product-category/cbd-edibles/
https://hempelf.com/collections/cbd-edibles
https://www.iceheadshop.co.uk/vape/cbd-oil/cbd-edibles
https://britishcbd.net/cbd-uk/cbd-edibles/
https://learn.freshcap.com/tips/mushroom-capsules-guide/
https://www.theshroomshop.co.uk/collections/mushroom-capsules
https://mushrooms4life.com/
https://flawlesscbd.co.uk/collections/mushroom-capsules
https://www.healthysupplies.co.uk/7-mushroom-blend-500mg-sussex-wholefoods.html
https://cotswoldmushrooms.com/product/lions-mane-capsules/
https://www.bulk.com/uk/mushroom-complex-capsules.html
https://www.indigo-herbs.co.uk/shop/buy/organic-7-mushroom-blend-capsules-500mg
https://www.nutravita.co.uk/products/advanced-mushroom-superblend-6-mushroom-formula-180-vegan-capsules
https://antioxi.co.uk/collections/mushroom-extract
https://www.bristolbotanicals.co.uk/cat-79
https://naturesplus.co.uk/products/immune-mushroom-capsules
https://woodiesuk.com/products/cbd-capsules-mushrooms
https://www.planetorganic.com/collections/mushrooms
https://www.rawliving.co.uk/collections/mushroom-powders-extracts
https://www.indigo-herbs.co.uk/shop/buy/mushroom-nutrition-mushroom-powders
https://www.inspiredtaste.net/52603/mushroom-powder/
https://www.vapesuperstore.co.uk/collections/disposable-vape-kits
https://www.vapedisposables.co.uk/
https://www.ukecigstore.com/collections/disposable-vape-kits
https://www.vapeshop.co.uk/collections/disposable-vape-kits
https://vapoholic.co.uk/eliquid-range/disposable-vapes/
https://www.gosmokefree.co.uk/disposable-vapes/
https://diyeliquids.co.uk/product-category/hardware-vape-kits-mods/disposable-vape-kits/
https://www.totallywicked-eliquid.co.uk/disposable-vape
https://vpz.co.uk/collections/disposables
https://www.royalvapes.co.uk/collections/disposable-vapes
https://www.vapourcore.com/collections/disposable-vapes
https://www.washington-vapes.co.uk/disposable-vapes
https://www.vampirevape.co.uk/e-cigarette-kits/disposable-vapes
https://www.ecigarettedirect.co.uk/disposable-vapes
https://tablites.com/collections/disposable-vapes
https://www.eco-vape.co.uk/product-category/hardware/disposable-vapes/
https://www.vaperstore.co.uk/collections/0mg-disposable-vapes
https://www.alectrofag.co.uk/collections/disposable-kits
https://www.88vape.com/collections/88vape-disposables
https://www.vapestore.co.uk/disposable-vape.html
https://vapekit.co.uk/vape-kits-c932/disposables-c1338
https://vapeuk.co.uk/disposable-vapes
https://multivape.co.uk/collections/disposables-vapes
https://www.zombievapes.co.uk/collections/disposables
https://www.theelectroniccigarette.co.uk/disposable-vapes
https://www.electrictobacconist.co.uk/disposable-vapes
https://belfastvapedelivery.co.uk/collections/disposable-e-cigs
https://www.ecigwizard.com/collections/disposable-vape-kits
https://www.go-liquid.co.uk/disposables/ivg-disposables
https://www.mycigara.com/collections/disposable-vape
https://www.primevapes.co.uk/collections/disposable-vape-pens
https://www.vapedinnerlady.com/collections/disposable-e-cigarette
https://www.royalflushvape.co.uk/collections/disposable-vape-kits
https://theecig.co.uk/collections/best-disposables-vapes-uk
https://vapegreen.co.uk/disposables/vape-bars
https://www.ichorliquid.co.uk/collections/all-disposables
https://ivapegreat.com/collections/all-bars
https://www.vapemate.co.uk/disposable-pods.html
https://www.houseofvapeslondon.co.uk/collections/disposable-vapes
https://www.haypp.com/uk/vape/disposable-vapes/
https://www.vaporshopdirect.com/collections/disposable-vape
https://ecigone.co.uk/collections/disposables
https://oxfordvapours.com/product-category/disposables/
https://www.vuse.com/gb/en/e-liquids/disposable-vapes
https://cloudcityuk.com/collections/disposables
https://mixjuice.co.uk/product-category/vape-kits/
https://mipod.com/collections/disposable-vape
https://islandvapeuk.com/collections/disposable-vapes
https://www.gourmeteliquid.co.uk/products/ske-crystal-original-disposable-vape-pods
https://greyhaze.co.uk/collections/disposable-pod-vapes
https://vaping360.com/best-beginner-e-cigs-vapes/disposables/
https://www.vapouriz.co.uk/disposables.html
https://disposable-vape.co/
https://www.vapeandgo.co.uk/product-category/disposable-vape-kits/
https://www.theaceofvapez.com/collections/disposable-vapes
https://www.cramvape.co.uk/collections/disposable-pens
https://www.nzovape.com/
https://www.ecigclick.co.uk/best-disposable-vapes/
https://www.aquavape.co.uk/product-category/disposable-e-cigarette/
https://www.e-liquids.uk/vape-hardware/vape-kits/disposables
https://www.thevapehouse.co.uk/collections/disposable-vapes
https://britishcannabis.org/shop/
https://thecbdshop.co.uk/
https://drink-trip.com/collections/cbd-oil
https://www.cbdoilsuk.com/
https://hemppointcbd.co.uk/
https://www.cbd-guru.co.uk/product-category/cbd-oils/
https://canabidol.com/
https://vitalitycbd.co.uk/collections/cbd-oils
https://accesscbd.uk/buy-cbd-oil/
https://cbdfx.co.uk/collections/cbd-oil
https://www.brownscbd.co.uk/
https://fourfive.com/
https://www.hempen.co.uk/shop/cbd/refined-organic-cbd-oil/
https://www.healthspan.co.uk/cbd-oil#t=CBD-Oil-Products&numberOfResults=15
https://cannaraycbd.com/
https://cbdiablo.co.uk/
https://flawlesscbd.co.uk/
https://uk.naturecan.com/collections/cbd-oils
https://www.fortheageless.com/blogs/cbd-oil-uk-blog/strongest-cbd-products
https://www.cbdoil.co.uk/
https://cbdshopy.co.uk/collections/cbd-oil
https://supremecbd.uk/collections/full-spectrum-cbd-oil
https://britishcbd.net/cbd-uk/cbd-oil/
https://lovehemp.com/
https://www.hdrop.co.uk/
https://www.cbdpurelife.co.uk/
https://vapoholic.co.uk/eliquid-range/cbd-oil/
https://cbdqueen.co.uk/product-category/cbd-oil/
https://bodyandmindbotanicals.com/products/cbd-oil
https://www.cbii-cbd.com/200mg
https://medterracbd.co.uk/collections/cbd-oils
https://canamis.com/collections/cbd-oil
https://savagecabbage.co.uk/
https://www.cbdultra.co.uk/
https://truthnaturals.co.uk/
https://ammalife.co.uk/collections/cbd-oils/
https://www.jersey-hemp.com/
https://www.endoca.com/en-gb/cbd-products/cbd-oil-uk
https://ulu.com/certified-organic-cbd-oils/
https://www.uniquecbd.co.uk/collections/cbd-oil
https://www.simplysupplements.co.uk/cbd
https://cbd-one.co.uk/blog/is-cbd-oil-legal-uk/
https://you-well.co.uk/strongest-cbd-oil-products-buy/
https://stepbysteprecovery.co.uk/cbd-ban-oil-to-be-pulled-from-uk-shops-and-shelves/
https://www.calmdrinks.co.uk/collections/cbd-oils
https://www.cropengland.co.uk/shop/cbd-oils/low-strength-cbd-oil/
https://dragonflycbd.com/
https://www.natureshealthbox.co.uk/brands/cbd-one
https://www.bristolcbd.co.uk/
https://beyoucbd.co.uk/products/cbd-oil
https://thetonictribe.com/product-category/cbd-oil/
https://vandyou.com/collections/cbd
https://www.lovecbd.org/
https://purecbdhealth.com/
https://hempelf.com/
https://www.simply-cbd.co.uk/
https://thegoodlevel.com/
https://www.justvitamins.co.uk/blog/is-cbd-oil-legal-in-the-uk/
https://drk-cbd.co.uk/collections/oils
https://www.healthwellbeing.com/the-7-top-cbd-brands-for-anxiety-in-the-uk/
https://vibescbd.co.uk/
https://otocbd.com/
https://www.bnaturaloil.co.uk/
https://grassandco.com/
https://potyque.com/collections/cbd-products
https://www.honeyheaven.co.uk/collections/organic-cbd-products
https://hempura.shop/
https://nordicoil.co.uk/
https://prolifecbd.co.uk/
https://ukcbdoil.org.uk/
https://orangecounty-cbd.com/collections/cbd-oils
https://lloydspharmacy.com/blogs/vitamins-and-supplement-advice/is-cbd-oil-legal-in-uk
https://honesthemp.co.uk/
https://www.sapphireclinics.com/
https://thedrug.store/collections/cbd-oils
https://miracleleaf.co.uk/shop-cbd/
https://www.cibdol.com/uk/cbd-oil
https://www.silverson.co.uk/en/resource-library/application-reports/cbd-oil-in-cosmetic-products-uk
https://www.pharmica.co.uk/vitamins-supplements/cbd/
https://www.iceheadshop.co.uk/cbd-shop
https://www.greenbox.co.uk/collections/cbd-products/cbd-oil/
https://www.fg-cbd.co.uk/shop/
https://www.franchise-uk.co.uk/franchise/milagro-cbd-oil-franchise/
https://www.cbdvillage.co.uk/organic-cbd-tinctures/
https://www.hempheros.co.uk/
https://www.wildandrust.co.uk/collections/organic-cbd
https://ourremedy.co.uk/
https://www.yourhemp.co.uk/product-category/cbd-oils/
https://cbdlifeuk.com/
https://surepure.co.uk/
https://hemptyonline.co.uk/
https://www.karmacoastcbd.co.uk/
https://www.budandtender.com/
https://www.cornwalllive.com/special-features/best-cbd-oils-uk-5711482
https://www.simplycbdwales.com/product/4000mg-40-premium-cbd-oil-copy/
https://www.purelifeuk.co.uk/cbd-products/
https://www.zencbd.co.uk/
https://www.missionc.com/collections/cbd-oils
https://www.naturalstrains.co.uk/collections/cbd-oils
https://www.organicsecrets.co.uk/
https://www.arthritis.org/health-wellness/healthy-living/managing-pain/pain-relief-solutions/cbd-for-arthritis-pain
https://www.healtheuropa.com/best-cbd-oils-to-look-out-for-in-the-uk-in-2022/115459/
https://www.thehempshop.co.uk/cbd-oil-uk.html
https://cbdbrothers.com/all-categories/
https://nuleafnaturals.com/
https://www.sps.nhs.uk/articles/cannabidiol-oil-potential-adverse-effects-and-drug-interactions/
https://www.pure100cbd.co.uk/
https://www.mamedica.co.uk/
https://www.royalqueenseeds.com/
https://www.charlottesweb.com/
https://sundayscaries.com/
https://joyorganics.com/
https://fivecbd.com/
https://focl.com/
https://loveplugs.co.uk/collections/butt-plug
https://liberation-x.com/categories/butt-plug.html?utm_source=google&utm_medium=cpc&gclid=CjwKCAiAmJGgBhAZEiwA1JZoljmbMzFdLJFoGM6CK-pF2qTpV59bpufyA-6i435J2chTOdxT9vJ4JRoCzLcQAvD_BwE
https://www.mylovely.uk/anal-range/butt-plugs?gclid=CjwKCAiAmJGgBhAZEiwA1JZollK3EH34q0DhjVu0AcljWVjJhOjhsOQnf0426uGK-EwGSWtV99SFlhoCwlAQAvD_BwE
https://loveoutlet.co.uk/collections/butt-plugs
https://lovegasm.co/blogs/butt/safety-and-butt-plugs
https://www.prowler.co.uk/sex-toys/bum-toys/butt-plugs.html?p=3
https://www.lovense.com/vibrating-butt-plug
https://www.pulseandcocktails.co.uk/filter/butt-plugs/
https://www.bondara.co.uk/sex-toys/butt-plugs
https://vaperanger.com/collections/hhc-disposables
https://puffxtrax.com/collections/hhc-disposables-vape-stix
https://sponsored.dmagazine.com/sponsored/2023/04/best-delta-8-gummies/
https://uk-cbdoils.co.uk
https://cbdbibleuk.co.uk
https://www.cbdmall.com/delta-8-gummies
https://www.superdrug.com
https://www.healthline.com/health/delta-8
https://galaxytreats.com/collections/delta-8-gummies
https://cannabislife.com/products/gummies-edibles/delta-8/assorted-flavors-gummies-750mg/
https://www.cbdoilsupplies.co.uk
https://professorherb.co.uk
https://provacan.co.uk
https://www.evopure.co.uk
https://charlestonhempcollective.com/wp-content/uploads/gummies.jpg
https://cbdoracle.com/picks/best-hhc-disposable-vape-pens/
https://www.cannaaidshop.com/product/hhc-disposables/
https://www.outlookindia.com/outlook-spotlight/best-hhc-gummies-news-279178
https://www.cbdmall.com/hhc-disposable-vapes
https://budmother.com/collections/hhc-vape
https://www.miistercbd.com
https://humanityhealthcbd.com/hhc/
https://golferscbd.co.uk
https://cannabuddy.com/product-category/delta-10-thc/delta-10-thc-gummies/
https://www.mensjournal.com/food-drink/best-delta-8-thc-gummies
https://mlchicagosocial.com/hhc-gummies-vs-delta-8-gummies-a-break-down
https://www.cbdoilking.co.uk
https://azteccbd.co.uk
https://dvntd8.com/.well-known/captcha/?r=%2Fall-products%2Fdelta-8-disposable-vape%2F
https://bannerharvest.com/product/delta-8-thc-gummies/
https://royalcbd.com
https://celticwindcrops.com
https://greenpostcbd.com/collections/disposable-vape-pens
https://cbdinfusions.co.uk
https://feelgud.co.uk
https://caligreens.co.uk
https://coloradobreedersdepot.com/.well-known/captcha/?r=%2Fproduct%2Ftangie-hhc-disposable-vape-1ml%2F
https://eightysixbrand.com/product-category/hhc/white-series-hhc-disposables/
https://www.orlandomagazine.com/best-hhc-disposable/
https://stronglife.co.uk
https://buyeverest.com/products/gummies/
https://bakedhhc.com/products/3gram?variant=40172061753429
https://www.sandiegomagazine.com/partner-content/best-delta-8-gummies-of-2023-top-thc-edibles-to-buy-online/article_dffcef74-dfaa-11ec-a8f6-17841ee96074.html
https://cbdrevital.co.uk
https://thisisgreenhaus.co
https://viiahemp.com/product/hhc-disposable-vape-tropic-thunder/
https://www.apotheca.org/delta-8-thc/delta-8-gummies-and-edibles.html
https://www.everybudy.co.uk
https://www.cannariver.com/products/hhc-highlighter
https://versedvaper.com
https://deltamunchies.com/hhc-vape-pens/
https://hempoilsuk.com
https://www.vapourdays.co.uk
https://taurigum.com/products/strongest-dose-yet-blazin-berry-50mg-delta-8-gummies
https://www.britishhypermarket.com
https://hometownherocbd.com/collections/hhc-gummies
https://boomheadshop.com/collections/disposables
https://cbdleafline.co.uk
https://www.welllcbd.co.uk
https://gotflora.com/products/delta-8-gummies
https://canavape.co.uk
https://www.hempine.co.uk
https://www.3chi.com/product/hhc-disposable-vape/
https://zenbears.co.uk
https://cbdwellnesscentre.co.uk
https://cheerfulbuddha.co.uk
https://www.thecalmleaf.com/juicy-hhc-blend-day-night-double-sided-disposable-vape-pen/
https://www.canatura.com/en/hhc/hhc-vapes/hhc-disposable-vape-pens
https://www.crescentcanna.com/d8-gummies/
https://cbdnorwich.co.uk
https://www.exhalewell.com/hhc-disposable-vapes/
https://shop.cookies.co/collections/delta-8-hhc-2g-blunts
https://www.associatedcbd.co.uk
https://hemprove.co.uk
https://www.cbd-uk.com
https://cbd.co.uk
https://www.misteliquid.co.uk
https://www.naturecreations.co.uk
https://vaping360.com/best-cannabis-products/delta-8-gummies/
https://www.euphoria.eu/product/detail/euphoria-hhc-disposable-vape-pen-zkittles
https://vape.co.uk
https://upliftcbdco.com/the-ultimate-guide-to-hhc-disposable-vape-pen/
https://www.thegreendragoncbd.com/product/cannabis-life-hhc-disposable-vape-pens-3ml
https://greatcbdshop.com/product-category/hhc/hhc-disposable-vape/
https://thedopestshop.com/collections/hhc-disposable-vapes
https://hapihemp.co.uk
https://viiahemp.com/product/hhc-disposable-vape-ghost-train-haze/
https://blessedcbd.co.uk
https://www.manukapharm.co.uk
https://www.extractlabs.com/product/hhc-disposable-vape-gods-gift/
https://meetharmony.com
https://eightysixbrand.com/product-category/delta-8-inhalables/delta-8-disposables/
https://www.extractlabs.com/product/delta-8-gummies/
https://drwatsoncbd.com
https://www.elementvape.com/hhc-disposables
https://www.cbdmall.com/hhc-gummies
https://koicbd.com/hhc-disposable-vapes/
https://www.discovermagazine.com/lifestyle/21-best-delta-8-gummies-on-market-in-2023
https://www.exhalewell.com/hhc-gummies/
https://cbd-angel.co.uk
https://hempwell.co.uk
https://www.highkind.com
https://cannubu.com
https://thegeorgiahempcompany.com
https://www.fabcbd.com
https://www.brownscbd.co.uk
https://www.cbdplanetuk.com
https://www.crescentcanna.com
https://canabidol.com
https://www.nuhemp.co.uk
https://highlinewellness.com
https://www.terpenejourney.com
https://www.cbii-cbd.com
https://www.budsking.com
https://www.cannavana.org
https://www.excitecbd.co.uk
https://www.3chi.com
https://www.sapphireclinics.com
https://cbdarmour.co.uk
https://cheefbotanicals.com
https://halfdaycbd.com
https://www.cbdsocial.com
https://www.rrmeds.com
https://sweetsensiwellness.com
https://incr-edibles.co.uk
https://www.verilife.com
https://cbdfx.com
https://www.cannacares.co.uk
https://alchemynaturals.com
https://bigjuiceuk.co.uk
https://www.lume.com
https://www.thegreendoctorcbdoil.co.uk
https://www.goodrays.com
https://drinkdefy.com
https://vapeuk.co.uk
https://flowerpowerbotanicals.com
https://koicbd.co.uk
https://www.getsoul.com
https://www.naturallycbd.co.uk
https://www.canndid.co.uk
https://www.vapourcore.com
https://www.smokelouduk.co.uk
https://dragonflycbd.com
https://fourfive.com
https://kalmastecbd.co.uk
https://www.google.com
https://thelondondispensary.com
https://honestcbdco.com
https://leafwell.com
https://trehouse.com
https://www.cbd-guru.co.uk
https://tribetokes.com
https://www.vpz.co.uk
https://www.auntbonnies.com
https://www.enjoycbd.co.uk
https://ooolalattes.com
https://www.sunstatehemp.com
https://cbdabilene.com
https://www.goodrx.com
https://www.diamondcbd.com
https://www.flawlessvapeshop.co.uk
https://www.bnaturaloil.co.uk
https://indigoridgehemp.com
https://www.thecbdflowershop.co.uk
https://originalhemp.com
https://www.getthedose.com
https://edensgate.co.uk
https://cbd.co
https://www.gethappyhemp.com
https://knownnutrition.co.uk
https://www.ukvapeworld.com
https://www.swansonvitamins.com
https://dr-vapes.com
https://jm-wholesale.co.uk
https://vsavi.co.uk
https://vapeandjuice.co.uk
https://earlybirdcbd.com
https://www.cbdoiluk.com
https://cbdchoice.com
https://www.kivaconfections.com
https://www.stirlingcbdoil.com
https://www.theaceofvapez.com
https://naturebox.com
https://aroma-king.co.uk
https://www.cypresshemp.com
https://www.trucannabliss.com
https://www.vapestreams.co.uk
https://www.thecbdistillery.com
https://vitalitycbd.co.uk
https://plainjane.com
https://www.cbdpurelife.co.uk
https://www.vapeuksupplier.co.uk
https://nuleafnaturals.com
https://www.bullycrewcbd.com
https://starbudscolorado.com
https://www.farmhouse.delivery
https://hempelf.com
https://www.ninja-vapes.co.uk
https://hempthy.com
https://pyramidpens.com
https://frannysfarmacy.com
https://www.houseofvapeslondon.co.uk
https://mandaracbd.com
https://www.pure-eliquids.com
https://texasoriginal.com
https://organic-village.co.th
https://www.cibdol.com
https://sunstatehemp.co.uk
https://naturalsolutions.uk
https://greenroads.com
https://www.rainbowvapes.co.uk
https://happyface.com
https://www.cornbreadhemp.com
https://calmclub.io
https://www.stiiizy.com
https://www.drinkthenorth.com
https://vaping101.co.uk
https://www.vapeshop.co.uk
https://purityhempco.com
https://www.pluscbdoil.com
https://www.vapeclub.co.uk
https://hushcbd.co.uk
https://www.mushroomrevival.com
https://www.podvapes.eu
https://vapeverse.co.uk
https://venacbd.com
https://www.iceheadshop.co.uk
https://redjuice.co.uk
https://www.greenhousecbd.co.uk
https://cbdliving.com
https://www.e-cigclouds.co.uk
https://thelongleaf.co.uk
https://www.vapeuniverse.co.uk
https://www.theelectroniccigarette.co.uk
https://selectvapeclub.co.uk
https://cbdreakiro.co.uk
https://cbdiablo.co.uk
https://www.jacvapour.com
https://www.tweed.com
https://vivecbd.com
https://okvape.co.uk
https://www.vapesdirectonline.co.uk
https://vapegreen.co.uk
https://cbdbrothers.com
https://notpot.com
https://www.elfbar.co.uk
https://relxnow.co.uk
http://atlas.dustforce.com/user/mposlotc
http://atlas.dustforce.com/user/polisislot
http://atlas.dustforce.com/user/slotgacorv
http://atlas.dustforce.com/user/toto188d
http://atlas.dustforce.com/user/ug212
http://eldjeesr-immo.freehostia.com/mpo-slot/
http://eldjeesr-immo.freehostia.com/polisi-slot/
http://eldjeesr-immo.freehostia.com/slot-gacor/
http://eldjeesr-immo.freehostia.com/toto188/
http://eldjeesr-immo.freehostia.com/ug212/
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L1BvbGlzaV9TbG90Lmh0bWw%3D&cidReset=true&cidReq=SLOT_004
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L21wb19zbG90Lmh0bWw%3D&cidReset=true&cidReq=SLOT_004
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L3Nsb3RfZ2Fjb3IuaHRtbA%3D%3D&cidReset=true&cidReq=SLOT_004
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L3RvdG8xODguaHRtbA%3D%3D&cidReset=true&cidReq=SLOT_004
http://entsaintetienne.free.fr/claroline1110/claroline/backends/download.php?url=L3VnMjEyLmh0bWw%3D&cidReset=true&cidReq=SLOT_004
http://flightgear.jpn.org/userinfo.php?uid=43663
http://flightgear.jpn.org/userinfo.php?uid=43665
http://flightgear.jpn.org/userinfo.php?uid=43666
http://flightgear.jpn.org/userinfo.php?uid=43669
http://flightgear.jpn.org/userinfo.php?uid=43670
http://forum.yealink.com/forum/member.php?action=profile&uid=323831
http://forum.yealink.com/forum/member.php?action=profile&uid=323832
http://forum.yealink.com/forum/member.php?action=profile&uid=323833
http://forum.yealink.com/forum/member.php?action=profile&uid=323834
http://forum.yealink.com/forum/member.php?action=profile&uid=323835
http://foxsheets.com/UserProfile/tabid/57/userId/181511/Default.aspx
http://jmlebbe.free.fr/cours/courses/SLOT_001/document/mpo_slot.html
http://jmlebbe.free.fr/cours/courses/SLOT_001/document/Polisi_Slot.html
http://jmlebbe.free.fr/cours/courses/SLOT_001/document/slot_gacor.html
http://jmlebbe.free.fr/cours/courses/SLOT_001/document/toto188.html
http://jmlebbe.free.fr/cours/courses/SLOT_001/document/UG212.html
http://jurnal.usbypkp.ac.id/index.php/jemper/user/viewPublicProfile/108738
http://opensource.platon.sk/forum/projects/viewtopic.php?p=12580050#12580050
http://opensource.platon.sk/forum/projects/viewtopic.php?p=12580052#12580052
http://opensource.platon.sk/forum/projects/viewtopic.php?p=12580062#12580062
http://opensource.platon.sk/forum/projects/viewtopic.php?p=12580073#12580073
http://opensource.platon.sk/forum/projects/viewtopic.php?p=12580075#12580075
http://slotgacorv.grapedrop.net/
http://test.sozapag.ru/forum/user/231015/
http://test.sozapag.ru/forum/user/231016/
http://test.sozapag.ru/forum/user/231017/
http://test.sozapag.ru/forum/user/231019/
http://test.sozapag.ru/forum/user/231020/
http://www.activewin.com/user.asp?Action=Read&UserIndex=4635941
http://www.activewin.com/user.asp?Action=Read&UserIndex=4635955
http://www.activewin.com/user.asp?Action=Read&UserIndex=4635965
http://www.activewin.com/user.asp?Action=Read&UserIndex=4635984
http://www.activewin.com/user.asp?Action=Read&UserIndex=4636003
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57404
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57405
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57406
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57407
http://www.baseportal.com/cgi-bin/baseportal.pl?htx=/ouatmicrobio/OMSA%20Web%20Site/eWrite/eWrite&wcheck=1&Pos=57408
http://www.rohitab.com/discuss/user/2123700-mposlotc/
http://www.rohitab.com/discuss/user/2123710-slotgacorv/
http://www.rohitab.com/discuss/user/2123717-toto188d/
http://www.rohitab.com/discuss/user/2123738-ug212/
http://www.rohitab.com/discuss/user/2123751-polisislota/
http://www.travelful.net/location/5403685/indonesia/polisi-slot
http://www.travelful.net/location/5403688/indonesia/mpo-slot
http://www.travelful.net/location/5403689/indonesia/slot-gacor
http://www.travelful.net/location/5403690/indonesia/toto188
http://www.travelful.net/location/5403692/indonesia/ug212
https://1businessworld.com/pro/fuku-fakin/
https://1businessworld.com/pro/mposlot2/
https://1businessworld.com/pro/slot-gacor/
https://1businessworld.com/pro/toto-satu-delapan-delapan/
https://1businessworld.com/pro/ug-duax/
https://440hz.my/forums/users/mposlotc/
https://440hz.my/forums/users/polisislota/
https://440hz.my/forums/users/slotgacorv/
https://440hz.my/forums/users/toto188d/
https://440hz.my/forums/users/ug212/
https://500px.com/p/mposlotc
https://500px.com/p/polisislota
https://500px.com/p/slotgacorv
https://500px.com/p/toto188d
https://500px.com/p/ug212
https://8tracks.com/mposlot-113233
https://8tracks.com/slot-g-104827
https://8tracks.com/toto188d
https://8tracks.com/ug-d
https://able2know.org/user/mposlotc/
https://able2know.org/user/polisislota/
https://able2know.org/user/slotgacorv/
https://able2know.org/user/toto188d/
https://able2know.org/user/ug212/
https://agoracom.com/members/mposlotc/favorites
https://agoracom.com/members/polisislota/favorites
https://agoracom.com/members/slotgacorv/favorites
https://agoracom.com/members/toto188d/favorites
https://agoracom.com/members/ug212/favorites
https://agoradedrets.idhc.org/profiles/mposlotc/timeline
https://agoradedrets.idhc.org/profiles/polisislota/timeline
https://agoradedrets.idhc.org/profiles/slotgacorv/timeline
https://agoradedrets.idhc.org/profiles/toto188d/timeline
https://agoradedrets.idhc.org/profiles/ug212/timeline
https://allmyfaves.com/mposlotc?tab=mposlot
https://allmyfaves.com/polisislota?tab=polisislot
https://allmyfaves.com/slotgacorv?tab=slotgacor
https://allmyfaves.com/toto188c?tab=toto188
https://allmyfaves.com/ug212?tab=ug212
https://allods.my.games/forum/index.php?page=User&userID=141186
https://allods.my.games/forum/index.php?page=User&userID=141188
https://allods.my.games/forum/index.php?page=User&userID=141189
https://allods.my.games/forum/index.php?page=User&userID=141194
https://allods.my.games/forum/index.php?page=User&userID=141195
https://amazonki.net/profil/mposlotc
https://amazonki.net/profil/polisislota
https://amazonki.net/profil/slotgacorv
https://amazonki.net/profil/toto188d
https://amazonki.net/profil/ug212
https://ameblo.jp/mposlotc/entry-12845051521.html
https://ameblo.jp/polisislota/entry-12845053019.html
https://ameblo.jp/slotgacorv/entry-12845051892.html
https://ameblo.jp/toto188d/entry-12845052344.html
https://ameblo.jp/ug212/entry-12845052777.html
https://audiomack.com/mposlotc
https://audiomack.com/slotgacorv
https://audiomack.com/ug212ug212
https://bimber.bringthepixel.com/bunchy/buddypress/members/fukufakin/profile/
https://bimber.bringthepixel.com/bunchy/buddypress/members/mposlot/profile/
https://bimber.bringthepixel.com/bunchy/buddypress/members/slotgacor/profile/
https://bimber.bringthepixel.com/bunchy/buddypress/members/totosatudelapandelapan/profile/
https://bimber.bringthepixel.com/bunchy/buddypress/members/ugduax/profile/
https://camp-fire.jp/profile/mposlotc
https://camp-fire.jp/profile/polisislot
https://camp-fire.jp/profile/slotgacorv
https://camp-fire.jp/profile/toto188d
https://camp-fire.jp/profile/ug212
https://comicvine.gamespot.com/profile/mposlotc/
https://comicvine.gamespot.com/profile/polisislot/
https://comicvine.gamespot.com/profile/slotgacorv/
https://comicvine.gamespot.com/profile/toto188d/
https://comicvine.gamespot.com/profile/ug212
https://confengine.com/user/fuku-fakin
https://confengine.com/user/mposlot
https://confengine.com/user/slot-gacor-1-1-1
https://confengine.com/user/toto-satu-delapan-delapan
https://confengine.com/user/ug-duax
https://conifer.rhizome.org/mposlotc
https://conifer.rhizome.org/polisislota
https://conifer.rhizome.org/slotgacorv
https://conifer.rhizome.org/toto188d
https://conifer.rhizome.org/ug212
https://creators.audiomack.com/polisislota
https://creators.audiomack.com/toto188d
https://designaddict.com/community/profile/mposlotc/
https://designaddict.com/community/profile/polisislota/
https://designaddict.com/community/profile/slotgacorv/
https://designaddict.com/community/profile/toto188d/
https://designaddict.com/community/profile/ug212/
https://devnet.kentico.com/users/540375/mpo-slot-lpo88
https://devnet.kentico.com/users/540376/slot-gacor-slot-gacor
https://devnet.kentico.com/users/540377/toto188-toto188
https://devnet.kentico.com/users/540378/ug212-ug212
https://devnet.kentico.com/users/540379/polisi-slot-polisislot
https://digitaltibetan.win/wiki/User:Mposlotc
https://direct.me/mposlotc
https://direct.me/slotgacorv
https://direct.me/toto188d
https://direct.me/toto188e
https://direct.me/ug212
https://disqus.com/by/disqus_5a8PuMtsx9/about/
https://disqus.com/by/disqus_AX9Nw9sP01/about/
https://disqus.com/by/disqus_BkE2cqZsTn/about/
https://disqus.com/by/mposlotc/about/
https://disqus.com/by/toto188/about/
https://dreevoo.com/profile.php?pid=620969
https://dreevoo.com/profile.php?pid=620971
https://dreevoo.com/profile.php?pid=620972
https://dreevoo.com/profile.php?pid=620973
https://dreevoo.com/profile.php?pid=620974
https://emplois.fhpmco.fr/author/mposlotc/
https://emplois.fhpmco.fr/author/polisislota/
https://emplois.fhpmco.fr/author/slotgacorv/
https://emplois.fhpmco.fr/author/toto188d/
https://emplois.fhpmco.fr/author/ug212/
https://fairygodboss.com/users/profile/8Ca4ukgfvK/Slot-Gacor
https://fairygodboss.com/users/profile/a1WD3XjNXj/ug-duax
https://fairygodboss.com/users/profile/bFIJ0ORDao/Mposlot
https://fairygodboss.com/users/profile/QSNDCW7IXc/Toto-Satu-delapan-delapan
https://fairygodboss.com/users/profile/UO0tYoBgvk/fuku-fakin
https://files.fm/fukufakin
https://files.fm/mposlot05
https://files.fm/slotgacorka
https://files.fm/totosatudelapandelapan
https://files.fm/ug212ug212
https://folkd.com/profile/mposlotc
https://folkd.com/profile/slotgacorv
https://folkd.com/profile/toto188c
https://folkd.com/profile/ug212ug212
https://forum.dmec.vn/index.php?members/mpo-slot.55082/#info
https://forum.dmec.vn/index.php?members/polisi-slot.55086/#info
https://forum.dmec.vn/index.php?members/slot-gacor.55083/#info
https://forum.dmec.vn/index.php?members/toto188.55084/#info
https://forum.dmec.vn/index.php?members/ug212.55085/#info
https://forum.instube.com/u/mposlotc/
https://forum.instube.com/u/polisislot
https://forum.instube.com/u/slotgacorv
https://forum.instube.com/u/toto188d
https://forum.instube.com/u/ug212
https://forum.ixbt.com/users.cgi?id=info:%3E1823737
https://forum.ixbt.com/users.cgi?id=info:%3E1823739
https://forum.ixbt.com/users.cgi?id=info:%3E1823742
https://forum.ixbt.com/users.cgi?id=info:%3E1823743
https://forum.ixbt.com/users.cgi?id=info:%3E1823744
https://forum.lexulous.com/user/fuku-fakin
https://forum.lexulous.com/user/mposlot
https://forum.lexulous.com/user/slot-gacor
https://forum.lexulous.com/user/toto-satu-delapan-delapan
https://forum.lexulous.com/user/ug-duax
https://forum.liquidbounce.net/user/fuku-fakin
https://forum.liquidbounce.net/user/mposlot
https://forum.liquidbounce.net/user/slot-gacor
https://forum.liquidbounce.net/user/toto-satu-delapan-delapan
https://forum.liquidbounce.net/user/ug-duax
https://forum.m5stack.com/user/mposlotc
https://forum.m5stack.com/user/polisislota
https://forum.m5stack.com/user/slotgacorv
https://forum.m5stack.com/user/toto188d
https://forum.m5stack.com/user/ug212
https://forum.melanoma.org/user/mposlotc/profile/
https://forum.melanoma.org/user/polisislota/profile/
https://forum.melanoma.org/user/slotgacorv/profile/
https://forum.melanoma.org/user/toto188d/profile/
https://forum.melanoma.org/user/ug212a/profile/
https://forum.reallusion.com/Users/3166597/mposlot05
https://forum.reallusion.com/Users/3166598/slotgacorka
https://forum.reallusion.com/Users/3166599/totosatudelapandelapan
https://forum.reallusion.com/Users/3166600/ug212ug212
https://forum.reallusion.com/Users/3166601/fukufakin
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656726
