Alberto Cisneros
Antibody Affinity Maturation in Antigen-Distal Residues
Project Abstract
The FV region of an antibody consists of the heavy chain (HC) and light chain (LC) variable domains whose association is maintained by a series of conserved, non-polar interactions. During chronic infections, somatic mutations are induced, often in the HC/LC interface. Sequence variation in these interactions allows the HC and LC domains to inhabit a range of orientations relative to one another. Thus, I hypothesize that these interface mutations are critical to orient and rigidify the HC/LC interface to arrange the paratope for optimal interaction with the antigen, thereby affecting antigen binding affinity allosterically. To test this hypothesis, I measured the HC/LC orientation of a set of broad and potent human HIV neutralizing antibodies. The HC/LC interface of these antibodies contained a large number of mutations and achieved unusual relative orientations compared to other human antibodies. I expressed and characterized a panel of recombinant HIV CD4 binding site antibodies as the fully matured variant and compared these with variants mutated to the HC/LC interface of the inferred unmutated common ancestor antibody. I found that HC/LC interface reverted antibodies have a reduced affinity, confirming that introduction of somatic mutations in the HC/LC interface was one of the critical steps in affinity maturation. I then used the Rosetta software suite to examine the mechanisms through which these mutations affect binding affinity. I determined to what extent the mutations were critical in altering the relative orientation of HC/LC domains to a conformation that is competent to bind the antigen. I further determined whether the mutations excluded alternative HC/LC conformations that would be incompetent to bind the antigen. These findings suggest that somatic mutations in the HC/LC interface, distant from the antigen/antibody contact region, play a critical role in affinity maturation of HIV antibodies by preconfiguring the bound conformation of the antibody in the orientation required for high affinity recognition of the antigen. Thus, optimization of HC/LC interface could serve as an important tool for maximizing antibody/antigen binding affinity without altering antigen contact residues.
Progress Report April 2019
The adaptive immune response (occasionally referred to as the “acquired immune response”), is the mechanism through which humans eliminate both bacterial and viral infections (Alberts, 2002) The strength of the adaptive immune response lies in its ability to recognize a vast number of foreign pathogens given a limited number of gene segments and options for gene segment recombination (Koonin, 2015). Prior to B cell activation, the diversity of the antibody repertoire is generated by V(D)J gene segment recombination (Roth 2014); using 69 VH, 27 DH, and 6 JH gene segments, the immune system generates over 11,000 unique VDJ recombination events for the heavy chain alone. Taking into account the 31 IGKV genes, 5 IGKJ genes, 45 IGLV genes, and 7 IGLJ genes that comprise light chains in conjunction with the junctional diversity that stems from recombination, an estimated 1011 antibodies can populate an individual’s antibody repertoire (Glanville, 2009). These antibodies, while dwarfed in number by the theoretical possible number of epitopes on pathogens that the immune system might encounter, provides compensatory protection through structural flexibility. Germline antibodies that have yet to undergo affinity maturation often are polyspecific and bind multiple targets at low affinity through a flexible binding surface – the paratope (Willis, 2013). However, antibody/antigen complexes are ternary structures (Sherrif, 1987). Since antibodies are formed by the combination of a heavy chain (HC) and a light chain (LC), the complex with the antigen constitutes a three-way interaction (Figure 1). This secondary interface in the Fv region of the antibody allows the variable HCs and variable LCs to take on a wide range of orientations relative to one another and is responsible for determining the geometry of the paratope (Chailyan, 2011; Abhinandan and Martin, 2011). Additionally, activated B cells undergo affinity maturation – an iterative process involving somatic hyper-mutation (Hwang, 2015), the process by which mutations are made in the rapidly proliferating B cell, diversifying the B cell receptor, and positive selection that ultimately leads to target-specific antibodies (Tiller, 2017). Antibodies that evolve in response to lifelong infections like HIV are often mutated beyond what we see in transient infections (Burton, 2005); upwards of 48% of amino acids in the VH gene for anti-HIV antibodies like VRC01 are mutated from their germline precursor (Georgiev, 2014). Fera et al. discerned that some anti-HIV CD4 binding-site (CD4BS) antibodies incorporate somatic mutations in the VH-VL interface to alter the geometry of the combining site, accommodating for the insertion of the HIV V5 loop (Fera, 2014) (Figure 1). This change in orientation is thought to mediate breadth of binding and neutralization.
