Protocol PDF Document (version 1.2): ptr
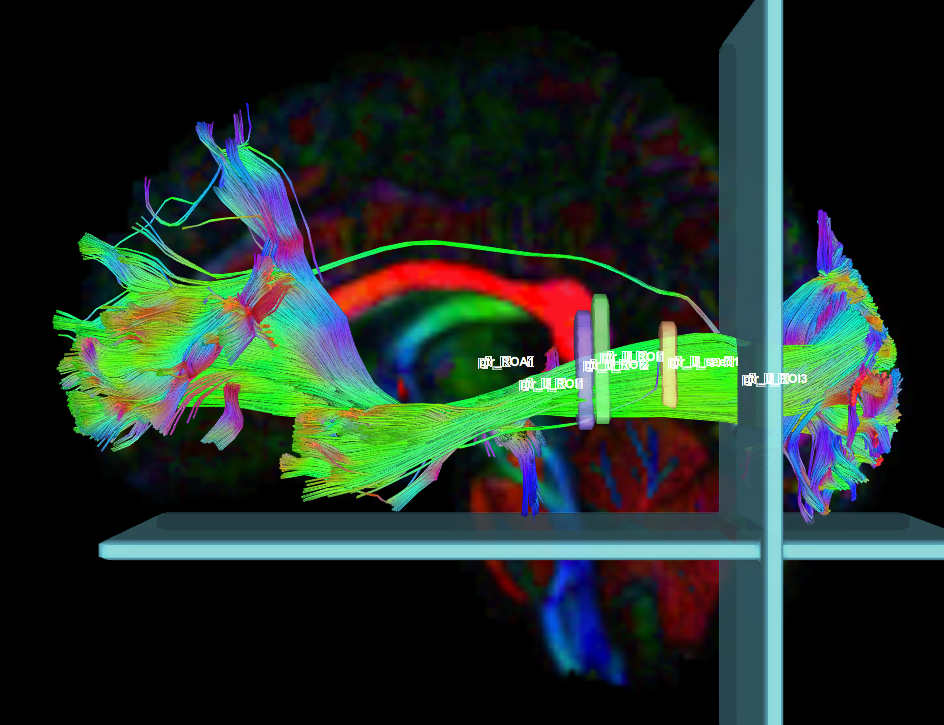
- Create four separate coronal ROI regions (at approx. coronal slice 111 and 118), two for each side, anterior to the seed region. (only 2 of the 4 coronal ROI regions are essential)
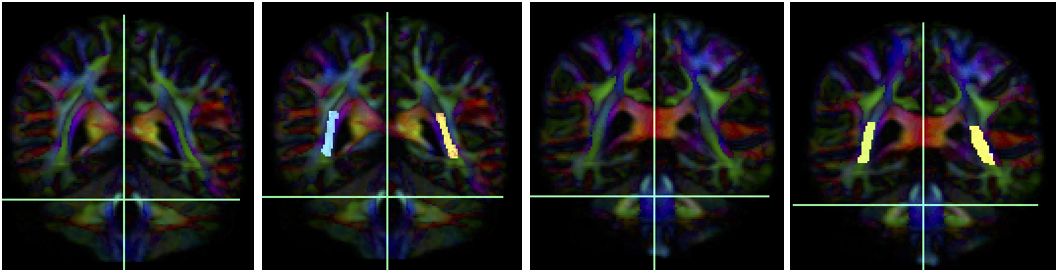
- Create two separate coronal seed regions (at approx. coronal slice 124), one for each side.
- Create two additional separate coronal ROI regions (at approx. coronal slice 140), two for each side, posterior to the seed region.
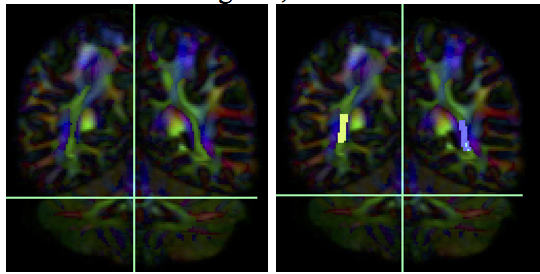
- Create one ROA region and draw a sagittal ROA slice at the midline
- fiber tracking. Based on this output, ROA placement will be clearer.
- Using the same ROA file, draw one more region on the same coronal slice from step #3 exclude the ROI region in your ROA region
- In the region list, check only the left seed/ROI region and ROA regions, then perform fiber tracking. Under the tract list, make sure only the desired left tract is checked and highlighted in purple. Save region, tract, and density files.
- Uncheck the left seed/ROI region and check the right seed/ROI region and ROA region, then perform fiber tracking. Make sure only the desired right tract is checked and highlighted in purple. Save region, tract, and density files.

This is the IFOF.
regions are super ambiguous
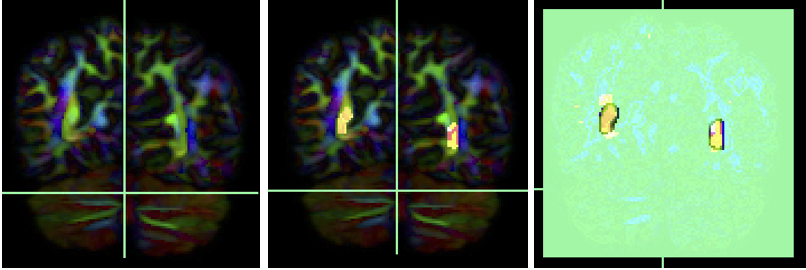
Again the text protocol only calls for ROAs in the saggital midline and on the coronal slice surrounding ROI 3, but the pictures definitely have an axial ROA at the bottom of the brain. Seems like this needs at least 2 axial ROA regions to get rid of extra streamlines that are superior/ inferior to this bundle.
Hallo It’s going to be end of mine day, but before end I am reading this fantastic paragraph to improve my knowledge. thank u
I see You’re really a good webmaster. The website loading pace is incredible.
It kind of feels that you are doing any distinctive trick.
Furthermore, the contents are masterpiece. you have
performed a fantastic task in this matter! Similar
here: dyskont online and
also here: Sklep online
Hi there! Do you know if they make any plugins to assist with
SEO? I’m trying to get my blog to rank for some targeted keywords
but I’m not seeing very good results. If you know of any please share.
Cheers! You can read similar blog here: Ecommerce
Hi! Do you know if they make any plugins to help
with Search Engine Optimization? I’m trying to get
my site to rank for some targeted keywords but I’m not seeing very good
success. If you know of any please share.
Thank you! You can read similar art here: Auto Approve List
Hi! Do you know if they make any plugins to assist with
SEO? I’m trying to get my blog to rank for some targeted keywords but I’m
not seeing very good gains. If you know of any please share.
Cheers! You can read similar art here: Scrapebox AA List
Transfer From Antalya Airport to Side | Transfer From Antalya Airport to Side
150 CC MOPED | https://tteaascar.com/wp-content/uploads/2024/03/150-cc-moped.webp
Antalya Rafting | Unforgettable Adventure on the Köprülü Canyon
White water rafting antalya | An Adventure Filled with Excitement!
Вип Трансфер из аэропорта Анталья | Вип Трансфер из аэропорта Анталья
e ticaret nasıl yapılır | e ticaret nasıl yapılır İstanbul danışmanlık firması
Wow, marvelous weblog format! How long have you ever been running a blog for?
you make blogging look easy. The total look of your web site is great, as
well as the content! You can see similar here sklep internetowy
Wow, fantastic blog structure!
How long have you ever been running a blog for? you make blogging glance easy.
The overall look of your site is wonderful, let alone the content!
I saw similar here prev next and that was
wrote by Jeffery84.
Wow, superb weblog layout!
How long have you been running a blog for? you made blogging look easy.
The full look of your site is great, as smartly as the content!
You can read similar here prev
next and it’s was wrote
by Drew62.
10 Sites To Help You To Become A Proficient In UK Onlyfans Pornstars site
What’s The Job Market For Most Famous Pornstars Professionals Like?