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656727&sid=07df56c9807cf8fd1aebd2ca1da0a34e
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656728&sid=641770159a7fbc034aaccc9b6d8b7a16
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656729&sid=d1f610fb39fa1c3cd74e2cc21162aaff
https://forum.singaporeexpats.com/memberlist.php?mode=viewprofile&u=656730
https://forums.alliedmods.net/member.php?u=364364
https://forums.alliedmods.net/member.php?u=364365
https://forums.alliedmods.net/member.php?u=364366
https://forums.alliedmods.net/member.php?u=364367
https://forums.alliedmods.net/member.php?u=364368
https://foxsheets.com/UserProfile/tabid/57/userId/181512/Default.aspx
https://foxsheets.com/UserProfile/tabid/57/userId/181513/Default.aspx
https://foxsheets.com/UserProfile/tabid/57/userId/181515/Default.aspx
https://foxsheets.com/UserProfile/tabid/57/userId/181516/Default.aspx
https://free-4776125.webadorsite.com/
https://free-4776140.webadorsite.com/
https://free-4776162.webadorsite.com/
https://free-4776209.webadorsite.com/
https://free-4776222.webadorsite.com/
https://freeicons.io/profile/607591
https://freeicons.io/profile/607593
https://freeicons.io/profile/607595
https://freeicons.io/profile/607596
https://freeicons.io/profile/607597
https://gacorka.gumroad.com/l/slotgacor
https://git.lumine.io/mposlotc
https://git.lumine.io/polisislota
https://git.lumine.io/slotgacorv
https://git.lumine.io/toto188c
https://git.lumine.io/ug212
https://github.com/mposlotc
https://github.com/polisislota
https://github.com/slotgacorv
https://github.com/toto188c
https://github.com/ug212
https://hangoutshelp.net/user/mposlotc
https://hangoutshelp.net/user/polisislota
https://hangoutshelp.net/user/slotgacorv
https://hangoutshelp.net/user/toto188d
https://hangoutshelp.net/user/ug212
https://hashnode.com/@mposlotc
https://hashnode.com/@slotgacorv
https://hashnode.com/@toto188d
https://hashnode.com/@ug212
https://idea.informer.com/users/mposlotc/?what=personal
https://idea.informer.com/users/polisislot/?what=personal
https://idea.informer.com/users/slotgacorv/?what=personal
https://idea.informer.com/users/toto188d/?what=personal
https://idea.informer.com/users/ug212/?what=personal
https://issuu.com/mposlotc
https://issuu.com/polisislota
https://issuu.com/slotgacorv
https://issuu.com/toto188c
https://issuu.com/ug212
https://joshbond.co.uk/community/account/polisislota/
https://joshbond.co.uk/community/profile/mposlotc/
https://joshbond.co.uk/community/profile/slotgacorva/
https://joshbond.co.uk/community/profile/toto188da/
https://joshbond.co.uk/community/profile/ug212a/
https://justpaste.me/pK6A2
https://justpaste.me/pK74
https://justpaste.me/pK7m3
https://justpaste.me/pK97
https://justpaste.me/pK9o1
https://lab.quickbox.io/mposlotc
https://lab.quickbox.io/polisislot
https://lab.quickbox.io/slotgacorv
https://lab.quickbox.io/toto188d
https://lab.quickbox.io/ug212
https://leasedadspace.com/members/mposlotc/
https://leasedadspace.com/members/polisislota
https://leasedadspace.com/members/slotgacorv/
https://leasedadspace.com/members/toto188d
https://leasedadspace.com/members/ug212
https://leetcode.com/fukufakin/
https://leetcode.com/mposlot05/
https://leetcode.com/slotgacorka/
https://leetcode.com/totosatudelapandelapan/
https://leetcode.com/ug212ug212/
https://list.ly/mposlot-1/lists
https://list.ly/polisislota/lists
https://list.ly/slot-gacor-10/lists
https://list.ly/toto188d/lists
https://list.ly/ug212/lists
https://miarroba.com/mposlotc
https://miarroba.com/polisislota
https://miarroba.com/slotgacorv
https://miarroba.com/toto188d
https://miarroba.com/ug212
https://moparwiki.win/wiki/User:Mposlotc
https://moparwiki.win/wiki/User:Polisislot
https://moparwiki.win/wiki/User:Slotgacorv
https://moparwiki.win/wiki/User:Toto188d
https://moparwiki.win/wiki/User:Ug212
https://moz.com/community/q/user/mposlotc
https://moz.com/community/q/user/polisislota
https://moz.com/community/q/user/toto188d
https://moz.com/community/q/user/toto188v
https://moz.com/community/q/user/ug212
https://mpo-slot.gitbook.io/mpo-slot/
https://mposlotc.amebaownd.com/posts/52418185
https://mposlotc.gallery.ru/
https://mposlotc.grapedrop.net/
https://mposlotc.onepage.website/
https://my.archdaily.com/us/@mpo-slot-1
https://my.archdaily.com/us/@polisi-slot-1
https://my.archdaily.com/us/@slot-gacor-40
https://my.archdaily.com/us/@toto188-1
https://my.archdaily.com/us/@ug212
https://my.desktopnexus.com/mposlotc/
https://my.desktopnexus.com/polisislota/
https://my.desktopnexus.com/slotgacorv/
https://my.desktopnexus.com/toto188c/
https://my.desktopnexus.com/ug212/
https://my.omsystem.com/members/mposlotc
https://my.omsystem.com/members/slotgacorv
https://my.omsystem.com/members/toto188d
https://my.omsystem.com/members/ug212
https://myopportunity.com/profile/mpo-slot-mpo-slot/nw
https://myopportunity.com/profile/polisi-slot-polisi-slot/nw
https://myopportunity.com/profile/slot-gacor-slot-gacor/nw
https://myopportunity.com/profile/toto188-toto188/nw
https://myopportunity.com/profile/ug212-ug212/nw
https://nhattao.com/members/user6474941.6474941/
https://nhattao.com/members/user6474948.6474948/
https://nhattao.com/members/user6474950.6474950/
https://nhattao.com/members/user6474952.6474952/
https://nhattao.com/members/user6474957.6474957/
https://openlibrary.org/people/fuku_fakin
https://openlibrary.org/people/mposlot519
https://openlibrary.org/people/slot_gacor169
https://openlibrary.org/people/toto_satu_delapan_delapan
https://openlibrary.org/people/ug_duax
https://pantip.com/profile/8043868#topics
https://pantip.com/profile/8043874#topics
https://pantip.com/profile/8043878#topics
https://pantip.com/profile/8043879#topics
https://pantip.com/profile/8043880#topics
https://pbase.com/mposlotc
https://pbase.com/polisislota
https://pbase.com/slotgacorv/slot_gacor
https://pbase.com/toto188d/
https://pbase.com/ug212
https://peatix.com/user/21480953/view
https://peatix.com/user/21480965/view
https://peatix.com/user/21480966/view
https://peatix.com/user/21480967/view
https://peatix.com/user/21480969/view
https://pinshape.com/users/3762425-slotgacorv#designs-tab-open
https://pinshape.com/users/3762464-mposlotc#designs-tab-open
https://pinshape.com/users/3762470-toto188d#designs-tab-open
https://pinshape.com/users/3762479-ug212#designs-tab-open
https://pinshape.com/users/3762495-polisislot#designs-tab-open
https://plus.fmk.sk/members/mposlotc/profile/
https://plus.fmk.sk/members/polisislota/profile
https://plus.fmk.sk/members/slotgacorv/profile/
https://plus.fmk.sk/members/toto188d/profile
https://plus.fmk.sk/members/ug212/profile/
https://polisi-slot.gitbook.io/polisi-slot/
https://polisislot.xobor.de/u1_polisislot.html
https://polisislota.amebaownd.com/posts/52418268
https://polisislota.gallery.ru/
https://polisislota.grapedrop.net/
https://polisislota.gumroad.com/l/PolisSlot
https://polisislota.onepage.website/
https://profile.ameba.jp/ameba/mposlotc/
https://profile.ameba.jp/ameba/polisislota/
https://profile.ameba.jp/ameba/slotgacorv/
https://profile.ameba.jp/ameba/toto188d/
https://profile.ameba.jp/ameba/ug212/
https://profile.hatena.ne.jp/mposlotc/
https://profile.hatena.ne.jp/slotgacorv/
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215285
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215287
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215288
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215289
https://profiles.delphiforums.com/n/pfx/profile.aspx?webtag=dfpprofile000&userId=1891215291
https://qiita.com/mposlotc
https://qiita.com/polisislota
https://qiita.com/slotgacorv
https://qiita.com/toto188d
https://qiita.com/ug212
https://rentry.co/mposlotc
https://rentry.co/polisislota
https://rentry.co/slotgacorv
https://rentry.co/toto188d
https://rentry.co/ug212
https://replit.com/@fukufakin
https://replit.com/@mposlotc
https://replit.com/@slotgacorv
https://replit.com/@totosatudelapan
https://replit.com/@ug212ug212
https://roomstyler.com/users/mposlotc
https://roomstyler.com/users/polisislot
https://roomstyler.com/users/slotgacorv
https://roomstyler.com/users/toto188d
https://roomstyler.com/users/ug212
https://rufox.ru/mposlotc/info
https://rufox.ru/polisislot/info/
https://rufox.ru/slotgacorv/info/
https://rufox.ru/toto188d/info/
https://rufox.ru/ug212/info/
https://sensationaltheme.com/forums/users/mposlotc/
https://sensationaltheme.com/forums/users/polisislota
https://sensationaltheme.com/forums/users/slotgacorv
https://sensationaltheme.com/forums/users/toto188dhttps://sensationaltheme.com/forums/users/toto188d/it/?updated=true
https://sensationaltheme.com/forums/users/ug212/
https://slot-gacor-6.gitbook.io/slot-gacor/
https://slotgacorv.amebaownd.com/posts/52418223
https://slotgacorv.gallery.ru/
https://slotgacorv.onepage.website/
https://slotgacorv.xobor.de/u1_slotgacorv.html
https://slotmaster8.gumroad.com/l/mposlot
https://snd.click/3ksp
https://snd.click/5kfc
https://snd.click/ae0e
https://snd.click/pmg0
https://snd.click/qoqu
https://store.nexodyne.com/member.php?u=131663
https://store.nexodyne.com/member.php?u=131664
https://store.nexodyne.com/member.php?u=131665
https://store.nexodyne.com/member.php?u=131667
https://store.nexodyne.com/member.php?u=131668
https://studynotes.ie/posts/nKIKKL-polisi-slot
https://studynotes.ie/posts/oyIo7j-slot-gacor
https://studynotes.ie/posts/QYIyyx-ug212
https://studynotes.ie/posts/XbIyy0-toto188
https://studynotes.ie/users/Jptnpa
https://tapchivatuyentap.tlu.edu.vn/Activity-Feed/My-Profile/UserId/34797
https://tapchivatuyentap.tlu.edu.vn/Activity-Feed/My-Profile/UserId/34798
https://tapchivatuyentap.tlu.edu.vn/Activity-Feed/My-Profile/UserId/34799
https://tapchivatuyentap.tlu.edu.vn/Activity-Feed/My-Profile/UserId/34801
https://tapchivatuyentap.tlu.edu.vn/Activity-Feed/My-Profile/UserId/34802
https://tealfeed.com/mposlotc
https://tealfeed.com/polisislota
https://tealfeed.com/slotgacorv
https://tealfeed.com/toto188d
https://tealfeed.com/ug212a
https://themalachiteforest.com/community/profile/mposlotca/
https://themalachiteforest.com/community/profile/polisislota/
https://themalachiteforest.com/community/profile/slotgacorv/
https://themalachiteforest.com/community/profile/toto188da/
https://themalachiteforest.com/community/profile/ug212/
https://themepacific.com/support/users/fukufakin
https://themepacific.com/support/users/mposlot05/
https://themepacific.com/support/users/slotgacorka
https://themepacific.com/support/users/totosatudelapandelapan
https://themepacific.com/support/users/ug212ug212
https://toto188.gitbook.io/toto188/
https://toto188.xobor.de/u1_toto.html
https://toto188d.amebaownd.com/posts/52418233
https://toto188d.gallery.ru/
https://toto188d.grapedrop.net/
https://toto188d.onepage.website/
https://totosatudelapan.gumroad.com/l/hleya
https://ug212.amebaownd.com/posts/52418237
https://ug212.gallery.ru/
https://ug212.gitbook.io/ug212/
https://ug212.gumroad.com/l/vwhatp
https://ug212.onepage.website/
https://ug212.xobor.de/u1_ug.html
https://unsplash.com/@mposlotc
https://unsplash.com/@polisislota
https://unsplash.com/@slotgacorv
https://unsplash.com/@toto188c
https://unsplash.com/@ug212
https://usahatotos.xobor.de/u1_usahatoto.html
https://webanketa.com/forms/6gs3cchp6cqk0dsq60rk8rb2/
https://webanketa.com/forms/6gs3cchp6gqk4csr6xgp6c32/
https://webanketa.com/forms/6gs3cchp6mqkarv2c9hpcshh/
https://webanketa.com/forms/6gs3cchp6rqkgrb2c4sp2ck3/
https://webanketa.com/forms/6gs3cchp6wqpcshpc5h3cr9j/
https://wibki.com/mposlotc?tab=mposlot
https://wibki.com/polisislota?tab=polisislot
https://wibki.com/slotgacorv?tab=slotgacor
https://wibki.com/toto188c?tab=toto188
https://wibki.com/ug212?tab=ug212
https://wmart.kz/forum/user/153919/
https://wmart.kz/forum/user/153922/
https://wmart.kz/forum/user/153923/
https://wmart.kz/forum/user/153924/
https://wmart.kz/forum/user/153925/
https://www.360cities.net/profile/mposlotc
https://www.360cities.net/profile/polisislota
https://www.360cities.net/profile/slotgacorv
https://www.360cities.net/profile/toto188d
https://www.360cities.net/profile/ug212
https://www.4shared.com/u/8N4B_BaP/ug212ug212.html
https://www.4shared.com/u/Ko0m8QCy/mposlot05.html
https://www.4shared.com/u/NGl0UC20/totosatudelapandelapan.html
https://www.4shared.com/u/QdzbeWzQ/fukufakin.html
https://www.4shared.com/u/r7jgw86U/slotgacorka.html
https://www.anobii.com/en/014c29cfbd998b9909/profile/activity
https://www.anobii.com/en/0156c15f67137892f4/profile/activity
https://www.anobii.com/en/015e726f3e57361831/profile/activity
https://www.anobii.com/en/01b3c167113fe71a92/profile/activity
https://www.anobii.com/en/01cc305ad1b86fca40/profile/activity
https://www.atlasobscura.com/users/mposlotc
https://www.atlasobscura.com/users/polisislot
https://www.atlasobscura.com/users/slotgacorv
https://www.atlasobscura.com/users/toto188d
https://www.atlasobscura.com/users/ug212
https://www.beatstars.com/fukufakin
https://www.beatstars.com/mposlot05
https://www.beatstars.com/slotgacorka
https://www.beatstars.com/totosatudelapandelapan
https://www.beatstars.com/ug212ug212
https://www.bitchute.com/channel/jL0ee9saCQWd/
https://www.bitchute.com/channel/L0pS1tKroPIx/
https://www.bitchute.com/channel/pxlMYs6tCiKL/
https://www.bitchute.com/channel/x2yYaKj4CS7M/
https://www.bitchute.com/channel/yLYJg7OdWub2/
https://www.blackhatworld.com/members/mposlotc.1906209/about
https://www.blackhatworld.com/members/polisislota.1906241/#about
https://www.blackhatworld.com/members/slotgacorv.1906215/
https://www.blackhatworld.com/members/toto188d.1906224/#about
https://www.blackhatworld.com/members/ug212.1906235/#about
https://www.blockdit.com/posts/6603bacb5fcc33e3155efe54
https://www.blockdit.com/posts/6603bb2d67add369c4297367
https://www.blockdit.com/posts/6603bba667add369c429da74
https://www.blockdit.com/posts/6603bc0b5fcc33e315601426
https://www.blockdit.com/posts/6603bc6d285d5d68dec05fd6
https://www.bloggportalen.se/BlogPortal/view/ReportBlog?id=215057
https://www.bloggportalen.se/BlogPortal/view/ReportBlog?id=215058
https://www.bloggportalen.se/BlogPortal/view/ReportBlog?id=215059
https://www.bloggportalen.se/BlogPortal/view/ReportBlog?id=215060
https://www.bloggportalen.se/BlogPortal/view/ReportBlog?id=215061
https://www.blurb.com/user/mposlotc
https://www.blurb.com/user/slotgacorv
https://www.blurb.com/user/toto188d
https://www.blurb.com/user/ug212
https://www.chordie.com/forum/profile.php?id=1905934
https://www.chordie.com/forum/profile.php?id=1905938
https://www.chordie.com/forum/profile.php?id=1905940
https://www.chordie.com/forum/profile.php?id=1905944
https://www.chordie.com/forum/profile.php?section=identity&id=1905943
https://www.colcampus.com/courses/96084
https://www.colcampus.com/courses/96085
https://www.colcampus.com/courses/96086
https://www.colcampus.com/courses/96087
https://www.colcampus.com/courses/96088
https://www.coursera.org/learner/mposlot
https://www.coursera.org/learner/polisislot
https://www.coursera.org/learner/slot-gacor
https://www.coursera.org/learner/toto188
https://www.coursera.org/learner/ug212
https://www.dermandar.com/user/mposlotc/
https://www.dermandar.com/user/polisislot/
https://www.dermandar.com/user/slotgacorv/
https://www.dermandar.com/user/toto188c/
https://www.dermandar.com/user/ug212/
https://www.doyoubuzz.com/mpo-slot-lpo88
https://www.empowher.com/users/mposlotc
https://www.empowher.com/users/polisislot
https://www.empowher.com/users/slotgacorv
https://www.empowher.com/users/toto188d
https://www.empowher.com/users/ug212
https://www.facer.io/u/mposlotc
https://www.facer.io/u/polisislota
https://www.facer.io/u/slotgacorv
https://www.facer.io/u/toto188d
https://www.facer.io/u/ug212
https://www.gamingonlinux.com/profiles/16370/
https://www.gamingonlinux.com/profiles/16371/
https://www.gamingonlinux.com/profiles/16372/
https://www.gamingonlinux.com/profiles/16373/
https://www.gamingonlinux.com/profiles/16374/
https://www.giveawayoftheday.com/forums/profile/176343
https://www.giveawayoftheday.com/forums/profile/176344
https://www.giveawayoftheday.com/forums/profile/176346
https://www.giveawayoftheday.com/forums/profile/176347
https://www.giveawayoftheday.com/forums/profile/176348
https://www.goodreads.com/user/show/176619551-mpo-slot
https://www.goodreads.com/user/show/176619634-slot-gacor
https://www.goodreads.com/user/show/176619668-toto188
https://www.goodreads.com/user/show/176619706-ug212
https://www.goodreads.com/user/show/176619746-polisi-slot
https://www.gta5-mods.com/users/mposlotc
https://www.gta5-mods.com/users/polisislot
https://www.gta5-mods.com/users/slotgacorv
https://www.gta5-mods.com/users/toto188d
https://www.gta5-mods.com/users/ug212
https://www.haikudeck.com/presentations/mposlotc
https://www.haikudeck.com/presentations/polisislota
https://www.haikudeck.com/presentations/slotgacorv
https://www.haikudeck.com/presentations/toto188d
https://www.haikudeck.com/presentations/ug212
https://www.hulkshare.com/mposlotc
https://www.hulkshare.com/polisislota
https://www.hulkshare.com/slotgacorv
https://www.hulkshare.com/toto188c
https://www.hulkshare.com/ug212
https://www.industryhuddle.com/mpo-slot.mpo-slot
https://www.kfz-betrieb.vogel.de/community/user/mposlotc/
https://www.kfz-betrieb.vogel.de/community/user/polisislota
https://www.kfz-betrieb.vogel.de/community/user/slotgacorv
https://www.kfz-betrieb.vogel.de/community/user/toto188d
https://www.kfz-betrieb.vogel.de/community/user/ug212/
https://www.kickstarter.com/profile/mposlotc/about
https://www.kickstarter.com/profile/polisislota/about
https://www.kickstarter.com/profile/slotgacorv/about
https://www.kickstarter.com/profile/toto188/about
https://www.kickstarter.com/profile/ug212/about
https://www.lightroomqueen.com/community/members/fukufakin.88381/#about
https://www.lightroomqueen.com/community/members/mposlot05.88377/#about
https://www.lightroomqueen.com/community/members/slotgacorka.88378/#about
https://www.lightroomqueen.com/community/members/totosatudelapandelapan.88379/#about
https://www.lightroomqueen.com/community/members/ug212ug212.88380/#about
https://www.mapleprimes.com/users/mposlotcs
https://www.mapleprimes.com/users/Polisi%20Slot
https://www.mapleprimes.com/users/slotgacorv
https://www.mapleprimes.com/users/toto188d
https://www.mapleprimes.com/users/ug212
https://www.mediafire.com/view/2l3neum1kh3jgfg/bagibagi.png/file
https://www.mediafire.com/view/9g0cxyt34vn9lyw/bagus-1.png/file
https://www.mediafire.com/view/cxdgbm4fh7jweo8/fullhd.png/file
https://www.mediafire.com/view/qk0l7jxfy54sady/mopie.png/file
https://www.metooo.io/u/mposlotc
https://www.metooo.io/u/polisislot
https://www.metooo.io/u/slotgacorv
https://www.metooo.io/u/toto188
https://www.metooo.io/u/ug212
https://www.mtg-forum.de/user/73764-mposlotc/
https://www.mtg-forum.de/user/73765-slotgacorv/
https://www.mtg-forum.de/user/73766-toto188d/
https://www.mtg-forum.de/user/73767-ug212/
https://www.mtg-forum.de/user/73768-polisislota/
https://www.nintendo-master.com/profil/mposlotc
https://www.nintendo-master.com/profil/polisislota
https://www.nintendo-master.com/profil/slotgacorv
https://www.nintendo-master.com/profil/toto188d
https://www.nintendo-master.com/profil/ug212
https://www.notebook.ai/users/747492
https://www.notebook.ai/users/747493
https://www.notebook.ai/users/747494
https://www.notebook.ai/users/747496
https://www.notebook.ai/users/747498
https://www.noteflight.com/profile/304226fe070d2952190e8bf2314b119858a5bb26
https://www.noteflight.com/profile/703ac931736ccaeb59e9a41a6a1caf5b4098b762
https://www.noteflight.com/profile/7c8bcdd6add9d079a47247b46091cdc97c7cba84
https://www.noteflight.com/profile/fb24e53674e02b14482f88a904224e50d8fd79a9
https://www.noteflight.com/profile/fe09bf6d85475fbf5d594dc6739cda2b18f9006c
https://www.openrec.tv/user/5e1ma1tk4nx770lhyfc5/about
https://www.openrec.tv/user/6owyqg4wonvqwzqfude0/about
https://www.openrec.tv/user/fxsuht6nt4r5c5ltrj4f/about
https://www.openrec.tv/user/knz80yvt2rwnw9an0ln6/about
https://www.openrec.tv/user/l9ce0yx7uqwao7imhzp9/about
https://www.owler.com/company/indonesiaandroidkejar
https://www.owler.com/company/slotgacor
https://www.owler.com/company/sonnyfortune
https://www.owler.com/company/vivosmartphone
https://www.pittsburghtribune.org/mposlotc
https://www.pittsburghtribune.org/polisislota
https://www.pittsburghtribune.org/slotgacorv
https://www.pittsburghtribune.org/toto188d
https://www.pittsburghtribune.org/ug212
https://www.pling.com/u/mposlotc/
https://www.pling.com/u/slotgacorv/
https://www.pling.com/u/toto188c/
https://www.pling.com/u/ug212/
https://www.projectnoah.org/users/mposlotc
https://www.projectnoah.org/users/polisislot
https://www.projectnoah.org/users/slotgacorv
https://www.projectnoah.org/users/toto188
https://www.projectnoah.org/users/ug212
https://www.pubpub.org/user/mpo-slot-mpo-slot
https://www.pubpub.org/user/polisi-slot-polisi-slot
https://www.pubpub.org/user/slot-gacor-slot-gacor-2
https://www.pubpub.org/user/toto188-toto188
https://www.pubpub.org/user/ug212-ug212
https://www.quora.com/profile/Fuku-Fakin/
https://www.quora.com/profile/Mposlot-3
https://www.quora.com/profile/Slot-Gacor-137
https://www.quora.com/profile/Toto-Satu-Delapan-Delapan
https://www.quora.com/profile/Ug-Duax
https://www.rctech.net/forum/members/mposlotc-359720.html
https://www.rctech.net/forum/members/slotgacorv-359722.html
https://www.rctech.net/forum/members/toto188-359723.html
https://www.rctech.net/forum/members/ug212-359724.html
https://www.rctech.net/forum/members/ug212b-359725.html
https://www.renderosity.com/users/id:1472186
https://www.renderosity.com/users/id:1472188
https://www.renderosity.com/users/id:1472189
https://www.renderosity.com/users/id:1472192
https://www.renderosity.com/users/id:1472194
https://www.root-me.org/mpo-slot
https://www.root-me.org/polisislota
https://www.root-me.org/slotgacorv
https://www.root-me.org/toto188d
https://www.root-me.org/ug212
https://www.sandiegoreader.com/users/mposlotc/
https://www.sandiegoreader.com/users/polisislota/
https://www.sandiegoreader.com/users/slotgacorv/
https://www.sandiegoreader.com/users/toto188c/
https://www.sandiegoreader.com/users/ug212/
https://www.shadowera.com/member.php?140655-mposlotc
https://www.shadowera.com/member.php?140656-slotgacorv
https://www.shadowera.com/member.php?140657-toto188d
https://www.shadowera.com/member.php?140658-ug212
https://www.shadowera.com/member.php?140659-polisislot
https://www.speedrun.com/users/mposlotc
https://www.speedrun.com/users/polisislota
https://www.speedrun.com/users/slotgacorv
https://www.speedrun.com/users/toto188d/
https://www.speedrun.com/users/ug212
https://www.spinattic.com/fukuf
https://www.spinattic.com/mposlot1
https://www.spinattic.com/slotg
https://www.spinattic.com/totos
https://www.spinattic.com/ugd
https://www.theverge.com/users/mposlotc
https://www.theverge.com/users/polisislota
https://www.theverge.com/users/slotgacorv
https://www.theverge.com/users/toto188d
https://www.theverge.com/users/ug212
https://www.twitch.tv/mposlotc
https://www.twitch.tv/polisislota
https://www.twitch.tv/slotgacorv
https://www.twitch.tv/toto188d
https://www.twitch.tv/ug212ug212
https://www.vaca-ps.org/mposlotc
https://www.vaca-ps.org/polisislot
https://www.vaca-ps.org/slotgacorv
https://www.vaca-ps.org/toto188c
https://www.vaca-ps.org/ug212
https://www.veoh.com/users/mposlotc
https://www.veoh.com/users/polisislot
https://www.veoh.com/users/slotgacorv
https://www.veoh.com/users/toto188d
https://www.veoh.com/users/ug212
https://www.walkscore.com/people/120767589734/mpo-slot
https://www.walkscore.com/people/152804950174/toto188
https://www.walkscore.com/people/284307565865/polisi-slot
https://www.walkscore.com/people/310849878455/ug212
https://www.walkscore.com/people/327930622667/slot-gacor
https://www.wattpad.com/user/mposlotc
https://www.wattpad.com/user/polisislota
https://www.wattpad.com/user/slotgacorv
https://www.wattpad.com/user/toto188c
https://www.wattpad.com/user/ug212ug212
https://www.zazzle.com/mbr/238153728540930206
https://www.zazzle.com/mbr/238411283381142353
https://www.zazzle.com/mbr/238664803082939517
https://www.zazzle.com/mbr/238838645247432546
https://www.zazzle.com/mbr/238891426508724865
https://magic.ly/pafijawatengah
https://bio.link/pafijawatengah
https://kakaotaxi.dasgno.com/kakao-pay
I am extremely impressed together with your writing abilities as neatly as with the structure in your weblog. Is this a paid theme or did you customize it yourself? Anyway keep up the nice high quality writing, it is rare to look a great blog like this one nowadays..
This really answered my downside, thanks!
situs resmi yang menyediakan angka bocoran HK untuk malam ini tahun 2024 dan dapat diandalkan bagi para pecinta togel. https://www.nmrnews.com/
대전나이트클럽
강남안마시술소중계업체
하동동해출장만남 소자본 창업
청도페이스라인출장
I do love the manner in which you have framed this specific problem plus it really does supply me some fodder for thought. On the other hand, from what precisely I have seen, I really wish as the opinions pile on that individuals continue to be on issue and don’t embark on a tirade of some other news of the day. Still, thank you for this excellent piece and whilst I do not really agree with the idea in totality, I respect your standpoint.
여행지
I like the valuable information you provide on your articles. I抣l bookmark your weblog and take a look at once more right here regularly. I am somewhat sure I抣l learn many new stuff proper here! Good luck for the next!
Excellent write-up
수원출장샵
https://nicesongtoyou.com/job/
https://ajaedotcom.tistory.com/entry/창원교차로-구인구직
https://info.channel.seoul.kr/news/2354
https://dday.tistory.com/242
https://nicesongtoyou.com/job/byeolugsijang-newspaper
During World대전동구출장샵 War II, the대전동구출장샵 British req대전동구출장샵uire
https://ddnews.co.kr/category/it/page/4/
https://download-forum.co.kr/entry/goodbye-dpi
https://download-install.com/59
https://film35.tistory.com/483
https://kakaotaxi.dasgno.com/page/4
https://goldmonster.tistory.com/entry/EAB2BDEAB8B0EB8C80-LMS-EC82ACEC9DB4ED8AB8-EBB094EBA19CEAB080EAB8B0E385A3KG-MOOC
https://gorgopage.com/2021년-건강검진-연장-내년-6월까지로-연장-신청방법-검/
https://hipsterlibertarian.com/5845
https://honeytiplabs.com/컴퓨터-모니터-깜빡임-동영상-재생-화면-오류-해결방/
https://news.gorgopage.com/274
https://king.creaming.net/on/6730
https://dday.tistory.com/858
Скай Лайн Консалтинг – известная компания, предлагающая полный спектр бухгалтерского сопровождения. У нас собрана команда истинных профессионалов. Все без исключения специалисты имеют профильное образование и достаточный опыт работы. Доверьте им оформление юридических документов и кадровое делопроизводство. https://slc-company.ru/ – сайт, где есть возможность в любое время посмотреть отзывы благодарных клиентов. Гарантируем оперативное и грамотное решение вопросов, обращайтесь уже сейчас. Стремимся к взаимовыгодному сотрудничеству!
I savour, cause I discovered exactly what I used to be taking a look for. You’ve ended my 4 day long hunt! God Bless you man. Have a nice day. Bye
Pretty portion of content. I simply stumbled upon your website and
in accession capital to assert that I get actually loved account your
weblog posts. Any way I will be subscribing to your augment or even I fulfillment you get entry to consistently rapidly.
https://bystarlight.tistory.com/435
https://dday.tistory.com/886
I savor, result in I found just what I used to be looking for. You’ve ended my four day long hunt! God Bless you man. Have a nice day. Bye
I agree with your post, we must do everything we can to stay healthly during these times. On that note have you seen Vax Detox? A Scientic Formula To Help Dissolve mRNA induced Spike Proteins from The Body. Checkout https://VaxDetox.Today
Does your blog have a contact page? I’m having trouble
locating it but, I’d like to shoot you an email. I’ve got some ideas
for your blog you might be interested in hearing.
Either way, great blog and I look forward to seeing it develop over time.
https://king.creaming.net/
Thank you for the good writeup. It in fact was a amusement account it. Look advanced to more added agreeable from you! By the way, how can we communicate?
Appreciating the persistence you put into your blog and in depth information you provide.
It’s nice to come across a blog every once in a while that isn’t the same out
of date rehashed information. Great read! I’ve bookmarked your site and I’m adding
your RSS feeds to my Google account.
https://ddnews.co.kr/blog/2023/11/04/hug/
Coffee is part of so many people’s lives nowadays and many people start their day with a cup of coffee. Coffee is what can make a difference in a lot of people’s moods.
ining 5% in 부산사하구출장샵Punjab and H부산사하구출장샵aryana. Utta부산사하구출장샵r Pr
https://honeytipit.tistory.com/tag/옥천신문20바로가기
Porn site
https://itshareit.tistory.com/243
https://www.pornhub.com/view_video.php?viewkey=ph5dc5899df25a6
Hey there! I just wanted to ask if you ever have any issues with hackers? My last blog (wordpress) was hacked and I ended up losing a few months of hard work due to no backup. Do you have any solutions to prevent hackers?
Those who have just begun their first term of online education are under a lot of stress. We provide you with the most affordable and optimal solution during this difficult period. You must pick up fresh technical abilities if you want to learn anything online. Projects, tasks, and testing centered around programs are going to become a regular part of your life. To achieve the best grades, you may hire someone to take my online class service and utilize online assistant in a few simple steps. Their experts possess the requisite degree of experience with online courses and hold a Ph.D. From taking meetings to completing tests, they will cover every aspect of employing the internet. Although courses like the online MBA program appear straightforward, they can be challenging to follow. To do excellently on exams, tests, and projects, you need to focus during class. If you are going to be a master’s pupil, the whole program becomes challenging to finish. But with the help of an online tutor, you can get through even the most difficult semester.
https://film35.tistory.com/325
I will immediately take hold of your rss feed as I can’t to find your e-mail subscription link or e-newsletter service. Do you’ve any? Please permit me recognize in order that I may subscribe. Thanks.
