Nathaniel Chapman
Mechanism of neutralization of Rift Valley Fever virus by human monoclonal antibodies
Project Abstract
The overall goal of this project is to provide a detailed understanding of the human humoral response to Rift Valley Fever Virus (RVFV) infection and to define the molecular and structural basisofneutralizationbyhumanantibodies. TheBunyavirusfamilyisadiverse–ssRNAbased family that can infect, cause disease and fatality in humans and animals. Specifically, RVFV has been listed as priority pathogens by the WHO and CDC. There are currently no approved vaccines, therapeutics, or prophylactics available to prevent or treat RVFV infection. Although work has been done on an experimental vaccine (MP-12) for RVFV, it has yet to be approved for general human use. Little is known about how the human antibody response is able to neutralize RVFV. To address this gap in knowledge, I have isolated antibodies to RVFV by the hybridoma platform with targeted screening strategies and have begun characterization. We are defining the protective epitopes and their mechanisms of neutralization. Preliminary data indicates that potently neutralizing monoclonal antibodies are able to block protein rearrangements necessary for the fusion process. We plan on further understanding rearrangement blockage in the context of in vivo protection.
Project Update April 2019
Overall, we have isolated a panel of neutralizing human monoclonal antibodies (mAbs) specific to RVFV glycoproteins, Gc and Gn, from naturally infected and vaccinated patients. In addition, we have begun to determine their epitopes and a novel mechanism of neutralization.
Using two assays in concert, we detected the presence of antibodies against RVFV glycoproteins. The first assay, allowed us to detect binding to potentially the heterodimer structure of the Gc/Gn surface of RVFV by transfecting the full-length M segment of the RVFV genome into 293F cells and detecting binding by high-throughput flow cytometry. In the second assay, we adapted a neutralization assay against RVFV MP-12 on vero cells for high-throughput capabilities based on foci formation upon infection.
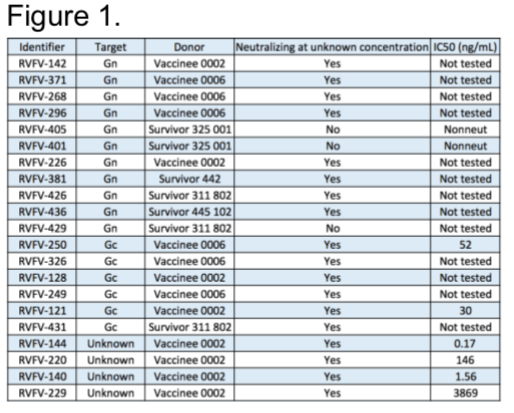
Our lab obtained PBMCs from USAMRIID Special Immunization Program from multiple donors that have been vaccinated with Rift Valley Fever Virus attenuated vaccine strain, MP-12 and PBMCs from wild-type infection survivors from Kenya. Thus far, we have transformed 200 million cells from multiple donors. We have successfully ended our pursuit of mAb from these donors with a final count of 21 mAbs, with 11 targeting Gn, 6 targeting Gc, and 4 targeting an unknown epitope. Furthermore, we are obtaining final inhibitory concertation of mAbs at 50% viral infection reduction relative to controls (IC50) (Figure 1).