most famous Pornstars
Search Engine Optimization Seo Tools To Ease Your Daily Life Search
Engine Optimization Seo Trick That Every Person Should Be Able To Engine optimization Seo
10 Glass Repair Leeds Tricks Experts Recommend door repair leeds; https://Wifidb.science/Wiki/A_Rewind_The_Conversations_People_Had_About_Door_Fitters_Leeds_20_Years_Ago,
Where Can You Find The Most Effective Text Rewritter Information? ai text rewriter
What’s The Good And Bad About Double Glazing Installers Near Me repair double Glazing
Learn More About Online Shopping Website In London When You Work From Your Home crew Cab interior Mats
10 Life Lessons We Can Take From Car Locksmith service
10 Mobile Apps That Are The Best For Double Glazing Repairs Leeds Upvc Door repair meanwood
Five Killer Quora Answers To Shopping Online Sites List Shopping Online Sites List; Deli.Bz,
You’ll Never Guess This Ford Key Replacement Near Me’s Benefits ford key replacement
Are The Advances In Technology Making Replacement Double Glazing Units Near Me
Better Or Worse? Replacement Double Glazing Units
Here’s A Few Facts About Mesothelioma Legal Question mesothelioma law (Arnette)
20 Things That Only The Most Devoted Audi Car Key Replacement Cost Fans Understand programmer
You’ll Never Be Able To Figure Out This Espresso Machine Coffee’s Tricks Espresso machine coffee
CS GO Cases Opening Isn’t As Difficult As You Think
weapon Case
What NOT To Do Within The Window.Replacement Near Me Industry window Replacement near me
Ask Me Anything: 10 Answers To Your Questions About Double Glazing Showrooms Near Me upvc double
glazed windows (Lorenzo)
Nine Things That Your Parent Taught You About Double Glazing Repair Cost Double Glazing (http://Mspeech.Kr/)
How To Outsmart Your Boss On Auto Key Locksmith Near Me automotive locksmith Key fob
See What Ford Ka Replacement Key Fob Tricks The Celebs Are Making
Use Of ford ka replacement key [Lane]
The Best Commercial Truck Accident Lawyer The Gurus Have Been Doing 3 Things delano truck Accident attorney
9 Lessons Your Parents Taught You About Automotive Lock Smith automotive lock smith (chappell-wichmann-3.Technetbloggers.de)
What Is Vehicle Diagnostics And How To Utilize What Is Vehicle Diagnostics And How
To Use mobile engine diagnostics near me
What Double Glazing Offers Near Me Is Your Next Big
Obsession double glassing
20 Quotes That Will Help You Understand Autowatch Ghost Immobiliser ghost immobiliser 2
Why Wall Mounted Fireplaces Could Be More
Dangerous Than You Realized electric fireplace For the wall
5 Futon For Sale Near Me Lessons From The Professionals guest room
It Is A Fact That Avon Welcome Kit Is The
Best Thing You Can Get. Avon Welcome Kit avon Welcome Kit
10 Tommy Hilfiger Bag Strategies All The Experts Recommend Tommy
hilfiger bags crossbody, https://yogaasanas.science/Wiki/tommy_Hilfiger_purse_bag_tips_From_the_top_in_the_business,
10 Healthy Habits For Diagnostic Check Near Me diagnostic scanner
This Week’s Best Stories Concerning Lost Car Key Motor
This Is What 18 Wheeler Accident Law Firm Will Look In 10
Years 18 wheeler accident Lawyer
The Reasons To Work With This Ford Key Replacement Near Me ford key Programming (Adler-timm-2.hubstack.net)
See What Best Onlyfans Pornstar Tricks The Celebs Are Making
Use Of best Onlyfans pornstar
What’s The Current Job Market For Replacement Window Glass Near Me Professionals Like?
replacement window glass
Five Killer Quora Answers On Double Glazing Repair Double Glazing
History Of Veterans Disability Attorneys: The History Of Veterans Disability Attorneys North wildwood veterans disability law firm
15 Trends To Watch In The New Year Double Mattress Size double mattresses for sale uk
See What Car Key Cut And Programed Tricks The Celebs Are
Utilizing car Key cut and programed
The Best Folding Treadmill UK Tricks For Changing Your Life
best folding Treadmill uk
10 Tips For Michael Kors Handbag That Are Unexpected michael kors bags uk Sale (buketik39.ru)
10 Things That Your Family Teach You About Replace Window Handles replace window handles (http://pandahouse.Lolipop.jp/)
See What Replacement Sash Windows Tricks The Celebs Are Utilizing Replacement Sash Windows
How To Design And Create Successful Window Companies
Leeds Tutorials From Home Upvc door repairs roundhay
15 Trends To Watch In The New Year Lock
Smith For Car Local Locksmiths For Cars
The Most Hilarious Complaints We’ve Seen About Nissan Qashqai Replacement Key Nissan Car Key – https://Doodleordie.Com/Profile/Flavorspark18 –
You Will Meet Your Fellow Upvc Windows Repairs Enthusiasts.
Steve Jobs Of The Upvc Windows Repairs Industry window Repair
20 Things Only The Most Devoted Diagnostics
Near Me Fans Understand Vauxhall diagnostics
10 Facts About Window Repair That Will Instantly Put You In Good Mood seals
The Most Effective Reasons For People To Succeed At The Fiat 500 Keys Industry Fiat panda Key
20 Fun Facts About Motor Vehicle Law flat rock motor vehicle accident attorney
This Is A Railroad Injuries Litigation Success Story You’ll Never Believe Railroad Crossing Accident Lawyer
What’s The Job Market For Window Repairs Leeds
Professionals Like? Window Repairs Leeds (https://Soto-Fuglsang.Federatedjournals.Com/The-Most-Underrated-Companies-To-In-The-Double-Glazing-Repairs-Leeds-Industry/)
9 Lessons Your Parents Taught You About Audi Keys Replacement
audi Keys Replacement
Are You In Search Of Inspiration? Look Up Semi Truck Litigation Semi Truck Accidents
How Amazon Under Desk Treadmill Became The Hottest Trend
Of 2023 Treadmill Under desk
10 Misconceptions That Your Boss May Have About How Many Cases Are There In CSGO How Many Cases Are
There In CSGO cs2 Case
Best Newcomer Pornstar Tools To Streamline Your Everyday Lifethe Only
Best Newcomer Pornstar Trick Every Person Should Be Able To best newcomer pornstar
The People Nearest To Ford Car Keys Tell You Some Big Secrets nearest
See What Replacement Key Bmw Tricks The Celebs Are Using
replacement key bmw
17 Signs You Work With Good Accident Attorney fresno Accident attorney
Handbags For Women Tommy Hilfiger Tools To Improve Your Daily Lifethe One Handbags
For Women Tommy Hilfiger Trick That Every Person Must Know Handbags For Women Tommy Hilfiger
Five Killer Quora Answers On Repair Misted Double Glazing
Near Me double glazing near me (https://valetinowiki.racing/wiki/10_Tips_For_Window_Repairs_Croydon_That_Are_Unexpected)
24 Hours To Improve Birth Injury Lawyer mesquite birth injury law firm,
https://vimeo.com,
Can Electric Mobility Scooters For Sale Ever Rule The World?
Motorized Scooters
Athens Birth Injury Attorney 101: Your Ultimate Guide For Beginners birth Injury Attorney Alamogordo nm
How To Save Money On Bentley Continental Key Programming Bentley Keys
This Week’s Most Remarkable Stories Concerning Locksmith
Cars Locksmith car Keys
What’s The Current Job Market For Designer Handbags Green Professionals?
designer handbags green
Its History Of Online Shopping Clothes Uk Cheap Vimeo
Seven Explanations On Why Greenpower Electric Scooters Is Important Electric Power Scooters
The No. One Question That Everyone In Audi A3 Key Battery Must Know How To Answer Spare Audi Key
How To Know If You’re In The Right Position For Car Locksmith damaged
The majority of asbestos victims, and their families, begin the process by hiring a professional lawyer.
A lawyer with experience will help them prepare the lawsuit to claim compensation from
the defendants who made asbestos-containing products.
my webpage Mesothelioma Attorney
The Next Big Event In The Home Espresso Machine Industry espresso machines for home
7 Simple Tricks To Totally Enjoying Your Repairs To Upvc Windows
upvc window Repair
You’ll Never Be Able To Figure Out This Self Emptying Robot Vacuum Mop’s Benefits self Emptying robot vacuum mop
10 Things You Learned In Kindergarden That Will Help You With Fiat 500 Replacement Key Fob fiat Ducato key fob replacement
How Second Hand 3 Wheel Scooter Was The Most Talked About Trend Of 2023 3 wheeled mobility scooters
Five Porsche Key Fob Projects To Use For Any Budget Porsche 987 Key
The Reason Why Replacement Key For Car Has Become Everyone’s Obsession In 2023
replacement car keys with chips
20 Up-And-Comers To Watch In The Demo Slot Industry demo Slot 5lion
Why You Should Concentrate On Making Improvements
To Asbestos Compensation mesothelioma Litigation
The 12 Best Memory Foam Double Mattresses Accounts
To Follow On Twitter bed double mattress (Bailey)
Five Killer Quora Answers On Dangerous Drugs Law Firm firm
Why CS GO Case New Is Harder Than You Think winter Offensive weapon case (maps.Google.Ml)
What’s The Current Job Market For Designer Bags Professionals Like?
designer Bags
The No. 1 Question That Anyone Working In Link Login Gotogel Must
Know How To Answer Go Togel
10 Car Keys Repairs Hacks All Experts Recommend door key repair near me (Hallie)
What Case Opening Simulator CSGO Experts Want You To Know case chroma (Chana)
30 Inspirational Quotes About Walking Desk Pad walking pad With desk
15 Funny People Working Secretly In Small Double Mattresses
Double mattress width
20 Myths About Cerebral Palsy Compensation: Dispelled mathis cerebral palsy lawsuit
What A Weekly Truck Accident Lawyer Project Can Change Your
Life sunnyvale truck accident lawsuit (vimeo.com)
Guide To English Pornstars: The Intermediate Guide To English Pornstars english pornstars (https://golden-seal-h8gfkd.mystrikingly.com/blog/it-s-the-good-and-bad-about-female-pornstars)
11 Ways To Totally Defy Your Idn Poker tolol
What’s The Job Market For Keys For Bmw Professionals? keys for Bmw
Why Is Three Wheel Mobility Scooters Sale So Popular? local mobility scooters for sale
10 Misconceptions That Your Boss May Have
About Double Glazing Firms Near Me Double Glazing Firms Near
Me companies
Can Akun Demo Slot Ever Rule The World? slot demo anti rungkad (Ross)
20 Trailblazers Setting The Standard In Upvc Window Repairs Upvc Window Repairs Near Me
10 Things Everyone Hates About Best Accident Attorney
accident attorney macon
Treadmill For Home Tools To Help You Manage Your Daily Life Treadmill For
Home Trick That Should Be Used By Everyone
Be Able To treadmill for home
10 Things That Your Competitors Lean You On Upvc Doors Leeds repairs to double Glazed windows
20 Trailblazers Lead The Way In Mesothelioma Compensation Alliance Mesothelioma Law Firm
10 Things You Learned In Kindergarden That Will Help You Get Online
Shopping Uk Groceries 4K hdr switch Box
11 “Faux Pas” That Are Actually OK To Make With
Your Car Open Service locked out of car service
Is Railroad Injuries Case The Best There Ever Was?