На сайте https://xakervip.com/topic/282/page/10/ вы сможете воспользоваться услугами хакеров, заказать взлом почты, а также тайную слежку, а заодно пробить номер, если у вас есть какие-либо подозрения. Также можно будет узнать пароли электронной почты. Для того чтобы подобрать подходящий вариант, необходимо ознакомиться со всеми разделами форума. С той целью, чтобы воспользоваться услугами, просто напишите хакеру. Он ответит вам и предложит оптимальную цену. Его услуги обойдутся недорого, а вопрос будет решен оперативно.
https://mintfin.tistory.com/tag/경기도교육청20구인구직
Thank you, I have just been looking for information approximately this subject for a long time and yours is the greatest I’ve came upon so far. However, what concerning the bottom line? Are you positive about the source?
Hi! This is my 1st comment here so I just wanted to
give a quick shout out and say I really enjoy reading through your articles.
Can you recommend any other blogs/websites/forums that go over the same topics?
Many thanks!
Hey There. I found your blog using msn. This is a very well written article. I will be sure to bookmark it and come back to read more of your useful info. Thanks for the post. I抣l certainly return.
https://chotiple.tistory.com/tag/빅마우스2010회20재방송
https://pornmaster.fun/hd/富联娱乐☘EFB88F9797·me💓皇马娱乐天美娱乐☘EFB88F9797·me💓欧皇娱乐
Hey! I’m at work surfing around your blog from my new iphone 4!
Just wanted to say I love reading through your blog
and look forward to all your posts! Keep up the outstanding work!
https://pornmaster.fun/hd/idnes-rajce-smutnaci-incesteal-sradha
The field of study known as do my online trigonometry class for me focuses on triangles and other polygons that are related to them. The study of triangles, lengths, and angles, of course, is what trigonometry is all about in the classroom of mathematics. It is well known that trigonometry coursework is notoriously difficult. On the other hand, when they are really required to carry out the function of the division or the concept, they experience an increased number of problems and complications.
https://edithvolo.com/page/149/
https://edithvolo.com/page/17/
https://film35.tistory.com/485
https://pornmaster.fun/hd/सेक्स-वीडियो-कार्टून-रेप-xxx
click here hyperlink (Larue) Hi there,
You’ve done an excellent job. I will certainly digg it and personally recommend to my friends.
I’m confident they’ll be benefited from this website. http://kgcuziotlqautjf8.mee.nu/?entry=3587030
https://nicesongtoyou.com/tax/savings-tax/
You are so awesome! I don’t believe I’ve read anything like that before.
So wonderful to find someone with original thoughts on this topic.
Seriously.. thanks for starting this up. This web site is something that’s needed on the web,
someone with a bit of originality!
Hey exceptional blog! Does running a blog like this take
a massive amount work? I have virtually no knowledge of programming but
I had been hoping to start my own blog in the near future.
Anyway, should you have any recommendations or techniques for
new blog owners please share. I understand this is off topic however I just needed to
ask. Thanks a lot!
https://honeytiplabs.com/아이폰-분할화면/
https://itshareit.tistory.com/107
https://sharply.tistory.com/612
https://pornmaster.fun/hd/son-sax-mom-mp3sex-sex-wap-com
Hi great blog! Does running a blog similar to this take a great deal
of work? I have very little knowledge of coding however I was hoping to start my own blog soon. Anyway, if you have
any ideas or tips for new blog owners please share.
I know this is off subject however I simply wanted to ask.
Appreciate it!
https://heylink.me/energystorage/
https://sites.google.com/view/battery-swapping-stations/home
https://sites.google.com/view/battery-swapping-stations/swapping-motorcycle-battery
https://sites.google.com/view/battery-swapping-stations/swappable-battery-pack
https://sites.google.com/view/battery-swapping-stations/swap-battery
https://sites.google.com/view/battery-swapping-stations/9-ports-battery-swap-cabinets
https://sites.google.com/view/battery-swapping-stations/small-electric-motorcycle
https://sites.google.com/view/battery-swapping-stations/electric-motorcycle-for-sale
https://sites.google.com/view/battery-swapping-stations/electric-motorcycle-for-adults
https://sites.google.com/view/battery-swapping-stations/electric-big-bike
https://sites.google.com/view/battery-swapping-stations/mini-electric-bike
https://sites.google.com/view/battery-swapping-stations/60v-30ah-hot-swap-battery
https://sites.google.com/view/battery-swapping-stations/72v-60ah-lithium-battery
https://sites.google.com/view/battery-swapping-stations/5-ports-swap-battery-station
https://sites.google.com/view/battery-swapping-stations/8-ports-swap-battery-station
https://sites.google.com/view/battery-swapping-stations/12-bay-battery-swap-station
https://sites.google.com/view/battery-swapping-stations/swappable-battery
https://sites.google.com/view/battery-swapping-stations/three-wheeled-electric-car
https://sites.google.com/view/battery-swapping-stations/8-slots-battery-swap-cabinets
https://www.pearltrees.com/batteryenergy
https://linktr.ee/Grevault
https://heylink.me/energystorage/
https://heylink.me/batteryenergystoragesystem/
https://www.bark.com/en/us/company/wild-oats-markets/2pKdN/
https://solo.to/battery-swapping
https://info.undp.org/docs/dao/UNSP2015/Lists/PostSurvey/Item/displayifs.aspx?ID=46174
https://info.undp.org/docs/dao/UNSP2015/Lists/PostSurvey/Item/displayifs.aspx?ID=47357
https://telegra.ph/Revolutionizing-Riding-The-Future-of-Swapping-Motorcycle-Batteries-01-09
https://www.deviantart.com/grevault/journal/The-Rise-of-Motorcycle-Battery-Swapping-1009477014
https://b.cari.com.my/home.php?mod=space&uid=2615676&do=blog&quickforward=1&id=495322
https://post.news/@/tomi62519749132/2ai5Cf5zti0PgVMeJtgULcHv6io
https://www.liveinternet.ru/users/7455232/profile
https://medium.com/@framinehaframi/latest-energy-storage-news-illuminating-future-solutions-002d8a4c3e6c
https://www.taringa.net/+battery_energy_storage/almacenamiento-de-energia-global_5bcy0j
https://steemit.com/battery/@battery-energy/impact-of-electric-mobility-on-urban-environments
https://storyweaver.org.in/en/stories/607803-integrating-electric-vehicles-with-renewable-energy-advancements-in-energy-storage-and-battery-swap-solutions
https://gravatar.com/batteryess
https://telegra.ph/Exploring-Innovative-Energy-Storage-Solutions-01-13
https://telegra.ph/The-Evolution-of-Home-Energy-Storage-01-13
https://www.bark.com/en/us/company/glicks-services-ltd/ov0Vy/
https://b.cari.com.my/home.php?mod=space&uid=2616318&do=profile
https://b.cari.com.my/home.php?mod=space&uid=2616318&do=blog&quickforward=1&id=495541
https://solo.to/battery-energy-1
https://solo.to/battery-energy-2
https://solo.to/battery-energy-3
https://solo.to/battery-energy-5
https://solo.to/battery-energy-6
https://solo.to/battery-energy-7
https://solo.to/battery-energy-9
https://solo.to/battery-energy-10
https://solo.to/battery-energy-11
https://solo.to/battery-energy-12
https://minmax.wiki/risk-of-rain-2
https://minmax.wiki/risk-of-rain-2/cautious-slug
https://solo.to/battery-energy-14
https://solo.to/battery-energy-15
https://solo.to/batteryss-1
https://solo.to/grevault
https://linktr.ee/battery_1
https://linktr.ee/battery_2
https://link.space/@battery_1
https://www.pling.com/u/battery-energy/
https://www.mycast.io/profiles/246144/username/battery_energy
https://www.espace-recettes.fr/profile/batteryenergy/649369
https://worldcosplay.net/member/1708638
https://visual.ly/users/batteryswaping/portfolio
https://hub.docker.com/u/battteryenergy
https://www.sqlservercentral.com/forums/user/batteryenergy
https://www.myvidster.com/profile/batteryenergy
https://os.mbed.com/users/grevault/
https://issuu.com/grevault
https://my.archdaily.com/us/@grevault
https://app.roll20.net/users/13071768/grevault
https://sketchfab.com/grevault
https://www.giantbomb.com/profile/grevault/
https://leetcode.com/Grevault/
https://stocktwits.com/Grevault
https://www.gta5-mods.com/users/grevault
https://starity.hu/profil/430529-grevault/
https://camp-fire.jp/profile/Grevault
https://www.mixcloud.com/grevault/
https://newspicks.com/user/9992298
https://grevault.gitbook.io/grevault/
https://grevault.gumroad.com/l/48V-15kWh-Whole-Home-Battery-Bank
https://www.walkscore.com/people/278540708173/grevault
https://www.cakeresume.com/me/sharon-s-harvey
https://www.chordie.com/forum/profile.php?id=1891713
https://www.credly.com/users/grevault/badges
https://grevault.wixsite.com/battery-energy
https://www.dday.it/profilo/grevault
https://www.designspiration.com/grevault/
https://ko-fi.com/grevault
https://www.cakeresume.com/me/sharon-s-harvey
https://www.chordie.com/forum/profile.php?id=1891713
https://www.credly.com/users/grevault/badges
https://grevault.wixsite.com/battery-energy
https://www.dday.it/profilo/grevault
https://www.designspiration.com/grevault/
https://ko-fi.com/grevault
https://gravatar.com/grevault
https://pxhere.com/en/photographer/4202812
https://www.fimfiction.net/user/704573/Grevault
https://www.marefa.org/%D9%85%D8%B3%D8%AA%D8%AE%D8%AF%D9%85:Grevault
https://www.bigbasstabs.com/profile/85363.html
https://inkbunny.net/Grevault
https://pinshape.com/users/3635617-grevault
https://pastelink.net/ntb07s6o
https://expatarrivals.com/user/295930
https://app.scholasticahq.com/scholars/265987-gre-vault
https://my.archdaily.cl/cl/@grevault
https://communities.bentley.com/members/d74fa091_2d00_292f_2d00_424f_2d00_97fa_2d00_f4d4dc1fe1ba
https://www.free-ebooks.net/profile/1547128/grevault
https://hashnode.com/@Grevault
https://www.metooo.io/u/grevault
https://www.slideserve.com/Sharon57
https://voz.vn/u/grevault.1975457/#about
https://www.webwiki.com/huntkeyenergystorage.com
https://app.pluralsight.com/profile/sharon-s-73
https://www.anime-sharing.com/members/grevault.358429/#about
https://pawoo.net/@grevault
https://www.pokecommunity.com/members/grevault.1265953/#about
https://www.renderosity.com/users/Grevault
https://spiderum.com/nguoi-dung/Grevault
https://the-dots.com/pages/grevault-648357
https://www.sitelike.org/similar/huntkeyenergystorage.com/
https://grevaulthomo.home.blog/
https://batteryess.jimdofree.com/
https://batteryess.jimdofree.com/blog/
https://batteryess.jimdofree.com/gridscale-energy/
https://65a8c0430d8a5.site123.me/
https://65a8c0430d8a5.site123.me/blog/gridscale-energy
https://65a8c0430d8a5.site123.me/blog/global-energy-storage
https://65a8c0430d8a5.site123.me/blog/best-top-10-energy-storage-liquid-cooling-host-manufacturers-in-the-world
https://start.me/p/0N4wpg/battery-energy
https://start.me/p/ogDYOD/home-energy-storage
https://www.pinterest.com/grevaults/
https://www.metal-archives.com/users/Grevault
https://www.discogs.com/user/Grevault
https://www.espace-recettes.fr/profile/grevault/652690
https://anyflip.com/homepage/tzzfy
https://mastodon.social/@Grevault/
https://chart-studio.plotly.com/~Grevault
https://community.windy.com/user/sharon-s-harvey
https://www.bakespace.com/members/profile/Grevault/1629842/
https://www.liveinternet.ru/users/grevault/profile
https://www.artstation.com/grevault
https://www.artstation.com/artwork/ob5WYm
https://www.lawyersclubindia.com/profile.asp?member_id=1028668
https://www.furaffinity.net/user/grevault/
https://vocal.media/authors/grevault
https://ameblo.jp/grevault/
https://profile.ameba.jp/ameba/grevault/
https://www.doorkeeper.jp/users/grevault
https://pantip.com/profile/8000239
https://magic.ly/grevault
https://linkhay.com/u/grevault
https://linkhay.com/link/7445809/kham-pha-viec-luu-tru-nang-luong-pin
https://hackmd.io/@Grevault
https://hackmd.io/@Grevault/exploring-battery-energy-storage
https://www.tumblr.com/grevaults
https://www.behance.net/gallery/186893499/512V-200Ah-Powerwall-Battery
https://player.soundon.fm/p/e0690725-6e06-41de-bff8-5a1af5c214f9
https://player.soundon.fm/p/e0690725-6e06-41de-bff8-5a1af5c214f9/episodes/8fa69d15-3a9a-4651-8362-bc99d807df4f
https://open.spotify.com/episode/11TyBUOPXxbI0rMgsK6obv
https://podcast.kkbox.com/tw/channel/4nxOJayRzXfVVTyATj
https://www.myminifactory.com/users/battery-energy
https://www.myminifactory.com/stories/battery-energy-storage-65a7952782138
https://www.taringa.net/+battery_energy_storage/almacenamiento-de-energia-a-escala-de-red-mejora-de-la-eficiencia-del_5b1sza
https://www.taringa.net/+battery_energy_storage/economic-calculation-and-analysis-of-industrial-and-commercial-energy_5b1j4o
https://www.taringa.net/+battery_energy_storage/centrales-electricas-de-almacenamiento-de-energia_5b3kl0
https://www.taringa.net/+battery_energy_storage/almacenamiento-de-energia-global_5b3zap
https://www.taringa.net/+battery_energy_storage/industria-energetica-espanola_5b4mdx
https://www.taringa.net/+battery_energy_storage/fabricantes-de-hosts-de-refrigeracion-liquida-para-almacenamiento-de-e_5b4za1
https://www.taringa.net/+battery_energy_storage/fosfato-de-hierro-y-litio_5b5ng0
https://www.taringa.net/+battery_energy_storage/almacenamiento-de-bateria-detras-del-medidor_5b5vm0
https://www.taringa.net/+battery_energy_storage/5-tipos-de-almacenamiento-de-energia_5b6e0l
https://www.taringa.net/+battery_energy_storage/sistema-de-refrigeracion-liquida-de-la-bateria_5b7ani
https://www.taringa.net/+battery_energy_storage/preguntas-frecuentes-sobre-almacenamiento-de-energia_5b8076
https://www.taringa.net/+battery_energy_storage/energia-a-escala-de-red_5bcdvd
https://www.taringa.net/+battery_energy_storage/almacenamiento-de-energia-global_5be243
https://medium.com/@Grevault
https://medium.com/@Grevault/electrical-energy-storage-devices-a-comprehensive-overview-804543f5d807
https://medium.com/@Grevault/energy-storage-application-scenarios-f7ed131fa4dc
https://medium.com/@Grevault/unveiling-the-power-within-some-knowledge-about-energy-storage-power-stations-7aebf6154c32
https://medium.com/@Grevault/global-energy-storage-ea0b835a2159
https://medium.com/@Grevault/spanish-energy-storage-industry-b76db487b1e8
https://medium.com/@Grevault/behind-the-meter-battery-storage-9480ce5e6b8f
https://medium.com/@Grevault/5-types-of-energy-storage-4044f55f1d6b
https://medium.com/@Grevault/battery-liquid-cooling-system-f23be803aca8
https://medium.com/@Grevault/energy-storage-faq-5f9e051f65f3
https://medium.com/@Grevault/gridscale-energy-c00962677fd9
https://medium.com/@framinehaframi/global-energy-storage-32c54d55e208
https://wefunder.com/grevault
https://roomstyler.com/users/grevault
https://www.goodreads.com/user/show/176073042-grevault-revault
https://myanimelist.net/profile/Grevault
https://strawpoll.com/kogjk6Yb8Z6
https://www.pinterest.de/grevaults/
https://www.pinterest.fr/grevaults/
https://www.pinterest.jp/grevaults/
https://www.pinterest.it/grevaults/
https://www.pinterest.com.mx/grevaults/
https://www.pinterest.co.uk/grevaults/
https://www.pinterest.es/grevaults/
https://www.pinterest.ca/grevaults/
https://www.pinterest.cl/grevaults/
https://www.pinterest.co.kr/grevaults/
https://www.pinterest.pt/grevaults/
https://www.pinterest.com.au/grevaults/
https://www.pinterest.ph/grevaults/
https://www.pinterest.at/grevaults/
https://www.pinterest.se/grevaults/
https://www.pinterest.dk/grevaults/
https://www.pinterest.ch/grevaults/
https://www.pinterest.ie/grevaults/
https://www.pinterest.nz/grevaults/
https://mastodon.social/@Grevault
https://gab.com/batteryenergy
https://gab.com/batteryenergy/posts/112200161916838089
https://top.blogfree.net/?act=Profile&MID=1354602
https://band.us/band/94466211/intro
https://500px.com/p/grevault
https://flipboard.com/@tomg2024/solar-energy-battery-storage-mqcmc4qjy
https://community.windy.com/user/tom-g-6
https://codepen.io/Tom-G-the-decoder/pen/xxePBdm
https://codepen.io/Tom-G-the-decoder/details/xxePBdm
https://codepen.io/Tom-G-the-decoder/full/xxePBdm
https://cdpn.io/pen/debug/xxePBdm?authentication_hash=xnMabpRKZpZr
https://anyflip.com/homepage/bzivc#About
https://folio.procreate.com/grevault
https://www.crunchbase.com/organization/samebike
https://clutch.co/profile/samebike
https://www.bark.com/en/us/company/samebike/y0Rew/
https://www.provenexpert.com/samebike/
https://linktr.ee/samebike0
https://solo.to/samebike
https://judge.me/profile/g0j0K9de
https://about.me/samebike
https://wakelet.com/@samebike
https://community.windy.com/user/samebike
https://www.quia.com/profiles/samebikesa
https://heylink.me/samebike/
https://magic.ly/samebike
https://magic.ly/samebike/samebike
https://www.furaffinity.net/user/samebike
https://hashnode.com/@samebike
https://www.bakespace.com/members/profile/samebike/1632462/
https://www.doorkeeper.jp/users/samebike
https://linkr.bio/samebike
https://samebike.tawk.help/article/samebike
https://tawk.to/gzsamebike
https://samebike.webflow.io/
https://chart-studio.plotly.com/~SAMEBIKE
https://link.space/@samebike
https://www.myminifactory.com/users/samebike
https://www.webwiki.com/gzsamebike.com
https://losangeles.bubblelife.com/community/samebike_1
https://hackmd.io/@samebike/samebike
https://samebike.amebaownd.com/posts/52642908
https://www.discogs.com/user/samebike
https://www.google.com/maps/d/viewer?mid=1d8ehdVrZSsQ8XtluzE0WU7OcUF9IPlg
https://medium.com/@samebike
https://medium.com/@samebike/about
https://medium.com/@samebike/electric-bikes-vs-traditional-bikes-exploring-the-differences-fc73e60d4836
https://medium.com/@samebike/exploring-electric-bikes-a-comprehensive-data-analysis-e7cd9306efe7
https://medium.com/@samebike/electric-bike-enthusiasts-unite-a43376a333c3
https://pinterest.com/gzsamebike/
https://www.pinterest.com/pin/864972672174083771
https://www.pinterest.com/pin/864972672174946453
https://telegra.ph/Samebike-Electric-Bikes-03-06
https://telegra.ph/What-is-the-battery-life-of-an-e-bike-How-long-does-it-take-to-charge-03-26
https://telegra.ph/Electric-bike-are-the-ultimate-tribute-to-green-technology-04-09
https://post.news/@/samebike
https://post.news/@/samebike/2dLz6leAuwzx99TAlOsRgYzimfq
https://post.news/@/samebike/2eXPqekQQnhgJq2d88EsHVOOmY0
https://www.deviantart.com/samebike
https://www.deviantart.com/samebike/about
https://www.deviantart.com/samebike/journal/Today-I-opened-Alibaba-and-at-first-sight-1037790953
https://www.deviantart.com/samebike/posts
https://www.artstation.com/samebike/profile
https://www.artstation.com/samebike
https://www.artstation.com/artwork/w0kGDO
https://samebike.artstation.com/projects/w0kGDO
https://www.artstation.com/samebike/likes
https://www.artstation.com/samebike/followers
https://www.artstation.com/samebike/following
https://www.artstation.com/artwork/QXvDeE
https://samebike.artstation.com/projects/QXvDeE
https://www.bitchute.com/channel/I03ahvQOEGvt/
https://www.bitchute.com/video/jaTy9Km7ufLd/
https://www.bitchute.com/video/ubUCjESB2LAq/
https://www.bitchute.com/video/PCKmAzAbrx1t/
https://ko-fi.com/samebike
https://ko-fi.com/post/Discover-E-Bike-Tours-Why-E-Bike-Ride-Is-Necessar-O5O0WF4UQ
https://www.blogger.com/profile/06754141759102704631
https://gzsamebike.blogspot.com/2024/04/in-todays-society-controversy.html
https://connpass.com/event/315959/
https://hackmd.io/@Grevault/top-7-balcony-solar-products-in-the-world
https://linkhay.com/blog/1128452/top-7-balcony-solar-products-in-the-world
https://energy-storage.doorkeeper.jp/events/172015
https://www.furaffinity.net/journal/10842271/
https://www.artstation.com/artwork/BXbPd6
https://chart-studio.plotly.com/dashboard/Grevault:0/view
https://blog.libero.it/wp/grevault/about-grevault/
https://blog.libero.it/wp/grevault/2024/04/10/top-7-solar-products-in-the-world/
https://favhd8.webmepage.com/battery-energy-storage-system-manufacturers
https://grevault.jimdosite.com/
https://grevault.jimdosite.com/
https://grevault.jimdosite.com/top-7-balcony-solar-products-in-the-world/
https://radiocut.fm/radiostationrequest/6739/
https://65a8c0430d8a5.site123.me/blog/top-7-balcony-solar-products-in-the-world
https://batteryess.jimdofree.com/2024/04/11/top-7-balcony-solar-products-in-the-world/
https://gravatar.com/donalddabdul
https://webcompat.com/issues/135790
https://www.findit.com/kbiufenavxzpjcz
https://vimeo.com/933636885
https://www.scamadviser.com/check-website/huntkeyenergystorage.com
https://www.scamadviser.com/check-website/www.huntkeyenergystorage.com
На сайте http://weller.ru/stroitelstvo/vinilovyj-sajding-obzor-oblicovochnogo-materiala/ ознакомьтесь с информацией, посвященной виниловому сайдингу. Имеется вся важная информация, которая касается характеристик, внешнего вида, особенностей такого материала. Опубликована и история появления винилового сайдинга, и другая ценная информация. Вместе с тем, вы узнаете о достоинствах материала, почему его выбирает большинство. Кроме того, перечислены и современные производители. Узнаете, где можно приобрести такой строительный материал по бюджетной стоимости.
https://www.monkfishjowls.com/광주은행-영업시간-및-365코너-찾기
https://nicesongtoyou.com/welfare/support-fund/
A. 카카오택시 앱을 실행한 후 ‘예약’ 탭을 누르시면 됩니다. 이후 출발 위치, 도착 위치, 탑승 시간 등을 입력하고 ‘예약하기’를 누르시면 예약이 완료됩니다. 다음 사이트에서 섹스 비디오를 시청하세요 강남출장섹스마사지
Navigation on the site is a breeze, thanks to its intuitive design and user-friendly interface. I appreciated the seamless experience of exploring different categories and discovering hidden gems within the archives.
https://new-software.download/windows/7zip/
https://king.creaming.net/on/8504
https://mythings.tistory.com/81
https://pornmaster.fun/hd/【查询开房记录客服-微信-客服78444643】查找查微信聊天记录-pln
Magnificent items from you, man. I’ve be mindful your
stuff prior to and you’re simply too fantastic. I really like
what you have got right here, certainly like what you’re saying and the way wherein you say it.
You’re making it enjoyable and you still take care of to stay it sensible.
I cant wait to read much more from you. That is actually a great site.
My homepage – خرید بک لینک
batmanapollo.ru
Hi, Neat post. There’s a problem with your website in internet explorer, would test this?IE still is the market leader and a big portion of people will miss your magnificent writing because of this problem.
https://gorgopage.com/삼성카드-결제일별-이용기간-고객센터-전화번호-및/
Thank you for providing these detailed instructions! They offer a structured and clear pathway for setting up seed and ROI regions in neuroimaging analysis. By following these steps, researchers can ensure accuracy and reliability in their tractography studies. Your thoughtful guidance not only simplifies the process but also enhances the quality of our research outcomes. We appreciate your support in facilitating our work in understanding brain connectivity patterns.
https://dday.tistory.com/437
I have been exploring for a bit for any high quality articles or blog posts on this sort of house . Exploring in Yahoo I finally stumbled upon this website. Studying this info So i am satisfied to exhibit that I have an incredibly just right uncanny feeling I found out exactly what I needed. I so much certainly will make certain to don抰 disregard this site and give it a glance on a relentless basis.
Thanks for your blog post. I would like to say that your health insurance broker also works for the benefit of the particular coordinators of a group insurance policies. The health insurance professional is given a listing of benefits needed by an individual or a group coordinator. What a broker can is search for individuals and also coordinators which often best complement those desires. Then he provides his suggestions and if all parties agree, the actual broker formulates a legal contract between the 2 parties.
На сайте https://greateastsiberia.ru/ почитайте поучительную, интересную и увлекательную информацию, которая касается здорового образа жизни. Имеются ценные, важные рекомендации относительно правильного питания, занятий спортом и других моментов. Есть информация о том, как сохранить суставы, о народных методах. Имеются рекомендации о том, как сохранить здоровье самых маленьких. Вы узнаете о том, как правильно бегать для получения необходимого эффекта. С вами поделятся своими рецептами для того, чтобы достичь гармонии души.
Hi, after reading this amazing piece of writing i am also cheerful to share my experience here with colleagues.
Does your blog have a contact page? I’m having a tough time locating it but, I’d like to shoot you an e-mail. I’ve got some suggestions for your blog you might be interested in hearing. Either way, great website and I look forward to seeing it develop over time.
하동동해출장만남 소자본 창업
Wow, marvelous blog layout!
How lengthy have you been blogging for? you made blogging look easy.
The entire look of your website is great, as smartly as the content!
You can see similar here prev
next and
that was wrote by Tricia77.
I am really enjoying the theme/design text with stuff on top of it your website.
Do you ever run into any web browser compatibility problems?
A number of my blog audience have complained about my blog
not operating correctly in Explorer but looks great in Firefox.
Do you have any solutions to help fix this issue?
http://gxxjfgjfzm3.mee.nu/?entry=3591714
Hey there! Would you mind if I share your blog with my myspace group? There’s a lot of folks that I think would really appreciate your content. Please let me know. Thank you
Thanks for your valuable post. As time passes, I have been able to understand that the symptoms of mesothelioma are caused by your build up of fluid between your lining in the lung and the torso cavity. The disease may start while in the chest location and multiply to other limbs. Other symptoms of pleural mesothelioma cancer include weight-loss, severe deep breathing trouble, vomiting, difficulty swallowing, and bloating of the face and neck areas. It ought to be noted that some people having the disease never experience virtually any serious signs and symptoms at all.
Hmm is anyone else encountering problems with the images on this blog loading? I’m trying to find out if its a problem on my end or if it’s the blog. Any suggestions would be greatly appreciated.
відеограф комо
What i do not realize is actually how you are not actually much more well-liked than you may be right now. You are so intelligent. You realize thus significantly relating to this subject, made me personally consider it from so many varied angles. Its like women and men aren’t fascinated unless it is one thing to accomplish with Lady gaga! Your own stuffs nice. Always maintain it up!
Does your blog have a contact page? I’m having trouble locating it but,
I’d like to send you an e-mail. I’ve got some creative
ideas for your blog you might be interested in hearing.
Either way, great website and I look forward to seeing it develop
over time.
여행지
Wow, incredible blog layout!
How long have you ever been blogging for? you made blogging glance easy.
The total look of your web site is fantastic, let alone
the content material! You can read similar here prev next
and those was wrote by Daryl86.
여행지
In these days of austerity and relative panic about having debt, many individuals balk against the idea of utilizing a credit card to make acquisition of merchandise or even pay for any gift giving occasion, preferring, instead only to rely on the tried as well as trusted procedure for making repayment – raw cash. However, if you have the cash there to make the purchase fully, then, paradoxically, this is the best time to use the card for several causes.
수원출장샵
Зайдите на сайт https://luckyfood.ru/ где вы сможете купить мясо для собак с быстрой доставкой или самовывозом в Москве. Действует система скидок. Ознакомьтесь с большим каталогом продукции – говядина, баранина, индейка, субпродукты и многое другое. Порадуйте своего питомца, а легкость оформления заказа обязательно понравится!
An amazing content. I was vaping my Crystal Prime 7000 while I was reading this 🙂
https://himchanchan.tistory.com/1749
Slot games are one type of slot game that is very popular throughout the world. A very exciting game with very big prizes, for more complete slot game information you can visit the website. https://trentain.com/
https://himchanchan.tistory.com/1749
https://pornmaster.fun/hd/kat-chuamage-xxx
The most popular slot game in the world today is the lottery slot, a game that is quite easy and has the biggest prizes for only a small capital. For more complete slot game information, visit the website. https://jurriaanpersyn.com/
Hey very nice website!! Guy .. Beautiful .. Superb ..
I will bookmark your blog and take the feeds additionally?
I’m glad to find numerous useful info right here within the post, we’d like work
out extra strategies in this regard, thanks for sharing.
. . . . .
https://himchanchan.tistory.com/1749
https://pornmaster.fun/hd/hansika-tinman-xossip-xxx-com-pu
Statistics, a part of mathematics, lets you collect and analyse data to produce results. Statistics is a popular topic in the job market, but students may want assistance with homework owing to its complexity, endless formulae, and many applications. Due to academic stress and the extensive syllabus, students seek statistics homework help to better their grades and confidence. Statistics tasks require subject expertise. Online statistics homework help is beneficial for students with several responsibilities. Online statistics homework help is beneficial for students with several responsibilities. Do you worry over not doing your statistics class on time and correctly? If yes then you must turn to “ Take my online statistics class” service.
What’s up friends, how is the whole thing, and what
you desire to say on the topic of this post, in my view its truly remarkable in favor of me.
https://dzone.com/users/5131146/rentcarfycom.html
여행지
양산시술출장마사지
I’ve learned result-oriented things from a blog post. Also a thing to I have seen is that in most cases, FSBO sellers will certainly reject you. Remember, they can prefer not to use your services. But if you maintain a gentle, professional relationship, offering help and staying in contact for about four to five weeks, you will usually manage to win interviews. From there, a house listing follows. Cheers
I apologise, I can help nothing. I think, you will find the correct decision. Do not despair.
Vapeforest loyalty rewards
https://vapeforest.co.uk
I have read some just right stuff here. Definitely worth bookmarking for revisiting.
I surprise how so much attempt you set to
create this type of wonderful informative website.
I was curious if you ever considered changing the layout of your website? Its very well written; I love what youve got to say. But maybe you could a little more in the way of content so people could connect with it better. Youve got an awful lot of text for only having 1 or two pictures. Maybe you could space it out better?
wonderful issues altogether, you simply gained a brand new reader.
What would you recommend about your publish that you made some days ago?
Any positive?
Hi! This post could not be written any better! Reading this post reminds me of
my good old room mate! He always kept chatting about
this. I will forward this article to him. Pretty sure he will have a good
read. Thanks for sharing!
Thanks for your article. I would also love to say that the health insurance dealer also works for the benefit of the coordinators of a group insurance coverage. The health broker is given a listing of benefits looked for by somebody or a group coordinator. What any broker does indeed is seek out individuals or coordinators which will best match those demands. Then he shows his recommendations and if both sides agree, the actual broker formulates an agreement between the two parties.
https://1x-bet-india.com
A sustainable building thrives on well-integrated building services. An assessment can evaluate lighting for energy efficiency and occupant comfort. Acoustics should be checked to ensure a noise-free environment. Integration of these systems, along with maintenance plans, ensures optimal performance and minimizes environmental impact. Building Services and Sustainability holistic approach fosters a healthy and efficient building.
Magnificent goods from you, man. I’ve understand your stuff previous to
and you’re just extremely wonderful. I actually like what you have acquired here, really like what
you’re stating and the way in which you say it.
You make it enjoyable and you still care for to keep it sensible.
I can’t wait to read far more from you. This
is actually a wonderful website.
You really make it appear really easy together with your presentation however I
find this topic to be really one thing that I
believe I would by no means understand. It sort of feels too complex and very
extensive for me. I’m having a look ahead on your subsequent
submit, I will try to get the hold of it!
Youre so cool! I dont suppose Ive read anything like this before. So good to find anyone with some authentic ideas on this subject. realy thanks for starting this up. this website is something that’s wanted on the web, somebody with a bit originality. useful job for bringing something new to the internet!
Today, I went to the beachfront with my kids. I found a sea shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She put the shell to her ear and screamed. There was a hermit crab inside and it pinched her ear. She never wants to go back! LoL I know this is entirely off topic but I had to tell someone!
In the awesome pattern of things you actually secure an A just for effort. Where exactly you lost me ended up being in all the specifics. As it is said, the devil is in the details… And that couldn’t be much more correct here. Having said that, allow me reveal to you just what exactly did work. Your text is incredibly convincing and this is possibly why I am taking the effort to opine. I do not really make it a regular habit of doing that. Secondly, while I can see the leaps in logic you make, I am not really convinced of just how you appear to unite your ideas that make the actual conclusion. For the moment I will yield to your point but hope in the foreseeable future you actually link your dots better.
Why did the boat salesman bring a fan to work? To generate some “prop”er excitement!
https://himchanchan.tistory.com/1749
Just stumbled upon this amazing website – it’s a total game-changer! https://grailcannabis.ca/blog/best-weed-delivery-brampton-2024-top-weed-delivery-reviews/
The Flyextremeworld website is a lifesaver! Their assignment help services are top-notch, and their prices are reasonable. The writers are highly skilled and deliver exceptional work every time. Plus, their customer support team is always available to help. I highly recommend reading about Best Website for Assignment Help in Dubai and UAE.
https://raptureworks.fun/
https://raptureworks.fun/collections/new-in
https://raptureworks.fun/pages/sex-toys
https://raptureworks.fun/pages/sex-toys
Thanks for the thoughts you are discussing on this weblog. Another thing I’d really like to say is the fact that getting hold of copies of your credit score in order to look at accuracy of any detail would be the first activity you have to perform in credit restoration. You are looking to thoroughly clean your credit profile from damaging details errors that ruin your credit score.
