Nagarajan Raju
Delineation of polyclonal antibody specificities in sera from donors infected by or vaccinated against dengue viruses
Project Abstract
The goal of this project is to develop and apply new technology for the delineation of polyclonal antibody specificities in sera from donors infected by or vaccinated against dengue viruses. The technology proposed here is built upon computational techniques for the generation and analysis of data related to monoclonal antibody and polyclonal serum interactions with the virus. Specifically, the technology will use large-scale antibody-virus neutralization matrices consisting of epitope-specific monoclonal antibody “neutralization fingerprints” (the potency-dependent pattern with which an antibody recognizes a set of diverse viral strains) to identify the types of antibodies present in a serum of interest. An analogous technology has been successfully applied to HIV-1 and should be generally extendible to other viruses, such as dengue, that exhibit sufficient levels of antigen sequence diversity. The polyclonal serum delineation approach described here has been a useful tool for efficient analysis of HIV-1 sera from infected and vaccinated individuals. Because of the substantial sequence diversity observed for dengue and the constantly increasing number of characterized monoclonal antibodies against this virus, we believe this fingerprinting-based approach should be extendible to dengue. Overall, the approach described here can be used for the analysis of sera both at the individual and population levels, and can thus be especially useful for high-throughput analysis of entire cohorts, such as for vaccine-elicited responses.
Progress Report
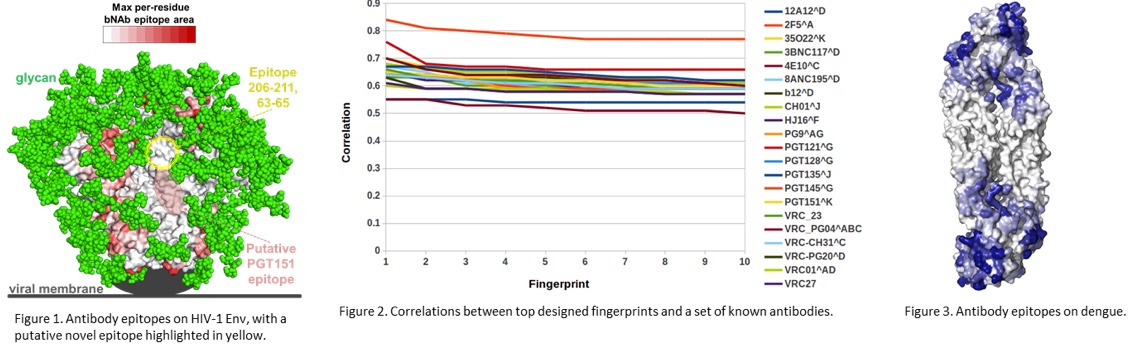
Conservation of dengue antibody epitopes on protein E. Maximum antibody-interactive area for each antigen residue is plotted as a heatmap (white-blue), capped at 100 Å2. Darker color corresponds to antigen residues that are a substantial part of at least one antibody epitope.
We have made progress toward developing novel technology for designing epitope-specific neutralization fingerprints for targeting new, previously uncharacterized, viral epitopes. Our initial studies have revolved around the HIV-1 Env glycoprotein, the target of broadly neutralizing antibodies. We have chosen HIV-1 Env due to the extensive characterization of antibody responses to this protein. However, our recent studies, also supported by findings from other groups, have indicated that there are still previously unseen antibody specificities elicited to HIV-1 infection. The identification and characterization of such novel epitope specificities can help build a more complete picture of the modes of HIV-1 recognition by antibodies and will guide further efforts for epitope-specific HIV-1 vaccine design. While our initial focus has been HIV-1, due to the large sets of functional and structural data available for this virus, the technology that we are developing will be generalizable to other pathogens. Specifically, we have started working on developing and applying this technology to dengue. In the case of HIV-1, we have been developing an algorithm that uses antigen sequence variability in order to design neutralization fingerprints targeting a novel epitope of interest. Initially, we created a dataset of published antibody-antigen structures and mapped their epitopes on the surface of the Env glycoprotein, identifying a potential novel epitope comprised of residues 63-65/206-211, not targeted by any known antibodies (Figure 1). We then assessed the ability of our new fingerprint design algorithm to recover known antibody specificities, and observed the initial implementations of this algorithm successfully generated fingerprints with high correlations to known antibodies (Figure 2). We also observed that some known antibodies are more easily recovered than others (Figure 2), so to address this, we are in the process of developing optimized versions of our algorithms with an improved ability to recover known antibody epitopes, as well as to uncover novel epitopes. Currently, we are applying the fingerprint design algorithm to generate fingerprints for the putative 63-65/206-211 epitope and will then screen HIV-infected donor sera for neutralization signals that map to this novel epitopes. We have also performed similar antibody-epitope analysis for the dengue E protein (Figure 3) and will be using these data in order to identify putative novel epitopes on this protein. We expect that an initial manuscript on the fingerprint generation technology will be prepared within the next six months, with additional manuscripts related to the application of this technology to actual novel epitope identification planned as part of the next steps for this study.
Mentors
Primary: Ivelin Georgiev
Secondary: James E. Crowe, Jr.
Type of Trainee
Postdoctoral Fellow

©2026 Vanderbilt University ·
Site Development: University Web Communications