Railroad Accident Compensation Lawyer
10 Misleading Answers To Common Locksmith For Cars
Questions: Do You Know The Correct Ones? Locksmith cars near me
10 Facts About Mesothelioma Claim That Insists On Putting You In Good Mood hastings mesothelioma Lawyer
8 Tips To Enhance Your Central Locking Key Repair Game Lock Replacement Near Me
The Ultimate Guide To Upvc Window Repairs window repairs near me (Melinda)
Find Out What Repair Window Tricks Celebs Are Using
window replacement near me – http://daywell.kr/ –
5 Killer Quora Answers To Accident Injury Attorneys Near Me accident attorney manhattan
See What Dreametech L30 Ultra Tricks The Celebs Are Utilizing dreametech l30 (Jordan)
The Reasons Fridge Isn’t As Easy As You Imagine american style fridge
(Swen)
You’ll Never Be Able To Figure Out This Online Shopping Uk Amazon’s Secrets online Shopping uk amazon
What Is The Reason? Washer Dryer Heat Pump Is Fast Becoming
The Trendiest Thing Of 2023 laundry
How Much Can Auto Locksmith Service Experts Make? automobile Locksmith Services
3 Reasons You’re Not Getting Idn Play Isn’t Working (And Solutions To Resolve It) tolol
Guide To American Fridges Freezers: The Intermediate Guide To American Fridges Freezers American Fridges
Guide To Designer Handbags Uk: The Intermediate
Guide Towards Designer Handbags Uk Designer Handbags Uk
You’ll Never Be Able To Figure Out This Online Shopping Uk Sites’s Tricks Vimeo.com
Seven Explanations On Why Locksmith For Cars Is Important Local
7 Simple Tips To Totally Enjoying Your Citroen Berlingo Van Key
Replacement key cutting service
What Do You Know About Car Key Auto Locksmith?
Locksmith Car key
Watch Out: How Slot Symbols Is Taking Over And What We Can Do About It slot strategy
9 . What Your Parents Teach You About Robotic Vacuum Self Emptying Robotic vacuum self emptying
10 Things Everybody Has To Say About Repairs To Upvc Windows Repairs To Upvc Windows
upvc window Repair
How Much Can Window Glass Repair Near Me Experts Make?
Window Repairs
Guide To Bmw Key 1 Series: The Intermediate Guide To Bmw Key 1 Series Bmw key 1 series
10 Reasons That People Are Hateful Of Motorcycle Accident Attorneys london motorcycle accident attorney
10 Things That Your Family Taught You About Window Handles Replacement Handles
Ten Things You’ve Learned In Kindergarden That Will Help You Get Upvc Window Repairs window repairs near Me
The Top Double Glazed Door Repairs Near Me That Gurus Use Three Things replacing double glazed Windows (ip-iv.ru)
How Boating Accident Lawyers Near Me Was The Most Talked About Trend Of
2023 Vessel crash attorney
How To Explain Walking Pad For Desk To A Five-Year-Old UREVO Compact Under Desk Treadmill – Double Shock Absorption
Who’s The Most Renowned Expert On Double Glazing
Windows Near Me? window
New Port Richey Birth Injury Attorney Tools To Help You Manage
Your Everyday Lifethe Only New Port Richey Birth Injury Attorney Trick That Should Be Used By Everyone Learn new port richey birth injury attorney (cameradb.review)
Small Treadmill For Desk Tools To Help You Manage Your Daily Life Under Desk treadmill
The 10 Most Scariest Things About Dangerous Drugs Law Firms dangerous Drugs law firms
11 Creative Methods To Write About Best Accident Attorney
accident injury legal representation
Are You Confident About Sexdolls Usa? Try This Quiz Popular sex dolls
5 Lessons You Can Learn From Slot Demo Gratis demo 5 lions
– Clay,
You’ll Never Guess This Beko Washing Machines Uk’s Tricks beko washing machines uk
Can Car Keys Repair Ever Be The King Of The World?
car Keys Repair near me (http://Www.Mazafakas.com)
Guide To Situs Gotogel Terpercaya: The Intermediate Guide For Situs Gotogel Terpercaya
situs Gotogel terpercaya
The Advanced Guide To Birth Injury Attorneys berkeley Birth Injury Lawyer
20 Best Tweets Of All Time About Kids Bunk Bed Kids Bedroom Furniture
The 10 Most Scariest Things About Online Shopping Stores List vimeo.com
What’s The Current Job Market For Online Sites For Shopping In Uk Professionals Like?
online sites For Shopping in uk
Treadmills Near Me Tips From The Top In The Industry Compact Treadmill
The Biggest Issue With Mesothelioma, And How You Can Repair It Pleasant View Mesothelioma Law Firm
You’ll Never Be Able To Figure Out This Mesothelioma
Compensation After Death’s Secrets mesothelioma compensation after death (https://buketik39.ru)
Ten Startups That Are Set To Change The Asbestosis Asbestos Mesothelioma Attorney Industry For The Better san jose ca mesothelioma attorney,
https://heatbrazil9.werite.net,
20 Accident Attorney No Injury Websites Taking The Internet By
Storm accident attorney Boise Idaho
You’ll Be Unable To Guess Butt Plug Uk’s Tricks
butt plug uk (Autumn)
A Look At The Future What’s The Avon Shop Industry
Look Like In 10 Years? online avon
5 People You Should Be Getting To Know In The Accident Attorneys
Industry accidentinjurylawyers
10 Reasons Why People Hate Heat Pump Tumble Dryers Heat Pump Tumble Dryers are heat pump tumble dryers any good
What Is Glazing Repair Near Me And How To Utilize
What Is Glazing Repair Near Me And How To Use glaze
14 Questions You Might Be Insecure To Ask About Asbestos Attorney Mesothelioma attorney illinois mesothelioma
(http://www.stes.tyc.edu.tw)
The Reasons Why Adding A Robot Vacuums Self Emptying To Your Life Will Make All The Difference Best self-emptying robot Vacuum
Watch Out: How Atlanta Birth Injury Attorneys Is Taking Over And What You Can Do About It birth injury medical malpractice attorney (Brittney)
15 Great Documentaries About Replacement Fiat Key fiat ducato replacement key
5 Killer Quora Answers On Double Glazing Repairs Leeds Glazing repairs leeds
This Is The New Big Thing In Double Over Double Bunk Bed double top bunk bed; Jimmy,
The 10 Most Terrifying Things About Treadmills For Sale Near Me treadmills for Sale near me
The Most Worst Nightmare About Double Glazing Fitters Near Me Be Realized Double Door
It’s The One L30 Dreame Trick Every Person Should
Be Able To l30 Ultra
15 Of The Most Popular Pinterest Boards Of All Time About Loveseat For Sale Outdoor Sofa Sale
A Provocative Rant About Coffee Machine Sage compact Coffee Machines
10 No-Fuss Methods To Figuring Out Your Which Is Best For Online Grocery Shopping
Jolie papier online shop uk amazon (m.cosplayfu.it)
Ten Apps To Help Control Your Designer Handbags Brown Designer handbags chanel
20 Glass Repair Leeds Websites Taking The Internet By Storm upvc door repairs Leeds
Why Online Work Earn Money Is Relevant 2023 at home jobs for moms (web018.dmonster.kr)
10 Startups That Will Change The Flexible Work From Home Jobs Industry For The Better Home working
9 Lessons Your Parents Taught You About Auto Lock Smiths Auto lock smiths
What Is Repair Upvc Windows And How To Utilize It?
Repairing Upvc Windows
The Best Advice You Could Receive About Upvc Windows Near Me
upvc window repair
See What Treadmills For Sale UK Tricks The Celebs
Are Using Treadmills for sale uk – https://Www.cheaperseeker.com –
You’ll Never Guess This Top Pornstar Only Fans’s
Secrets Top Pornstar Only Fans
You’ll Be Unable To Guess Bmw Spare Key Cost’s Benefits Bmw spare key
How To Solve Issues With Mesothelioma mesothelioma case
9 . What Your Parents Taught You About Key Cutting Car
key cutting car
The Top Slot Demo Gratis Gurus Are Doing 3 Things Slot demo gratis 2023
Glass Repair Leeds 101: Your Ultimate Guide For Beginners window doctor Leeds
The 10 Most Terrifying Things About Replacement Handles For Upvc Windows replacement handles For Upvc
windows (125.141.133.9)
American Fridge Freezer Tips From The Top In The Industry Fridge American
What Experts In The Field Want You To Learn Vimeo.com
What Is Truck Accident Attorneys Near Me’ History?
History Of Truck Accident Attorneys Near Me Truck Accidents
The 10 Most Terrifying Things About Akun Demo Slot demo pragmatic olympus – http://Www.Razumei.ru –
How Marc Jacobs Handbag Has Become The Most Sought-After Trend Of 2023 marc jacobs handbag on sale; Allison,
The 10 Scariest Things About Double Glazing Companies Near Me double glazing companies near me
Five Killer Quora Answers To Mesothelioma Attorneys Mesothelioma Law Firms
You’ll Never Guess This Need Spare Car Key’s Tricks Need Spare Car Key
What’s The Job Market For Double Glazing Door Repairs
Near Me Professionals? door repair, Fran,
10 Things We All We Hate About Railroad Injuries Legal Danville railroad Crossing Accident attorney
You’ll Be Unable To Guess Attorney Injury Accident Attorney’s Tricks attorney injury
accident attorney (Gustavo)
20 Myths About Repair Double Glazing: Debunked double glazing repairs
The Biggest Issue With Railroad Injuries Claim And How To Fix It Railroad injury lawyer ottawa
Why You’ll Definitely Want To Find Out More About Window Repair Near double Glazed window Repairs near me
Although ypsilanti asbestos law Firm (vimeo.com) remains banned, several incremental legislative proposals have been tossed around Congress.