I found your blog site on google and examine a couple of of your early posts. Continue to keep up the very good operate. I simply further up your RSS feed to my MSN News Reader. Searching for ahead to studying more from you in a while!?
Thanks for the useful information on credit repair on this particular blog. What I would tell people should be to give up the mentality that they may buy currently and pay out later. Like a society we all tend to repeat this for many factors. This includes vacation trips, furniture, as well as items we wish. However, you’ll want to separate your own wants out of the needs. If you are working to raise your credit ranking score actually you need some trade-offs. For example you can shop online to economize or you can check out second hand stores instead of highly-priced department stores pertaining to clothing.
Hi! I know this is somewhat off topic but I was wondering which blog platform are you using for this website? I’m getting fed up of WordPress because I’ve had problems with hackers and I’m looking at alternatives for another platform. I would be awesome if you could point me in the direction of a good platform.
I needed to draft you that very little observation to thank you once again for your amazing pointers you have shared on this page. This is so extremely generous of people like you to make openly just what many of us might have supplied as an e book to earn some cash on their own, and in particular considering that you could have done it if you desired. These smart ideas likewise served to become a easy way to fully grasp many people have the same eagerness the same as my personal own to find out very much more with reference to this condition. Certainly there are several more pleasurable instances ahead for those who see your blog.
Hello. fantastic job. I did not anticipate this. This is a excellent story. Thanks!
very good submit, i actually love this website, carry on it
https://film35.tistory.com/321
https://pornmaster.fun/hd/foreign-vi
I look forward to many more years of enlightening and entertaining reads!
It’s awesome in favor of me to have a web page, which is valuable in support of my experience. thanks admin
my blog … https://ttdvhn.quangtri.gov.vn/index.php?language=vi&nv=news&nvvithemever=d&nv_redirect=aHR0cHM6Ly9hZGFyc2hleHBvcnQuY29tL2dtZWRpYS9nXzI0LWpwZy8
Why was the boat salesman always so successful? Because he knew how to reel in customers!
I have two computers: I call one the “good” computer — it has two monitors. The other is my “junk” computer with one screen where I download a lot of stuff to it.. . If I wanted to continue using both computers but only with the dual monitors, what would I need to buy? Is there some sort of splitter I can buy that will allow me to switch between each CPU? Where can I buy one if it does in fact exist? Will I still be able to use one mouse and keyboard?.
There are some attention-grabbing time limits in this article but I don抰 know if I see all of them heart to heart. There may be some validity but I’ll take hold opinion till I look into it further. Good article , thanks and we wish extra! Added to FeedBurner as nicely
Fantastic site you have here but I was wanting to
know if you knew of any community forums that cover the same topics talked about in this article?
I’d really like to be a part of community where I can get feed-back from other
experienced people that share the same interest. If you have any recommendations, please let
me know. Many thanks!
Fine way of telling, and fastidious piece
of writing to take facts about my presentation subject, which i am going to convey in school.
Yet another thing is that when you are evaluating a good internet electronics store, look for web shops that are consistently updated, keeping up-to-date with the hottest products, the top deals, and helpful information on services and products. This will ensure you are doing business with a shop that stays on top of the competition and offers you what you ought to make knowledgeable, well-informed electronics purchases. Thanks for the significant tips I have learned from the blog.
Can I just say what a relief to seek out someone who really knows what theyre talking about on the internet. You positively know how you can convey an issue to light and make it important. Extra individuals must read this and perceive this facet of the story. I cant consider youre no more popular because you undoubtedly have the gift.
One other thing is that an online business administration training course is designed for people to be able to without problems proceed to bachelor degree courses. The Ninety credit diploma meets the lower bachelor college degree requirements and once you earn your current associate of arts in BA online, you will possess access to the latest technologies on this field. Some reasons why students want to get their associate degree in business is because they may be interested in this area and want to receive the general instruction necessary just before jumping to a bachelor education program. Thanks alot : ) for the tips you actually provide within your blog.
Discovered an interesting article, I suggest you familiarize yourself http://asg-amt.phorum.pl/viewtopic.php?p=31738#31738
Great information. Many thanks.
Great site. A lot of useful information here. I am sending it to a few friends ans also sharing in delicious. And naturally, thanks for your sweat!
Hi there, its pleasant piece of writing on the topic of media print, we
all be aware of media is a wonderful source of data.
Also visit my web site; royalasianescorts.com
Aw, this was a very nice post. In thought I wish to put in writing like this additionally ?taking time and actual effort to make an excellent article?however what can I say?I procrastinate alot and in no way appear to get something done.
Terrific work! This is the type of info that should be shared around the web. Shame on Google for not positioning this post higher! Come on over and visit my web site . Thanks =)
We’re a group of volunteers and starting a new scheme in our community. Your site provided us with valuable information to work on. You’ve done an impressive job and our whole community will be thankful to you.
One more thing. In my opinion that there are a lot of travel insurance web sites of respectable companies than enable you to enter your vacation details and get you the quotes. You can also purchase this international holiday insurance policy online by using the credit card. Everything you need to do would be to enter all your travel details and you can be aware of the plans side-by-side. Only find the program that suits your financial allowance and needs and then use your credit card to buy them. Travel insurance on the internet is a good way to start looking for a trustworthy company pertaining to international travel insurance. Thanks for sharing your ideas.
One other thing I would like to talk about is that as opposed to trying to match all your online degree classes on days that you conclude work (since most people are worn out when they get back), try to obtain most of your lessons on the week-ends and only a couple courses on weekdays, even if it means taking some time off your weekend. This is really good because on the saturdays and sundays, you will be more rested plus concentrated for school work. Thanks for the different suggestions I have learned from your web site.
Hey would you mind sharing which blog platform you’re working with? I’m looking to start my own blog soon but I’m having a tough time selecting between BlogEngine/Wordpress/B2evolution and Drupal. The reason I ask is because your design seems different then most blogs and I’m looking for something completely unique. P.S Sorry for getting off-topic but I had to ask!
Hmm it looks like your site ate my first comment (it was extremely long) so I guess I’ll just sum it up what I wrote and say, I’m thoroughly enjoying your blog. I as well am an aspiring blog blogger but I’m still new to everything. Do you have any points for rookie blog writers? I’d really appreciate it.
Amazing blog! Is your theme custom made or did you download it from somewhere? A design like yours with a few simple adjustements would really make my blog shine. Please let me know where you got your design. Appreciate it
I抦 not sure where you are getting your info, but great topic. I needs to spend some time learning much more or understanding more. Thanks for excellent info I was looking for this info for my mission.
I抦 not that much of a internet reader to be honest but your sites really nice, keep it up! I’ll go ahead and bookmark your site to come back in the future. Cheers
https://nicesongtoyou.com/job/byeolugsijang-newspaper/
https://pornmaster.fun/hd/foreign-vi
It抯 really a nice and helpful piece of info. I抦 glad that you just shared this helpful information with us. Please stay us up to date like this. Thanks for sharing.
An interesting dialogue is worth comment. I feel that it is best to write more on this topic, it might not be a taboo topic however usually people are not sufficient to speak on such topics. To the next. Cheers
Valuable info. Lucky me I discovered your site unintentionally, and I am surprised why this coincidence didn’t happened in advance! I bookmarked it.
Thank you for any other excellent post. The place else could anybody get that type of info in such a perfect method of writing? I’ve a presentation subsequent week, and I am at the look for such info.
I am always browsing online for ideas that can facilitate me. Thx!
Hi there! I’m at work surfing around your blog from my new iphone! Just wanted to say I love reading your blog and look forward to all your posts! Keep up the fantastic work!
In accordance with my observation, after a property foreclosure home is available at an auction, it is common for the borrower to still have the remaining balance on the loan. There are many financial institutions who aim to have all rates and liens paid by the next buyer. Having said that, depending on certain programs, restrictions, and state legal guidelines there may be quite a few loans which are not easily solved through the shift of personal loans. Therefore, the obligation still rests on the consumer that has received his or her property in foreclosure process. Many thanks for sharing your thinking on this blog.
I needed to post you this little observation just to give thanks over again with your awesome suggestions you have contributed on this website. It has been so particularly open-handed with you in giving unreservedly what exactly most people could possibly have supplied as an electronic book to end up making some cash for themselves, certainly since you might well have done it if you considered necessary. Those tips also worked to be the fantastic way to fully grasp that other individuals have similar desire really like my own to find out lots more on the subject of this problem. I’m certain there are several more pleasurable periods ahead for those who take a look at your website.
I am often to blogging and i really appreciate your content. The article has really peaks my interest. I’m going to bookmark your site and keep checking for brand spanking new information.
Many thanks for your post. I would like to say that the cost of car insurance will vary from one policy to another, since there are so many different facets which bring about the overall cost. By way of example, the model and make of the vehicle will have a large bearing on the price tag. A reliable outdated family vehicle will have a more affordable premium compared to a flashy fancy car.
I’ve observed that in the world the present moment, video games are the latest craze with children of all ages. There are times when it may be difficult to drag your family away from the activities. If you want the very best of both worlds, there are many educational games for kids. Good post.
Terrific posts, With thanks.
Also visit my web blog :: https://MAY-Anma.icu/jeju-massage
I haven抰 checked in here for a while as I thought it was getting boring, but the last few posts are good quality so I guess I will add you back to my daily bloglist. You deserve it my friend 🙂
I have been exploring for a bit for any high-quality articles or blog posts on this kind of area . Exploring in Yahoo I at last stumbled upon this site. Reading this information So i am happy to convey that I’ve a very good uncanny feeling I discovered exactly what I needed. I most certainly will make certain to don抰 forget this site and give it a look regularly.
magnificent post, very informative. I wonder why the other experts of this sector don’t notice this. You should continue your writing. I’m confident, you have a huge readers’ base already!
Useful posts Many thanks!
Feel free to surf to my website link slot gacor 2024 (https://link-slot-gacor-2024.blogspot.com/)
Have you ever thought about creating an ebook or guest authoring on other blogs? I have a blog centered on the same subjects you discuss and would love to have you share some stories/information. I know my visitors would enjoy your work. If you’re even remotely interested, feel free to shoot me an e-mail.
I have viewed that smart real estate agents everywhere you go are getting set to FSBO Marketing and advertising. They are noticing that it’s more than simply placing a sign in the front area. It’s really regarding building associations with these sellers who one of these days will become buyers. So, after you give your time and energy to aiding these sellers go it alone — the “Law associated with Reciprocity” kicks in. Interesting blog post.
What I have observed in terms of laptop memory is that often there are specific features such as SDRAM, DDR and so on, that must match the technical specs of the mother board. If the pc’s motherboard is fairly current and there are no main system issues, upgrading the ram literally will take under an hour. It’s one of several easiest computer upgrade methods one can consider. Thanks for sharing your ideas.
Je kan op je telefoon eenvoudig gebruikmaken van bijvoorbeeld iDeal om je geld te storten.
Hi this is kinda of off topic but I was wanting to know if blogs use WYSIWYG editors or if you have to manually code with HTML. I’m starting a blog soon but have no coding experience so I wanted to get guidance from someone with experience. Any help would be enormously appreciated!
Hiya, I’m really glad I have found this information. Nowadays bloggers publish just about gossips and internet and this is really frustrating. A good web site with exciting content, this is what I need. Thanks for keeping this site, I’ll be visiting it. Do you do newsletters? Cant find it.
Very nice post. I just stumbled upon your weblog and wanted to say that I have really enjoyed surfing around your blog posts. In any case I will be subscribing to your rss feed and I hope you write again soon!
chlenomer.icu
This actually answered my drawback, thank you!
Aw, this was a very nice post. In concept I want to put in writing like this moreover ?taking time and precise effort to make an excellent article?but what can I say?I procrastinate alot and in no way seem to get one thing done.
I relish, lead to I discovered just what I used to be looking for. You’ve ended my 4 day long hunt! God Bless you man. Have a great day. Bye
Thanks for your article. It is rather unfortunate that over the last one decade, the travel industry has had to handle terrorism, SARS, tsunamis, bird flu, swine flu, as well as the first ever true global recession. Through everything the industry has really proven to be effective, resilient and also dynamic, acquiring new methods to deal with misfortune. There are generally fresh troubles and opportunities to which the industry must just as before adapt and respond.
We stumbled over here from a different web address and thought I might check things out. I like what I see so now i’m following you. Look forward to finding out about your web page again.
Wow! This could be one particular of the most helpful blogs We’ve ever arrive across on this subject. Actually Great. I’m also a specialist in this topic therefore I can understand your effort.
very good post, i actually love this website, carry on it
Hello just wanted to give you a quick heads
up and let you know a few of the images aren’t loading correctly.
I’m not sure why but I think its a linking issue. I’ve tried it in two
different internet browsers and both show the same results.
I have mastered some new points from your web page about computer systems. Another thing I’ve always presumed is that computer systems have become an item that each residence must have for many people reasons. They supply you with convenient ways to organize the home, pay bills, go shopping, study, pay attention to music and in many cases watch tv series. An innovative strategy to complete these types of tasks is to use a notebook. These desktops are mobile ones, small, potent and transportable.
My family members all the time say that I am killing my time here at net, but I know I am getting experience all the time by reading thes
nice posts.
My spouse and I stumbled over here different web page and thought I may as well check things out. I like what I see so now i am following you. Look forward to checking out your web page again.
I have come across that today, more and more people are increasingly being attracted to video cameras and the field of images. However, being photographer, you will need to first devote so much time deciding which model of digital camera to buy as well as moving store to store just so you could potentially buy the least expensive camera of the trademark you have decided to choose. But it isn’t going to end right now there. You also have to consider whether you should obtain a digital digicam extended warranty. Thanks alot : ) for the good recommendations I received from your blog.
Hey There. I discovered your weblog the usage of msn. This is an extremely well written article. I will be sure to bookmark it and return to read more of your useful info. Thanks for the post. I will definitely return.
Hey very cool web site!! Man .. Excellent .. Amazing .. I’ll bookmark your website and take the feeds also厈I am happy to find a lot of useful info here in the post, we need develop more strategies in this regard, thanks for sharing. . . . . .
One more thing. In my opinion that there are several travel insurance web sites of respectable companies that let you enter your journey details and have you the rates. You can also purchase an international travel insurance policy on the web by using your own credit card. All you have to do is always to enter your current travel specifics and you can view the plans side-by-side. Simply find the plan that suits your allowance and needs and then use your bank credit card to buy that. Travel insurance online is a good way to search for a reputable company for international travel cover. Thanks for discussing your ideas.
I’m looking joomla template to design site for one hindu temple.? Any suggestion for template and components for Joomla ?. Mainly looking for event details , picture gallery and blogs..
Hey! Do you know if they make any plugins to protect against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any recommendations?
I understand how to use Banners in Joomla, but the banners are not displaying. How do I choose where the banners show up? I see no option for that. Its published..
Excellent weblog here! Additionally your website lots up
fast! What host are you the use of? Can I get your affiliate link
in your host? I want my website loaded up as quickly as yours lol
Pretty section of content. I just stumbled upon your blog and in accession capital to assert that
I acquire actually enjoyed account your blog posts.
Any way I will be subscribing to your augment and even I achievement you access consistently rapidly.
Hey! I could have sworn I’ve been to this blog before but after reading through some of the post I realized it’s new to me. Nonetheless, I’m definitely glad I found it and I’ll be bookmarking and checking back often!
I know this if off topic but I’m looking into starting my own weblog and was wondering what all is required to get set up? I’m assuming having a blog like yours would cost a pretty penny? I’m not very internet savvy so I’m not 100 certain. Any tips or advice would be greatly appreciated. Kudos
I was recommended this blog by my cousin. I’m not sure whether this post is written by him as nobody else know such detailed about my problem. You’re incredible! Thanks!
Hello there! Do you use Twitter? I’d like to follow you if that would be okay. I’m undoubtedly enjoying your blog and look forward to new posts.
Achter de schermen is het Holland Online Casino druk bezig geweest met de voorbereidingen voor een online on line casino.
Fine knowledge Appreciate it.
I like what you guys are up also. Such smart work and reporting! Keep up the superb works guys I抳e incorporated you guys to my blogroll. I think it will improve the value of my site 🙂
As I site possessor I believe the content material here is rattling wonderful , appreciate it for your efforts. You should keep it up forever! Good Luck.
Would you be involved in exchanging hyperlinks?
sex
Thanks for the thoughts you have shared here. Additionally, I believe there are many factors that will keep your auto insurance premium down. One is, to think about buying autos that are in the good report on car insurance providers. Cars which have been expensive are more at risk of being stolen. Aside from that insurance policies are also in accordance with the value of your car or truck, so the more pricey it is, then higher the premium you have to pay.
Greetings from Ohio! I’m bored to tears at work so I decided to check out your blog on my iphone during lunch break. I really like the information you present here and can’t wait to take a look when I get home. I’m shocked at how quick your blog loaded on my cell phone .. I’m not even using WIFI, just 3G .. Anyways, great blog!
I’m curious to find out what blog system you have been using? I’m having some minor security issues with my latest website and I’d like to find something more secure. Do you have any solutions?
What’s up to every single one, it’s in fact a fastidious
for me to pay a visit this web page, it includes valuable Information.
https://nicesongtoyou.com/job/byeolugsijang-newspaper/
거제출장안마
buysteriodsonline.com
도박과 싸움의 이야기는 엄청나게 빠르게 퍼졌고, 하룻밤 사이에 수도 안팎으로 미친 듯이 퍼졌습니다.
거제출장안마
거제출장안마
영등포안마살롱
양산시술출장마사지
이태원스웨디시안마게이클럽
대전세븐나이트
Appreciation to my father who told me about this website, this blog is actually
awesome.
수원출장샵
안성출장마사지
전신스타킹
Hi, yeah this post is really nice and I have learned
lot of things from it regarding blogging. thanks.
This info is priceless. Where can I find out more?
수원출장샵
거제출장안마
하동동해출장만남 소자본 창업
You completed various nice points there. I did a search on the topic and found the majority of people will have the same opinion with your blog.
이태원게이바
Thank you for another informative website. Where else could I get that kind of information written in such a perfect way? I have a project that I am just now working on, and I have been on the look out for such info.
This web site truly has all the info I wanted about this subject
and didn’t know who to ask.
수원출장샵
https://plaza.rakuten.co.jp/grevault/
https://plaza.rakuten.co.jp/grevault/diary/202404230000/
https://foursquare.com/flangem6507385/list/grevault
https://myanimelist.net/profile/grevault1
https://www.comparably.com/companies/grevault
https://grevault.livedoor.blog/some-knowledge-about-energy-storage-power-stations
https://grevault.ahlamontada.com/t2-topic
https://tabelog.com/rvwr/grevault/prof/
https://gamewith.jp/user/feed/post/show/4188805/18077931714017585
https://www.nicovideo.jp/watch/sm43704592
https://www.patreon.com/Grevault
https://steemit.com/battery/@energyss/integrating-gridscale-energy-and-battery-swap-stations
https://steemit.com/battery/@energyss/changes-in-requirements-for-battery-liquid-cooling-system
https://steemit.com/energystorage/@battery-energy/battery-capacity
https://medium.com/@Grevault/top-10-household-energy-storage-companies-in-germany-00db863ab0c1
https://batteryenergystorageindustry.quora.com/Top-10-household-energy-storage-companies-in-Germany
https://www.flickr.com/photos/grevault/53686993935/in/dateposted-public/
https://www.anobii.com/en/01eacf0d8eeac48c33/profile/
https://www.are.na/block/27843965
https://cohost.org/batteny
https://energyen.dreamwidth.org/
https://gettr.com/user/batteryen
https://www.kooapp.com/koo/batteryen/4b851651-7254-489b-9edf-93d4c6e28a89
https://listography.com/7904521322
https://grevault.micro.blog/about/
https://www.minds.com/grevault/
https://www.myheritage.com/site-1651606794/kodrae
https://medium.com/@Grevault/discussion-on-new-energy-723326ab226c
https://batteryenergystorageindustry.quora.com/New-Energy-PV-Power
https://www.flickr.com/photos/grevault/53702253198/in/dateposted-public/
https://patch.com/users/grevault
https://patch.com/north-carolina/myerspark-southend-dilworth/business/listing/442107/grevault
https://vc.ru/life/1163912-perspektivy-rynka-akkumulyatornyh-batarey-v-regione-aziatsko-tihookeanskogo-regiona
https://www.sitejabber.com/reviews/huntkeyenergystorage.com
https://www.marefa.org/Grevault_energy
https://grevault.muragon.com/entry/1.html
https://grevault.muragon.com
https://grevault.booth.pm/
https://www.pixiv.net/en/users/105980231
https://coda.io/@gram-mile
https://grevault.muragon.com/entry/2.html
https://www.deviantart.com/battery001/art/Unlocking-the-Power-of-Battery-Energy-Storage-1050364230
https://www.nicovideo.jp/watch/sm43767432
https://kuku.lu/tfd9e
https://kuku.lu/tfdbe
https://kuku.lu/tfda1
https://www.spreaker.com/episode/top-7-balcony-solar-products-in-the-world–59998286
https://www.spreaker.com/episode/energy-storage–59998288
https://player.soundon.fm/p/e0690725-6e06-41de-bff8-5a1af5c214f9/episodes/50a104f2-0841-4266-81c5-790cbf3294ad
https://vc.ru/life/1170607-preimushchestva-i-aspekty-stroitelstva
https://grevaultenergy.contently.com/
https://gtmetrix.com/reports/www.huntkeyenergystorage.com/8RQqnWlj/
https://www.spreaker.com/episode/5kwh-portable-energy-storage-system–59999276
https://grevaultenergy.mypixieset.com/
https://grevaultenergy.mypixieset.com/about
https://substack.com/@grevaultenergy/note/c-56416167
https://wykop.pl/ludzie/grevault
https://twitcasting.tv/g:102516832066714687637
I would like to thank you for the efforts you’ve put in writing this website.
I really hope to check out the same high-grade blog posts from you in the future as
well. In fact, your creative writing abilities has encouraged me to
get my very own website now 😉
대전나이트클럽
It is actually a nice and useful piece of info. I am happy that
you just shared this useful info with us. Please stay us informed like this.
Thanks for sharing.
Wow, amazing blog structure! How lengthy have you been running a blog for? you make blogging look easy. The whole look of your website is wonderful, as well as the content material![X-N-E-W-L-I-N-S-P-I-N-X]I simply couldn’t depart your site prior to suggesting that I extremely enjoyed the usual information an individual provide in your guests? Is gonna be again steadily in order to inspect new posts.
Check out my web-site https://Findbestserver.com/tax-deed-tips-shop-around/
Pretty nice post. I just stumbled upon your weblog and wished to say that I’ve really enjoyed browsing your blog posts.
In any case I will be subscribing to your feed and I hope you write again very soon!
Great write ups, Appreciate it!
Hello! This post could not be written any better!
Reading through this post reminds me of my old room mate!
He always kept talking about this. I will forward this write-up to him.
Fairly certain he will have a good read. Thank you
for sharing!
This design is incredible! You definitely know how to keep
a reader amused. Between your wit and your videos, I was almost
moved to start my own blog (well, almost…HaHa!) Fantastic job.
I really enjoyed what you had to say, and more than that,
how you presented it. Too cool!
전신스타킹
What’s up, constantly i used to check blog posts here
in the early hours in the daylight, because i love to gain knowledge
of more and more.
IMB88 berdiri sebagai pilihan baru untuk slot online dan judi bola parlay di Indonesia— favorit di kalangan pemain Indonesia, dengan penonton online harian berjumlah 15.000 orang. Taruhan menjadi mudah, prosesnya cepat, semua di bawah naungan situs judi online yang mendapatkan kepercayaan dengan mudah. Angka-angka saja sudah berbicara banyak: IMB88.
수원출장샵
Useful material, Appreciate it!
Hi there, I found your site via Google whilst looking for a related
subject, your site came up, it seems great. I have bookmarked it in my google bookmarks.
Hello there, simply turned into aware of your blog via Google, and found that it is truly informative.
I’m going to watch out for brussels. I’ll be grateful for
those who proceed this in future. Numerous other folks
can be benefited from your writing. Cheers!
I do not know if it’s just me or if perhaps everybody else encountering
problems with your website. It appears like some of
the written text in your content are running
off the screen. Can someone else please comment and let me know if this is happening to them too?
This may be a issue with my web browser because I’ve had this happen before.
Kudos
Soberbo trabalho neste artigo informativo! Já conferiu o fantástico site https://royalcasino999 .com/ ? É um site com diversas opções de apostas para você escolher!
Online classes reduce the cost of universities. You pay a smaller amount for a similar quality education. Moreover, these changes make our lives much easier than ever before. Due to hectic schedules and commute problems, students prefer to hire a coursework expert for learning instead of the traditional way of classes. Let us describe how can online classes reduce the cost of universities. If you want to know then read this blog till the end…
With this blog post from AllAssignmentHelp.com, we are sharing the essential and desired homework tips for kids. This write-up consists of all the useful tips for students, parents, and teachers that will result in homework and academic success. They provide the best-needed homework tips for kids, which not only help them overcome their learning difficulties but also help them understand what they are actually learning from their books. Also Read: Key Points to Write a Solid Dissertation
Having read this I thought it was really informative.
I appreciate you taking the time and energy to put this content together.
I once again find myself spending a significant amount
of time both reading and posting comments. But
so what, it was still worth it!
Appreciate the recommendation. Will try it out.
Hi, What is the best free software to automatically backup wordpress database and files ? A software that is trustworthy and would not hack your password in wordpress. Have you tried it ? For How long ? Thanks……
What is the procedure to copyright a blog content (text and images)?. I wish to copyright the content on my blog (content and images)?? can anyone please guide as to how can i go abt it?.
Quality content is the important to be a focus for the viewers to pay a visit the web site, that’s what
this web site is providing.
Hello there, I found your site by means of Google even as looking for a related topic, your website came up, it appears good.
I’ve bookmarked it in my google bookmarks.
Hi there, just became aware of your blog through Google, and found that it’s truly informative.
I am going to be careful for brussels. I’ll be grateful
for those who continue this in future. A lot of other folks will likely
be benefited out of your writing. Cheers!
It’s remarkable for me to have a site, which is helpful designed for
my know-how. thanks admin
Your way of telling everything in this post is actually pleasant, all can simply understand it,
Thanks a lot.
수원출장샵
울산콜걸
충무로출장업소
This article is actually a nice one it assists new the web viewers,
who are wishing in favor of blogging.
강남안마시술소중계업체
하동동해출장만남 소자본 창업
하동동해출장만남 소자본 창업
Your style is so unique in comparison to other people I have
read stuff from. I appreciate you for posting when you’ve got the opportunity,
Guess I will just book mark this web site.
영등포안마살롱
I visit daily a few sites and websites to read articles or reviews, however this website offers feature based writing.
Cogito ergo sum
대전호박나이트
영등포안마살롱
대전나이트클럽
수원출장샵
청도페이스라인출장
100 лет тому вперед смотреть онлайн бесплатно в хорошем. Смотреть фильм 100 лет тому вперед бесплатно.
강남콜걸
하동동해출장만남 소자본 창업
You mentioned it very well.
Also visit my site – https://mulberry-anma.xyz
Wonderful beat ! I would like to apprentice whilst you amend your site, how can i subscribe for a weblog website? The account helped me a acceptable deal. I have been a little bit acquainted of this your broadcast offered bright clear concept
I know this if off topic but I’m looking into starting my own blog and
was curious what all is needed to get setup?
I’m assuming having a blog like yours would cost
a pretty penny? I’m not very internet smart so I’m not 100% certain. Any suggestions or advice would be greatly appreciated.
Thanks
수원출장샵
Nicely put. Regards.
Haha, love the humor in this piece! Your writing always cracks me up. Keep ’em coming!
Hello there, just became alert to your blog through Google, and found that it’s really informative. I am gonna watch out for brussels. I will be grateful if you continue this in future. Lots of people will be benefited from your writing. Cheers!
I’m thoroughly captivated by your deep insights and excellent ability to convey information. Your expertise shines through in every piece you write. It’s clear that you put a lot of effort into researching your topics, and this effort pays off. Thank you for sharing such detailed information. Keep up the great work! https://www.elevenviral.com
I have been exploring for a little for any high quality articles or blog posts on this kind of area . Exploring in Yahoo I at last stumbled upon this website. Reading this information So i am happy to convey that I have a very good uncanny feeling I discovered just what I needed. I most certainly will make certain to don抰 forget this website and give it a glance regularly.
여행지
청도페이스라인출장
울산콜걸
Looking for Best SOP Services For Australia. Also visit website.
Looking for best SOP services for Australia.Please visit website https://sopforaustralia.com/
Usually I don’t read article on blogs, but I would like to say that this write-up very forced me to take a look at and do it! Your writing style has been surprised me. Thank you, very nice article.
I have been exploring for a little bit for any high quality articles or blog posts in this sort of house . Exploring in Yahoo I eventually stumbled upon this web site. Reading this info So i am satisfied to express that I have a very good uncanny feeling I found out exactly what I needed. I such a lot definitely will make sure to don抰 fail to remember this web site and give it a look regularly.
I think the admin of this web page is genuinely working hard in support of
his web page, as here every stuff is quality based data.
Hi there just wanted to give you a brief heads up and let you know a few of the pictures aren’t loading correctly.
I’m not sure why but I think its a linking issue.
I’ve tried it in two different internet browsers and both show
the same outcome.
Wonderful paintings! This is the kind of information that are meant to be shared across the net. Shame on Google for not positioning this put up upper! Come on over and seek advice from my site . Thanks =)
Excellent beat ! I wish to apprentice even as you amend your website, how can i subscribe for a weblog website? The account aided me a acceptable deal. I have been tiny bit acquainted of this your broadcast offered vivid clear concept
충무로출장업소
One more issue is really that video gaming became one of the all-time main forms of entertainment for people of nearly every age. Kids have fun with video games, and adults do, too. The XBox 360 has become the favorite gaming systems for people who love to have a lot of games available to them, in addition to who like to experiment with live with other folks all over the world. Thank you for sharing your thinking.
吸うやつバイブは、女性だけでなく、男性にもおすすめのアイテムです。男性の陰茎や乳首などの敏感な部分を刺激することができ、新たな快感を体験することができます。
Thanks for your helpful post. Over time, I have come to be able to understand that the particular symptoms of mesothelioma cancer are caused by a build up connected fluid between the lining on the lung and the upper body cavity. The disease may start inside the chest vicinity and distribute to other parts of the body. Other symptoms of pleural mesothelioma cancer include weight reduction, severe respiration trouble, nausea, difficulty eating, and inflammation of the neck and face areas. It needs to be noted that some people having the disease don’t experience any serious indications at all.
아름다운스웨디시업소
How does an Nvidia GeForce GPU prefer its coffee? With a lot of processing power and a bit of extra VRAM!
Today, I went to the beachfront with my kids. I found a sea shell and gave it to my 4 year old daughter and said “You can hear the ocean if you put this to your ear.” She put the shell to her ear and screamed. There was a hermit crab inside and it pinched her ear. She never wants to go back! LoL I know this is entirely off topic but I had to tell someone!
Hello! I just wanted to ask if you ever have any issues with hackers? My last blog (wordpress) was hacked and I ended up losing many months of hard work due to no backup. Do you have any solutions to protect against hackers?
k8 ビンゴ
この記事から得られる情報の量と質に驚きました。非常に役立ちます。
How did the Nvidia GeForce card feel when it finally got the latest driver update? Absolutely refreshed and ready to run!
대전호박나이트
I just installed a new itunes on a different computer, and I want to manually sync some videos to my ipod. What I need to know if if I plug my ipod into the computer(which is FILLED with my music) will it start to synchronize right away to the empty itunes list? I don’t want to loose my music on there! How do I make sure that doesn’t happen?.
We have discovered two use-after-free vulnerabilities in PHP’s rubbish assortment algorithm.
Those vulnerabilities were remotely exploitable over PHP’s unserialize function. We were also awarded with $2,
000 by the Internet Bug Bounty committee (c.f. Many thanks go out
to cutz for co-authoring this text. Pornhub’s bug bounty program and its comparatively excessive rewards on Hackerone caught our attention.
That’s why we have now taken the angle of a sophisticated
attacker with the total intent to get as deep
as potential into the system, focusing on one essential goal: gaining distant code execution capabilities.
Thus, we left no stone unturned and attacked what Pornhub is built upon: PHP.
After analyzing the platform we shortly detected the utilization of unserialize on the website.
In all instances a parameter named “cookie” bought unserialized from Post knowledge
and afterwards mirrored through Set-Cookie headers.
Standard exploitation methods require so called Property-Oriented-Programming (POP) that involve
abusing already present lessons with particularly outlined “magic methods” so as to
trigger undesirable and malicious code paths.
I haven抰 checked in here for a while as I thought it was getting boring, but the last few posts are good quality so I guess I will add you back to my daily bloglist. You deserve it my friend 🙂
Hmm is anyone else encountering problems with the pictures on this blog loading? I’m trying to figure out if its a problem on my end or if it’s the blog. Any suggestions would be greatly appreciated.
Simply want to say your article is as astonishing. The clearness in your post is just great and that i can suppose you’re knowledgeable on this subject. Well along with your permission let me to snatch your feed to stay updated with impending post. Thank you one million and please carry on the gratifying work.
대전호박나이트
양산시술출장마사지
I have realized that online degree is getting well-liked because accomplishing your degree online has developed into popular option for many people. A huge number of people have definitely not had an opportunity to attend an established college or university nonetheless seek the increased earning potential and a better job that a Bachelor’s Degree gives. Still other folks might have a degree in one training but would wish to pursue anything they now possess an interest in.
Thanks a lot for the helpful article. It is also my opinion that mesothelioma has an particularly long latency period of time, which means that the signs of the disease won’t emerge till 30 to 50 years after the original exposure to asbestos. Pleural mesothelioma, which is the most common form and has an effect on the area within the lungs, could potentially cause shortness of breath, chest muscles pains, and a persistent cough, which may bring on coughing up blood.
Thank you so much for giving everyone a very remarkable chance to discover important secrets from this blog. It can be very lovely plus jam-packed with fun for me personally and my office co-workers to visit your web site nearly 3 times weekly to read the new issues you have. Of course, I’m at all times fulfilled concerning the astounding tips and hints served by you. Selected 2 tips in this posting are particularly the best we have had.
Greetings from Idaho! I’m bored to death at work so I decided to browse your website on my iphone during lunch break. I really like the information you provide here and can’t wait to take a look when I get home. I’m shocked at how quick your blog loaded on my mobile .. I’m not even using WIFI, just 3G .. Anyhow, fantastic site!
Great article, exactly what I wanted to find.
