Alex Sevy
Abstract
Many antibodies have been identified that can bind and neutralize diverse viral variants of influenza or HIV-1. This broad recognition can be achieved when an antibody generated against a primary strain of the virus undergoes somatic hypermutation to increase affinity for variant strains. Although this is a well-known phenomenon for increasing neutralization breadth among variants of a single viral protein, there is little data showing this mechanism can occur across surface proteins from distinct viral classes. We have observed a high degree of structural similarity between antibodies targeting conserved regions of influenza hemagglutinin or HIV gp41. Based on this structural similarity, we hypothesize that a common antibody conformation may accommodate binding to both of these targets, and that an antibody can shift specificity between influenza hemagglutinin and HIV gp41 via somatic hypermutation. This unexpected mechanism of cross-reactivity would expand the pool of potential clones available to respond to an HIV infection and would represent the first details of an antibody maturation pathway involving maturation against multiple viral antigens. The first specific aim will test these hypotheses by using computational design to simulate affinity maturation and generate antibody variants with altered specificity from HIV to influenza. We will determine the biophysical characteristics of these complexes using ELISA and bio-layer interferometry, and low-resolution structural characterization using hydrogen-deuterium exchange mass spectrometry. Another goal of this project is to generate optimized antibody variants capable of recognizing both influenza and HIV, which would represent intermediates in the affinity maturation process. To this end, the second aim of the proposal is to develop improved multi-specificity design methods capable of designing a cross-reactive sequence. This approach requires algorithm development to enable sequence optimization for binding to multiple proteins, which would be widely applicable to the field of protein design. The third specific aim is then to generate cross-reactive antibodies capable of recognizing both HIV and influenza, using a combination of existing multi-specificity design algorithms, rational design, and the method developed in Aim 2. These antibody variants then will be expressed and characterized biophysically and structurally.
Project Update
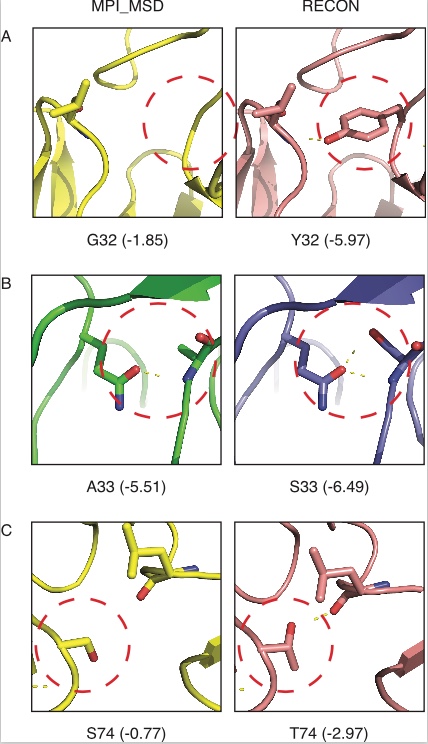
The RECON algorithm for multi-specificity was implemented and benchmarked on a set of promiscuous proteins. In these cases, this algorithm was more effective at recovering natural sequence variation than a competing algorithm, MPI_MSD. The RECON method was able to recover low energy sequences that MPI_MSD failed to sample (Figure 1).
In addition, we have successfully developed a protocol for hydrogen-deuterium exchange (HDX). We characterized the antigen binding domain (paratope) of antibody 14N4 targeting respiratory syncytial virus (RSV) F protein. I was able to obtain an HDX map of 14N4 using deuterium labeling time points of 15, 30, and 60 min. The HDX profile of this antibody indicated that the CDRH3 loop is the primary mediator of antigen binding, with limited engagement by the CDRL2 and, to a lesser extent, CDRH1 (Figure 2). This HDX data was compared to a low resolution (4.1 Å) crystal structure of the 14N4-RSV F protein co-complex, and the HDX data matches very well to the paratope as determined by the co-crystal structure. We plan to use this HDX protocol to characterize a number of antibody-antigen interactions.
Mentors:
Primary: Jens Meiler and James Crowe
List of publications:
Mousa JJ, Sauer MF, Sevy AM, et al. Structural basis for nonneutralizing antibody competition at antigenic site II of the respiratory syncytial virus fusion protein. Proc Natl Acad Sci USA. 2016;113(44):E6849-E6858.
*Sevy AM, *Bender, BJ et al. Protocols for Molecular Modeling with Rosetta3 and RosettaScripts. Biochemistry 55, 4748–4763 (2016).
*Authors contributed equally to this work
Sevy AM, Jacobs TM, Crowe JE Jr, Meiler J. Design of Protein Multi-specificity Using an Independent Sequence Search Reduces the Barrier to Low Energy Sequences. PLoS Comput. Biol. 11, e1004300 (2015).
Sevy AM, Meiler J. Antibodies: Computer-Aided Prediction of Structure and Design of Function. Microbiol Spectr 2, 173–190 (2014).doi:10.1128/microbiolspec.AID-0024-2014.


©2024 Vanderbilt University ·
Site Development: University Web Communications