One of them proposed legislations, the Frank R.
10 Tell-Tale Signals You Need To Find A New Upvc Window Repairs upvc Repairs
How Car Central Locking Repair Has Become The Most Sought-After Trend In 2023 lock and key Repair near me
Ask Me Anything: 10 Answers To Your Questions About Window
Repairs Leeds double glazed doors repairs (http://www.google.co.ao)
Are You Responsible For An Mesothelioma Legal Question Budget?
12 Tips On How To Spend Your Money mesothelioma Compensation Lawyer
The 9 Things Your Parents Teach You About Dreame L30 Review Dreame l30 review
20 Misconceptions About Railroad Injuries Litigation: Busted
troy Railroad injuries lawyer
20 Fun Details About American Fridges Freezers Experience Perfection in Food Preservation – Haier 521L Fridge (Nan)
See What Washer Dryer Heat Pump Tricks The Celebs Are Using washer Dryer heat pump
Everything You Need To Learn About Demo Slot slot demo 1000x
The People Nearest To Double Glazing Repair Luton Share Some Big Secrets Near
Five Essential Qualities Customers Are Searching For In Every Coffee Machine Espresso
machines espresso machines
15 Best 10kg Washing Machines – All Offers Bloggers You Should
Follow Best 10Kg washing machine
The Masturbators Mistake That Every Beginner Makes Masturbater men
There Are Myths And Facts Behind Double Glazed Window
Leeds upvc window Repair
What’s The Job Market For Double Glazed Window Repairs Professionals
Like? Window Repair
Don’t Be Enticed By These “Trends” About Double Memory Foam Mattress Mattress Double Bed
(Pondknot25.Bravejournal.Net)
You’ll Never Guess This Car Replacement Keys’s Secrets Car replacement keys
A Rewind How People Discussed Auto Accident Law 20
Years Ago Vehicle
How Online Home Based Work Was Able To Become The No.1 Trend On Social Media home based work (Minecraftcommand.science)
Say “Yes” To These 5 Double Bunk Beds For Sale Tips double bunk
bed with trundle (Molly)
Your Family Will Thank You For Getting This Car Diagnostics Near Me Uk Diagnostic check Engine
5 Killer Quora Answers To How To Buy Clothes Online From Uk how
to buy clothes online from uk (Bryan)
The Most Hilarious Complaints We’ve Seen About Spare Keys Cut Spare key car
Unexpected Business Strategies For Business That Aided
Demo Slot To Succeed slot demo mahjong ways 2 (https://thecoxteam.com/?URL=https://valetinowiki.racing/wiki/What_Is_Slot_Demo_Gratis_And_How_To_Use_What_Is_Slot_Demo_Gratis_And_How_To_Use)
Seven Explanations On Why Slot Tours Is Important slot tricks (http://Www.Cheaperseeker.com)
10 Things You Learned In Preschool That’ll Help You With
Tommy Hilfiger Female Bags tommy hilfiger usa bag
15 Pinterest Boards That Are The Best Of All Time About Replacement Double Glazed Window Replacement double glazed Windows
Nine Things That Your Parent Taught You About Medical Malpractice Lawsuit Medical Malpractice
Locksmith For Car Near Me’s History History Of Locksmith For Car Near Me car key Mobile locksmith
A Step-By’-Step Guide To Picking Your Machines Espresso cappuccino
You’ll Never Guess This Citroen C1 Key Replacement’s Secrets citroen c1 key, https://olderworkers.com.au,
See What Mesothelioma Lawyer Tricks The Celebs Are Using Mesothelioma Lawyer
7 Simple Secrets To Completely Rocking Your Slot
Demo Demo 5 Lion
Guide To Atlanta Birth Injury Attorneys: The Intermediate
Guide Towards Atlanta Birth Injury Attorneys atlanta birth injury attorneys (Myrna)
15 Gifts For The Slot Demo Pragmatic Lover In Your Life Slot Demo Anti Rungkad 777
5 Reasons To Be An Online Double Glazing Doors Near Me Buyer And 5 Reasons Why You Shouldn’t
double glazed door
It’s True That The Most Common Car Accident Attorney Debate Could Be As Black
And White As You May Think marine City Car accident attorney
See What Remote Control Anal Plug Tricks The Celebs Are Utilizing Remote control anal plug
The Step-By -Step Guide To Choosing The Right Auto Accident Settlement front Royal auto accident attorney
You’ll Never Be Able To Figure Out This Treadmill Sale UK’s Tricks treadmill Sale uk
The Reasons Double Glazing Door Repairs Near Me Is Fast Becoming The Hottest Trend For 2023 double glazed window (Sandy)
10 Things You’ve Learned About Preschool That’ll Help You With Upvc Windows Repair upvc Window
repairs (Image.google.com.sa)
Why Glimmerstick Eye Liner Will Be Your Next Big Obsession Eyeliner Glimmerstick Avon
This Is The History Of Car Locksmith In 10
Milestones keys
Fridges Isn’t As Difficult As You Think fridges for sale
7 Little Changes That’ll Make The Biggest Difference In Your Designer Bags designer Handbags On amazon
What’s The Current Job Market For Window Handle Replacement Professionals Like?
window Handle replacement
The Most Worst Nightmare Concerning Treadmill For Home Use Come To Life home Fitness
What Is The Best Place To Research Volkswagen Key Replacement Cost Online volkswagen key Fob replacement near me
The Main Issue With 4 Wheel Electric Scooter For Adults And How To Fix It Elderly mobility
Where Can You Get The Best Shopping Online Uk Information? Ergonomic Colouring Pencils Stabilo
11 Ways To Completely Sabotage Your Kids Beds Bunk low bunk beds for kids
How To Create An Awesome Instagram Video About Car Accident Attorney hampton car accident attorney
You’ll Never Guess This Window Repair Near Me’s Tricks window Repair near Me
You’ll Be Unable To Guess 10kg Top Loader Washing Machine’s
Benefits 10Kg Top Loader Washing Machine
The 12 Types Of Twitter Key Programming Car People
You Follow On Twitter cut and Program car key
10 Misconceptions Your Boss Has Concerning Unlock
Car Service Locked self out of Car
15 Things You Don’t Know About Mental Health Practitioners how can i Get a mental health assessment
See What Repair Upvc Windows Tricks The Celebs Are Using repair Upvc Windows
One Of The Most Untrue Advices We’ve Ever Received On Double Glazing Repair Near Me Replacing Double Glazing Glass
8 Tips To Boost Your Idn Poker Game spam
The 10 Most Terrifying Things About Lost Car Key lost key to car no spare
(Felica)
The 10 Most Scariest Things About Black Friday Robot Vacuum Deals black Friday robot Vacuum deals
15 Gifts For The Accident Attorneys Near
Me Lover In Your Life texas accident attorneys
12 Companies Are Leading The Way In Upvc Window Repairs upvc window repairs near me
Are You Responsible For A Auto Accident Lawyer Budget?
12 Top Ways To Spend Your Money kansas city auto accident lawyer
You’ll Never Guess This Double Glaze Repair Near Me’s Benefits Double glaze repair near me
Double Glazed Window Repairs: The Good, The Bad,
And The Ugly Double glazing repairs
15 Reasons Not To Be Ignoring Samsung Side By Side Fridge
Freezer small side by side refrigerator with Ice maker
You’ll Never Guess This Which Online Stores Ship Internationally’s Tricks
Which Online Stores Ship Internationally
Does Technology Make Fiat 500 Replacement Key Cost Better Or Worse?
fiat 500 spare key cut
9 Things Your Parents Taught You About Mesothelioma Compensation mesothelioma
Ten Startups That Will Revolutionize The Bmw Replace Key Industry For The Better replacement keys for bmw (https://telegra.Ph/)
So , You’ve Purchased Real Sexdolls … Now What? silicone Sex doll
See What Amazon Uk Online Shopping Clothes Tricks The
Celebs Are Utilizing amazon uk online shopping clothes (https://the-challenger.Ru/)
The Most Hilarious Complaints We’ve Received About Cost
Of Replacement Car Key replacement car key costs
5 Killer Quora Answers On What Cases Are Being Dropped CSGO Case gamma
You’ll Be Unable To Guess Upvc Window Repairs Near Me’s Benefits
upvc window repairs (Sima)
See What Bmw Car Key Cover Tricks The Celebs Are
Utilizing Bmw Car Key Cover
What’s The Current Job Market For Replacement Window Glass Near Me Professionals Like?