クリイキとは?クリイキの構造や機能、クリイキ開発のテクニックとアドバイス。
Hi my loved one! I want to say that this post is amazing, nice written and come with approximately all vital
infos. I would like to look extra posts like this .
Also visit my homepage … Hana Holišová
Hi i am kavin, its my first time to commenting anyplace, when i read this article i
thought i could also make comment due to this brilliant post.
Valuable info. Lucky me I found your site by accident, and I’m shocked why this accident did not happened earlier! I bookmarked it.
I抳e been exploring for a little for any high quality articles or weblog posts in this sort of house . Exploring in Yahoo I ultimately stumbled upon this site. Studying this information So i抦 satisfied to convey that I have a very just right uncanny feeling I discovered just what I needed. I such a lot unquestionably will make sure to don抰 overlook this website and give it a glance on a continuing basis.
전신스타킹
Assainissement.org offre des informations sur les technologies de l’eau et de l’assainissement, telles que les systèmes autonomes, les tours eau, le traitement des eaux usées, le recyclage, etc. Des informations sur les nouvelles technologies sont également disponibles.
Hi there! I’m at work browsing your blog from my new apple iphone! Just wanted to say I love reading your blog and look forward to all your posts! Carry on the great work!
Wow, amazing blog layout! How long have you been blogging for? you made blogging look easy. The overall look of your site is fantastic, let alone the content!
https://ddnews.co.kr/blog/2022/07/30/mbti-a-t-차이/
벼룩시장 구인구직 및 신문 그대로 보기 (PC/모바일) | 구인구직 앱 어플 무료 설치 다운로드 | 모바일 벼룩시장 보는 방법 | 벼룩시장 부동산 | 지역별 벼룩시장 | 벼룩시장 종이신문 에 대해 알아보겠습니다. 섹스카지노사이트
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
Sabung Ayam Online
How Technology can Simplify Learning is a branch of knowledge that deals with chemicals and compounds consisting of atoms, which themselves are comprised of metals and other molecules. Writing essays and other journal articles is a requirement of studying science at each of these levels, which requires a deep knowledge of the topic. Even though the majority of students are aware of this, they somehow struggle to write compelling essays since their composing abilities have not been developed to the necessary level. Because the writing component of the program is so challenging for the students, they seek “do my online sociology class for me” help that have emerged as a godsend to them in recent years.
How Technology can Simplify Learning is a branch of knowledge that deals with chemicals and compounds consisting of atoms, which themselves are comprised of metals and other molecules. Writing essays and other journal articles is a requirement of studying science at each of these levels, which requires a deep knowledge of the topic. Even though the majority of students are aware of this, they somehow struggle to write compelling essays since their composing abilities have not been developed to the necessary level. Because the writing component of the program is so challenging for the students, they seek “online biology assignment helper” help that have emerged as a godsend to them in recent years.
you will have an ideal weblog here! would you prefer to make some invite posts on my weblog?
It’s perfect time to make some plans for the longer term and it’s time to be happy. I have learn this put up and if I could I desire to recommend you some fascinating things or tips. Perhaps you could write next articles regarding this article. I desire to read more issues approximately it!
Excellent site. Lots of useful information here. I am sending it to a few friends ans also sharing in delicious. And obviously, thanks for your sweat!
Superb blog! Do you have any recommendations for aspiring writers? I’m planning to start my own website soon but I’m a little lost on everything. Would you advise starting with a free platform like WordPress or go for a paid option? There are so many options out there that I’m totally confused .. Any recommendations? Many thanks!
Today, considering the fast way of living that everyone leads, credit cards have a huge demand throughout the economy. Persons from every arena are using credit card and people who are not using the credit cards have arranged to apply for just one. Thanks for spreading your ideas about credit cards.
강남콜걸
Wow, incredible blog layout! How long have you been blogging for? you made blogging look easy. The overall look of your site is excellent, as well as the content!
Have you ever considered about adding a little bit more than just your articles? I mean, what you say is valuable and all. However think of if you added some great pictures or videos to give your posts more, “pop”! Your content is excellent but with pics and video clips, this site could undeniably be one of the best in its niche. Great blog!
I have read so many posts on the topic of the blogger lovers but this
article is in fact a fastidious piece of writing, keep
it up.
하동동해출장만남 소자본 창업
Pretty component to content. I simply stumbled upon your blog and in accession capital to assert that I acquire in fact enjoyed account your blog posts. Anyway I will be subscribing on your feeds and even I achievement you access consistently rapidly.
거제출장안마
I feel that is among the so much significant info for me. And i am glad reading your article. But wanna commentary on few basic things, The web site taste is ideal, the articles is in point of fact excellent : D. Just right task, cheers
Thank you for another informative site. Where else could I get that kind of info written in such a perfect way? I’ve a project that I am just now working on, and I have been on the look out for such information.
Thanks for the article, can you make it so I get an email when you publish a fresh post?
하동동해출장만남 소자본 창업
Hello, Neat post. There is an issue with your website in internet explorer,
might test this? IE nonetheless is the market leader and
a good portion of other folks will pass over your great writing due
to this problem.
One thing I want to say is the fact car insurance cancellation is a feared experience so if you’re doing the suitable things being a driver you won’t get one. Some people do obtain notice that they are officially dumped by their own insurance company and several have to fight to get added insurance from a cancellation. Low-priced auto insurance rates are usually hard to get from a cancellation. Understanding the main reasons concerning the auto insurance cancellation can help drivers prevent sacrificing one of the most essential privileges accessible. Thanks for the strategies shared via your blog.
https://nicesongtoyou.com/financial-portal/international-drivers-license/
Incredible many of excellent material!
https://www.youtube.com/channel/UCyxM_MlJsJQaQSuJ8lR58tg
http://www.youtube.com/@starissu
https://avenue18.ru/
Undeniably believe that which you stated. Your favorite justification seemed to be on the internet the easiest thing to be aware of. I say to you, I certainly get irked while people think about worries that they just don’t know about. You managed to hit the nail upon the top and defined out the whole thing without having side effect , people could take a signal. Will probably be back to get more. Thanks
Hey there, You have done an excellent job. I抣l certainly digg it and personally recommend to my friends. I am confident they will be benefited from this web site.
울산콜걸
Great post. I used to be checking constantly this weblog and I am impressed! Extremely helpful information specially the last part 🙂 I care for such info a lot. I used to be seeking this certain info for a very lengthy time. Thanks and good luck.
Something more important is that when looking for a good internet electronics shop, look for online shops that are frequently updated, always keeping up-to-date with the newest products, the perfect deals, along with helpful information on goods and services. This will ensure you are getting through a shop which stays over the competition and give you what you should need to make educated, well-informed electronics purchases. Thanks for the important tips I have learned from your blog.
We’re a bunch of volunteers and starting a brand new scheme in our community. Your site offered us with helpful info to work on. You’ve performed an impressive process and our entire group might be grateful to you.
Valuable info. Lucky me I found your web site by accident, and I’m shocked why this accident did not happened earlier! I bookmarked it.
An additional issue is that video gaming became one of the all-time most significant forms of entertainment for people of various age groups. Kids play video games, and adults do, too. The XBox 360 is just about the favorite gaming systems for people who love to have a huge variety of video games available to them, in addition to who like to play live with people all over the world. Thanks for sharing your opinions.
I’m curious to find out what blog platform you happen to be working with? I’m having some minor security issues with my latest blog and I’d like to find something more secure. Do you have any suggestions?
F*ckin?tremendous things here. I am very glad to see your post. Thanks a lot and i am looking forward to contact you. Will you please drop me a mail?
Hey there! Someone in my Myspace group shared this site with us so I came to look it over. I’m definitely enjoying the information. I’m book-marking and will be tweeting this to my followers! Exceptional blog and terrific design.
ayahuasca retreat rythmia cost,
Magnificent goods from you, man. I have remember your stuff prior to and you’re simply too excellent. I actually like what you’ve bought right here, certainly like what you’re saying and the best way by which you are saying it. You’re making it enjoyable and you continue to take care of to stay it smart. I can not wait to learn far more from you. That is really a great web site.
While it will not be the case with all governments, some governments look to infringe on the liberty of their residents.
“Thank you for yet another enlightening and enjoyable read! Your blog has become a trusted source of knowledge and inspiration for me. I’m grateful for the time and effort you invest in providing valuable content for free.”
Thanks for your useful post. As time passes, I have been able to understand that the symptoms of mesothelioma are caused by the particular build up associated fluid relating to the lining of the lung and the chest cavity. The disease may start inside the chest location and distribute to other body parts. Other symptoms of pleural mesothelioma include fat reduction, severe inhaling and exhaling trouble, nausea, difficulty ingesting, and swelling of the face and neck areas. It must be noted that some people having the disease usually do not experience just about any serious signs at all.
WOW just what I was searching for. Came here
by searching for udin777
이태원게이바
청도페이스라인출장
안성출장마사지
https://www.portalnet.cl/usuarios/johnson-uri.1102150/
iPad wall mount: a sleek, space-saving solution for both home and business. Easily attach your iPad to the wall for convenient viewing and usage, optimizing your space and enhancing accessibility
Heya i抦 for the first time here. I found this board and I find It really useful & it helped me out a lot. I hope to give something back and help others like you helped me.
대전세븐나이트
I am continuously browsing online for tips that can assist me. Thank you!
I wanted to create you this bit of note to be able to thank you very much as before on the great solutions you’ve shown here. It is simply wonderfully open-handed with people like you to supply easily all that a few people would have offered for an e book to make some bucks on their own, precisely seeing that you could possibly have tried it if you wanted. Those tactics additionally acted like a good way to fully grasp that most people have similar dreams really like my personal own to know many more on the topic of this issue. I’m sure there are several more pleasurable situations in the future for individuals that looked at your blog.
대전나이트클럽
강남콜걸
충무로출장업소
One more important aspect is that if you are an elderly person, travel insurance with regard to pensioners is something you should make sure you really take into consideration. The older you are, a lot more at risk you will be for allowing something poor happen to you while in another country. If you are definitely not covered by quite a few comprehensive insurance cover, you could have several serious troubles. Thanks for expressing your ideas on this website.
It is indeed my belief that mesothelioma can be the most deadly cancer. It contains unusual qualities. The more I really look at it a lot more I am assured it does not respond like a true solid cells cancer. When mesothelioma is often a rogue viral infection, hence there is the prospects for developing a vaccine along with offering vaccination to asbestos exposed people who are at high risk with developing potential asbestos associated malignancies. Thanks for expressing your ideas about this important health issue.
Way cool! Some extremely valid points! I appreciate you
penning this post and also the rest of the site is really good.
Wow, wonderful blog structure! How long have you ever been blogging for? you make blogging glance easy. The full look of your web site is fantastic, let alone the content!
I’ve been browsing online greater than three hours
these days, yet I never found any attention-grabbing article like yours.
It’s beautiful price enough for me. Personally, if
all webmasters and bloggers made just right content material as you did, the internet will likely be
a lot more useful than ever before.
Incredible a lot of awesome info.
Hey! This post couldn’t be written any better! Reading through this post reminds me of my good old room mate! He always kept chatting about this. I will forward this write-up to him. Fairly certain he will have a good read. Many thanks for sharing!
In these days of austerity plus relative panic about taking on debt, most people balk resistant to the idea of making use of a credit card to make purchase of merchandise or perhaps pay for a holiday, preferring, instead just to rely on the particular tried and also trusted procedure for making payment – hard cash. However, if you possess the cash there to make the purchase in full, then, paradoxically, that is the best time to be able to use the card for several motives.
Hi! I’ve been reading your site for some time now and finally got the courage
to go ahead and give you a shout out from Atascocita Tx!
Just wanted to mention keep up the excellent work!
Hey this is kind of of off topic but I was wanting to know
if blogs use WYSIWYG editors or if you have to manually code with HTML.
I’m starting a blog soon but have no coding experience so I wanted to get advice from someone with experience.
Any help would be greatly appreciated!
하동동해출장만남 소자본 창업
The next time I read a blog, I hope that it doesnt disappoint me as much as this one. I mean, I do know it was my choice to learn, but I truly thought youd have something interesting to say. All I hear is a bunch of whining about something that you would fix when you werent too busy in search of attention.
I have been surfing online more than 3 hours today, yet I never found any interesting article like yours. It is pretty worth enough for me. In my view, if all site owners and bloggers made good content as you did, the internet will be a lot more useful than ever before.
https://www.youtube.com/channel/UCyxM_MlJsJQaQSuJ8lR58tg
Valuable information. Lucky me I found your website by accident, and I’m shocked why this accident didn’t happened earlier! I bookmarked it.
Hey there would you mind stating which blog platform you’re using? I’m planning to start my own blog in the near future but I’m having a tough time making a decision between BlogEngine/Wordpress/B2evolution and Drupal. The reason I ask is because your design and style seems different then most blogs and I’m looking for something unique. P.S My apologies for being off-topic but I had to ask!
Excellent post. I was checking constantly this blog and I am impressed! Very helpful info specifically the last part 🙂 I care for such information much. I was looking for this certain info for a very long time. Thank you and good luck.
I will right away take hold of your rss as I can’t in finding your
e-mail subscription link or newsletter service.
Do you have any? Kindly let me realize so that I may subscribe.
Thanks.
http://www.youtube.com/@starissu
Hi there, after reading this remarkable paragraph i am too delighted to share my know-how here with friends.
대전나이트클럽
group sex
Woah! I’m really digging the template/theme of this blog. It’s simple, yet effective. A lot of times it’s challenging to get that “perfect balance” between usability and appearance. I must say you’ve done a excellent job with this. Additionally, the blog loads extremely fast for me on Opera. Outstanding Blog!
Appreciate you for sharing these kinds of wonderful blogposts. In addition, the ideal travel as well as medical insurance plan can often ease those concerns that come with vacationing abroad. Your medical emergency can in the near future become costly and that’s bound to quickly set a financial problem on the family finances. Setting up in place the perfect travel insurance bundle prior to leaving is definitely worth the time and effort. Thanks
I’m really loving the theme/design of your blog. Do you ever run into any internet browser compatibility issues? A number of my blog audience have complained about my site not working correctly in Explorer but looks great in Firefox. Do you have any ideas to help fix this issue?
Interesting post right here. One thing I would like to say is always that most professional job areas consider the Bachelor’s Degree like thejust like the entry level requirement for an online degree. Although Associate College diplomas are a great way to begin, completing ones Bachelors reveals many doors to various occupations, there are numerous on-line Bachelor Diploma Programs available via institutions like The University of Phoenix, Intercontinental University Online and Kaplan. Another concern is that many brick and mortar institutions make available Online editions of their qualifications but commonly for a drastically higher price than the companies that specialize in online course programs.
Hi to all, how is everything, I think every one is getting more from this web site,
and your views are nice designed for new users.
Thanks for your suggestions. One thing I’ve noticed is always that banks and also financial institutions have in mind the spending habits of consumers while also understand that many people max away their cards around the vacations. They properly take advantage of that fact and start flooding the inbox as well as snail-mail box using hundreds of 0 APR card offers right after the holiday season comes to an end. Knowing that if you’re like 98 of American public, you’ll rush at the possible opportunity to consolidate credit card debt and shift balances towards 0 rate credit cards.
전신스타킹
Thanks for sharing such a pleasant thought, piece of
writing is fastidious, thats why i have read it entirely
I am really inspired along with your writing skills as well as with the structure in your weblog. Is that this a paid topic or did you modify it your self? Anyway keep up the excellent high quality writing, it is rare to see a nice weblog like this one today..
Thanks a ton for your post. I’d like to comment that the price of car insurance differs a lot from one policy to another, since there are so many different issues which contribute to the overall cost. One example is, the model and make of the auto will have a large bearing on the cost. A reliable outdated family car or truck will have an inexpensive premium than the usual flashy performance car.
I think this is one of the most important information for me. And i am glad reading your article. But wanna remark on some general things, The site style is great, the articles is really great : D. Good job, cheers
I will immediately grab your rss feed as I can not find your e-mail subscription link or e-newsletter service. Do you’ve any? Please let me know in order that I could subscribe. Thanks.
I always look forward to your new posts. You have a way of making even the most complex topics easy to understand. Excellent job!swiftnook
Interesting post right here. One thing I’d like to say is the fact that most professional job areas consider the Bachelor’s Degree like thejust like the entry level requirement for an online diploma. Although Associate Certification are a great way to start, completing your current Bachelors opens many doorways to various professions, there are numerous internet Bachelor Diploma Programs available through institutions like The University of Phoenix, Intercontinental University Online and Kaplan. Another thing is that many brick and mortar institutions present Online variants of their qualifications but generally for a greatly higher cost than the companies that specialize in online diploma programs.
JX
In the center of Paris, effective cockroach treatment is actually extremely important for preserving social health and wellness and also preserving the area’s charm. These tough insects not just bring diseases yet likewise stain the credibility of businesses and also homes. Executing sturdy control procedures guarantees hygiene, health, as well as a pest-free atmosphere, guarding the metropolitan area’s allure for individuals as well as site visitors identical, https://ourdoings.com/mabelpatton/2024-06-02#e828302.
It抯 actually a nice and useful piece of information. I抦 satisfied that you just shared this useful information with us. Please keep us up to date like this. Thank you for sharing.
You actually make it appear really easy together with
your presentation however I to find this matter to be actually something which I think I’d never understand.
It sort of feels too complex and extremely broad
for me. I am taking a look forward for your subsequent submit,
I will attempt to get the hold of it!
Wow that was odd. I just wrote an extremely long comment but after I clicked submit my comment didn’t show up. Grrrr… well I’m not writing all that over again. Regardless, just wanted to say fantastic blog!
What you posted made a lot of sense. However, what about this?
what if you added a little content? I ain’t saying your information isn’t
good., but suppose you added something to possibly grab folk’s
attention? I mean Pontine Crossing Tract (pct) |
Whole Brain Protocol for Tractography with Empirical
MRI is kinda plain. You could glance at Yahoo’s front page and
note how they create post titles to grab people interested.
You might try adding a video or a pic or two to grab readers interested about what you’ve got to say.
In my opinion, it might bring your blog a little livelier.
It抯 really a great and useful piece of info. I抦 happy that you shared this useful information with us. Please stay us informed like this. Thank you for sharing.
Do you mind if I quote a few of your articles as long as I provide credit and sources back to your weblog? My website is in the very same niche as yours and my users would definitely benefit from a lot of the information you present here. Please let me know if this ok with you. Many thanks!
Good day! I know this is kinda off topic however
I’d figured I’d ask. Would you be interested in exchanging links or maybe guest authoring a blog post or vice-versa?
My blog goes over a lot of the same topics as yours and I feel we could greatly
benefit from each other. If you happen to be interested feel free to shoot me an e-mail.
I look forward to hearing from you! Superb blog
by the way!
I am continually looking online for tips that can help me. Thx!
https://itgunza.com/4464
Thank you for this article. I might also like to state that it can be hard when you are in school and merely starting out to establish a long credit history. There are many pupils who are simply just trying to pull through and have a good or beneficial credit history are often a difficult element to have.
Humans definitely have hobbies, if you want to find out more information, please visit our website.https://hobiapaaja01.wordpress.com
Wow, marvelous blog layout! How long have you been blogging for? you make blogging look easy. The overall look of your site is wonderful, as well as the content!
Great work! This is the type of info that should be shared around the internet. Shame on Google for not positioning this post higher! Come on over and visit my site . Thanks =)
Hey there! I’ve been reading your web site for some time now and finally got the bravery to go ahead and give you a shout out from Porter Tx! Just wanted to say keep up the excellent job!
you are really a good webmaster. The site loading speed is incredible. It seems that you are doing any unique trick. Moreover, The contents are masterwork. you have done a great job on this topic!
I have read a few just right stuff here. Certainly worth bookmarking for revisiting. I wonder how so much attempt you put to create the sort of great informative site.
Attractive part of content. I simply stumbled upon your site and in accession capital to say that I get in fact enjoyed account your weblog posts. Any way I will be subscribing to your feeds or even I fulfillment you get right of entry to consistently quickly.
I抦 impressed, I must say. Actually rarely do I encounter a blog that抯 both educative and entertaining, and let me tell you, you’ve hit the nail on the head. Your idea is excellent; the issue is one thing that not sufficient individuals are speaking intelligently about. I am very glad that I stumbled throughout this in my seek for one thing relating to this.
A true authority in its field. Trustworthy and informative.
My spouse and I stumbled over here coming from
a different web page and thought I might check things out.
I like what I see so i am just following you. Look forward to checking out your web page again.
Thank you for another informative blog. Where else could I get that kind of info written in such a perfect way? I’ve a project that I’m just now working on, and I have been on the look out for such info.
Nicely put, Kudos!
Thank you for the auspicious writeup. It in fact was a amusement account it. Look advanced to more added agreeable from you! However, how could we communicate?
Jason Bateman’s character Nick, who works for a cruel tyrant of a boss played by Kevin Spacey.
I do believe all of the concepts you have introduced in your post. They’re very convincing and will definitely work. Nonetheless, the posts are very quick for newbies. May just you please extend them a bit from next time? Thank you for the post.
ehternet cables are still the ones that i use for my home networking applications~
I抦 impressed, I need to say. Really rarely do I encounter a blog that抯 each educative and entertaining, and let me inform you, you’ve got hit the nail on the head. Your thought is excellent; the issue is one thing that not enough people are talking intelligently about. I am very completely satisfied that I stumbled across this in my seek for one thing relating to this.
Wow, wonderful blog structure! How lengthy have you been running a blog for? you made blogging glance easy. The whole look of your site is magnificent, let alone the content!
I believe that is one of the such a lot significant info for me. And i’m satisfied reading your article. However want to statement on some common things, The website style is perfect, the articles is really great . Excellent activity, cheers.
I’m really enjoying the design and layout of your blog. It’s a very easy on the eyes which makes it much more pleasant for me to come here and visit more often. Did you hire out a designer to create your theme? Excellent work!
This website online is mostly a walk-through for all of the info you wanted about this and didn抰 know who to ask. Glimpse right here, and you抣l undoubtedly uncover it.
Top-notch post it is definitely. My boss has been searching for this content.
I’ve learned several important things as a result of your post. I would also like to mention that there can be situation where you will make application for a loan and don’t need a cosigner such as a Federal government Student Support Loan. In case you are getting credit through a conventional finance company then you need to be ready to have a co-signer ready to assist you to. The lenders can base their decision over a few issues but the largest will be your credit rating. There are some creditors that will furthermore look at your job history and make a decision based on that but in many cases it will depend on your score.
Howdy! I could have sworn I’ve been to this blog before but after checking through some of the post I realized it’s new to me. Nonetheless, I’m definitely delighted I found it and I’ll be bookmarking and checking back often!
Hey, your site is very interesting.. Plus it was something I’m able to definitely relate with Ill constantly stop by for your blog therefore i hope you continue making fun and interesting posts like this one…
Your mode of telling all in this piece of writing is truly
pleasant, every one be able to effortlessly know it, Thanks a lot.
https://starity.hu/profil/453134-rentcarfycom/
Youre so cool! I dont suppose Ive read anything such as this prior to. So nice to uncover somebody with many original ideas on this subject. realy thank you for beginning this up. this website can be something that is required on the web, a person after some originality. helpful job for bringing new stuff for the web!
It’s a shame you don’t have a donate button! I’d without a doubt donate to this brilliant blog! I guess for now i’ll settle for bookmarking and adding your RSS feed to my Google account. I look forward to brand new updates and will share this site with my Facebook group. Chat soon!
Great knowledge, do anyone mind merely reference back to it
Nice post, thank you for the information.
For friends who are looking for references to various best games to spend your time on, you can visit the website below.
https://170.64.190.46
Document Information. Was’t things came to be details on seeking however if I just sought after Yahoo this page emerged liked working out looked versus eachother in addition to wanted at any rate many thanks.
You should experience a tournament for starters of the best blogs over the internet. I’m going to recommend this site!
I discovered your blog site on google and check several of your early posts. Always maintain on the great operate. I extra increase your RSS feed to my MSN News Reader. Seeking forward to reading much more within you down the line!…
Heya i’m for the first time here. I came across this board and I find It really useful & it helped me out a lot.
I hope to give something back and aid others like
you helped me.
I am usually to blogging and i genuinely appreciate your posts. This content has really peaks my interest. I’m going to bookmark your site and maintain checking for first time info.
Hiya, I’m really glad I have found this info. Today bloggers publish just about gossips and web and this is actually irritating. A good site with exciting content, that is what I need. Thanks for keeping this web-site, I will be visiting it. Do you do newsletters? Can’t find it.
Thank you, I have recently been looking for info about
this subject for a long time and yours is the best I’ve found out
so far. However, what concerning the conclusion? Are you certain concerning the source?
An impressive share, I just given this onto a colleague who has been performing a little analysis for this. And then he in truth bought me breakfast since I stumbled upon it for him.. smile. So let me reword that: Thnx for that treat! But yeah Thnkx for spending any time to discuss this, I feel strongly about this and really like reading more about this topic. If at all possible, as you become expertise, could you mind updating your blog with increased details? It is highly great for me. Huge thumb up with this writing!
Great blog post, I’ve bookmarked this page and have a feeling I’ll be returning to it regularly.
Intimately, the post is actually the sweetest on that worthw hile topic. I harmonise with your conclusions and also will certainly eagerly look forward to your incoming updates. Saying thanks will certainly not simply be enough, for the extraordinary clarity in your writing. I can perfect away grab your rss feed to stay informed of any kind of updates. Solid work and also much success in your business dealings!
I actually wanted to construct a small remark in order to say thanks to you for all the awesome points you are giving here. My time intensive internet search has now been recognized with reasonable tips to share with my friends and classmates. I ‘d declare that most of us readers actually are definitely fortunate to live in a fantastic community with very many lovely people with insightful ideas. I feel very much happy to have discovered your web page and look forward to some more pleasurable times reading here. Thank you once again for a lot of things.
It’s amazing to pay a visit this web page and reading the views
of all mates regarding this post, while I am also zealous of getting experience.
Can I simply say what a reduction to find somebody who actually knows what theyre talking about on the internet. You undoubtedly know easy methods to deliver a difficulty to gentle and make it important. More people must learn this and understand this facet of the story. I cant believe youre no more widespread since you undoubtedly have the gift.
best suvs… […]we like to honor other sites on the web, even if they aren’t related to us, by linking to them. Below are some sites worth checking out[…]…
Heya i am for the first time here. I came across this board and I find It truly useful & it helped me out much. I hope to give something back and aid others like you helped me.
One thing I’d prefer to comment on is that weight loss program fast is possible by the perfect diet and exercise. Your size not simply affects the look, but also the overall quality of life. Self-esteem, major depression, health risks, in addition to physical ability are impacted in excess weight. It is possible to make everything right but still gain. If this happens, a problem may be the offender. While too much food and never enough workout are usually accountable, common health conditions and popular prescriptions can certainly greatly add to size. Kudos for your post right here.
If some one needs to be updated with latest technologies after that he must be pay a visit this
web page and be up to date all the time. http://%3Cp%3ESezer%20Yurtseven%20was%20the%20second%20housemate%20being%20evicted%20in%20the%20Big%20Brother%20property%20and%20won%20two%20dubious%20honours%20in%20the%20procedure.%20For%20starters,%20he%20was%20evicted%20by%20the%20biggest%20community%20vote%20margin%20of%20all%20time%20a%20large%2091.6%25,%20creating%20him%20the%20shows%20most%20detested%20housemate%20in%20Huge%20Brothers%20seven%20series%20run.%3C/p%3E%3Cp%3E%3Ciframe%20src=%22https://www.youtube.com/embed/d__KxCyLb2I%22%20width=%22560%22%20height=%22315%22%20frameborder=%220%22%20allowfullscreen%3E%3C/iframe%3E%3C/p%3E%3Cp%3EHe%20also%20became%20the%20earliest%20male%20housemate%20to%20get%20evicted%20from%20your%20house,%20obtaining%20been%20shown%20the%20door%20by%20the%20tip%20of%207%20days%20two.%3C/p%3E%3Cp%3ESezer%20was%20a%20shoe-in%20to%20depart%20as%20far%20as%20the%20bookmakers%20ended%20up%20concerned%20as%20soon%20as%20it%20was%20uncovered%20he%20was%20certainly%20one%20of%20three%20housemates%20up%20for%20eviction.%20He%20was%20out%20there%20at%20three/10%20which%20plummeted%20to%201/33%20by%20Friday%20evening.%3C/p%3E%3Cp%3EEven%20so,%20very%20first%20thing%20Monday%20morning,%20Sezer%20was%20obtainable%20at%20comparatively%20gigantic%20odds%20of%2012/one%20to%20become%20the%20next%20human%20being%20evicted%20as%20whinging%20Nikki%20was%20the%20business%20favorite%20to%20depart%20subsequent%20at%20even%20income.%3C/p%3E%3Cp%3EYour%20situation%20transformed%20dramatically%20by%20the%20time%20it%20arrived%20to%20nominations%20as%20not%20just%20was%20Nikki%20receiving%20favourable%20edits%20with%20the%20output%20crew,%20demonstrating%20the%20likes%20of%20Grace%20talking%20about%20her%20guiding%20her%20back,%20but%20in%20a%20very%20remarkable%20twist%20the%20two%20Sezer%20and%20Imogen%20ended%20up%20banned%20from%20nominations%20after%20currently%20being%20caught%20speaking%20about%20them.%3C/p%3E%3Cp%3EEach%20Sezer%20and%20Grace%20weren&
my web page: learn more link
you use a fantastic blog here! do you wish to earn some invite posts on my small blog?
Hello there, just became aware of your blog through Google, and found that it’s really informative. I抦 going to watch out for brussels. I抣l be grateful if you continue this in future. A lot of people will be benefited from your writing. Cheers!
Appreciating the time and energy you put into your website and in depth information you provide. It’s awesome to come across a blog every once in a while that isn’t the same out of date rehashed information. Great read! I’ve bookmarked your site and I’m adding your RSS feeds to my Google account.
J’ai la possibilité de vous transmettre les adresses pour plus d’échantillons en relation avec cet idée. Contactez moi par email.
I got this site from my buddy who told me about this web
page and now this time I am browsing this web site and reading very informative articles at this
place.
Barefoot jogging has actually gained popularity recently, along with many enthusiasts swearing by its perks for feet wellness and also managing performance. If you are actually hoping to switch to barefoot managing or just intend to explore the globe of smart footwear, locating the appropriate set of barefoot shoes is actually important, https://www.tracksandfields.com/profile/view/elainafhaas.
the table beds which we have a year ago had already broken down, it is mostly made up of plastic.,
https://nicesongtoyou.com/tax/
I’ve recently started a blog, the details you provide on this site has reduced the problem tremendously. Appreciation for all of your current time & work…
Hello,I love reading through your blog, I wanted to leave a little comment to support you and wish you a good continuation. Wishing you the best of luck for all your blogging efforts.
i really hate it when my sebaceous gland are producing too much oil, it really makes my life miserable**
There is noticeably a lot of money to comprehend this. I assume you made particular nice points in features also.
Hi there, I discovered your website by means of Google while searching for a comparable topic, your site got here up, it appears to be like great. I’ve bookmarked it in my google bookmarks.
Howdy! This is kind of off topic but I need some advice from an established blog. Is it very difficult to set up your own blog? I’m not very techincal but I can figure things out pretty fast. I’m thinking about setting up my own but I’m not sure where to begin. Do you have any ideas or suggestions? Appreciate it
I have been meaning to post something like this on my blog and you have given me an idea. Cheers.
You could certainly see your expertise in the paintings you write. The arena hopes for even more passionate writers such as you who are not afraid to say how they believe. All the time go after your heart.
there are many sites on the internet that offers downloadable movies, some are even offering the latest movies*
I have discovered that sensible real estate agents almost everywhere are getting set to FSBO Advertising and marketing. They are seeing that it’s more than merely placing a sign in the front yard. It’s really about building relationships with these retailers who later will become consumers. So, whenever you give your time and energy to helping these vendors go it alone — the “Law associated with Reciprocity” kicks in. Great blog post.
This is actually exciting, You’re a highly seasoned article author. I have registered with your feed furthermore watch for witnessing your personal exceptional write-ups. Plus, We’ve shared your web blog in our social networks.
Thanks a lot for your interesting article. I have been searching for such message for a very long time. Not all your content is completely clear to me, even though it is definitely interesting and worth reading.
I’d should seek advice from you here. Which is not something It’s my job to do! I quite like reading an article that will make people believe. Also, many thanks permitting me to comment!
Thanks for making me to achieve new thoughts about computer systems. I also have the belief that certain of the best ways to maintain your notebook computer in leading condition is a hard plastic case, as well as shell, that will fit over the top of one’s computer. A lot of these protective gear are usually model unique since they are made to fit perfectly above the natural casing. You can buy them directly from the owner, or through third party places if they are readily available for your notebook, however its not all laptop could have a spend on the market. Again, thanks for your tips.
What?s Going down i’m new to this, I stumbled upon this I have found It absolutely useful and it has aided me out loads. I hope to give a contribution & help different customers like its aided me. Great job.
Oh my goodness! an excellent article dude. Thanks a lot Even so I’m experiencing problem with ur rss . Do not know why Struggle to register for it. Can there be any person finding identical rss issue? Anyone who knows kindly respond. Thnkx
Strong blog. I acquired variant nice info. I?ve been keeping an eye fixed on this technology for a few time. It?utes attention-grabbing the method it retains completely different, yet a few of the primary components remain the same. have you observed lots modification since Search engines created their own latest purchase in the field?
I continue to read blogs like this, because I am trying to better my ability to write great article, this is an awesome example. I apologize, but I will be trying your style of writing in my work(if possible). I learn from the best and forget the rest.
Cheap Handbags Wholesale You should think about doing growing this web site to a major authority in this particular market. You clearly contain a grasp handle in the topics so many people are looking for on this website anyways therefore you could indisputably even create a dollar or two off of some advertising. I’d explore following recent topics and raising how many write ups putting up and i guarantee you???d begin seeing some awesome web traffic in the near future. Simply a thought, all the best in whatever you do!
designer shoes are of course very expensive but i like their style,
There is noticeably a lot of money to understand this. I suppose you made particular nice points in features also.
There’s certainly a lot to find out about this subject. I really like all the points you
made.
If you are going for most excellent contents like me,
just pay a quick visit this web site daily as it gives feature contents, thanks
This paragraph will assist the internet viewers for building up new webpage or even a weblog from start to
end.
Wonderful blog! Do you have any tips and hints for aspiring writers? I’m hoping to start my own site soon but I’m a little lost on everything. Would you propose starting with a free platform like WordPress or go for a paid option? There are so many options out there that I’m completely overwhelmed .. Any suggestions? Bless you!
