Clara Schoeder
Computational Design of Epitope-Focused Ebola and Marburg Immunogens
Project Abstract
Filoviruses Marburg (MARV), Ravn (RAVV), Ebola (EBOV), Sudan (SUDV) and Bundibugyo (BDBV) cause periodic outbreaks of a highly lethal disease in humans. There are no licensed vaccines against any of these viruses. Clara’s project “Computational design of vaccine candidates based on marburgvirus and ebolavirus membrane-proximal external region and receptor binding domain” is aimed at development and downselection of two vaccine antigens: one for marburgviruses MARV and RAVV and another for ebolaviruses EBOV, SUDV and BDBV. The project involves modeling the three-dimensional structures of human antibodies (Abs) as they engage viral surface proteins. It leverages innovative computational tools developed in the Meiler laboratory for the structure-based design of improved human Abs and immunogens. We will use the Rosetta software platform techniques to design novel structure-based vaccine candidate antigens, using high-resolution structures of antigen-antibody complexes in the GP receptor-binding domain (RBD) and membrane-proximal external region (MPER). We will collaborate with James Crowe for the structural and functional characterization of novel human Abs and vaccines against marburgviruses and ebolaviruses. Further, we will collaborate with Ivelin Georgiev to generate rationally designed epitope-based vaccines. Their protective efficacy against marburgviruses and ebolaviruses in vivo via an external collaboration.
Progress Report – April 2019
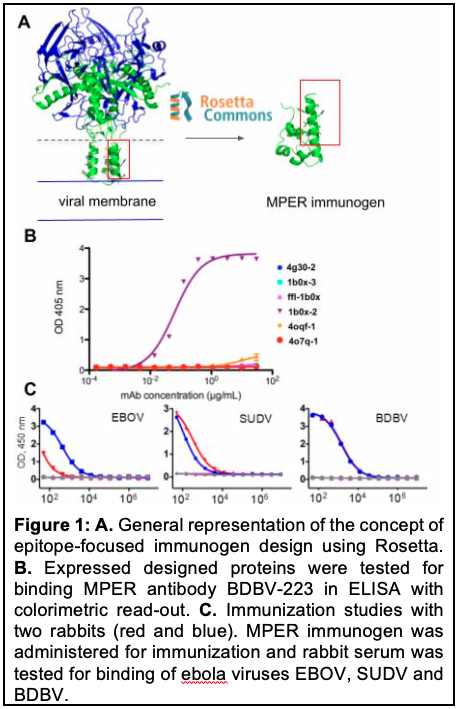
Small immunogens were sampled using epitope-focused vaccine design protocols in the Rosetta protein design software suite. The MPER-epitope of the BDBV glycoprotein was chosen due to its structural favorable organization and the availability of well-characterized anti-MPER antibodies, namely BDBV-223, BDBV-317 and BDBV-340 (Flyak, A., et al. Nat. Microbiol. 2018), which will be used to confirm the correct folding, positioning and function of small MPER immunogen designs. Of eleven chosen designs, six were successfully expressed in E. coli. and were further investigated in antibody binding versus BDBV-223, one of the control antibodies. Out of the six expressed designs, one protein was able to bind the BDBV-223 antibody (Figure 1B). In an initial animal study the small protein MPER immunogen design showed a moderate induction of antibodies in two rabbits, which bound to all three filoviruses BDBV, EBOV and SUDV, although the response to EBOV was very weak (Figure 1C). Consequently, the designed immunogen displayed already breadth and induced the desired immune response, however, the reaction was relatively weak and the breadth especially against EBOV required optimization.
Two different design strategies were further pursued. The lack of breadth can be addressed on a molecular level by redesigning the epitope on the small protein immunogen by considering the amino acid sequence differences of the three filoviruses and fine tuning the binding affinity to reflect the natural behavior of the MPER epitope most closely. We therefore employed site-directed mutagenesis to generate immunogen designs with modifications in the amino acid sequence (currently undergoing functional characterization). The other technique we established in our lab is the utilization of self-assembling cage proteins that can effectively be decorated with immunogens and reach sizes of 20-30 nm in diameter. These self-assembling protein cages have the advantage of being easily purified and produced and they form reliably particles (Bale et al. Science, 2016). They have successfully been employed as immunogen carrier system for the respiratory syncytial virus prefusion surface glycoprotein (Marcandalli et al. Cell 2019). In collaboration with the King lab at the University of Washington, we were able to establish the system in our lab and produced assembly particles displaying our MPER immunogen. With ELISA antibody binding against BDBV223, we further confirmed that our immunogen’s epitope was maintained, while an assembly particle without the epitope did not bind the control antibody at all (Figure 2B).
Overall, the MPER immunogen design could be further developed both on a molecular level as also its macromolar display to the immune system through introducing the self-assembly protein platform. Both projects are currently progressing further to optimize the chosen immunogen and characterize its interaction with the control antibodies, which will lead to a small protein immunogen that carries the MPER epitope in the most native-like conformation.
Notable accolades
Clara is 1 of 6 speakers at our annual Symposium on Modeling Immunity May 1, 2019. She will present “Epitope-focused vaccine design based on Ebola virus glycoprotein membrane-proximal external region”
Mentors
Primary: Jens Meiler
Secondary: James E. Crowe, Jr.
Type of Trainee
Postdoctoral Fellow

©2024 Vanderbilt University ·
Site Development: University Web Communications