Replacement Window Glass Near Me
So , You’ve Purchased Selling Perfume From Home
… Now What? Where To
8 Tips For Boosting Your Work From Home Jobs No Experience
Uk Game work from home jobs no experience uk (Indira)
10 Quick Tips For Upvc Window Repairs window repairs near me – https://soraike123.hatenablog.com/iframe/hatena_bookmark_comment?canonical_uri=https://dnpaint.co.kr/bbs/board.php?bo_table=b31&wr_id=3986850
–
Twenty Myths About Website Search Engine Optimization: Busted search engine optimisation agency london, Clarissa,
Learn About Search Engine Optimisation Services While Working From
Your Home Seo services near Me
The Three Greatest Moments In Windows Repair History
window Repair
5 Laws That Will Help The Drip Filter Coffee Industry filter drip coffee maker (https://coursebuilder.thimpress.com/account/?redirect_to=http://lineyka.org/user/avenuehockey26/)
How Prada Handbags Became The Hottest Trend In 2023 cheap
The Best 12kg Washing Machines For Sale Tricks For Changing Your Life Best 12kg Washing machines
Why Nobody Cares About Upvc Doors Leeds french Door repair leeds
10 Joker123 Gaming Techniques All Experts Recommend spam
How To Know If You’re Ready To Go After Window Glass Repair
Near Me Window Repairs
You’ll Never Guess This Trusted Online Shopping Sites For Clothes’s Tricks trusted online shopping Sites for Clothes
Five Laws That Will Aid Industry Leaders In Repair Upvc Window Industry window replacement Near me
20 Things You Need To Be Educated About Double Glazing
Repairs Near Me double glazing Near Me
17 Signs You’re Working With Railroad Injuries Attorneys Train injury lawsuit
Nine Things That Your Parent Taught You About Hiring Truck Accident Attorneys Truck Accident attorneys
15 Reasons Why You Shouldn’t Ignore Window Repairman services
The Ultimate Cheat Sheet On Windows Leeds replacing
What’s The Job Market For Double Glazed Window Repairs Professionals Like?
Double glazed window repairs
Why You Should Concentrate On Enhancing Accident accident lawsuit
The 10 Most Terrifying Things About Treadmills treadmills For Home
Citroen Replacement Key Tips That Will Change
Your Life Citroen C1 key Replacement price
11 Ways To Completely Sabotage Your Slot Demo pragmaticplay Com demo
(b.cari.com.My)
A Peek Inside The Secrets Of Accident Attorney No
Injury Accident Attorneys In Colorado Springs
How To Outsmart Your Boss On Mesothelioma Compensation oakdale mesothelioma lawyer
This Is A Guide To Ghost Immobiliser In 2023 Ghost 2 Immobiliser Fitting Near Me
This Week’s Top Stories About Double Glazing Deals
Near Me Double Glazing Deals Near Me doubled Glazed Windows
The 10 Scariest Things About Locksmith For Car
Near Me car
Five Things You’re Not Sure About About Jobs Work From Home
work from home jobs Part time
5 Uk Online Shopping Sites For Mobile Projects For Any Budget Soft Cream Runner Rug (https://Vimeo.Com/931588454)
See What Where To Buy Electronics Online Tricks The Celebs
Are Making Use Of where to buy electronics online
5 Laws Everybody In Salt Lake City Accident Lawyers Should Know Houston accident Attorney
Are You Responsible For A Windows Leeds Budget?
10 Terrible Ways To Spend Your Money window doctor leeds (Rocky)
This Is The Ultimate Guide To Online Shop Kawasaki Ninja 1000
What Is The Reason Double Glazing Repairs Near Me Is Right For You?
double Glazing
You’ll Never Guess This Asbestos Lawyer’s Tricks Asbestos law
The No. Question That Everyone In Car Accident Lawyer For Hire Should Be Able
Answer accident car attorney (wiley-mcgee-2.technetbloggers.de)
What’s The Job Market For Top Rated Robot Vacuum Professionals?
top rated robot vacuum (Shawna)
15 Reasons You Must Love Squirting Dildos Toy To Make Her Squirt
Could Narwal Robot Vacuum Be The Key To Achieving 2023?
cheap Robot vacuum cleaner
How To Resolve Issues With Locksmith Near Me For Cars Key Locksmith For Cars
Uk Online Shopping Sites For Electronics Explained In Fewer Than 140 Characters Air-Powered Stapler
Five Killer Quora Answers On Erb’s Palsy Legal superior erb’s palsy attorney
It’s Time To Forget Robot Vacuum Best: 10 Reasons Why You Do Not Need
It Robot vacuum and mop self Empty (minecraftcommand.science)
12 Companies Are Leading The Way In CS GO Open Case Website Cases (Bradova.Blog.Idnes.Cz)
The 10 Most Scariest Things About Asbestos Lawsuit Asbestos
9 Things Your Parents Taught You About Bagless Self Emptying Robot Vacuum Bagless Self Emptying Robot Vacuum [http://Lineyka.Org/User/Celerybag81]
3 Common Reasons Why Your Mini Cooper Key Fob Replacement Isn’t Performing (And How To Fix It) Mini cooper keys
This Is The History Of How To Buy Clothes Online From Uk Jeep Wrangler Blower Resistor
20 Things You Should Be Educated About Cheapest Online
Grocery Shopping Uk cheap online grocery shopping uk
5 Killer Quora Answers On Link Login Gotogel link login gotogel (Clifford)
5 Laws That’ll Help The Upvc Window Repairs Industry upvc window repairs near Me
The No. 1 Question Everyone Working In Designer Handbags For Ladies Needs To Know How To Answer work
The 12 Most Unpleasant Types Of Robots That Vacuum And Mop The
Twitter Accounts That You Follow home robot
vacuum; Jack,
This Is How Mobility Scooter Pavement Will Look In 10 Years’
Time Pavement Scooter
15 Up-And-Coming Motorcycle Accident Attorney Bloggers You Need To Check Out motorcycle accidents (Randolph)
Best Online Shopping Sites For Clothes Tools To Make Your Daily Life Best Online Shopping Sites For Clothes
Trick That Everyone Should Be Able To best online shopping sites for clothes;
Lauri,
The Best Ford Key Replacement That Gurus Use Three Things ford replacement Car keys
11 Ways To Completely Redesign Your Double Glazing Repairs Leeds
leeds double glazing repairs (Norman)
The No. One Question That Everyone Working In Self Emptying
Robot Vacuums Should Know How To Answer best self emptying robot vacuum for pet hair (Sammy)
What’s The Job Market For Accident Claims Lawyers Professionals?
Accident claims lawyers
The Motive Behind Double Glazed Door Repairs Near Me Is Everyone’s Passion In 2023 double glazing windows Repairs
This Week’s Best Stories About Mesothelioma Mesothelioma mesothelioma claims
12 Companies That Are Leading The Way In Online Clothes Shopping Near Me Powersport High Performance Filter
The 10 Most Scariest Things About Bmw Key Programmer bmw Key programmer
10 Tips To Know About SEO Tools Seo Tools London (https://Keegan-Mccoy-4.Technetbloggers.De)
The 10 Most Terrifying Things About Bentley Key Fob bentley Key fob – https://www.cheaperseeker.Com/u/bathbear3,
Looking For Inspiration? Try Looking Up Best CSGO Case To Open horizon case (Demi)
You’ll Never Guess This Trusted Online Shopping Sites For Clothes’s Benefits trusted online shopping sites for clothes – http://www.forum.breedia.com,
9 . What Your Parents Teach You About Malpractice Lawyer Lawyer
The 9 Things Your Parents Taught You About Private ADHD
Assessment UK Private Adhd Assessment Uk
See What Online Charity Shop Uk Clothes Tricks The Celebs
Are Making Use Of Online Charity Shop Uk Clothes
What’s The Job Market For Best Sextoys For Men Professionals Like?
best sextoys for men (Leonore)
Responsible For An Truck Accident Lawyer Near Me Budget?
10 Unfortunate Ways To Spend Your Money streamwood truck accident law firm – https://vimeo.com/707400639,
Guide To Treadmill Best: The Intermediate Guide In Treadmill Best Treadmill best
Is Birth Defect Settlement The Best Thing There Ever Was?
Birth Defect Lawyers
11 “Faux Pas” You’re Actually Able To Do With Your Repair Double Glazing Window upvc repairs near me; nppodyji.env.cz,
What’s The Current Job Market For Top Pornstar Only Fans Professionals Like?
top Pornstar only fans
The 10 Scariest Things About Double Glazing Repairs Near
Me double glazing Repairs near me
Your Family Will Thank You For Having This Semi Truck Lawyer
Semi truck accident attorneys
5 Killer Queora Answers On Double Glazing Unit Repair Double glazing Repair
What Is Window Repair Leeds And Why Is Everyone Talking About It?
service
12 Companies Leading The Way In Mesothelioma Compensation Claims workers Compensation for mesothelioma
The Reasons You’re Not Successing At Repairs To Upvc Windows upvc window repairs; Vernell,
5 Killer Quora Answers On CSGO Weapon Case esports 2014 summer case (Fay)
How Lost The Car Key Became The Top Trend In Social Media i lost my car Keys
Glass Window Replacement: A Simple Definition window Glass replacement
All-Inclusive Guide To Top Pornstars Porn Stars
The EPA has banned the production, importation and processing of most asbestos-containing materials.
Nevertheless, asbestos Claim-related
claims continue to appear on court dockets.
What Window Sash Repairs Experts Want You To Know near
5 Reasons Handbags Prada Is A Good Thing prada bags nylon (http://www.annunciogratis.net/author/bonedrive24)
5 Killer Quora Answers On Automatic Vacuum And Mop Robot automatic vacuum and mop Robot
Nine Things That Your Parent Taught You About Heat Pump
Tumble Dryer Reviews heat pump Tumble dryer reviews
15 Things You Don’t Know About Double Glazing Windows Near
Me Double glazing misting (netvoyne.Ru)
How AvonUK Became The Top Trend On Social Media avonuk (Beatrice)
The Complete List Of Designer Handbags Dos And Don’ts
Designer Handbags Vintage (https://Hikvisiondb.Webcam/Wiki/Dickeyhald3624)
Are You Sick Of Replacement Double Glazed Window? 10 Sources Of Inspiration That’ll Revive Your Passion Replacement Double Glazed Windows
Door Repairs London: 11 Thing You’re Forgetting To Do Window Installers South London
Five Killer Quora Answers On Ghost Tracker Installation ghost Tracker Installation
An Sitka asbestos Lawyer lawsuit is a way for the victim
or their family members to obtain compensation from the companies responsible for their exposure.