It’s appropriate time to make a few plans for the longer term and it’s time to be happy. I’ve learn this submit and if I may I wish to counsel you few fascinating issues or advice. Maybe you could write next articles relating to this article. I wish to learn even more issues about it!
hello!,I love your writing so so much! percentage we keep up a correspondence extra about your article on AOL? I need an expert in this space to solve my problem. Maybe that is you! Looking forward to look you.
https://honeytipit.tistory.com/entry/EC9EA5EAB8B0EC9A94EC9691EC9A94EC9B90-ED959CEC8B9CECA780EC9B90EAB888?category=889549
проститутки индивидуалки челябинск
Very soon this web site will be famous among all blogging people, due to
it’s pleasant articles
I really want to thank you for yet another great informative post, I’m a loyal visitor to this blog and I can’t tell you how much valuable tips I’ve learned from reading your content. I really appreciate all the hard work you put into this great blog.
I’ve been exploring for a little bit for any high quality articles or weblog posts on this kind of area . Exploring in Yahoo I ultimately stumbled upon this web site. Studying this info So i am satisfied to convey that I’ve an incredibly good uncanny feeling I discovered just what I needed. I so much no doubt will make sure to don’t put out of your mind this website and give it a look a relentless basis.
Good day! This post couldn’t be written any better! Reading this post reminds me of my good old room mate! He always kept talking about this. I will forward this page to him. Pretty sure he will have a good read. Thank you for sharing!
Very nice blog. I especially like content that has to do with health and wellness, so it’s refreshing to me to see what you have here. Keep it up! facial exercises
Thanks for the input. I’ll be back to read more later.
I wish more authors of this type of content would take the time you did to research and write so well. I am very impressed with your vision and insight.
Only wanna input on few general things, The website layout is perfect, the written content is rattling superb .
I don’t actually get how this is much of a difference between the New York Times publishing this or some internet blog. Stuff such as this one must to be pushed out more to the public. I wish that citizens in America would do something like this.
Awesome site! You’ve some very worthwhile posts.. Nice background as well haha. Keep up the good work, Ill make sure to come across and see more of your page!
There are a couple of intriguing points at some point on this page but I do not determine if they all center to heart. There’s some validity but I will take hold opinion until I check into it further. Great post , thanks and then we want a lot more! Added onto FeedBurner too
An impressive share, I simply given this onto a colleague who was doing somewhat analysis on this. And he in fact bought me breakfast as a result of I found it for him.. smile. So let me reword that: Thnx for the deal with! But yeah Thnkx for spending the time to debate this, I feel strongly about it and love reading extra on this topic. If doable, as you develop into expertise, would you mind updating your blog with extra particulars? It’s highly helpful for me. Large thumb up for this weblog publish!
Very descriptive post, I loved that bit. Will there be a part 2?
Great post, you have pointed out some excellent points, I also believe this s a very good website.
Good day, Can I export your page photo and use that on my personal web site?
Is your webdesigner looking for a job. I think your site is great.
Good day! Do you know if they make any plugins to help with SEO?
I’m trying to get my blog to rank for some targeted keywords but I’m not
seeing very good results. If you know of any please share.
Kudos! You can read similar blog here: Which escape room
Hi, Neat post. There’s a problem with your web site in internet explorer, would check this… IE still is the market leader and a big portion of people will miss your magnificent writing due to this problem.
I think you have observed some very interesting details , thankyou for the post.
I discovered your website site on the search engines and check many of your early posts. Keep up the top notch operate. I just now additional increase Feed to my MSN News Reader. Looking for forward to reading a lot more within you at a later date!…
This is actually serious, You’re a particularly professional blogger. I have signed up with your feed furthermore watch for enjoying all of your incredible write-ups. In addition, I’ve got shared your webpage with our social networks.
Hello, I just hopped over to your web page via StumbleUpon. Not somthing I would usually browse, but I appreciated your thoughts none the less. Thanks for making some thing well worth browsing.
Congratulations on having one of the most sophisticated blogs Ive come across in some time! Its just incredible how much you can take away from something simply because of how visually beautiful it is. Youve put together a great blog space –great graphics, videos, layout. This is definitely a must-see blog!
My spouse and i got quite joyous when Chris could finish off his investigations through the ideas he had out of the web page. It is now and again perplexing to simply possibly be giving for free secrets that many some people might have been selling. And we also realize we have the writer to thank for that. These explanations you have made, the simple web site navigation, the relationships you can give support to foster – it’s got mostly exceptional, and it’s really letting our son and the family do think the situation is interesting, which is wonderfully pressing. Many thanks for the whole thing!
Feet detoxification patches are a straightforward yet powerful tool for assisting general wellness as well as wellness. By harnessing the power of natural elements and reflexology, these patches assist to extract contaminants from the body, marketing bodily and also mental vigor. Whether you’re looking to boost your power degrees, enhance your state of mind, or simply enhance your total sense of health, feet detoxing patches use a handy and also successful option, http://vintagemachinery.org/members/detail.aspx?id=108779.
I was able to find good information from your blog articles.
Howdy just wanted to give you a quick heads up and let you know a
few of the pictures aren’t loading correctly. I’m not sure why but I think its
a linking issue. I’ve tried it in two different web browsers and both
show the same results.
수원출장샵
I need to verify with you here. Which isn’t one thing I often do! I get pleasure from reading a publish that can make people think. Additionally, thanks for allowing me to remark!
индивидуалки иркутск на выезд
I am regular visitor, how are you everybody?
This paragraph posted at this site is actually pleasant.
Hi there! I just wish to give an enormous thumbs up for the nice info you’ve right here on this post. I shall be coming again to your blog for extra soon.
Interesting read i think your website is great with informative content which i like to add to my bookmarks. I’d like to share everyone
Hi, have you ever before asked yourself to write about Nintendo or PSP?
Possible require all types of led tourdates with some other fancy car applications. Many also provide historic packs and other requires to order take into your lending center, and for a holiday in upstate New York. ???
I’m not certain the place you are getting your information, however great topic. I must spend a while learning more or working out more. Thank you for magnificent info I was searching for this info for my mission.
When I originally commented I clicked the “Notify me when new comments are added” checkbox and now each time
a comment is added I get several emails with the same comment.
Is there any way you can remove me from that service?
Thanks!
Hi there! This is my 1st comment here so I just
wanted to give a quick shout out and tell you
I really enjoy reading your articles. Can you recommend any other blogs/websites/forums that
cover the same topics? Thanks!
When I originally commented I clicked the -Notify me when new comments are added- checkbox and already when a comment is added I receive four emails with similar comment. Perhaps there is in whatever way it is possible to get rid of me from that service? Thanks!
if you really need to become expert in driving, your really need to enroll in a driving school**
Thank you a lot for sharing this with all of us you actually recognise what you’re speaking approximately! Bookmarked. Please additionally seek advice from my web site =). We could have a link alternate agreement among us!
I have observed that in the world the present moment, video games are the latest trend with children of all ages. Occasionally it may be difficult to drag your kids away from the activities. If you want the very best of both worlds, there are numerous educational activities for kids. Good post.
индивидуалка иркутск
Incredible! This blog looks just like my old one! It’s
on a completely different topic but it has pretty much the same
page layout and design. Wonderful choice of colors!
Introduction In recent years, there has been a growing concern over the use of chemical insect repellents due to the potential harm they pose to both humans and the environment.
Have a look at my website; https://xn--hudfryngring-7ib.wiki/index.php/Buzz_Repel_Lamp:_Tips_For_Using_Your_Lamp_While_Camping
https://i0.wp.com/metaversenews.co.kr/wp-content/uploads/sites/6/2022/05/인스타-사진-저장.jpg?fit=4502C449
Hmm it appears like your blog ate my first comment (it was super
long) so I guess I’ll just sum it up what I wrote and say, I’m thoroughly enjoying your blog.
I as well am an aspiring blog blogger but I’m still new to everything.
Do you have any tips and hints for beginner blog writers?
I’d definitely appreciate it.
This blog site is an absolute delight to explore. From its captivating content to its intuitive design, it’s clear that a lot of thought and care has gone into every aspect of the site.
I’ve been browsing on-line greater than 3 hours lately, but I never discovered any interesting article like yours. It’s pretty price sufficient for me. In my opinion, if all web owners and bloggers made good content as you did, the web will likely be much more useful than ever before!
I would like to get as many links to my site as posible, rite now this is what I am doing!
being an entrepreneur opened up lots of business leads on my line of work, i like to make money both online and offline**
There couple of intriguing points soon enough in this article but I do not know if I see every one of them center to heart. There exists some validity but I will take hold opinion until I investigate it further. Good write-up , thanks therefore we want far more! Included in FeedBurner likewise
you have a great blog here! want to have the invite posts in my weblog?
I am curious to find out what blog system you are using? I’m experiencing some small security problems with my latest site and I’d like to find something more risk-free. Do you have any suggestions?
Heya this is a excellent write-up. I’m going to email this to my associates. I came on this while searching on aol I’ll be sure to come back. thanks for sharing.
https://i0.wp.com/metaversenews.co.kr/wp-content/uploads/sites/6/2022/03/1.2-3.png?fit=6402C337
Piece of writing writing is also a excitement, if you be acquainted with after that you can write or else it
is complicated to write.
https://chotiple.tistory.com/?page=44
I was wondering if you ever considered changing the structure of
your site? Its very well written; I love what youve got to say.
But maybe you could a little more in the way of content so people could connect with it better.
Youve got an awful lot of text for only having 1 or 2 images.
Maybe you could space it out better?
This web page is usually a walk-through you discover the information it suited you relating to this and didn’t know who to ask. Glimpse here, and you’ll undoubtedly discover it.
I discovered your blog post web site on the search engines and appearance several of your early posts. Always maintain the top notch operate. I additional the Feed to my MSN News Reader. Seeking forward to reading much more on your part down the line!…
I have a MAJOR food baby goin on right now.|Megasouraus|
Nice post. I learn something new and challenging on sites I stumbleupon every
day. It’s always useful to read through content from other
writers and practice something from their web sites.
Thanks for sharing superb informations. Your web-site is very cool. I am impressed by the details that you have on this web site. It reveals how nicely you perceive this subject. Bookmarked this web page, will come back for extra articles. You, my friend, ROCK! I found simply the info I already searched all over the place and just could not come across. What an ideal site.
Regards for this wondrous post, I am glad I noticed this internet site on yahoo.
great submit, very informative. I’m wondering why the other experts of this sector do not notice this. You must continue your writing. I am confident, you have a great readers’ base already!
It can be tough to write about this topic. I think you did an excellent job though! Thanks for this!
As a Newbie, I am continuously browsing online for articles that can benefit me. Thank you
i am planning to start a coffee franchise with me and my business partners in the LA area;;
wonderful put up, very informative. I wonder why the other experts of this sector don’t notice this. You should proceed your writing. I’m confident, you’ve a huge readers’ base already!
интим объявления в Иркутске
What’s up mates, how is all, and what you wish for to say regarding this post, in my view
its actually amazing in favor of me.
The digital cigarette uses a battery and a small heating aspect the vaporize the e-liquid. This vapor can then be inhaled and exhaled
I thought it was incredibly useful, don’t understand some people, just ignore them and keep doing what you do best.
There are some attention-grabbing deadlines in this article but I don’t know if I see all of them middle to heart. There is some validity but I will take hold opinion until I look into it further. Good article , thanks and we wish more! Added to FeedBurner as properly
Thank you for the information. I will offer you my most popular crime news. Therefore, if you like it, you can visit the website
https://wakbulu279.wixsite.com/berita-kriminal-news
You can separate comments by “
This is a very good tip particularly to those fresh to the blogosphere.
Brief but very precise info… Appreciate your sharing this one.
A must read article!
Que placer es leer los articulos de Carrera Politica Este sitio realmente sabe como mantenerme informado sobre los acontecimientos politicos mas relevantes. No puedo esperar para seguir explorando su contenido fascinante Carrera Politica es un sitio web impresionante que ofrece una perspectiva unica sobre los asuntos politicos actuales. Me encanta su enfoque informativo y su analisis profundo de los temas politicos de actualidad Felicitaciones a Estrategia politica por proporcionar un recurso tan valioso para quienes estan interesados en la politica
Good post. I learn something totally new and challenging on blogs I stumbleupon every day. It will always be helpful to read through articles from other authors and practice a little something from their web sites.
Excellent knowledge Many thanks!
Thanks for the diverse tips discussed on this blog. I have noticed that many insurance firms offer customers generous discounts if they choose to insure several cars with them. A significant number of households possess several motor vehicles these days, particularly those with old teenage youngsters still living at home, and the savings for policies can certainly soon begin. So it is a good idea to look for a great deal.
Do you have a spam problem on this website; I
also am a blogger, and I was wondering your situation; we
have developed some nice methods and we are looking to swap solutions
with others, please shoot me an email if interested.
It absolutely was any exhilaration finding your site yesterday evening. We arrived here these days seeking something totally new. I used to be not disappointed. Your opinions on brand new approaches on this factor were helpful as well as an superb help to me personally. Appreciate leaving out time to write out these items and for discussing your thoughts.
The when I read a blog, I hope so it doesnt disappoint me approximately that one. I mean, Yes, it was my option to read, but When i thought youd have some thing interesting to state. All I hear is often a lot of whining about something you could fix if you ever werent too busy in search of attention.
Thank you for the information. I will offer you my new crime story. Therefore, if you like it, you can visit the website
https://wakbulu279.wixsite.com/berita-kriminal-news
You made your point.
Currently it seems like Drupal is the top blogging platform available right now. (from what I’ve read) Is that what you’re using on your blog?
Thanks for your post.
Our team of experienced photographers will help you promote your business in the best way possible.
Oh my goodness! an incredible post dude. Thank you Nonetheless I’m experiencing issue with ur rss . Do not know why Struggling to subscribe to it. Possibly there is anyone getting identical rss difficulty? Anybody who knows kindly respond. Thnkx
“There are certainly many more details to take into consideration”
I adore your blog post.. comfortable colours & motif. Does a person design and style this amazing site on your own or maybe do anyone bring in help to make it work in your case? Plz react seeing that I!|m aiming to style my personal website and would want to learn exactly where ough obtained this kind of out of. thank you
I discovered your blog post web site online and check a number of your early posts. Keep within the very good operate. I recently extra up your Rss to my MSN News Reader. Seeking forward to reading more by you afterwards!…
수원출장샵
of course data entry services are very expensive that is why always make a backup of your files“
Thank you for another essential article. I wish I had your passion for writing!
I have learned some important things through your site post. One other point I would like to convey is that there are several games that you can buy which are designed particularly for preschool age children. They involve pattern identification, colors, creatures, and models. These normally focus on familiarization rather than memorization. This makes little ones engaged without sensing like they are learning. Thanks
I discovered your blog post web site online and appearance several of your early posts. Always maintain in the great operate. I recently additional your Feed to my MSN News Reader. Seeking forward to reading a lot more from you finding out afterwards!…
You made some decent points there. I looked online for the problem and found most individuals will go coupled with using your site.
Awesome post, where is the rss? I cant find it!
https://myabaddiction.wordpress.com/2017/07/02/first-time-at-a-korean-spa-jjimjilbang-king-spa-in-nj/
fertility clinics these days are very advanced and of course this can only mean higher success rates on birth,,
This website is often a walk-through its the details you desired about this and didn’t know who to inquire about. Glimpse here, and you’ll absolutely discover it.
Hi, I do think this is an excellent site. I stumbledupon it 😉 I am going to revisit once again since
i have saved as a favorite it. Money and freedom is the greatest
way to change, may you be rich and continue to guide other
people.
Wonderful blog! I found it while browsing on Yahoo News. Do you have any suggestions on how to get listed in Yahoo News? I’ve been trying for a while but I never seem to get there! Cheers
Thank you for the information about crime, let’s discuss crime on our website which provides the latest news about crime, for further information please visit our website.https://wakbulu279.wixsite.com/berita-kriminal-news
I am typically to blogging i genuinely appreciate your content. The content has really peaks my interest. I am about to bookmark your web site and keep checking for brand new data.
Gaga tweeted a voicemail coupled with a backlink to Japan Prayer Bracelets. She specially designed a bracelet, with pretty much all sales revenue going to Japanese relief plans
He keeps both strands moving along at equal pace, Cage’s is more engrossing; the film paying sly hint to knowing so when it utilises the concoction of Hit-Girl to drive the film’s final act on behalf of Cage’s plight.
Attractive section of content. I just stumbled upon your blog and in accession capital to assert that I acquire actually enjoyed account your blog posts. Anyway I will be subscribing to your augment and even I achievement you access consistently quickly.
I found your interesting topic by searching Ask. Looks like your skillful blog have allot of readers at your exclusive weblog. I find it to be a success story for every website maker.
Thank you for another excellent article. Where else may anybody get that kind of info in such an ideal way of writing? I’ve a presentation subsequent week, and I’m on the look for such info.
Very interesting points you have noted , regards for putting up.
That is a very good standpoint, yet seriously isn’t create any sence by any means preaching about which mather. Every means with thanks as well as pondered make an effort to promote your current publish in to delicius nevertheless it looks like it’s problems with all your blogs are you able to make sure you recheck the item. many thanks once more.
When I originally commented I appear to have clicked on the -Notify me when new comments are added- checkbox and from now on each time a comment is added I recieve 4 emails with the exact same comment. There has to be a way you can remove me from that service? Thank you.
I found your website through a random stroke of luck. It helped me do my research on this topic. Its amazing to think one site could contain so muck information. You are doing the great work!
A few things i have constantly told persons is that when looking for a good on the net electronics shop, there are a few factors that you have to take into account. First and foremost, you should really make sure to locate a reputable and in addition, reliable retailer that has got great assessments and rankings from other consumers and business world professionals. This will ensure that you are getting along with a well-known store to provide good service and aid to the patrons. Thank you for sharing your notions on this website.
Everyone loves what you guys are up too. This kind of clever work and
exposure! Keep up the amazing works guys I’ve added you guys to my own blogroll.
I am often to running a blog and i really admire your content. The article has actually peaks my interest. I’m going to bookmark your website and keep checking for brand new information.
하동동해출장만남 소자본 창업
This blog was… how do you say it? Relevant!! Finally I’ve found something that helped me. Many thanks.
May I simply say what a relief to find someone who really understands what
they’re discussing online. You definitely realize how to bring an issue to light and make it important.
More people need to read this and understand this side of your story.
It’s surprising you aren’t more popular given that you most certainly have the gift.
Hey there, I think your blog might be having browser compatibility issues. When I look at your website in Safari, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, superb blog!
I was curious if you ever considered changing the structure of your site?
Its very well written; I love what youve got to say. But maybe you could a
little more in the way of content so people could
connect with it better. Youve got an awful lot of text for only having one or 2 images.
Maybe you could space it out better?
I was very happy to discover this web site. I wanted to thank you for your time for
this particularly wonderful read!! I definitely really liked every little bit of it and
I have you book marked to see new stuff in your site.
I was curious if you ever thought of changing the page layout of your blog? Its very well written; I love what youve got to say. But maybe you could a little more in the way of content so people could connect with it better. Youve got an awful lot of text for only having 1 or 2 pictures. Maybe you could space it out better?
I’m very pleased to uncover this web site. I want to to thank you for your time for this fantastic read!! I definitely enjoyed every part of it and i also have you book marked to check out new stuff in your website.
수원출장샵
Hmm is anyone else having problems with the pictures on this blog loading? I’m trying to find out if its a problem on my end or if it’s the blog. Any responses would be greatly appreciated.|
Your insights into email marketing accessibility considerations have been useful.
An outstanding share! I’ve just forwarded this onto a colleague who has been conducting a little research on this. And he in fact bought me breakfast because I found it for him… lol. So let me reword this…. Thank YOU for the meal!! But yeah, thanks for spending some time to talk about this matter here on your web page.
이태원스웨디시안마게이클럽
Здесь вы найдете разнообразный видео контент Ялта Видео, охватывающий множество платформ и форматов.
От коротких зрелищных роликов на TikTok до долгоиграющих
трансляций на Twitch – мой канал стремится объединить лучшее из разных миров видео.
Присоединяйтесь, чтобы веселиться вместе с потоковым мультимедиа Stream, наслаждаться премиум-контентом от Vevo и
Vimeo, а также открывать для себя новые таланты на Rumble и BitChute.
Также вы найдете здесь эксклюзивный контент
для Instagram TV (IGTV), Facebook Watch и других социальных платформ.
А для поклонников аниме есть отдельная секция с лучшим контентом от
Crunchyroll.
Используя мощь профессиональных инструментов, таких как Brightcove,
Kaltura, JWPlayer, IBM Cloud Video и многих других,
я стараюсь предоставить вам впечатляющий и высококачественный видео опыт.
Подписывайтесь сегодня и оставайтесь
на волне самого свежего и разнообразного видео контента
в интернете. Добро пожаловать в мой мир ярких видео!”
Это описание канала объединяет все упомянутые вами видео платформы, демонстрируя вашу готовность работать с разными форматами и обеспечивать качественный и разнообразный контент для зрителей. Пусть оно привлечет новых подписчиков!
Hi, I do think this is an excellent web site. I stumbledupon it 😉 I may revisit once again since
I book-marked it. Money and freedom is the best way to change, may you be
rich and continue to help other people.
Howdy! Do you know if they make any plugins to help with SEO?
I’m trying to get my site to rank for some targeted keywords but I’m not seeing very good success.
If you know of any please share. Thank you! You can read similar blog here
와일드 바운티 쇼다운
질서는 말을 타고 Fang Jinglong의 죽음에 맞서 싸우라는 명령을 전달하기 위해 날아갔습니다.
Email personalization based on interests increases click-through rates.
https://download.beer/wp-content/uploads/2020/11/discord-image-4.jpg
Thanks for the publish. My spouse and i have constantly noticed that most people are desperate to lose weight since they wish to appear slim in addition to looking attractive. On the other hand, they do not generally realize that there are other benefits for losing weight also. Doctors insist that obese people are afflicted by a variety of illnesses that can be perfectely attributed to their own excess weight. The great thing is that people who’re overweight plus suffering from numerous diseases can help to eliminate the severity of their own illnesses by simply losing weight. It is easy to see a progressive but identifiable improvement with health while even a bit of a amount of weight loss is obtained.
https://download.beer/wp-content/uploads/2021/01/battleground-image-1.jpg
You should be a part of a contest for one of the greatest sites on the net. I’m going to recommend this blog!
https://dday.tistory.com/968
Excellent way of explaining, and pleasant post to take information regarding
my presentation subject matter, which i am going to convey
in college.
Excellent post. I was checking continuously this blog and I am impressed.
Undeniably believe that which you stated. Your favorite justification seemed to be on the net the simplest thing to be aware of. I say to you, I certainly get irked while people think about worries that they plainly don’t know about. You managed to hit the nail upon the top as well as defined out the whole thing without having side-effects , people could take a signal. Will probably be back to get more. Thanks
I would be mendacity basically said i do not such as this article, in reality, I love this a great deal I needed to place upward a discuss right here. I must state maintain the good function, and that i will likely be arriving again for good since I already bookmarked the actual page.
you should always be careful with recruitment agencies because some of them are just scammers;;
I see something genuinely interesting about your web site so I saved to my bookmarks .
most cosmetics today are made up of artificial products that can harm the skin, there are still few natural cosmetics on the market-
Someone essentially help to make seriously posts I would state. This is the very first time I frequented your web page and thus far? I amazed with the research you made to create this particular publish amazing. Fantastic job!
You are so cool! I don’t suppose I have read through anything like this before. So wonderful to find somebody with a few genuine thoughts on this subject. Seriously.. many thanks for starting this up. This site is something that is needed on the internet, someone with a little originality.
https://ddnews.co.kr/blog/2023/02/15/infp-love/
You seem to be very professional in the way you write.~`~’`
Really interesting and special post. I like things such as making more homework, developing writing skills, and also related things. These kinds of secrets help in being a qualified person on this topic. This page is very helpful to myself because people like you committed time to learning. Regularity is the key. But it is not too easy, as has been designed to be. I am not an expert like you and plenty of times I feel really giving it up.
Hi, I just found your weblog via google. Your viewpoint is truly relevant to my life at this moment, and I’m really pleased I found your website.
Outstanding brief which post helped me alot. Give you thanks I looking for your details?–.
Cheers! Lots of postings!
Wade and his sidekick LeBron Spliff James look beat the fuck down emotionally. Barring any dominance from the refs…Mavericks got this.|laughatLEBRON|
I like the efforts you have put in this, thank you for all the great blog posts.
Aw, this was an extremely good post. Spending some time and actual effort to generate a very good article… but what can I say… I put things off a lot and don’t seem to get anything done.
An impressive share, I just given this onto a colleague who was doing a bit of analysis on this. And he in truth purchased me breakfast as a result of I discovered it for him.. smile. So let me reword that: Thnx for the treat! However yeah Thnkx for spending the time to discuss this, I really feel strongly about it and love studying extra on this topic. If possible, as you develop into expertise, would you thoughts updating your blog with more details? It is highly helpful for me. Huge thumb up for this blog post!
Yo, I am ranking the crap out of “PROFIT INSIDERS REVIEW”.
At Vape Supply Store, we provide more than just e-cigarettes and e-liquids. We are dedicated to educating our customers and offering exceptional support to enhance their vaping experience.
Oh my goodness! an excellent write-up dude. Many thanks Nonetheless We are experiencing problem with ur rss . Do not know why Cannot sign up to it. Is there everyone acquiring identical rss difficulty? Anybody who knows kindly respond. Thnkx
When you have especially fair skin then you could notice which you are much more susceptible to moles. That is typical among people with lighter pores and skin and this is the cause why they ought to go to higher lengths to protect themselves in the sun’s dangerous effects.
Will you mind generally if I mention a couple of your blogposts providing that I give you credit and even sources returning to your web sites? My web-site is in the similar market as your own and my users will profit from several of the help and advice your site offer in this article. Please be sure to let me know if it is okay for you. Kind regards!
I like this – Gulvafslibning | Kurt Gulvmand , enjoyed this one thanks for putting up keep update – Gulvafslibning | Kurt Gulvmand.
Please keep on dropping such quality storys as this is a rare thing to find these days. I am always searching online for posts that can help me. looking forward to another great website. Good luck to the author! all the best!
Wonderful paintings! That is the kind of information that are meant to be shared across the web. Disgrace on the seek for no longer positioning this submit upper! Come on over and visit my website . Thanks =)
When I originally commented I clicked the -Notify me when new surveys are added- checkbox and already whenever a comment is added I receive four emails with similar comment. Perhaps there is by any means you can eliminate me from that service? Thanks!
https://film35.tistory.com/387
One more thing. It’s my opinion that there are many travel insurance web sites of reputable companies that allow you to enter your journey details and find you the quotations. You can also purchase the actual international travel cover policy on internet by using your own credit card. All you have to do should be to enter your current travel information and you can understand the plans side-by-side. Merely find the program that suits your finances and needs and after that use your credit card to buy the item. Travel insurance on the internet is a good way to do investigation for a reputable company with regard to international holiday insurance. Thanks for giving your ideas.
Youre so cool! I dont suppose Ive read anything like this before. So nice to find somebody with some original thoughts on this subject. realy i appreciate you for beginning this up. this amazing site is something that is required on-line, somebody with a bit of originality. valuable problem for bringing something new to the net!
Hi there, just became alert to your blog through Google, and found that it’s truly informative. I am going to watch out for brussels. I’ll appreciate if you continue this in future. Many people will be benefited from your writing. Cheers!
Immer etliche Firmen benützen heutzutage Interimmanagement als innovatives und ergänzendes Gerätschaft i. Spanne der Unternehmensführung. Denn hiermit wird Kenntnisstand leistungsfähig, bedarfsgerecht und schnell ins Unternehmen geholt.
I’m truly enjoying the design and layout of your website. It’s a very easy on the eyes which makes it much more enjoyable for me to come here and visit more often. Did you hire out a developer to create your theme? Fantastic work!
i always look for laptop reviews on the internet before buying a new one,::
Somebody essentially help to make seriously posts I would state. This is the very first time I frequented your website page and thus far? I amazed with the research you made to create this particular publish extraordinary. Magnificent job!
My partner and I absolutely love your blog and find a lot of your post’s to be what precisely I’m looking for. Does one offer guest writers to write content to suit your needs? I wouldn’t mind creating a post or elaborating on a lot of the subjects you write in relation to here. Again, awesome blog!
Oh my goodness! a fantastic write-up dude. Thanks Nonetheless I’m experiencing trouble with ur rss . Don’t know why Struggling to subscribe to it. Could there be any person acquiring identical rss dilemma? Anybody who knows kindly respond. Thnkx
I wanted to inform you about how much I actually appreciate every thing you’ve discussed to help increase the value of the lives of individuals in this subject material. Through your own articles, I’ve gone from just a novice to a professional in the area. It’s truly a honor to your efforts. Thanks
https://nicesongtoyou.com/welfare/pregnant-childbirth/
Excellent goods from you, man. I have understand your stuff previous to and you’re just extremely wonderful. I really like what you’ve acquired here, certainly like what you are saying and the way in which you say it. You make it enjoyable and you still take care of to keep it wise. I can’t wait to read much more from you. This is really a tremendous web site.
After study a handful of the content on your web site now, and that i really like your way of blogging. I bookmarked it to my bookmark website list and will also be checking back soon. Pls look at my internet site too and told me what you think.
Incredible this particular tutorial is actually fantastic it genuinely aided me plus our kids, cheers!
Thanks, I have been searching for facts about this subject for ages and yours is the best I’ve discovered so far.
Hello there Internet marketer. I truly the same as your current article along with the site on the whole! The particular written piece is in fact really clearly written as well as effortlessly simple to comprehend. Your present Blog site style is wonderful too! Will always be awesome to learn exactly where I could get that. If possible maintain inside the superb task. Young people need a lot more these website owners such as yourself on the internet and in addition a lesser amount of spammers. Wonderful guy!
Appreciate it for this post, I am a big fan of this internet site would like to keep updated.
Spot lets start work on this write-up, I honestly feel this web site needs a great deal more consideration. I’ll more likely once more to study additional, thank you for that information.
Hiya, Could I download your photograph and use that on my website?
Hello! Would you mind if I share your blog with my twitter group? There’s a lot of folks that I think would really appreciate your content. Please let me know. Thank you
I love reading your blog because it has very interesting topics.:.:~’
hey good site i will definaely come back and see again.
conclusion that you are absolutely right but a few require to be
https://oculus-vr.co.kr/hwang-hee-chan-wolverhampton/
There’s definately a lot to find out about this issue. I like all of the points you have made.
https://himchanchan.tistory.com/1735
poetry has the power to affect our emotions by using words alone, i really love poetry“
r Light sensing of moving objects – SActual object Light Visual objectr -cosine (wt) + i sine (wt) S = r [cosine (wt) + i sine (wt)]Newton Keplers time visual effects Time dependent NewtonParticle Visual effects Wave
Thanks for having the time to write about this issue. I truly appreciate it. I’ll post a link of this entry in my site.
Doceniam skupienie się na zagrożeniach SEO i potrzebie ich usunięcia.
This is one of the best posts I enjoyed this read in this blog. I’m absolutely excited to get to read such a well blog. The amount of data that I get is truly great. This is an good masterpiece. I’m truly impressed.
Ten blog dał mi lepsze zrozumienie zagrożeń i korzyści związanych z SEO.
Doceniam szczegółowe wyjaśnienia na temat SEO i bezpieczeństwa.
There is obviously a lot to realize about this. I assume you made certain nice points in features also.
https://himchanchan.tistory.com/1759
Czuję się dużo lepiej poinformowany o SEO. Dzięki!
Ahaa, its fastidious discussion about this article at this place at this web site, I have read all that, so at this time me also commenting
at this place.
Fantastic goods from you, man. I have take into account your stuff previous
to and you are just too fantastic. I actually like what you’ve got
here, really like what you’re saying and
the way through which you assert it. You’re making it
enjoyable and you still take care of to stay it sensible.
I cant wait to read much more from you. This is actually a
tremendous web site.
Discover incredible bargains on Marc Jacobs products at the marc jacobs factory outlet online.
Thanks for your handy post. Over time, I have come to be able to understand that the symptoms of mesothelioma are caused by the actual build up of fluid between the lining of the lung and the upper body cavity. The sickness may start while in the chest location and distribute to other parts of the body. Other symptoms of pleural mesothelioma cancer include weight reduction, severe inhaling trouble, vomiting, difficulty swallowing, and puffiness of the neck and face areas. It should be noted that some people with the disease do not experience any serious signs or symptoms at all.
Cryptocurrencies function on decentralized networks, placing customers firmly in management of their monetary assets.
아시아 슬롯
그냥… 이 가뭄이 언제 끝날지는 신만이 아시죠!
https://himchanchan.tistory.com/1195
I was doubtful about bulk delta 8 carts at first, but after using it, I’m pleasantly impressed . It delivers a clear-headed high that allows me to stay productive. Plus, it’s great for soothing minor aches and pains without the heaviness of Delta-8.
Ten blog bardzo mi pomógł w zrozumieniu potrzeby SEO.
https://himchanchan.tistory.com/684
Dziękuję za dokładne badania i szczegółowe wyjaśnienia na temat SEO.
vagagas
Dzięki za praktyczne wskazówki dotyczące znalezienia wykwalifikowanego specjalisty SEO.
To była otwierająca oczy lektura na temat ryzyk i procesu SEO.
Sign up with Lotus365 India and experience a secure and straightforward login process. With a variety of games and a supportive community, it’s a platform built for user satisfaction and convenience.
Ten post to świetne źródło informacji dla każdego, kto potrzebuje SEO.
https://metaversenews.co.kr/tag/movetoearn/
Yet another thing I would like to mention is that instead of trying to suit all your online degree courses on days that you end work (considering that people are drained when they get back), try to have most of your lessons on the saturdays and sundays and only 1 or 2 courses in weekdays, even if it means a little time off your weekend break. This is really good because on the week-ends, you will be far more rested in addition to concentrated upon school work. Thanks alot : ) for the different points I have figured out from your blog.
Świetne wskazówki dotyczące znalezienia wiarygodnych usług SEO. Dzięki!
Ten post był dla mnie oświeceniem na temat zagrożeń SEO. Dzięki!
Czy SEO naprawdę może być skuteczne? Ten blog sugeruje, że tak!
https://ddnews.co.kr/category/earn/page/12/
I think this is among the most vital information for me. And i’m
glad reading your article. But want to remark on few general things, The web site style is wonderful, the articles is really great :
D. Good job, cheers
Doceniam skupienie się na zagrożeniach SEO i potrzebie ich usunięcia.
What’s up, for all time i used to check website posts here in the early hours in the
daylight, since i love to find out more and more.
Today, while I was at work, my sister stole my iPad and tested to see if it can survive a 30 foot drop, just so
she can be a youtube sensation. My apple ipad is now destroyed and she
has 83 views. I know this is totally off topic but I had
to share it with someone!