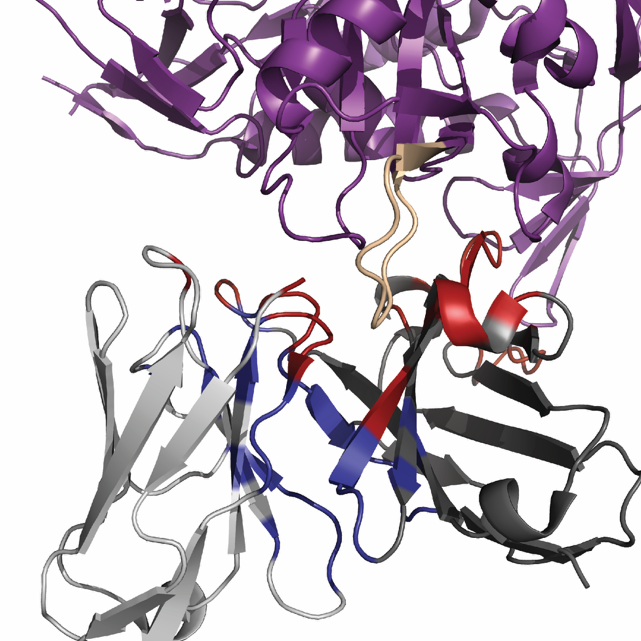
Figure 1. The ternary nature of antibody/Antigen interfaces. Cartoon representation of antibody-antigen and HC/LC interface using VRC03 and 93TH057 gp120 (PDBID:3SE8). The gp120 is shown in purple, the V5 loop is shown in yellow, the heavy chain is shown in dark grey, and the light chain is shown light grey. The paratope was defined as residues within a Cβ-Cβ cutoff of 8 Å or a pair of non-hydrogen atoms within 5.5 Å across the antibody/antigen interface, and is shown in red. The HC/LC interface was defined using the same interface parameters but excludes residues that are included in the paratope and is shown in blue.
Previous studies have inferred that non-contact residues contribute to overall binding affinity and activity of HIV antibodies, however they were not focused on the unique role of heavy chain/light chain interactions in governing binding affinity or the thermodynamic implications behind such a mechanism (Klein, 2013). In this work I apply a novel pipeline to observe and interpret the changes in orientation that can be attributed to mutations in the HC/LC interface by computational modeling and docking. The models generated for the HC/LC interface reversion exhibited a shift in the range of HC/LC orientations sampled during docking, which is consistent with the concept of mutations in the HC/LC interface as a means of mitigating entropic loss upon binding and increasing the enthalpy for the bound conformation. The results show that highly mutated HIV-specific CD4 binding site antibodies achieve unusual orientation features that are distinguishable from most human antibody heavy chain/light chain orientations in order to bind their epitope and that mutations in the HC/LC interface govern the overall orientation of the CDRs by modulating the range of accessible orientations.
The mechanisms involved in B cell development and subsequently in affinity maturation require a conserved HC/LC interface in order to achieve HC/LC pairing. B cells undergo somatic hypermutation, incorporating random mutations in their BCRs in order to increase the affinity of their receptors for antigen and compete for survival. Traditionally, mutations in the HC/LC interface have been seen as disruptive to the aforementioned process, as they may prevent efficient HC/LC pairing (Koenig, 2017). The work presented in this project challenges this conventional notion of antibody affinity maturation. The findings show that antigen-distal somatic mutations in the HC/LC interface indirectly affect binding affinity through mitigation of entropic loss and pre-configuration for the bound conformation of the antibody, and extends the known molecular determinants of antibody/antigen binding and neutralization to include non-contact residues, thus discerning an additional mechanism through which binding affinity is mediated.
Additionally, while an increasing number of co-crystal structures become available, it is still unknown whether the antibodies in complex are optimal in sequence and structure in terms of affinity for the target. The work presented in this project suggests that antibodies devoid of mutations in HC/LC interface can be improved using their bound conformation as a template for in silico affinity maturation.
Cisneros 3rd A, Nargi RS, Parrish EH, Haliburton CM, Meiler J, Crowe Jr. JE. Role of antibody heavy and light chain interface residues in affinity maturation of binding to HIV envelope glycoprotein. Mol. Syst. Des. Eng., 2019; Advance Article
Primary: James E. Crowe, Jr.
Graduate student

©2024 Vanderbilt University ·
Site Development: University Web Communications