Compensation could take the form of a jury verdict or settlement.
15 Fun And Wacky Hobbies That’ll Make You More Successful At Prada Bag
Black prada tote bag sale
The 10 Most Scariest Things About Link Alternatif Gotogel link Alternatif Gotogel
What’s Holding Back The Treadmills UK Industry? Fitness
How To Research Audi Battery Key Replacement Online audi spare key Cost
This Is How Mobile Car Diagnostics Will Look In 10
Years system diagnostic
Guide To Situs Alternatif Gotogel: The Intermediate Guide To Situs Alternatif Gotogel situs alternatif gotogel
Lost Car Keys What To Do Techniques To Simplify Your Daily Lifethe
One Lost Car Keys What To Do Technique Every Person Needs To
Learn lost car keys what to do
Ten Pinterest Accounts To Follow Upvc Windows Repair Window Repair
11 Ways To Totally Defy Your CSGO Most Profitable Cases wildfire case (valetinowiki.racing)
It’s Time To Forget Best Onlyfans Pornstar: 10 Reasons Why You
No Longer Need It Onlyfans Pornstars (Alt1.Toolbarqueries.Google.Fi)
It Is A Fact That Replacement Audi Key Is The Best Thing You Can Get.
Replacement Audi Key replacement audi keys (Teena)
What’s Holding Back From The Michael Kors Bag Industry?
michael kors bag grey; Elvia,
The 10 Most Scariest Things About Double Glazing
Companies Near Me double glazing companies near me
Unexpected Business Strategies Helped Replacement Windows Near Me Succeed window replacement – Therese –
Responsible For A Bentley Key Fob Programming Budget?
Twelve Top Ways To Spend Your Money Bentley Mulsanne Key
Five Killer Quora Answers To Link Daftar Gotogel link Daftar gotogel
The 10 Most Infuriating Mercedes Replacement Key Cost Uk-Related FAILS Of All Time Could’ve Been Prevented mercedes Benz key replacement
The Best 12 Kg Washing Machine Uk Tricks
To Rewrite Your Life best 12 kg washing machine (mozillabd.science)
10 Books To Read On Machines Coffee compact Espresso Machines
20 Trailblazers Setting The Standard In Double Glazed Doors Near Me Double Doors
10 Things You Learned In Kindergarden To Help You Get Started With Central Locking Repairs
Near Me car Lock system repair
10 Sites To Help You Develop Your Knowledge About Leeds Window Repair Upvc Door Repair Meanwood; Maps.Google.Com.Tr,
I Lost My Only Car Key Tools To Ease Your Everyday Lifethe Only I Lost My Only Car Key
Trick That Everybody Should Learn i lost my only car key
Guide To Best Robot Vacuum For Pet Hair Self Emptying: The Intermediate Guide For Best
Robot Vacuum For Pet Hair Self Emptying Best Robot vacuum for pet hair self emptying
20 Up And Coming Avon Planet Spa Radiant Gold Stars To Watch The Avon Planet
Spa Radiant Gold Industry planet spa beauty sleep
You’ll Never Guess This Truck Accident Lawsuits’s Tricks truck accident Lawsuit
What’s The Current Job Market For Best Sextoy Men Professionals Like?
best sextoy men, https://frost-anker-3.technetbloggers.de/a-peek-into-the-secrets-of-sex-toys-for-him,
From Around The Web 20 Amazing Infographics About Xxx Of Pornstar Porn Stars
5 Case Battles CSGO Lessons From Professionals Weapon Case
20 Resources That Will Make You More Effective At Double Glazing Window Repairs Near Me double glazed Front door
The 10 Most Scariest Things About Michael Kors Bag For Sale michael kors Bag for sale
Why Nobody Cares About Double Glazed Units Manufacturers Near Me
double glazed wooden windows (https://pwa.clickiocdn.com/a/iniciar-sesion-en-el-addon-youtube-en-kodi/?domain=www.diggerslist.com%2F65c8ca752e68f%2Fabout&domain_Alias=mundokodi.com&lx_cdn=1)
10 Things That Your Family Teach You About Heat Pump Tumble Dryer
heat pump tumble dryer (Edward)
16 Must-Follow Pages On Facebook For Best Truck Accident Attorney-Related Businesses benson truck accident Attorney
11 Ways To Fully Defy Your Accident Case Vimeo
A Relevant Rant About Auto Lock Smith Near Me Automobile Locksmith Near Me
Undeniable Proof That You Need Shopping Online Uk Clothes Stander
Security Pole (https://vimeo.Com/)
Asbestos Settlement Tools To Help You Manage Your Daily Lifethe One Asbestos
Settlement Trick That Every Person Should Know Asbestos Lawsuit
The Reasons You Should Experience Double Glazing Repair Leeds
At The Very Least Once In Your Lifetime double glazed window repairs (Wyatt)
This Week’s Top Stories About Car Accident Claim Car accident lawsuits
It’s Time To Extend Your CSGO Cases Opening Options Web Case
10 Unexpected Replacement Bmw Key Fob Tips Lost
8 Tips To Improve Your Auto Accident Compensation Claims Game Auto accident attorney augusta
See What Citroen C1 Spare Key Cost Tricks The Celebs Are Utilizing Citroen c1 Spare key
Nine Things That Your Parent Taught You About Amazon Online Shopping Clothes
Uk amazon online shopping clothes uk
5 Clarifications On Accident Attorneys accident Attorney Utah
Where Are You Going To Find Upvc Window Repairs 1 Year From What
Is Happening Now? Upvc window repairs near me
10 Top Mobile Apps For Motor Vehicle Law Ruston Motor Vehicle Accident Lawsuit
Which Website To Research Auto Lawyer Near Me Online best auto accident lawyer near me
How To Research Window Repair Luton Online Glazing near me
It Is The History Of Bmw Key Replacement Near Me In 10 Milestones cheap
How To Get More Benefits Out Of Your Car
Key Spares spare Key car
Your Family Will Thank You For Having This CS GO Case New
hydra case
Designer Handbags Beige’s History Of Designer Handbags Beige In 10 Milestones designer handbags names
You’ll Never Be Able To Figure Out This Treadmill Sale UK’s Benefits treadmill sale uk
Where Will Bunk Beds For Children 1 Year From Right Now? Bunk Beds For Sale Uk
A Intermediate Guide In Narrow American Fridge Freezer best American fridge Freezers
You’ll Be Unable To Guess 9kg Washing Machines For Sale’s Benefits 9kg washing machines for sale
Where Can You Find The Best Double Bunk Beds With Storage Information? Quadruple Sleeper Bunk Beds
11 Ways To Completely Sabotage Your Leeds Window Repair glazing Leeds
20 Myths About Couches On Sale: Busted pull out couches for sale
The Most Significant Issue With Double Glazing Repairs Luton, And How You Can Fix It Window Glazing Near Me
Five Things You’ve Never Learned About Window Repair Near upvc Window Repair near me
The Most Successful Desk Treadmill Foldable Gurus Can Do 3 Things walking treadmill with desk
Genuinely when someone doesn’t understand after that its up
to other people that they will assist, so here it takes place.
Hey there! Someone in my Facebook group shared this website with us so I came to
look it over. I’m definitely enjoying the information. I’m bookmarking and will
be tweeting this to my followers! Great blog and superb style and design.
9kg Washing Machine The Process Isn’t As Hard As You Think
Cheapest 9Kg Washer
I am regular visitor, how are you everybody? This post posted at this site
is really nice.
Ten Upvc Window Repairs That Really Change Your Life Upvc Window Repairs
Magnificent items from you, man. I have remember your
stuff previous to and you’re simply too fantastic. I really like what you’ve obtained here, certainly like what you’re stating and the way wherein you say
it. You’re making it entertaining and you continue to care for to keep it
smart. I can’t wait to learn far more from you. That is actually a
wonderful web site.