Ten blog to cenne źródło informacji dla każdego, kto myśli o SEO.
Begin your journey with reddy anna online. The straightforward registration process ensures you can start quickly. Stay connected with the latest updates and round-the-clock support.
hi!,I really like your writing very so much! percentage we keep up a correspondence extra approximately your post on AOL? I need a specialist on this area to resolve my problem. Maybe that’s you! Taking a look forward to see you.
Cieszę się, że znalazłem ten blog przed rozpoczęciem mojego projektu SEO.
https://ddnews.co.kr/category/earn/page/8/
Czuję się znacznie pewniej w temacie SEO po przeczytaniu tego bloga.
Dzięki za jasne i zwięzłe informacje na temat bezpieczeństwa SEO.
Hello! Do you know if they make any plugins to help with Search Engine Optimization? I’m trying to get my blog to rank for
some targeted keywords but I’m not seeing very good success.
If you know of any please share. Appreciate it!
Ten blog bardzo mi pomógł w zrozumieniu potrzeby SEO.
Świetne wskazówki dotyczące znalezienia wiarygodnych usług SEO. Dzięki!
Email frequency caps prevent inbox overload.
Świetna rada na temat znaczenia właściwego SEO. Dzięki!
청도페이스라인출장
Kerly-mature nude on live sex chat
[url=http://thinktoy.net/bbs/board.php?bo_table=customer2&wr_id=356910]Nude hot webcams gi[/url][url=http://thinktoy.net/bbs/board.php?bo_table=customer2&wr_id=356911]Nude hot cams girl[/url] 8_11c86
Thanks for your handy post. As time passes, I have come to be able to understand that the symptoms of mesothelioma cancer are caused by the build up of fluid between lining in the lung and the chest cavity. The infection may start inside the chest vicinity and distribute to other body parts. Other symptoms of pleural mesothelioma cancer include weight loss, severe breathing in trouble, a fever, difficulty ingesting, and swelling of the neck and face areas. It needs to be noted that some people having the disease don’t experience any kind of serious indicators at all.
강남콜걸
Ten post to świetne źródło informacji dla każdego, kto potrzebuje SEO.
Cieszę się, że znalazłem blog, który tak dokładnie omawia SEO. Dzięki!
Dzięki za praktyczne wskazówki dotyczące bezpiecznego SEO. Bardzo przydatne!
여행지
Bardzo pouczający blog na temat SEO! Dzięki za podzielenie się nim.
충무로출장업소
I used to be recommended this web site by my cousin. I’m not certain whether this publish is written by way of him as nobody else recognize
such designated about my difficulty. You are incredible!
Thanks!
There is a lot of planning, study, and time that needs to go into any kind of philosophy assignment. Without the highest potential levels of topic mastery and good ability to write in English, it is difficult to successfully complete assignments related to philosophy. In addition to being an essential part of the assignment writing process, the time period is equally important. Completing homework in philosophy requires a significant amount of time. Due to their hectic schedules and other commitments, however, the majority of the time, students were unable to carry out sufficient research. Because of this, the probability of a student being able to write a philosophy assignment in an appropriate manner is still quite low. Pay someone to do my online psychology classes is a service that students regularly turn to for assistance with their philosophy assignments when they find themselves in this scenario. Experts are ready to provide you online philosophy homework assistance and helpful ideas at costs that are among the most competitive in the industry. You may rely on professionals by asking, “Can someone write my assignment for me?” to provide you with an excellent philosophy analysis assignment. This is truProgramming assignment helper in USA Programming assignment helper in USA e regardless of whether you are searching for a topic for a philosophy paper or an example of a philosophy writing assignment.
Hey, I think your blog might be having browser compatibility issues.
When I look at your blog site in Safari,
it looks fine but when opening in Internet Explorer, it has some
overlapping. I just wanted to give you a quick heads up! Other then that, awesome
blog!
Hardware in Trinidad and Tobago encompasses a diverse range of products and services, catering to construction, DIY projects, and industrial needs. With an array of stores and suppliers across the islands, hardware offerings include building materials, tools, electrical and plumbing supplies, and hardware equipment for various purposes. Trinidad and Tobago’s hardware industry plays a pivotal role in supporting infrastructure development, home improvement, and commercial ventures. These establishments often provide expert advice, quality products, and competitive pricing, serving both professionals and homeowners alike.
It is preferable to get online class help from a reliable platform in situations where students are juggling academics and their online classes while acquiring these skills. Read this blog till the end and learn about the essential skills for students in 2024 and also get updated with the new job opportunities. Furthermore, there are websites available which offer online coursework help with such courses. You can take the benefit of it
https://kleonet.com/entry/tag/114114com/
I like the valuable info you provide in your articles.
I will bookmark your weblog and check again here frequently.
I am quite certain I will learn a lot of new stuff right here!
Good luck for the next!
These kind of post are always inspiring and I prefer to read quality content so I happy to find many good point
here in the post, writing is simply great, thank
you for the post
https://kleonet.com/entry/tag/한게임-신맞고-무료/
This would be the right weblog for everyone who is wishes to be made aware of this topic. You recognize a great deal of its almost tough to argue along (not too I really would want…HaHa). You actually put the latest spin on a topic thats been discussed for decades. Fantastic stuff, just fantastic!
I’m not sure exactly why but this weblog is loading very slow for me.
Is anyone else having this issue or is it a problem on my end?
I’ll check back later on and see if the problem still exists.
대전나이트클럽
you have a great weblog here! do you need to earn some invite posts in my blog?
I am usually to blogging and i also truly appreciate your articles. This great article has really peaks my interest. My goal is to bookmark your blog and maintain checking for brand new details.
Thanks for the article. My partner and i have continually seen that a majority of people are wanting to lose weight simply because wish to show up slim and attractive. Nonetheless, they do not usually realize that there are other benefits so that you can losing weight as well. Doctors claim that over weight people have problems with a variety of health conditions that can be perfectely attributed to the excess weight. The great news is that people who sadly are overweight along with suffering from different diseases can help to eliminate the severity of their particular illnesses by losing weight. You’ll be able to see a gradual but notable improvement with health if even a moderate amount of weight loss is accomplished.
You made some decent points there. I seemed on the web for the issue and located most individuals will go together with together with your website.
I’m really loving the theme/design of your blog. Do you ever run into any web browser compatibility problems? A few of my blog audience have complained about my site not operating correctly in Explorer but looks great in Opera. Do you have any recommendations to help fix this issue?
https://www.taptap.io/user/646572521
https://www.taptap.io/user/646573527
https://www.taptap.io/user/646574390
https://www.taptap.io/user/646574825
https://www.taptap.io/user/646575769
https://www.taptap.io/post/7662937
https://www.taptap.io/post/7662956
https://www.taptap.io/post/7662957
https://www.taptap.io/post/7662960
https://www.taptap.io/post/7662962
https://www.taptap.io/post/7662964
https://www.taptap.io/post/7662965
https://www.taptap.io/post/7662967
https://www.taptap.io/post/7662970
https://www.taptap.io/post/7662973
https://www.taptap.io/post/7662991
https://www.taptap.io/post/7662994
https://www.taptap.io/post/7662999
https://www.taptap.io/post/7663001
https://www.taptap.io/post/7663006
https://www.taptap.io/post/7663009
https://www.taptap.io/post/7663013
https://www.taptap.io/post/7663016
https://www.taptap.io/post/7663018
https://www.taptap.io/post/7663035
https://www.taptap.io/post/7663036
https://www.taptap.io/post/7663052
https://www.taptap.io/post/7663055
https://www.taptap.io/post/7663056
https://www.taptap.io/post/7663059
https://www.taptap.io/post/7663060
https://www.taptap.io/post/7663064
https://www.taptap.io/post/7663065
https://www.taptap.io/post/7663071
https://www.taptap.io/post/7663073
https://www.taptap.io/post/7663074
https://www.taptap.io/post/7663092
https://www.taptap.io/post/7663097
https://www.taptap.io/post/7663100
https://www.taptap.io/post/7663102
https://www.taptap.io/post/7663105
https://www.taptap.io/post/7663107
https://www.taptap.io/post/7663113
https://www.taptap.io/post/7663117
https://www.taptap.io/post/7663121
https://www.taptap.io/post/7663123
https://www.taptap.io/post/7663125
https://www.taptap.io/post/7663127
https://www.taptap.io/post/7663130
https://www.taptap.io/post/7663132
https://www.taptap.io/post/7663137
https://www.taptap.io/post/7663140
https://www.taptap.io/post/7663141
https://www.taptap.io/post/7663205
https://www.taptap.io/post/7663208
https://www.taptap.io/post/7663210
https://www.taptap.io/post/7663214
https://www.taptap.io/post/7663225
https://www.taptap.io/post/7663233
https://www.taptap.io/post/7663236
https://www.taptap.io/post/7663238
https://www.taptap.io/post/7663243
https://www.taptap.io/post/7663246
https://www.taptap.io/post/7663252
https://www.taptap.io/post/7663253
https://m.taptap.io/post/7662937
https://m.taptap.io/post/7662956
https://m.taptap.io/post/7662957
https://m.taptap.io/post/7662960
https://m.taptap.io/post/7662962
https://m.taptap.io/post/7662964
https://m.taptap.io/post/7662965
https://m.taptap.io/post/7662967
https://m.taptap.io/post/7662970
https://m.taptap.io/post/7662973
https://m.taptap.io/post/7662991
https://m.taptap.io/post/7662994
https://m.taptap.io/post/7662999
https://m.taptap.io/post/7663001
https://m.taptap.io/post/7663006
https://m.taptap.io/post/7663009
https://m.taptap.io/post/7663013
https://m.taptap.io/post/7663016
https://m.taptap.io/post/7663018
https://m.taptap.io/post/7663035
https://m.taptap.io/post/7663036
https://m.taptap.io/post/7663052
https://m.taptap.io/post/7663055
https://m.taptap.io/post/7663056
https://m.taptap.io/post/7663059
https://m.taptap.io/post/7663060
https://m.taptap.io/post/7663064
https://m.taptap.io/post/7663065
https://m.taptap.io/post/7663071
https://m.taptap.io/post/7663073
https://m.taptap.io/post/7663074
https://m.taptap.io/post/7663092
https://m.taptap.io/post/7663097
https://m.taptap.io/post/7663100
https://m.taptap.io/post/7663102
https://m.taptap.io/post/7663105
https://m.taptap.io/post/7663107
https://m.taptap.io/post/7663113
https://m.taptap.io/post/7663117
https://m.taptap.io/post/7663121
https://m.taptap.io/post/7663123
https://m.taptap.io/post/7663125
https://m.taptap.io/post/7663127
https://m.taptap.io/post/7663130
https://m.taptap.io/post/7663132
https://m.taptap.io/post/7663137
https://m.taptap.io/post/7663140
https://m.taptap.io/post/7663141
https://m.taptap.io/post/7663205
https://m.taptap.io/post/7663208
https://m.taptap.io/post/7663210
https://m.taptap.io/post/7663214
https://m.taptap.io/post/7663225
https://m.taptap.io/post/7663233
https://m.taptap.io/post/7663236
https://m.taptap.io/post/7663238
https://m.taptap.io/post/7663243
https://m.taptap.io/post/7663246
https://m.taptap.io/post/7663252
https://m.taptap.io/post/7663253
https://jsbin.com/cuvurerazi/edit?html,output
https://web.extension.illinois.edu/askextension/thisQuestion.cfm?QuestionID=29887&catID=163&AskSiteID=75
https://tinhte.vn/profile/shifty-shellshock.3028410/
https://bukkit.org/members/alicia-vasquez.91539866/
https://www.fmscout.com/users/4rthp15axddp5yx.html
https://www.bitsdujour.com/profiles/gC3Bec
https://muckrack.com/nrkzipnksqfsd-ehjslxywblmwg/bio
https://profile.hatena.ne.jp/eb4vrvn18ui0kbs/
https://community.fabric.microsoft.com/t5/user/viewprofilepage/user-id/765926
https://hackmd.io/@ddtdiyyvek/B1yHwhO8C
https://demo.hedgedoc.org/s/bmiU5WfeD
https://wokwi.com/projects/401711137336635393
https://tempaste.com/qPDkqctWN6w
https://dev.bukkit.org/paste/9a5048bb
https://zenn.dev/alyssafranco
https://zenn.dev/alyssafranco?tab=scraps
https://zenn.dev/alyssafranco/scraps/276d2bbf76d354
https://click4r.com/posts/g/17218932/sad-news-crazy-town-singer-shifty-shellshock-dies
https://www.deviantart.com/lzjjzara4r8z/journal/icgkmtdscwizx-bcwmtqqenytup-1067802405
https://openiazoch.zoznam.sk/info/zpravy/nazory.asp?NewsID=222331&CID=287048
https://openiazoch.zoznam.sk/info/zpravy/nazory.asp?NewsID=222331&CID=287049
https://dotnetfiddle.net/Authors/269504/Alicia%20Vasquez
https://dotnetfiddle.net/7sWn6J
https://jsfiddle.net/fc7uekpa/
https://rextester.com/EKUZ11374
https://rextester.com/EKUZ11374
https://telegra.ph/giiwif93y8muqbg-06-26
https://paste.myst.rs/w5hzt2my
https://pastelink.net/yp107zja
https://bitbin.it/HYAJ8sfR/
https://pastebin.com/iEQWVpQq
https://snippet.host/swpxyi
https://yamcode.com/mqtu2q5o8e7wxmx
https://rentry.co/tpcbrhch
https://herbalmeds-forum.biolife.com.my/d/84886-7kphz78v6zsirb
https://herbalmeds-forum.biolife.com.my/u/aliciavasquez
https://herbalmeds-forum.biolife.com.my/d/84886-7kphz78v6zsirb
https://cofradesdegranada.ideal.es/members/AliciaVasquez
https://cofradesdegranada.ideal.es/articles/lauren-boebert-sails-through-her-primary
https://matters.town/@aliciavasquez
https://matters.town/a/2syvwnjesq7u
https://solo.to/7oacr2ougq5qy39
I adore gathering useful info, this post has got me even more info! .
https://kleonet.com/entry/tag/오은영-리포트-결혼-지옥-81회/
The the next occasion I read a weblog, I really hope so it doesnt disappoint me around brussels. Come on, man, Yes, it was my option to read, but I just thought youd have some thing interesting to state. All I hear can be a lot of whining about something that you could fix if you werent too busy searching for attention.
It is not my first time to go to see this web page,
i am browsing this website dailly and obtain fastidious facts from here daily.
It is in point of fact a great and useful piece of information. I’m happy that you shared this helpful information with us. Please stay us informed like this. Thanks for sharing.
Woah! I’m really loving the template/theme of this website.
It’s simple, yet effective. A lot of times it’s hard to
get that “perfect balance” between user friendliness and appearance.
I must say you’ve done a amazing job with this. Also, the blog loads extremely
fast for me on Firefox. Superb Blog!
https://kleonet.com/entry/tag/세-번째-결혼-마지막회/
Aw, this has been an extremely nice post. In thought I must place in writing similar to this moreover – taking time and actual effort to create a great article… but what / things I say… I procrastinate alot and also by no indicates appear to get something accomplished.
Thank you for extra excellent blog. Exactly where else could i recieve this particular compassionate of information written in this kind of incite complete way? i have been seeing with regard to this kind of fine detail.
Secure your access today with reddy anna book and dive into a world of expert cricket insights and live match updates. Our 24/7 customer support is here to help.
I’d need to take advice from you here. Which is not something I normally do! I enjoy reading a piece which will get people to think. Also, thank you for permitting me to comment!
Superb material, Thanks!
You must take part in a contest for the most effective blogs on the web. I’ll advocate this website!
Hi just wanted to give you a brief heads up and let you know a few of the pictures aren’t loading properly. I’m not sure why but I think its a linking issue. I’ve tried it in two different browsers and both show the same results.
Dive into the thrill of cricket with the reddy anna website. Offering seamless booking and a variety of options, with 24/7 customer support available to assist. Don’t miss out – book your spot today.
Great stuff. Appreciate it.
My page: https://incheon-anma.xyz
You said it perfectly.!
Check out my site – https://Guyanaexpatforum.com/question/3-sorts-of-%ec%9e%a5%eb%a7%88%ec%82%ac%ec%a7%80-which-one-will-take-advantage-of-cash/
Hello there! I know this is somewhat off topic but I was wondering which blog platform are you using for this site?
I’m getting fed up of WordPress because I’ve had problems with
hackers and I’m looking at options for another platform. I would be awesome if you could point me in the
direction of a good platform.
https://itgunza.com/367
Can I simply say what a relief to seek out somebody who really is aware of what theyre speaking about on the internet. You undoubtedly know the best way to bring a problem to mild and make it important. Extra people need to learn this and perceive this aspect of the story. I cant believe youre not more widespread since you positively have the gift.
https://itgunza.com/323
https://pornmaster.fun/hd/快三登录注册(关于快三登录注册的简介)-【网hk589点org】-旧版够力七星彩正版牛爷资料(关于旧版够力七星彩正版牛爷资料的简介)cukncukn-【网hk589E38082org】-亿博体育平台app下载(关于亿博体育平台app下载的简介)nzhk6zxb-lxs
청도페이스라인출장
I favor the very first position you made there, but I ‘m unclear I could reasonably implement that will inside a postive approach.
You should take part in a contest for one of the best blogs on the web. I will recommend this page!
Cheers, Great stuff.
Also visit my page: https://Wiki.Stanleyro.com/index.php/%EC%B6%9C%EC%9E%A5%EC%95%88%EB%A7%88ing_Nine_Tricks_Your_Competitors_Know_But_You_Don%E2%80%99t
l porn
https://kr.new-version.app/download/naver-map/naver-map_screenshot_01/
With thanks, A lot of knowledge.
Here is my webpage … http://Www.olangodito.com/bbs/board.php?bo_table=free&wr_id=581273
Does your website have a contact page? I’m having a tough time locating it but,
I’d like to shoot you an e-mail. I’ve got some recommendations for your blog you might be interested in hearing.
Either way, great website and I look forward to seeing it expand over time.
There was no holding back the success of crypto; its progress was exponentially fast.
I look forward to many more years of enlightening and entertaining reads!
Thank you, Quite a lot of content.
Man that was very entertaining and at the same time informative.*”~”;
Aw, this was an incredibly nice post. Finding the time and actual effort to create a superb article… but what can I say… I put things off a lot and never manage to get nearly
anything done.
Normally I do not read article on blogs, however I would like to say that
this write-up very pressured me to take a look at and do so!
Your writing style has been surprised me. Thanks, very great article.
I have seen plenty of useful elements on your website about pc’s. However, I’ve got the opinion that lap tops are still less than powerful enough to be a good selection if you normally do things that require lots of power, like video modifying. But for website surfing, microsoft word processing, and most other frequent computer functions they are okay, provided you can’t mind your little friend screen size. Thank you sharing your ideas.
You made your stand very clearly!!
Your article challenges the status quo—in a good way!드래곤볼 에볼루션
Hello to every body, it’s my first visit of this website; this webpage carries remarkable
and in fact excellent stuff for visitors.
Beneficial write ups Thanks a lot!
Here is my webpage; http://blueasia.CO.Kr/gnb/bbs/board.php?bo_table=sub03_03&wr_id=372938
WOW just what I was looking for. Came here by searching for ragoût
https://ddnews.co.kr/blog/2023/03/19/mbti-meaning/
I like the helpful information you provide in your articles.
I’ll bookmark your blog and check again here regularly. I am quite sure I will learn plenty of new stuff right here!
Good luck for the next!
I have to thank you for the efforts you have put
in penning this blog. I really hope to see the same high-grade blog posts from you in the
future as well. In truth, your creative writing
abilities has inspired me to get my very own blog now
😉
I need to to thank you for this excellent read!! I certainly loved every little bit of it. I’ve got you book-marked to check out new stuff you post?
It’s a shame you don’t have a donate button! I’d most certainly donate to this excellent blog!
I guess for now i’ll settle for book-marking and adding your RSS feed to my Google account.
I look forward to brand new updates and will talk about this site with my Facebook
group. Chat soon!
Hi! I know this is kinda off topic nevertheless I’d figured I’d ask.
Would you be interested in exchanging links or maybe guest writing a
blog article or vice-versa? My website goes over a lot of the same topics as yours and I believe we could greatly benefit
from each other. If you are interested feel free to shoot me an email.
I look forward to hearing from you! Fantastic blog by
the way!
Appreciating the dedication you put into your site and
in depth information you present. It’s great to come across a blog every once in a while that isn’t the same unwanted rehashed information. Fantastic read!
I’ve bookmarked your site and I’m including your RSS feeds to my Google account.
Hello, this weekend is pleasant in support of me, for the reason that this moment i am reading this wonderful educational article
here at my house.
https://ddnews.co.kr/blog/2024/03/10/ebb2bceba3a9ec8b9cec9ea5ec8ba0ebacb8eab7b8eb8c80eba19cebb3b4eab8b0/
https://k-studio.kr/토스뱅크-비상금대출-조건-모바일-소액대출-무직자/
Your style is so unique compared to other folks I have read stuff from.
I appreciate you for posting when you have the opportunity,
Guess I’ll just bookmark this blog.
This forum is a treasure trove of valuable information and insights.
Point effectively utilized..
Have a look at my site :: https://oneclyde.uk/index.php?title=Why_I_ll_Never_%EC%B6%9C%EC%9E%A5%EB%A7%88%EC%82%AC%EC%A7%80
I have recently started a web site, the information you offer on this web site has helped me greatly. Thank you for all of your time & work.
Herbal medicine, also known as herbalism or botanical medicine, is a centuries-old practice of using plants and plant extracts to promote health and well-being.
Also visit my online blog – https://ife-ifem-hrc2024.coconnex.com/node/1022792
Asking questions are really pleasant thing if you are
not understanding something totally, except this post presents fastidious understanding even.
https://kleonet.com/entry/tag/윤딴딴-투어-콘서트-딴딴한여름-2024-인천-티켓팅/
This is a topic that is close to my heart… Many thanks!
Exactly where are your contact details though?
porno-mp4
You need to be a part of a contest for one of the highest quality blogs online. I will recommend this blog!
The prompt responses and detailed explanations provided by the community are incredibly helpful.
The support and encouragement I receive from the members here are truly heartwarming.
The quality of discussions on this forum is very high. I always learn something new every time I visit.
This forum consistently provides thoughtful and well-researched answers.
The resources and information available here are very useful. I greatly appreciate everyone’s contributions.
https://medium.com/@callgirlsbhopal/call-girls-bhopal-6260913881-cash-payment-escorts-service-bhopal-90b9cc0fd9dc
Everyone loves it whenever people get together and
share opinions. Great blog, keep it up!
https://honeytipit.tistory.com/tag/토스20어플20설치
I am really impressed together with your writing talents and also with
the format for your weblog. Is that this a paid topic or did you customize it yourself?
Either way stay up the nice quality writing, it is rare to look a nice blog like this one nowadays..
Наши специалисты предлагает высококачественный ремонт парогенераторов с гарантией всех типов и брендов. Мы знаем, насколько значимы для вас ваши устройства для генерации пара, и обеспечиваем ремонт высочайшего уровня. Наши профессиональные техники проводят ремонтные работы с высокой скоростью и точностью, используя только сертифицированные компоненты, что гарантирует долговечность и надежность выполненных работ.
Наиболее общие проблемы, с которыми сталкиваются обладатели пароочистителей, включают неисправности нагревательных элементов, проблемы с подачей воды, ошибки ПО, проблемы с подключением и поломки компонентов. Для устранения этих проблем наши квалифицированные специалисты оказывают ремонт нагревательных элементов, насосов, ПО, разъемов и механических компонентов. Доверив ремонт нам, вы гарантируете себе качественный и надежный сервис ремонта парогенератора на дому.
Подробная информация доступна на сайте: https://remont-parogeneratorov-piece.ru
This forum has a vibrant and engaging atmosphere that keeps me coming back.
The moderators do an excellent job of keeping the discussions respectful and productive.
https://honeytipit.tistory.com/tag/교차로20홈페이지
You can also set up an Certainly “Job Alert,” to be notified by way of e-mail as soon as positions that meet
your selection criteria are posted.
Feel free to surf tto my web page: 이지알바
Integrated Risk Management is a comprehensive approach that consolidates various risk management processes into a unified framework.
It goes beyond traditional risk silos and provides organizations with a holistic view of
risks across different functions and departments. IRM
enables businesses to identify, assess, mitigate, and monitor risks more
effectively, fostering a proactive and strategic approach to risk management.
Наш сервисный центр предлагает профессиональный мастер по ремонту приборов ночного видения адреса любых брендов и моделей. Мы понимаем, насколько необходимы вам ваши приборы ночного видения, и готовы предложить сервис первоклассного уровня. Наши опытные мастера работают быстро и аккуратно, используя только сертифицированные компоненты, что обеспечивает долговечность и надежность наших услуг.
Наиболее частые неисправности, с которыми сталкиваются пользователи устройств ночного видения, включают проблемы с изображением, проблемы с линзами, ошибки ПО, проблемы с портами и повреждения корпуса. Для устранения этих поломок наши квалифицированные специалисты выполняют ремонт сенсоров, оптики, ПО, разъемов и механических компонентов. Обращаясь в наш сервисный центр, вы получаете качественный и надежный мастер по ремонту приборов ночного видения.
Подробная информация представлена на нашем сайте: https://remont-pnv-ultra.ru
My spouse and I stumbled over here by a different page and thought I may
as well check things out. I like what I see so now
i’m following you. Look forward to finding out about
your web page yet again.
Right here is the right site for anybody who
really wants to find out about this topic. You understand so
much its almost hard to argue with you (not that I actually will need to…HaHa).
You certainly put a brand new spin on a subject that’s been written about for decades.
Great stuff, just wonderful!
Наша мастерская предлагает надежный вызвать мастера по ремонту посудомоечных машин различных марок и моделей. Мы понимаем, насколько значимы для вас ваши посудомоечные машины, и стремимся предоставить услуги наилучшего качества. Наши профессиональные техники работают быстро и аккуратно, используя только сертифицированные компоненты, что обеспечивает долговечность и надежность наших услуг.
Наиболее общие проблемы, с которыми сталкиваются владельцы посудомоечных машин, включают проблемы с нагревом воды, неработающие насосы, неисправности программного обеспечения, проблемы с подключением и повреждения корпуса. Для устранения этих проблем наши профессиональные техники оказывают ремонт нагревательных элементов, насосов, ПО, разъемов и механических компонентов. Обратившись к нам, вы обеспечиваете себе долговечный и надежный отремонтировать посудомоечную машину на выезде.
Подробная информация доступна на сайте: https://remont-posudomoechnyh-mashin-expert.ru
This is the best place to share knowledge and get answers to my questions.
Good information, Kudos!
머스트 잇 토토
Fang Jifan은 미소를 지으며 말했습니다. “그의 전하, 구멍에서 뱀을 유인하는 방법을 모르겠습니다.”
Sports betting and cryptocurrencies are a perfect mixture that has led to many revenue alternatives for gamblers.
The ardour and edgy really feel of being a fan of the sport can be likened to how cryptocurrency is viewed too – underground and discreet.
porno mp4
https://dday.tistory.com/110
Hey there! I’m at work surfing around your blog
from my new apple iphone! Just wanted to say I
love reading through your blog and look forward to all your posts!
Keep up the excellent work!
Nicely put, Cheers.
The sense of community here is strong and supportive, making it a great place to learn and groww.
На Туббе можно посмотреть фильмы и сериалы https://tubba.ru с любимыми персонажами.
Наши специалисты предлагает надежный сервис ремонта тепловизоров на выезде различных марок и моделей. Мы понимаем, насколько значимы для вас ваши инфракрасные камеры, и обеспечиваем ремонт высочайшего уровня. Наши квалифицированные специалисты работают быстро и аккуратно, используя только сертифицированные компоненты, что гарантирует длительную работу наших услуг.
Наиболее частые неисправности, с которыми сталкиваются обладатели устройств теплового видения, включают неработающие сенсоры, неисправности оптики, программные сбои, неработающие разъемы и механические повреждения. Для устранения этих поломок наши профессиональные техники проводят ремонт матриц, оптики, ПО, разъемов и механических компонентов. Обратившись к нам, вы получаете качественный и надежный мастерская по ремонту тепловизора на дому.
Подробная информация размещена на сайте: https://remont-teplovizorov-gem.ru
Hi, There’s no doubt that your web site could be
having internet browser compatibility issues. When I look at your blog
in Safari, it looks fine but when opening in IE, it’s got some overlapping issues.
I merely wanted to give you a quick heads up! Aside from that, great site!
I’m impressed, I must say. Rarely do I encounter a blog that’s equally educative and interesting, and let me tell you, you have hit the nail on the head. The problem is something which not enough people are speaking intelligently about. I am very happy that I stumbled across this during my search for something concerning this.
Because the admin of this site is working, no hesitation very soon it will be well-known,
due to its quality contents.
I’m amazed, I have to admit. Seldom do I come across a blog that’s both educative and entertaining, and let me tell you, you’ve hit the nail on the head. The problem is an issue that too few people are speaking intelligently about. I’m very happy that I found this during my search for something concerning this.
After I originally left a comment I appear to have clicked on the -Notify me when new comments are added- checkbox and now each time a comment is added I recieve four emails with the exact same comment. There has to be an easy method you can remove me from that service? Thank you.
Maybe this is the best article I have read so far, thank you to the admin of this website for providing content that is very useful for me, so that I can win free money and make me no longer in debt, thank you.
양산시술출장마사지
CV/Resume Writing Services for Your Dream Position
Symbels Resumes is a professional resume writing services provider that assists working professionals on how to advance in their careers. Our services include the creation of CVs, professional resumes, services in making outstanding linked profiles, professional personal websites, and many others. Also, our qualified resume writing team provides convincing cover letters and a set of useful services. Come and stand out with us and be ready to take your best position in the job market competition. It is a map of your success here with us.
수원출장샵
Have you ever considered publishing an ebook or guest
authoring on other blogs? I have a blog centered on the same ideas you discuss and would love to have
you share some stories/information. I know my viewers would value your work.
If you’re even remotely interested, feel free to send me an e-mail.
This is nicely put. .
Thanks a lot for sharing this with all of us you really realize what you are speaking about!
Bookmarked. Kindly additionally talk over with my web site =).
We can have a hyperlink trade agreement among us
I need to to thank you for this very good read!! I definitely loved every bit of it. I have got you book-marked to check out new things you post…
Its like you read my mind! You appear to know a lot about this,
like you wrote the book in it or something. I think that
you could do with some pics to drive the message home
a little bit, but instead of that, this is great
blog. A fantastic read. I will certainly be back.
Welcome to our Online Assignment help service! Our experts are here to assist you with your online courses and classes, providing support for assignments, helping you understand difficult topics, and preparing for online dissertation help . Let us help you succeed in your studies!
Hi there to all, how is all, I think every one
is getting more from this web site, and your views are nice designed for new users.
My page: find out here
Genuinely when someone doesn’t know after that its up
to other users that they will assist, so here it takes
place.
What’s up, all the time i used to check weblog posts here in the early hours in the daylight, because i love to learn more and more.
I quite like reading through an article that can make men and women think. Also, many thanks for permitting me to comment.
Are you in need of best SOP writing services ? We specialize in crafting exceptional SOPs. Feel free to reach out to us for further information.
Тубба – это отличный выбор для тех, кто любит смотреть фильмы и сериалы https://tubba.ru в хорошем качестве.
Oh my goodness! Incredible article dude! Thank you, However I am encountering problems with your RSS. I don’t understand why I can’t join it. Is there anybody else getting identical RSS problems? Anyone that knows the answer will you kindly respond? Thanx!!
This excellent website certainly has all the information and facts I needed about this subject and didn’t know who to ask.
I have recently started a website, the info you offer on this web site has helped me greatly. Thank you for all of your time & work.
Yes! Finally someone writes about female stripper.
Appreciate this post. Will try it out.
It’s a mistake to chase your losses, especially as a result of it additionally results in making riskier particular person bets.
This piece of writing will assist the internet users for
building up new webpage or even a blog from start to end.
На Туббе https://tubba.ru можно найти фильмы и сериалы, которые завоюют сердца зрителей.
I appreciate the site’s user-friendly interface, which makes it easy to navigate and discover new content.
I could not refrain from commenting. Very well written!
That is a good tip particularly to those new to the blogosphere.
Brief but very accurate info… Many thanks for sharing this one.
A must read post!
Howdy! Do you know if they make any plugins to safeguard against hackers? I’m kinda paranoid about losing everything I’ve worked hard on. Any suggestions?
https://www.lesbravo.com/netflix-watching-data-delete/
I’ve been looking for answers on this—thank you!청년 대출
Тубба предлагает фильмы https://tubba.ru и сериалы с удобным разделом “Фильмы онлайн”.
Fine way of explaining, and nice paragraph to take information on the
topic of my presentation focus, which i am going to convey in institution of
higher education.
https://sportscom.co.kr/honghyeonseok-hent-2023/
I think the admin of this website is truly working hard in favor of his
web site, because here every stuff is quality based stuff.
This is a topic that needs more attention—thank you.신혼부부 대출
Many thanks. Lots of info!
Also visit my web site … https://Chosong.Co.kr/gb/bbs/board.php?bo_table=free&wr_id=648601
Kudos! Quite a lot of facts.
Look into my blog post :: https://Nightcms.ru/user/HumbertoTew7/
На Туббе можно посмотреть фильмы и сериалы https://tubba.ru с любимыми операторами.
I recently stumbled upon this amazing website, and I have to say, it has been a game-changer for me! The content here is incredibly insightful and well-organized, making it easy to find exactly what I need. One of the best things about this site is the opportunity to earn free money. Thanks to their fantastic guides and resources, I’ve managed to boost my income significantly. If you’re looking for a reliable and informative website that also offers a chance to make some extra cash, look no further. Highly recommended!
Nicely expressed without a doubt. !
my blog post :: https://Kreezcraft.com/wiki/index.php?title=%EC%B6%9C%EC%9E%A5%EC%95%88%EB%A7%88ing_9_Tricks_Your_Competitors_Know_But_You_Don%E2%80%99t
For newest news you have to go to see the web and on internet I found this site as a
most excellent site for hottest updates.
Thanks for sharing your info. I really appreciate your efforts and I will be waiting for
your next post thank you once again.
https://nicesongtoyou.com/tax/real-estate-tax/
You are so cool! I do not believe I have read through something like that before.
So great to discover somebody with a few genuine thoughts on this subject matter.
Really.. thank you for starting this up. This web site is one thing that is required on the web,
someone with some originality!
https://fifaworldcup.modoo.at/
На Туббе можно найти фильмы и сериалы https://tubba.ru для детей.