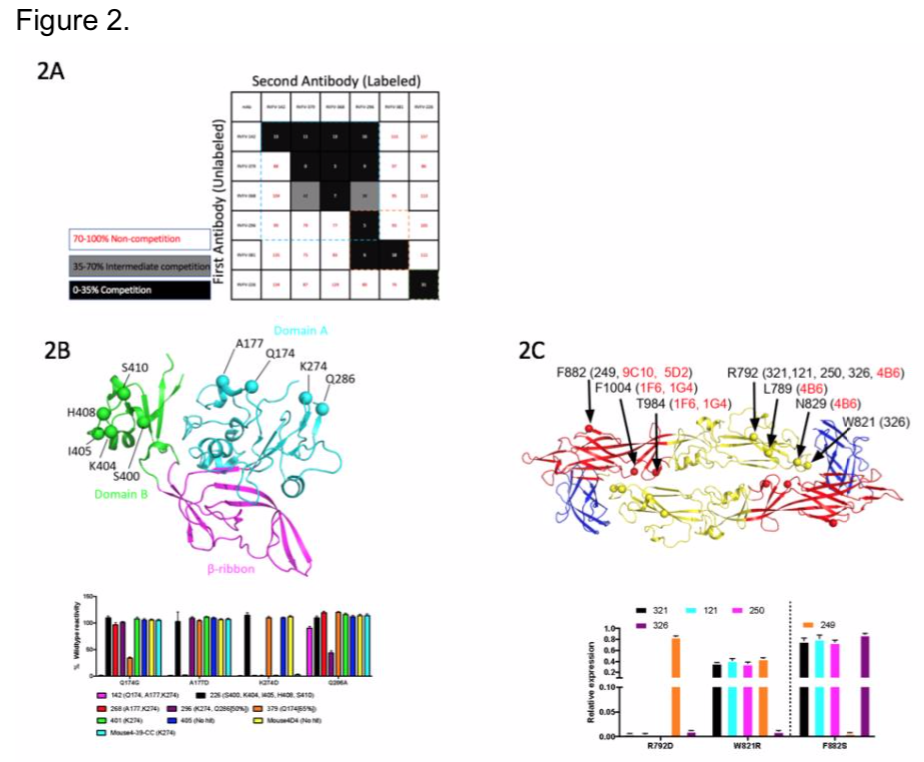
We then wanted to determine epitopes of the mAbs for a subset we have in hand to begin preliminary characterization. We began by competing our panel of Gn-binding mAbs to determine the number of competition groups on the protein. We identified 3 major groups recognized in our panel, indicating there are 3 major regions that correlate with neutralization (Figure 2A). In collaboration with Dr. Fremont and Dr. Haiyan Zhao at Washington University at St. Louis, we performed a loss-of-binding screen using a rationally designed mutant library that replaces surface exposed residues by integrating opposing charges. We observed that the Gn mAbs recognized domain A or domain B (Figure 2B), and Gc mAbs recognize domain I and II (Figure 2C). We also wanted to identify major mechanisms of neutralization using these mAbs. Two major points of the viral life cycle, attachment and fusion, are potential targets of these mAbs.
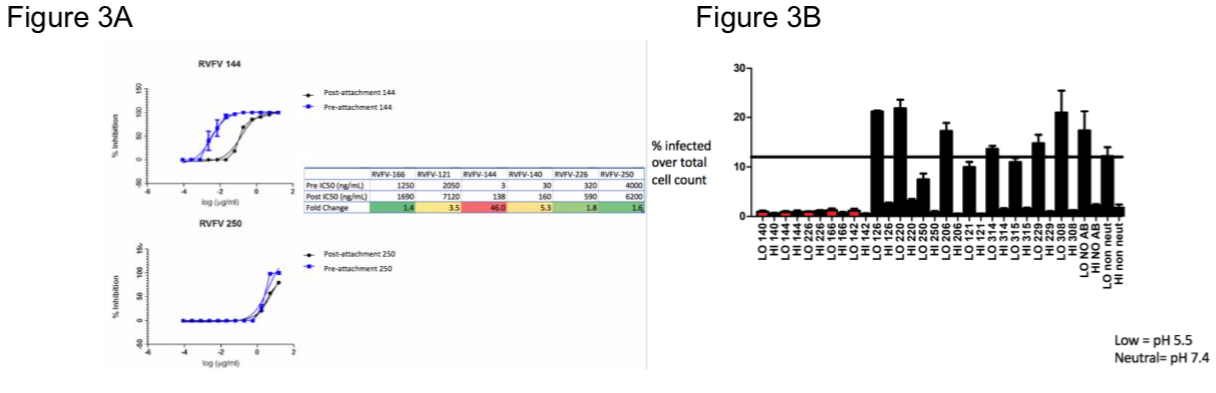
We first began by performing a pre- and post- attachment assay and found that most of the mAbs tested do not neutralize by blocking attachment (Figure 3A). Although RVFV-144 and RVFV-140 may play a role by blocking attachment to the cellular receptor. We also performed a fusion from without assay (FFWO), and saw that the most potent mAbs block the fusion process from occurring, and all antibodies involved at this point either bind the unknown epitope or Gn (Figure 3B).
Given this, I hypothesize that Gn mAbs block fusion, not by blocking the fusion loop hidden in Gc, but by blocking the allosteric alterations of Gn necessary to uncap the Gc fusion loop for it to interact with the endosomal membrane (Figure 4). Also, Figure 4 summarizes the process thus far. More studies to elucidate the mechanism include animal studies, Cyro-EM, epitope mapping by HDX-MS and X-ray crystallography of antibody-antigen complexes.
Applying for F31 in August 2019
Mentors
Primary: James E. Crowe, Jr
Secondary: Jens Meiler
Type of Trainee
Graduate student

©2026 Vanderbilt University ·
Site Development: University Web Communications