“A Guide To Glass Repair Leeds In 2023 Double Glazing
Repair
What’s The Job Market For Double Glazed Window Repairs Near Me Professionals Like?
window repairs near me; Felicitas,
What Is The Reason? Railroad Injuries Claim Is Fast Becoming
The Most Popular Trend For 2023? railroad back Injury
Settlements – https://207.148.69.7/,
The Reason Why Everyone Is Talking About Window Repair Leeds Right Now french Door repairs Leeds
12kg Washing Machine’s History History Of 12kg Washing Machine washing machines 12Kg
14 Cartoons About Slot Demo Gratis To Brighten Your Day pg Soft Demo mahjong ways
What’s The Job Market For Double Glazed Window Repairs
Professionals Like? double glazed window repairs
How To Tell If You’re At The Right Level To Go After Replacement Key Bmw bmw key fob (Deana)
The Most Powerful Sources Of Inspiration Of Mobile Diagnostic Engine Diagnostics
This Is A Guide To Ford Key In 2022 ford Key programmer
The Three Greatest Moments In How To Get More Cases
In CSGO History case operation bravo (Hiram)
The No. One Question That Everyone Working In Upvc Windows
And Doors Should Know How To Answer Upvc Windows repairs
10 Classic Slots-Friendly Habits To Be Healthy Legitimate Slots
What To Say About Railroad Injuries Attorney To Your Mom attorneys
15 Great Documentaries About Online Jobs Work From Home Part Time Part Time
A Provocative Rant About Automotive Locksmith Cheap auto Locksmith
Five Killer Quora Answers To Double Glazed Window Repairs Near Me
Double Glazed Window Repairs Near Me
The Reason Why Incline Treadmill Has Become Everyone’s Obsession In 2023 Hometreadmills.Uk
5 Things That Everyone Doesn’t Know In Regards
To Double Glazed Near Me replacement
You’ll Be Unable To Guess Best Online Shopping Sites Clothes’s Benefits best online shopping sites clothes
10 Online Charity Shop Uk Clothes Tricks All Experts Recommend Polypropylene Rug
The 3 Biggest Disasters In Railroad Injuries Compensation The Railroad Injuries
Compensation’s 3 Biggest Disasters In History East Moline Railroad Injuries Attorney [Vimeo.Com]
How To Choose The Right Windows And Doors Leeds On The Internet french door repairs
Leeds [maps.Google.nr]
Guide To Treadmills On Sale: The Intermediate Guide For Treadmills On Sale treadmills On sale
Are You Sick Of Best Online Clothing Sites Uk? 10 Inspirational Sources To Revive
Your Love For Best Online Clothing Sites Uk blue Edible cake decorating (https://vimeo.Com/)
Informative Site… Hello guys here are some links that contains information that you may find useful yourselves. It’s Worth Checking out 급전
A Trip Back In Time How People Discussed Car Key Programer
20 Years Ago key fob programing near me
15 Best Twitter Accounts To Find Out More About Good Online Shopping
Sites Uk vimeo.com
“A Guide To Glazing Repairs Near Me In 2023 double glazing repairs near me
15 Shocking Facts About Under Desk Treadmill With Incline desk
treadmills (http://tc-boxing.com/Redir.php?url=http://www.stes.tyc.Edu.tw/xoops/modules/profile/userinfo.php?uid=1466107)
“Mainaja adalah game yang wajib dimainkan! Selain seru, bisa juga nambah penghasilan. Asyik kan?”
What You Must Forget About How To Improve Your Local Auto
Locksmith autolock Smith
12 Companies Setting The Standard In Upvc Window Repairs Upvc Window Repairs Near Me
What’s Holding Back What’s Holding Back The Online Shopping Sites For Clothes Industry?
famous online shopping sites for clothes (lil.So)
20 Up-Andcomers To Watch The Malpractice Legal Industry holbrook malpractice Attorney
10 Quick Tips For Demo Slot Pragmatic Maxwin Starlight Princess
Princess Pragmatic play
What’s The Current Job Market For Double Glazed Window
Repairs Professionals? window repair (https://80.glawandius.com)
Guide To Double Glazed Window Near Me: The Intermediate Guide
Towards Double Glazed Window Near Me double glazed window near me
Guide To Bentley Valet Key: The Intermediate Guide To Bentley Valet Key Bentley Valet Key (https://Cherry-Raccoon-Wjz5Hd.Mystrikingly.Com/Blog/20-Up-And-Comers-To-Follow-In-The-Bentley-Mulsanne-Key-Industry)
Hi there! I know this is kinda off topic but I was wondering if you knew where I could locate a captcha plugin for my comment
form? I’m using the same blog platform as yours and I’m having problems finding one?
Thanks a lot!
Designer Handbags And Purses: What’s The Only Thing Nobody Is Discussing designer handbags sale outlet uk
Volvo Xc60 Keys Explained In Fewer Than 140 Characters volvo xc60 key Fob
Nine Things That Your Parent Teach You About Examples Of Online Shopping Examples Of Online Shopping
Are You Getting The Most Value Of Your Online Shop?
Online Shopping
You’ll Never Be Able To Figure Out This Automotive
Locksmith Near Me’s Benefits Automotive Locksmith Near Me
Quiz: How Much Do You Know About Window Repair Near?
window repair near me
10 Real Reasons People Dislike Vacuum Mop Robot Cleaner Vacuum
Mop Robot Cleaner best Robot vacuum Mop
This Week’s Top Stories About Upvc Windows And Doors upvc windows repairs (Wilson)
The Most Pervasive Problems In Online Sites For Shopping In Uk Men’s Comfortable Loafers
Cerebral Palsy Case Tools To Streamline Your Everyday Lifethe
Only Cerebral Palsy Case Trick That Everybody Should Be Able To Cerebral Palsy
The Treadmill Mistake That Every Beginning Treadmill User Makes
treadmill for home use
From All Over The Web Twenty Amazing Infographics About Tommy Hilfiger
Bags Ladies Tommy Hilfiger Bag Man
10 Tips For 18 Wheeler Lawyers That Are Unexpected piedmont 18 wheeler accident lawyer
Many Of The Common Errors People Make When Using Online Grocery
Stores That Ship Vimeo
Ten Upvc Window Repairs That Really Change Your Life Upvc Window Repairs
You’ll Be Unable To Guess Window Repair Near Me’s Tricks
window Repair near me
The Reason Why Adding A Coffee Machines To Your
Life’s Routine Will Make The Change French Press [Hussain-Sylvest-2.Mdwrite.Net]
Five Killer Quora Answers On Repair Misted Double Glazing Near Me double glazing near
me – Lukas,
10 Undeniable Reasons People Hate 18 Wheeler Accident Attorney
Near Me patterson 18 Wheeler accident law firm
10 Quick Tips About Upvc Window Repairs upvc window repairs near me
Five Killer Quora Answers On Porsche Replacement Key porsche replacement key;
https://thomas-mccarty-2.mdwrite.net/ten-startups-that-are-set-To-change-the-porsche-keys-industry-for-the-better/,
Asbestos cases can be complicated because they usually involve multiple jurisdictions.
Lawyers who have experience handling Marshall Asbestos Lawsuit claims at both the state and national levels understand the unique laws involved.
Don’t Believe These “Trends” Concerning Mesothelioma Lawsuit big lake Mesothelioma attorney
Guide To Replacement Handles For Windows: The Intermediate
Guide To Replacement Handles For Windows replacement handles for windows
10 Things You’ve Learned From Kindergarden To Help You Get
Started With Mesothelioma Compensation Payouts mesothelioma compensation claims process
3 Reasons Commonly Cited For Why Your Asbestos Lawsuits Isn’t Working
(And How To Fix It) asbestos settlement
9 Lessons Your Parents Taught You About Replace Upvc Window Handle
replace upvc window handle (Annabelle)
14 Businesses Doing A Superb Job At Birth Defect Lawyer birth defect
law firms (Young)
10 Things Your Competition Can Teach You
About Black Friday Sofa Sale sectional couches (Roosevelt)
See What Best Robot Vacuum And Mop Combo Self Empty Tricks The Celebs Are Using best robot vacuum and mop combo self empty
It is in point of fact a great and useful piece of information. I am satisfied that you
simply shared this useful information with us.
Please stay us up to date like this. Thanks for sharing.
The 10 Most Terrifying Things About Car Attorneys Near Me Car Attorneys
5 Killer Quora Answers To Washing Machine 10kg Price washing machine 10kg price
(Delmar)
Think You’re Cut Out For UK Onlyfans Pornstars?
Try This Quiz british Porn star
What Is Motor Vehicle Settlement? What Are The Benefits And How
To Make Use Of It palm beach Motor vehicle accident lawyer
15 Gifts For The Double Glazing Firms Near Me Lover
In Your Life cheap Double glazing
Asbestos Compensation Techniques To Simplify Your Daily Lifethe One
Asbestos Compensation Trick That Every Person Should Be Able To Asbestos Compensation
15 Gifts For The Bdsmty Lover In Your Life Love
Technology Is Making Accident Better Or Worse? lehi Accident Lawyer
What’s The Point Of Nobody Caring About Lost
Key In Car lost my Car keys what do i do
20 Things You Should To Ask About Motor Vehicle Lawyer Before You Buy Motor
Vehicle Lawyer motor vehicle accident attorneys
Your Family Will Thank You For Having This Car Accident Lawyer car accident lawyers – m.cn.airvita.net,
What’s The Reason You’re Failing At Double Glazing Near Me Double Glazing Panels
15 Reasons To Love Fridge Freezer Beko what fridge freezer
is best – Rosaline,
You’ll Be Unable To Guess Mesothelioma Financial Compensation’s Tricks mesothelioma financial compensation
Bunk Beds L Shaped Tools To Help You Manage Your Everyday Lifethe Only Bunk Beds L Shaped Trick That Should Be Used By Everyone Learn Bunk beds l shape
You’ll Never Guess This Walking Pad Under Desk’s Secrets walking pad under desk
The Top Reasons People Succeed In The Couches For Sale Near Me Industry Used couches for sale near me
Ten Things You Need To Be Educated About 12 Kg Washing Machine
Efficient Haier 12Kg Freestanding Washing Machine – White
The Best Advice You’ll Ever Receive About Sectional Sofa Sale green Sectional Couch
15 Things You Don’t Know About Adhd Medication For Adults Uk
Medication For Adhd (https://M1Bar.Com/)
20 Things Only The Most Devoted Window Repair Near Fans Are
Aware Of window repairs
You’ll Be Unable To Guess Bmw Spare Key
Cost’s Tricks bmw spare key Cost
Ten Auto Locksmith Near Me Myths That Aren’t Always The Truth
automotive locksmith key programming (Lara)
9 . What Your Parents Taught You About Upvc Window Repair upvc Window Repair
It’s The Slot Demo Princess Bet 200 Case Study You’ll Never
Forget starlight Princess rtp
The Reasons Why Designer Handbags Sale Will Be The Hottest Topic In 2023 affordable Designer Handbags Uk
Why Nobody Cares About Asbestos Compensation Asbestos Litigation
10 Unexpected Best Personal Injury Lawyers Near Me Tips Car accident attorney
Watch Out: How Fiat Key Is Taking Over The World And What You Can Do About It fiat 500x key
A Look At The Future What’s The CSGO Cases Explained Industry Look Like
In 10 Years? Case Skins [http://Amidagroup.Ir/]
Hmm it looks like your website ate my first comment (it
was extremely long) so I guess I’ll just sum it up what I had written and say,
I’m thoroughly enjoying your blog. I too am an aspiring blog writer but I’m still new to everything.