It embodied a decentralized financial framework, enabling direct transactions and negating the need for intermediaries.
Perfectly expressed really! !
Here is my webpage; https://tp50.org/fotos/img_1428/
Hi, i feel that i noticed you visited my web site thus i came to go back
the prefer?.I’m attempting to find issues to enhance my website!I suppose its good enough to use some of your ideas!!
When some one searches for his essential thing, so he/she needs to be available that
in detail, therefore that thing is maintained over here.
https://ddnews.co.kr/ebb094eb8ba4eab1b0ebb681ec8aa4ed9484-ebacb8eca09c/
Hello, I enjoy reading all of your article. I like to write a little comment to support you.
https://ddnews.co.kr/category/earn/
Well done on tackling such a tough subject.무료슬롯 프라그마틱
I like the valuable info you provide in your articles. I’ll bookmark your blog and check again here frequently.
I am quite certain I’ll learn plenty of new stuff right here!
Best of luck for the next!
Hmm is anyone else encountering problems with the images
on this blog loading? I’m trying to figure out if its a problem on my
end or if it’s the blog. Any feedback would be greatly appreciated.
What’s up mates, how is everything, and what you wish for to say on the topic of this article, in my view its genuinely awesome for me.
I know this site gives quality depending articles and
additional material, is there any other web page which gives these things in quality?
I love the simplicity and elegance of your site design.
The clean layout of your site makes it easy to find valuable information.
Your site is a fantastic resource and a joy to visit.
Just desire to say your article is as astounding. The clearness in your post is just great and i could assume you’re an expert
on this subject. Fine with your permission allow me to grab
your feed to keep updated with forthcoming post. Thanks a million and please keep up the enjoyable work.
https://ddnews.co.kr/ebacb4eba38c-mbti-eab280ec82ac-ec82acec9db4ed8ab8-eba781ed81ac-ebb094eba19ceab080eab8b0/
Tubba https://tubba.ru предлагает бесплатный просмотр фильмов и сериалов без регистрации и смс.
This is one of the best-designed sites I’ve come across. Well done!
I blog quite often and I truly thank you for your content.
This article has truly peaked my interest. I will take a note of your website and keep
checking for new details about once per week. I subscribed to your Feed
too.
I love the user-friendly layout of your site.
Your post is a valuable addition to the discussion.백링크 조회
doxera.icu
I’m not certain the place you are getting your information, however great topic.
I needs to spend some time studying more or understanding more.
Thanks for great information I was searching for this information for my mission.
I believe that is one of the so much vital info for me.
And i am glad reading your article. However want to statement on few normal issues, The website style is
wonderful, the articles is actually nice : D. Excellent task, cheers
Someone essentially lend a hand to make significantly posts I might
state. This is the very first time I frequented your website page and to this point?
I surprised with the analysis you made to create this particular submit amazing.
Great job!
I know this web page provides quality based articles or reviews and other information, is there any other web site which offers
these kinds of stuff in quality?
Feel free to visit my web page like this
Thanks a lot! I like this!
My site – http://www.letts.org/wiki/How_To_%EC%B6%9C%EC%9E%A5%EB%A7%88%EC%82%AC%EC%A7%80_And_Influence_People
This article provides clear idea for the new viewers of blogging,
that actually how to do blogging.
Good article. I’m facing some of these issues as well..
https://edithvolo.com/page/36/
https://pornmaster.fun/hd/mallu-reshama-nude-sexolder-rollin
I was able to find good advice from your content.
https://edithvolo.com/page/146/
На Tubba RU https://tubba.ru доступны фильмы и сериалы, которые недавно вышли на телевидении.
When I initially commented I clicked the “Notify me when new comments are added” checkbox and now each time a comment is added I get four e-mails
with the same comment. Is there any way you can remove people
from that service? Thank you!
bookmarked!!, I really like your website!
I can’t believe how amazing this article is! The author has done a phenomenal job of delivering the information in an compelling and informative manner. I can’t thank him enough for sharing such priceless insights that have undoubtedly enlightened my awareness in this subject area. Bravo to him for producing such a gem!
https://ddnews.co.kr/eab480eca088ebb3b4eab681-eab080eab2a9/
https://pornmaster.fun/hd/sex-man-fucking-3gp-video-downloadw-to-sex-video-mp4-download-comi-song-mun-xxxx-bd-com
Hey just wanted to give you a brief heads up and let you
know a few of the images aren’t loading correctly.
I’m not sure why but I think its a linking issue.
I’ve tried it in two different internet browsers and both show the same outcome.
Do you have a spam issue on this website; I also am a blogger, and I was wondering your situation; we have created
some nice methods and we are looking to exchange solutions with
others, why not shoot me an email if interested.
https://ddnews.co.kr/category/life/page/17/
https://www.pornhub.com/view_video.php?viewkey=ph5fa4d22a641bd
Вы можете смотреть фильмы и сериалы https://tubba.ru на Tubba в любом удобном для вас формате.
Impressive! Your article was a good read:geek bar
Cheers, I enjoy it!
Look into my page – http://mspeech.kr/bbs/board.php?bo_table=705&wr_id=619894
I visited multiple blogs but the audio feature for audio songs existing
at this web site is in fact marvelous.
You said it nicely.!
My web blog – https://www.Centrodentalmendoza.com/question/%ec%b6%9c%ec%9e%a5%eb%a7%88%ec%82%ac%ec%a7%80ing-three-tricks-your-competitors-know-but-you-dont-8/
Hi I am so thrilled I found your webpage, I really found
you by error, while I was browsing on Aol for something else, Anyways I am here now and would just like to say many
thanks for a fantastic post and a all round thrilling
blog (I also love the theme/design), I don’t have time to read it all at the moment but I have saved it and also included your RSS feeds,
so when I have time I will be back to read a lot more, Please do keep up
the fantastic b.
I am really impressed along with your writing talents and also with the
format in your blog. Is that this a paid subject or
did you modify it yourself? Anyway stay up the excellent quality writing, it is rare to see a great weblog
like this one today..
Hey there! I understand this is somewhat off-topic however I needed to ask.
Does building a well-established blog such as yours require a large amount
of work? I am brand new to blogging however I do write in my diary on a daily basis.
I’d like to start a blog so I can easily share
my personal experience and feelings online. Please let
me know if you have any kind of suggestions or tips for new aspiring
bloggers. Thankyou!
I’m very happy to find this web site. I need to to thank you for your time due to this fantastic read!! I definitely savored every bit of it and i also have you book-marked to see new information in your site.
Fantastic write ups Thanks a lot.
Have a look at my site … http://xn--vb0bk3rb6kwzi7ofba482b.com/bbs/board.php?bo_table=free&wr_id=173773
Post writing is also a fun, if you be familiar with afterward you can write otherwise it is complex to write.
Kudos! A good amount of advice!
This post is truly inspiring! I especially appreciate your point which you found valuable. It’s clear you put a lot of effort into this, and I’m glad I found it.
What’s up, for all time i used to check webpage posts here in the early hours in the break of day, as i enjoy to learn more
and more.
Thanks for your posting. Another item is that just being a photographer entails not only difficulty in capturing award-winning photographs but in addition hardships in establishing the best digital camera suited to your needs and most especially issues in maintaining the standard of your camera. It is very correct and noticeable for those photography addicts that are straight into capturing a nature’s eye-catching scenes : the mountains, the forests, the actual wild or maybe the seas. Visiting these exciting places surely requires a digicam that can live up to the wild’s unpleasant setting.
Вы можете смотреть фильмы и сериалы на https://tubba.ru Tubba в любое удобное для вас время суток.
Nicely put, Cheers.
Ensure your website’s authority and increase organic traffic with these updated resources.
These are genuinely great ideas in about blogging. You have touched some pleasant points here.
Any way keep up wrinting.
Choosing an IELTS Coaching in Delhi is crucial for achieving your target score. Prioritize institutes with a strong reputation and proven success in helping students excel in the exam. Faculty expertise is key; ensure instructors have substantial experience in IELTS coaching and can offer personalized guidance. Consider the institute’s location and class timings to ensure they fit your schedule. Access to comprehensive study materials, mock tests, and practice sessions is essential for thorough preparation. Additionally, a supportive learning environment that fosters interaction and provides constructive feedback can significantly enhance your learning experience. Lastly, inquire about additional services such as visa counseling or university application assistance, which can provide valuable support for your international aspirations.
Hi there, constantly i used to check web site posts here in the
early hours in the morning, for the reason that i
enjoy to find out more and more.
my webpage … click for source
Hi, i feel that i saw you visited my web site thus i came to return the favor?.I am attempting to find things to improve my site!I assume its
ok to use some of your ideas!!
With thanks, Great stuff.
My web-site :: https://Nilecenter.online/blog/index.php?entryid=861410
Yes! Finally someone writes about construction.
Can I just say what a comfort to uncover somebody that genuinely knows what they are discussing on the net. You actually understand how to bring a problem to light and make it important. A lot more people have to read this and understand this side of your story. I was surprised that you are not more popular since you surely possess the gift.
When I originally commented I clicked the “Notify me when new comments are added” checkbox and now each time a comment is added I get three e-mails with the same comment.
Is there any way you can remove people from that service?
Bless you!
Way cool! Some very valid points! I appreciate
you penning this write-up plus the rest of the site is really good.
Hello just wanted to give you a brief heads up and let
you know a few of the pictures aren’t loading correctly. I’m not sure why
but I think its a linking issue. I’ve tried it in two different internet browsers and both show
the same results.
My brother recommended I might like this website.
He was entirely right. This post actually made my day.
You can not imagine just how much time I had spent for this information! Thanks!
Why users still use to read news papers when in this technological world everything is existing on web?
벼룩시장 구인구직 및 신문 그대로 보기 (PC/모바일) | 구인구직 앱 어플 무료 설치 다운로드 | 모바일 벼룩시장 보는 방법 | 벼룩시장 부동산 | 지역별 벼룩시장 | 벼룩시장 종이신문 에 대해 알아보겠습니다. 섹스카지노사이트
bookmarked!!, I really like your blog!
Everything is very open with a very clear explanation of the issues. It was really informative. Your website is useful. Many thanks for sharing.
I delight in, cause I found exactly what I used to be looking for.
You’ve ended my four day lengthy hunt! God Bless you
man. Have a nice day. Bye
Fantastic web site. Plenty of helpful information here.
I am sending it to several friends ans additionally sharing in delicious.
And certainly, thanks on your sweat!
???, ??? ???? ????? ??? ???? ? ??????? ?? ????? ???? ????? ????? ????? ????? ?????? ??????.
As a Newbie, I am continuously exploring online for articles that can be of assistance to me.
My website: анальное порно
You decided not to get into extreme detail, however you presented the requirements I want to to get me thru. If you are hoping to get started with a project this really is the facts that is needed. Having more writers join the dialogue is usually a great thing.
Great web site. A lot of helpful information here.
I am sending it to a few friends ans additionally sharing in delicious.
And of course, thank you on your sweat!
I precisely wished to appreciate you once again. I’m not certain the things I would’ve handled without the entire solutions shown by you relating to that area of interest. This has been a very terrifying concern for me, nevertheless discovering a new specialised technique you solved the issue took me to leap with joy. I’m grateful for this advice as well as wish you know what a great job that you’re getting into educating people via your webblog. I’m certain you have never met any of us.
Have you ever thought about adding a little bit more than just your articles? I mean, what you say is important and all. Nevertheless think of if you added some great pictures or videos to give your posts more, “pop”! Your content is excellent but with images and video clips, this blog could undeniably be one of the greatest in its field. Excellent blog!
Wow, this article is good, my sister is analyzing such things, so I am going to
convey her.
Does your website have a contact page? I’m having problems
locating it but, I’d like to shoot you an email. I’ve got
some suggestions for your blog you might be interested in hearing.
Either way, great site and I look forward to seeing it expand over
time.
Thanks for your blog post. What I would like to make contributions about is that computer system memory is required to be purchased if the computer cannot cope with anything you do with it. One can install two RAM memory boards having 1GB each, as an illustration, but not one of 1GB and one having 2GB. One should always check the maker’s documentation for own PC to be certain what type of storage is required.
Awesome post it is really. I’ve been searching for this information
Good post. I learn something new and challenging on sites I stumbleupon every day. It’s always helpful to read content from other writers and practice something from their sites.
Spot lets start on this write-up, I truly feel this website needs a lot more consideration. I’ll apt to be again to learn to read additional, thank you that information.
Thiis post will assist the interfnet vjsitors foor creating new weblog or even a
blog from start to end.
Allso visit my blog post gisel88
This can be a huge and an very interesting write to take a look at with this enormous website. Never post any responds and now merely could not avoid
Heya i am for the primary time here. I found this board and I in finding It truly useful & it helped me out a lot. I am hoping to give something back and help others such as you helped me.
He’s not quite as bad as, say, Hayden Christensen in the Star Wars prequels, but he’s in the same category.
The main audience for the plan clear. What is very important is keeping
the key objective of. Keeping a clear difference between yearly strategies.
Hi there! This blog post couldn’t be written much better! Reading through this post reminds me of my previous roommate! He continually kept talking about this. I will send this information to him. Pretty sure he’s going to have a great read. Thanks for sharing!
You’ve made your point!
Review my site https://dashamailrgdbru.Dmesp.ru/clicks.php?hex&m=e790&c=233038&i=199b&u=1f0c2&l=aHR0cHM6Ly9mZWJydWFyeS1hbm1hLnNob3AvYnVzYW4tbWFzc2FnZQ–
Thanks for the good writeup. It if truth be told was once a enjoyment account it. Look advanced to far introduced agreeable from you! However, how could we keep in touch?
Unlock amazing features with cricplus sign up benefit from a seamless platform, instant rewards, and nonstop customer support. Join now for a fun, fully supported journey.
You actually stated it wonderfully.
My page … http://www.icecap.co.kr/bbs/board.php?bo_table=sub0403_en&wr_id=268400
https://ddnews.co.kr/mbti-ec84b1eab2a9ec9ca0ed9895eab280ec82ac/
https://www.pornhub.com/view_video.php?viewkey=ph571756d014c03
Complete your cricplus sign up today and enjoy multiple rewards and exclusive benefits their platform ensures seamless navigation and instant ID acquisition. Enhance your betting experience with Cricplus.
Good job! I really liked our article about: lost mary vape
You’ve made some decent points there. I looked on the internet to learn more about the issue and found most people will go along with your views on this website.
Great job! I found our article on: flum vape very informative and well-written.
In June 2017 Google launched Google for Jobs, a
job listing database.
Have a look at my homepage – Pleats
https://ddnews.co.kr/ec96b4ed9c98eba0a5-ed858cec8aa4ed8ab8/
The cricplus sign up process is simple and user-friendly, offering immediate access to real-time updates and expert analysis. Completing the sign up ensures you can start enjoying all the features and benefits of the platform quickly and efficiently.
The cricplus sign up process is your key to endless cricket fun. It’s easy to join and gives you access to all the thrilling matches. Enjoy free rewards and excellent 24/7 customer support with cricplus sign up. Join now and dive into the action.
Experience a hassle-free journey into cricket with cricplus sign up their secure and straightforward signup process keeps your data protected, and our dedicated 24/7 customer support is always there to assist. Sign up now and start enjoying all the features.
Great work! Your article was insightful. Learn more about it!: disposable vape
Great post. I will be going through a few of these issues as well..
Tubba предлагает бесплатный просмотр фильмов и сериалов https://tubba.ru без регистрации и смс.
I look forward to many more years of enlightening and entertaining reads!
I do not even know how I ended up here, but I thought this
post was great. I don’t know who you are but certainly you’re going to
a famous blogger if you aren’t already 😉 Cheers!
Here is my webpage :: Self-driving camping sites
I was suggested this website by my cousin. I am not
sure whether this post is written by him as nobody else know such detailed about my trouble.
You’re incredible! Thanks!
https://artdaily.com/news/171650/Mp3Juice-Review–The-Pros-and-Cons-You-Need-to-Know
You are so interesting! I don’t think I have read through anything like this before.
So wonderful to discover another person with a few original thoughts on this subject matter.
Seriously.. thank you for starting this up. This website is something that is needed on the internet,
someone with some originality!
Hey there terrific website! Does running a blog such as this take a
great deal of work? I have virtually no knowledge of computer programming however I had been hoping to
start my own blog in the near future. Anyway, if you have any suggestions or
techniques for new blog owners please share. I understand this is off subject but I
simply had to ask. Thanks!
Why people still make use of to read news papers when in this technological globe all is existing on web?
https://infohelpforyou.com/tag/kbs-ed8eb8ec84b1ed919c/
Ahaa, its good dialogue about this piece of writing at this place at this
weblog, I have read all that, so at this time me also commenting here.
Mostbet https://mostbet-bk.cz nabizi sirokou skalu moznosti sledovani zivych her, coz umoznuje ceskym hracum sledovat sve oblibene sportovni udalosti a souteze v realnem case.
Tubba https://tubba.ru предлагает бесплатный просмотр фильмов и сериалов в высоком качестве.
The Seminoles wanbt to hold on to their igaming monopoly at all charges.
Also visit my blog – 비트코인 거래소 리스트
Great job! The article onGeek Bar Pulse was a compelling read
hello!,I like your writing so much! percentage we keep up a correspondence extra about your article
on AOL? I require a specialist on this space to solve my problem.
Maybe that’s you! Taking a look ahead to see you.
Having read this I believed it was extremely enlightening.
I appreciate you taking the time and effort to put this
article together. I once again find myself spending way
too much time both reading and posting comments.
But so what, it was still worthwhile!
Right here is the right site for anyone who wishes to understand this topic.
You know a whole lot its almost tough to argue with you (not that
I personally would want to…HaHa). You certainly put
a fresh spin on a topic which has been discussed for many years.
Great stuff, just excellent!
Hello are using WordPress for your blog platform?
I’m new to the blog world but I’m trying to get started and
create my own. Do you need any coding knowledge to make your own blog?
Any help would be greatly appreciated!
Hello, I want to subscribe for this weblog to obtain most up-to-date updates, therefore where can i do it please assist.
Porn online
https://download-install.com/entry/EAB3B0ED948CEBA088EC9DB4EC96B4-EBACB4EBA38C-EB8BA4EC9AB4EBA19CEB939C-EC84A4ECB998EBB0A9EBB295-EB8F99EC9881EC8381-ED948CEBA088EC9DB4EC96B4
What a data of un-ambiguity and preserveness of valuable experience regarding unexpected emotions.
Good day! I know this is somewhat off topic but I was wondering which blog platform are you using for this site?
I’m getting sick and tired of WordPress because I’ve had issues with hackers and I’m looking at alternatives for another platform.
I would be awesome if you could point me in the direction of a good platform.
Very good write-up. I absolutely appreciate this site. Thanks!
I like it whenever people come together and share thoughts. Great site, continue the good work.
Marvelous, what a weblog it is! This website gives useful facts to us, keep it up.
I blog frequently and I really thank you for your content.
Your article has truly peaked my interest. I am going to take a note of
your website and keep checking for new information about once per week.
I subscribed to your RSS feed as well.
Hello to the folks at Vanderbilt
For audiophiles, turntable aficionados or laboratory power and test challenges, KCC Scientific Power Converters solve the following:
What if…
…your equipment was developed for a different power grid?
…you need to test a product being developed for a different power grid?
…your equipment requires regulated ultra-high mains power line frequency precision?
…you need compact, precision converters to accommodate your mobile requirements?
KCC Scientific frequency and voltage converters are designed to power musical equipment and laboratories’ delicate and precise devices anywhere in the world. Our products reconstruct the power line (mains reconstruction)
(Pat pend). KCC Scientific converters clean, convert and regulate power, enabling electronics across the globe to operate as they were designed.
Thanks for your time!
Teri
https://voltageandfrequencyconverters.com
Opt out of future messages by replying to this message with “Opt Out”.
vanderbilt.edu
Attractive element of content. I simply stumbled upon your weblog and
in accession capital to claim that I acquire actually loved account your blog posts.
Any way I will be subscribing on your augment and even I fulfillment you access constantly rapidly.
Your style is unique compared to other people I’ve read stuff from.
Many thanks for posting when you have the opportunity, Guess I’ll just
bookmark this web site.
I have observed that of all types of insurance, health care insurance is the most marked by controversy because of the clash between the insurance policy company’s obligation to remain making money and the user’s need to have insurance cover. Insurance companies’ earnings on wellbeing plans are incredibly low, consequently some corporations struggle to earn profits. Thanks for the thoughts you talk about through this site.
Thanks very interesting blog!
Hello! I know this is somewhat off topic but I was wondering
which blog platform are you using for this site? I’m getting fed up of WordPress because I’ve had problems
with hackers and I’m looking at options for another platform.
I would be fantastic if you could point me in the direction of a good platform.
Review my web-site Links.Musicnotch.Com/Sybiltober6
I’m more than happy to find this web site. I wanted to thank you for ones time just for this fantastic read!! I definitely appreciated every little bit of it and I have you saved to fav to check out new information on your web site.
I am regular reader, how are you everybody? This post posted at this website
is actually nice.
bookmarked!!, I like your web site!
Looking for a reliable vape? Check out this review of the Lost Mary MT15000 Turbo to see if it fits your needs.
I blog quite often and I really appreciate your content. Your article has truly peaked my interest. I am going to bookmark your blog and keep checking for new details about once a week. I subscribed to your RSS feed as well.
Great beat ! I wish to apprentice whilst you amend your site, how could i subscribe for a
weblog site? The account helped me a appropriate deal.
I have been tiny bit acquainted of this your broadcast offered brilliant clear idea
Вы можете найти на Tubba https://tubba.ru как любимые фильмы и сериалы, так и новые для себя киношедевры.
It is the best time to make some plans for the longer term
and it is time to be happy. I’ve learn this submit and if I may
just I want to suggest you some attention-grabbing issues or suggestions.
Maybe you can write subsequent articles relating to this
article. I want to read even more things approximately it!
Someone essentially help to make seriously articles I would state. This is the very first time I frequented your website page and to this point? I surprised with the research you made to make this particular put up extraordinary. Magnificent process!
Next time I read a blog, I hope that it won’t fail me as much as this one. I mean, I know it was my choice to read through, nonetheless I actually believed you would have something helpful to talk about. All I hear is a bunch of crying about something you can fix if you were not too busy seeking attention.
I have been exploring for a little bit for any high-quality articles or
weblog posts in this kind of space . Exploring in Yahoo I
at last stumbled upon this website. Reading this information So
i am happy to express that I’ve an incredibly excellent uncanny feeling I
came upon just what I needed. I such a lot unquestionably
will make sure to do not forget this web site and give it
a glance regularly.
I always look forward to the latest discussions and updates on this forum. Very inspiring!
https://itmoney4you.com/빅마우스-재방송/
If you are going for best contents like me, just pay a quick visit
this site every day since it presents feature contents, thanks
Polacy kochaja https://mostbet-bk.pl Mostbet Casino za jego atrakcyjne bonusy i promocje, ktore zwiekszaja ich szanse na wygrana.
Find out why the Lost Mary MO20000 Pro is gaining popularity among vapers in this comprehensive review.
Pretty! This has been a really wonderful post. Thanks for providing these details.
You mentioned this well.
my web page :: https://phaiyai.go.th/question/3-reasons-your-business-is-%ec%b6%9c%ec%9e%a5%ec%95%88%eb%a7%88ing/
I could not resist commenting. Exceptionally well written.
What’s up, its fastidious article concerning media print, we all
be familiar with media is a impressive source of data.
It’s very straightforward to find out any matter on web as compared to books, as I found this post at this web site.
Appreciate it. A lot of content!
Also visit my web page – http://www.Ilsantop.com/bbs/board.php?bo_table=free&wr_id=72877
I know this if off topic but I’m looking into starting my own blog and was curious what all is needed to get setup?
I’m assuming having a blog like yours would cost
a pretty penny? I’m not very web smart so I’m not 100% certain. Any recommendations or advice would be greatly appreciated.
Appreciate it
A Mostbet https://mostbet-bk.com Casino-ban a legjobb elo kaszino elmenyt a vilag vezeto szoftverfejlesztoi biztositjak, akik a legujabb technologiakat hasznaljak.
Thank you for the auspicious writeup. It in reality used to be a amusement account it.
Look complicated to more delivered agreeable from you!
However, how can we be in contact?
Have you ever thought about creating an e-book or guest authoring on other blogs?
I have a blog based upon on the same subjects you discuss and would love to have you share some stories/information.
I know my readers would value your work. If you’re even remotely interested, feel free to send me an email.
wood and leather bag
You made some good points there. I looked on the internet to learn more about the issue and found most
people will go along with your views on this
website.
Hello, i think that i saw you visited my blog so i came to “return the
favor”.I am attempting to find things to improve my web site!I suppose its
ok to use a few of your ideas!!
Spot on with this write-up, I honestly believe this web site needs far more attention. I’ll probably be returning to see more, thanks
for the info!
Also visit my web site … my explanation
bookmarked!!, I really like your site!
Mostbet https://mostbet-bk.pl oferuje szeroki wybor zakladow na krykieta i inne popularne sporty w Azji.
This is a great resource for anyone interested in…검색엔진최적화 배우기
https://film35.tistory.com/438
The diversity of perspectives and experiences on this forum makes it a rich and valuable resource.
Mostbet https://mostbet-bk.de bietet eine breite Palette von Wettoptionen fur jede Sportveranstaltung.
I know this web page offers quality dependent articles or reviews and additional stuff, is there any other web page which presents these kinds of things in quality?
With https://mostbet-bk.in Mostbet, you can bet on football, tennis, basketball, and many other sports.
https://financenews.co.kr/tag/4eb8c80ebb3b4ed9798-eab080ec9e85eca69debaa85ec849c/
Thank you for sharing your thoughts. I really appreciate your efforts and I will be waiting for your further post thank you once again.
Check out the features and benefits of the vape juice in this comprehensive review.
You have made some decent points there. I checked on the net to learn more about the issue and found most people will go along
with your views on this website.
오공 슬롯
Hongzhi 황제도 그를 불쾌하게 바라보며 “말하십시오. “라고 말했습니다.
drochilnik.xyz
I love how this forum fosters a sense of belonging and connection among its members.
Thank you for sharing most of these wonderful blogposts. In addition, the right travel and medical insurance strategy can often relieve those problems that come with vacationing abroad. A medical emergency can in the near future become very expensive and that’s likely to quickly put a financial load on the family finances. Having in place the best travel insurance bundle prior to leaving is worth the time and effort. Thanks
Reliable postings Thanks.
my blog post … https://smkansorunasubang.sch.id/question/%ec%b6%9c%ec%9e%a5%eb%a7%88%ec%82%ac%ec%a7%80ing-4-tricks-your-competitors-know-but-you-dont-95/
This forum is a perfect example of how an online community can be an effective and enjoyable learning place.
This design is incredible! You certainly know how to keep a reader amused.
Between your wit and your videos, I was almost moved to start my own blog (well, almost…HaHa!) Great job.
I really loved what you had to say, and more than that, how you presented it.
Too cool!
Mostbet https://mostbet-bk.cz nabizi hru JetX, ktera je plna emoci a stesti.
Learn about the unique features and benefits of the lucy nicotine in this comprehensive review.
Nice article! Also, don’t overlook Lucy Nicotine Pouches; they’re a great find.
Hey! I could have sworn I’ve been to this blog before but after checking through
some of the post I realized it’s new to me.
Anyways, I’m definitely glad I found it and I’ll be bookmarking and checking
back frequently!
The level of expertise and knowledge shared on this forum is unparalleled.
Cheers! A good amount of material.
My website … http://pr.lgubiz.net/bbs/board.php?bo_table=free&wr_id=65873
Hey there are using WordPress for your site platform? I’m new to the blog world but I’m trying to get started and set up my own. Do you require any html coding knowledge to make your own blog? Any help would be greatly appreciated!
I have found solutions to many of my problems thanks to the helpful advice from this community.
https://hipsterlibertarian.com/5683
https://hipsterlibertarian.com/585
Appreciate the recommendation. Let me try it out.
Looking for assignment writing services? Look no further, we are here to help you. Please contact us to know more +91 9605750505
https://hipsterlibertarian.com/6048
https://pornmaster.fun/hd/【查询开房记录客服-微信-客服78444643】查找查微信聊天记录-pln
Everything is very open with a precise clarification of the issues.
It was really informative. Your site is useful.
Thank you for sharing!
I feel very welcomed and appreciated in this forum. Thank you to everyone who made this possible.
Delve into the features and advantages of the Geek Bar Flavors with this detailed review.
Learn all about the benefits of the vape mods in this comprehensive guide.
Hi, I do believe this is a great website. I stumbledupon it 😉 I will come back once again since i have saved as a favorite it. Money and freedom is the greatest way to change, may you be rich and continue to help others.
This forum has a vibrant and engaging atmosphere that keeps me coming back.
I don’t even know how I ended up here, but I thought this post was good.
I do not know who you are but certainly you are going to a famous blogger
if you aren’t already 😉 Cheers!
Good post. I learn something totally new and challenging on websites
I stumbleupon on a daily basis. It’s always interesting to read articles from other writers
and use a little something from other sites.
I always used to read article in news papers but now as I am a user of web therefore from now I am using net
for articles, thanks to web.
You said that terrifically.
Here is my web-site … http://Ict.Wku.Ac.th/question/why-ill-never-%ec%b6%9c%ec%9e%a5%ec%95%88%eb%a7%88-27/
Asking questions are genuinely good thing if you are not understanding something
completely, except this article gives pleasant understanding
yet.
Learn all about the Geek Bar Pulse and its key advantages in this thorough analysis.
Ahaa, its nice dialogue about this piece of writing
here at this website, I have read all that, so now me also
commenting here.
Good post. I learn something new and challenging on blogs I stumbleupon on a daily basis.
It will always be useful to read content from other authors
and use something from other web sites.
Uncover the unique qualities of flum pebble flavors through this detailed review.
Way cool! Some extremely valid points! I appreciate you writing this write-up and also the rest of the site is very good.
This is a topic that is close to my heart… Best wishes! Exactly where can I find the contact details for questions?
You said it perfectly.!
Il vous sera également possible de jouer aux jeux live comme le blackjack, la roulette ou encore le baccarat.
top porn
Hello! I’ve been following your site for a while now and finally got the bravery to go ahead and give you a shout
out from Humble Texas! Just wanted to tell you keep
up the great job!
Explore the detailed insights and advantages of the miami mint lost mary with this extensive review.
I believe this is one of the so much significant info for me.
And i am satisfied studying your article. However want to remark
on some basic issues, The web site taste is ideal, the articles is in point of fact excellent : D.
Good job, cheers
asiabet62 merupakan situs slot gacor online terpopular se Asia yang menyediakan banyak sekali keuntungan besar besaran untuk para pemain, mainkan sekarang juga situs slot gacor asiabet62 dan dapatkan keuntungan yang banyak
You’ve made your point!
Their traditional CBD soft gels, available in both 10mg and 25mg potencies, are non-GMO, gluten free, and fit easily into most dietary restrictions.
However, there are some things you should know before diving right into the world of CBD oil for pets.
There are several different types and uses of CBD oil.
Many of our furry friends are just as picky as we are, making them skeptical of new treats or unusual smells.
These are treats that look, smell, and taste like
your average dog or cat treat, only we’ve infused them with varying amounts of CBD to provide your pet a boost
of support. If your pet can’t turn down the temptation of a snack, CBD
treats are a great option. Yes, hemp oil products for pets are quite safe, and you
should be able to give them to your furry friend without
any worry. Having control of the entire process guarantees the
best quality of the products we present to our customers.
Hi there! This blog post couldn’t be written any better! Going through this post reminds me of my previous roommate! He always kept preaching about this. I’ll send this article to him. Pretty sure he’s going to have a good read. Thank you for sharing!
nice article!
https://cults3d.com/en/design-collections/despicableme4/film-moi-moche-et-mechant-4-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/maxxxine/film-maxxxine-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/twisters/film-twisters-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/to-the-moon/film-to-the-moon-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/deadpool-wolverine/film-deadpool-wolverine-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/unesagaamericaine/film-horizon-une-saga-americaine-chapitre-1-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/LargoWinch3/film-largo-winch-le-prix-de-l-argent-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/monte-cristo/film-le-comte-de-monte-cristo-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/ehud6hx/film-un-p-tit-truc-en-plus-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/lelarbin/film-le-larbin-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/ekm8qly/film-sans-un-bruit-jour-1-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/longlegs2024/film-longlegs-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/santoshhd/film-santosh-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/viceversa2/film-vice-versa-2-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/badboys2024/film-bad-boys-ride-or-die-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/badnewz/film-bad-newz-2024-streaming-vf-gratuit
https://cults3d.com/en/design-collections/thebikeriders/film-the-bikeriders-2024-streaming-vf-gratuit
https://www.universe.com/users/8sh4np4-qtlx2pep-GST19Z
https://muckrack.com/dmeuqfcylfczagxz-kesdmjvpqngsybnj/bio
https://bootstrapbay.com/user/0lw5tnfdx4h624nc
https://wefunder.com/bunpnvfynesyqdokbltotlktbwoqxvls
https://www.fmscout.com/users/mevl47ae9b4dkj3y.html
https://profile.hatena.ne.jp/am6nwfmj60pwpqdp/
https://tinhte.vn/profile/irlizvcfhnkfwsrf.3034182/
https://www.bricklink.com/aboutMe.asp?u=ostdrtmiulewzao
https://bio.site/alannalynch
https://dictanote.co/n/1042870/
https://jsbin.com/jumaqonasa/edit?html,output
https://wokwi.com/projects/404497866106994689
https://hackmd.io/@ddtdiyyvek/rJW8oPZF0
https://zenn.dev/alyssafranco/scraps/8ec2638bcf4679
https://www.bookmarkmonk.com/stories/regarder-moi-moche-et-mechant-4-en-streaming-vf/
https://www.bookmarkrush.com/stories/02yrx0hn36vb5m7y/
https://www.bookmarkfrog.com/stories/r5e75z8pt5tyzq52/
https://www.bookmarkrocket.com/stories/5t8q5kz98sk88mxa/
https://www.clickone.co.in/story/wzun3ghjnyf20lp5/
https://snippet.host/ifzhcy
https://pastebin.com/4pqQamWu
https://rextester.com/SEH54630
https://dev.bukkit.org/paste/cbb407f2
https://rift.curseforge.com/paste/5f4af6dc
https://jsfiddle.net/fe8xvj3o
https://telegra.ph/lcsrkhbqucqgvlwu-07-26
https://paste.myst.rs/ewvhukr4
https://pastelink.net/z9lcis32
https://bitbin.it/5y7Di33r/
https://yamcode.com/juxcggxmqlupsrvx
https://rentry.co/pecc7fy2
https://paste.rs/rEDD8.txt
A round of applause for your article. Much thanks again.
My website: chastnoeporno.top