About
This program builds on an existing collaboration between PIs Jens Meiler and James Crowe but transforms it substantially, expands scope, and shifts focus to the next frontier: the integration of big data and structural biology into vaccine design. It adds a comprehensive training program, eight mostly early/mid-career faculty, and a series innovative technologies to form the Vanderbilt Program for Next Generation Vaccines. We embrace a comprehensive vision for educating at the interface of big data, structural biology, and vaccine development at all levels – undergraduate, graduate, and post-graduate. Through re-investment we can implement a two-pronged approach: as an institution, VU becomes the leader in an emerging, highly dynamic and translational research field and we simultaneously educate the next generation of scientific leaders, creating a multiplier effect where trainees from VU continue to shape this emerging field.
The ten faculty members will co-mentor the trainees as teams of two or three, thereby creating the kernel for a research and education center that has the potential to develop into a comprehensive Program for Next Generation Vaccines.
In parallel this program aggressively expands the technologies that trainees are educated to use, technologies that are critical to advance development of next generation vaccines. For this purpose, our proposal integrates eight mostly junior and mid-career faculty members who contribute innovative technologies. New is the integration of experimental techniques of structural biology such as electron microscopy (EM, Melanie Ohi), X-ray crystallography (Benjamin Spiller), deuterium exchange and cross-linking mass spectrometry (DXMS and XLMS, Kevin Schey), and small angle scattering (SAS, Walter Chazin). New are also innovative computational techniques to cope with big data (Daniel Fabbri) that enters the field of vaccine discovery through next generation sequencing, game theoretic approaches to model viral escape and design antibodies which cannot be easily escaped by viral mutations (Yevgeniy Vorobeychik), a rigorous biostatistics analysis of computational protocols (James C. Slaughter), as well as novel computational approaches to engineer vaccines (Ivelin Georgiev).
The human immune system creates antibodies in response to viral threats such as influenza (Flu), human immunodeficiency virus (HIV), or Ebola/Marburg viruses. These antibodies recognize proteins on the surface of the virus, so-called antigens. In the best-case scenario, human antibodies neutralize viruses by blocking the interaction of viruses with the surface of the human cell. Viral vaccines mimic the effect of infection to provoke our immune system to elicit neutralizing antibodies, thereby preventing infection during later exposures. Unfortunately, not all humans create neutralizing antibodies quickly enough upon infection, and there are no effective vaccines available for many pathogens.
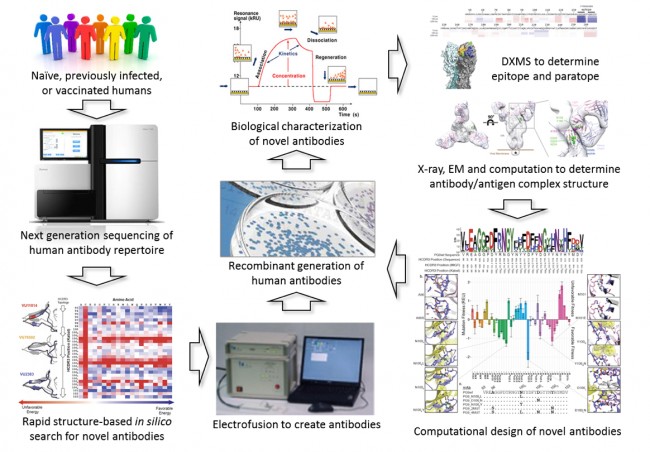
In the VVC, researchers in the laboratory of James Crowe discover novel human antibodies that are broadly neutralizing against the above-mentioned and many additional viruses. In collaboration with others, the Crowe laboratory continues to determine the three-dimensional structure of interesting antibodies in complex with viral proteins, typically using X-ray crystallography. This approach is a resource-intensive process with the ultimate goal that a better understanding of the interaction of the viral protein with broadly neutralizing antibodies will allow for the design of new, effective vaccines.
With the advent of next-generation high-throughput sequencing (using technologies now available in the Vanderbilt VANTAGE core), the number of human antibody sequences available is exploding from thousands to billions. This explosion of sequence data provides a potential resource for quickly identifying novel, broadly neutralizing antibodies and thereby fueling the development of next-generation vaccines – i.e., vaccines against HIV or Ebola or a Flu vaccine that do not require annual vaccination. Further, the knowledge gleaned from these large antibody databases will enable us to engineer novel and improved antibodies not (yet) observed in humans. Ultimately such antibodies will provide an improved template for developing vaccines. However, before the promise of this research can be fully achieved, it is essential that the resource-intensive step of moving from antibody sequence to 3D structures of antibody/antigen complexes is streamlined in order to take advantage of the rapidly increasing antibody sequence databases to aid in vaccine development.
©2024 Vanderbilt University ·
Site Development: University Web Communications