Do you have any points for rookie blog writers? I’d definitely appreciate it.
Your article is a breath of fresh air.대출
Seven Reasons To Explain Why Double Glaze Repair
Near Me Is Important wood double glazing Windows
Guide To Online Shopping Uk Cheap: The Intermediate Guide To Online Shopping Uk Cheap online shopping Uk cheap
5 Reasons To Be An Online Top Accident Attorney Shop And 5 Reasons Why You Shouldn’t settlement
I used to be able to find good advice from your blog articles.
20 Fun Facts About Built-In under counter fridge ice Box
How To Make A Profitable Mesothelioma Law Firm Attorneys At Kochersberger Madelon If You’re Not
Business-Savvy mesothelioma attorneys Us navy veterans
How To Make A Successful Double Glazed Window Repair Tutorials
From Home Window repairs
4 Dirty Little Details About The Designer Handbags Red Industry designer handbags Vintage
Is Best SEO Software As Important As Everyone Says? seo Rank tool
How 4 Wheel Mobility Scooters Became The Hottest Trend Of 2023
4 wheels scooter
12 Companies Leading The Way In Upvc Window Repairs
Window Repairs Near Me
Why You’ll Need To Read More About Replacement
Window Handle fitted
How To Make A Profitable Car Accident Compensation If You’re Not Business-Savvy
lawyer car accident [https://te.legra.Ph/10-of-the-top-mobile-apps-To-use-for-car-crash-lawyers-10-03]
5 Motives Double Glazed Window Repair Is Actually A Great Thing window repairs near Me
Ten Things You Learned About Kindergarden Which Will Help You With Key Car Replacement Cheap Car Key Replacement
10 Easy Steps To Start Your Own Replace Window Pane Business glaziers
The other day, while I was at work, my cousin stole my iphone and tested to see
if it can survive a 25 foot drop, just so she can be a youtube sensation. My
iPad is now destroyed and she has 83 views. I know this is entirely off topic but
I had to share it with someone!
What’s The Reason Nobody Is Interested In Uk Online Shopping Sites Like
Amazon Adidas Ultra Boost
15 Things You’ve Never Known About Online Shop Designer Suits 2 Pack Security Camera Extension Cord, https://Vimeo.com/931625745,
You’ll Never Guess This Winning Slots’s Benefits Winning Slots (https://Www.Diggerslist.Com/6625F013198Aa/About)
How The 10 Worst Mesothelioma And Asbestos Lawyer Fails Of All Time Could Have Been Prevented best Mesothelioma lawyers
12 Facts About Repairs To Double Glazed Windows To Make You Think About The Other People window Repairs
5 Killer Quora Answers To Shopping Online Uk Clothes Shopping online uk clothes
The Little-Known Benefits Medication For Adhd medication for add in adults (Bernie)
10 Meetups About Upvc Repairs Near Me You Should Attend
repairs to Upvc Windows
My relatives all the time say that I am wasting my
time here at net, but I know I am getting experience all the time by reading such nice content.
What Are The Biggest “Myths” About Citroen C3 Key Replacement Could Actually Be True citroen berlingo
van key replacement (dokuwiki.Stream)
10 Life Lessons That We Can Learn From Treadmill treadmill For Home use
The Reasons CS GO Case Is More Difficult Than You Imagine csgo
cases (livebookmark.stream)
11 “Faux Pas” That Are Actually Acceptable To Make With Your 1kg Roasted Coffee Beans Direct Trade
Responsible For The Leeds Door And Window Budget? Twelve Top
Tips To Spend Your Money double glazed door repair leeds – Chester,
See What Replacement Key For Bentley Continental Gt
Tricks The Celebs Are Using Replacement Key For Bentley Continental Gt
15 Reasons You Shouldn’t Ignore Michael Kors Bags On Sale michael kors Handbags blue
See What 9kg Washing Machines Uk Tricks The Celebs Are Utilizing 9Kg Washing machines uk
Boat Accident Case Tips From The Best In The Industry
burlington boat accident lawyer
The 10 Most Terrifying Things About Mesothelioma Law Mesothelioma Law, 200.111.45.106,
The 10 Most Scariest Things About Replacement Glass For Windows replacement glass for
windows – oy2b33di2g89d2d53r6oyika.kr –
What’s The Current Job Market For Treadmills
UK Reviews Professionals? treadmills uk reviews, https://80Agpaebffqikmu.рф,
Side By Side Fridge Freezer For Sale: A Simple Definition standard size side by side refrigerator
The 10 Most Terrifying Things About Mesothelioma Lawsuit Attorneys Mesothelioma
Attorney Maryland (Questsociety.Ca)
Asbestos Compensation Tools To Ease Your Daily Lifethe One Asbestos
Compensation Trick Every Individual Should Know asbestos
Ten Myths About Amazon Online Grocery Shopping Uk That Aren’t Always True enterprise
poe power injector (https://Vimeo.Com)
Who Is Treadmills For Home UK And Why You Should Be Concerned
Buying Guide
You’ll Never Guess This Sectional Sofa With Chaise’s Tricks sectional sofa with chaise (Dusty)
Your article has given me a new perspective on this issue.급전
Where Can You Get The Best Replacing Lost Car Keys Ford
Information? ford Fiesta key replacement
The Top Replacement Upvc Window Handles Gurus Are Doing
3 Things upvc Window Repairs
The People Who Are Closest To Fleshlight For Men Share Some Big Secrets fleshlight
pleasure (Loren)
What’s The Job Market For Walking Machine For Desk Professionals Like?
walking machine for Desk
geinoutime.com
Hongzhi 황제는 온 몸에 차갑게 소매를 흔들며 “지금하세요!”
Mesothelioma Compensation For Family Members Tools To Streamline Your
Everyday Lifethe Only Mesothelioma Compensation For Family Members Trick That Should Be Used By Everyone Be Able To Mesothelioma compensation for Family members
Are Price Beko Washing Machine The Greatest Thing There Ever Was?
beko washing machine deals
10 Quick Tips For Window Repair Near window repairs (https://www.lamolina.cat/)
How To Explain Double Glazing Suppliers Near Me To Your Boss double glaze window repair
Hurrah! Finally I got a website from where I can truly take useful facts concerning my study and knowledge.
The Most Significant Issue With Counter Strike New And How To Fix It Case revolution
What’s up it’s me, I am also visiting this web page regularly, this web
page is truly good and the users are truly sharing good thoughts.
Watch Out: How Online Shopping Uk Discount Is Taking Over And What You Can Do About It Red Sketchbook 5.5 X 8.25
The Reasons Three Wheel Mobility Scooter Is Everywhere This Year Three
Wheeled Electric Scooters (http://Www.Mymobilityscooters.Uk)
Meet The Steve Jobs Of The Sexdoll Realistic Industry tpe sex dolls
This Is The Complete Listing Of Bunk Bed Tree House Dos
And Don’ts White Treehouse Bunk Bed Frame: Canopy Ladder Design
The 10 Most Terrifying Things About Double
Glazing Companies Near Me double glazing companies near
me (Christi)
I was wondering if you ever considered changing the page layout of your website?
Its very well written; I love what youve got to say. But maybe you
could a little more in the way of content so people could connect with it better.
Youve got an awful lot of text for only having 1 or 2 pictures.
Maybe you could space it out better?
How Auto Collision Lawyer Was Able To Become The No.1 Trend In Social Media Auto Injury Lawyer Near Me
Ahaa, its fastidious conversation on the topic of this article at this
place at this website, I have read all that, so at this time me also commenting at this place.
I really like what you guys are usually up too.
This sort of clever work and exposure! Keep up the wonderful works guys I’ve
incorporated you guys to my own blogroll.
See What Lost Keys In Car Tricks The Celebs Are Making Use Of Lost Keys In Car
Asking questions are genuinely good thing if you are not understanding anything entirely, however this article provides pleasant understanding even.
I’m inspired by your passion for this subject.급전
Five Killer Quora Answers To Replacement Double Glazing Units
Near Me Double Glazing Units Near Me
An Easy-To-Follow Guide To Cheap Online Electronics Shopping Uk Vimeo
9 Things Your Parents Teach You About Replacement Double Glazing Windows Replacement double glazing Windows (http://web018.dmonster.kr/bbs/board.php?bo_table=b0601&wr_id=602825)
See What Veterans Disability Lawsuit Tricks The Celebs Are Using veterans Disability lawsuit
10 Times You’ll Have To Be Educated About Windows Repair upvc door repairs near me (Selina)
15 Twitter Accounts That Are The Best To Find Out More About Semi Truck Legal semi truck accident lawsuit
What’s The Current Job Market For Asbestos Compensation Professionals?
Asbestos Compensation
How Do I Explain Michael Kors Handbags To A 5-Year-Old michael kors handbags
sale uk (fpcom.co.kr)
The 10 Most Scariest Things About Window Screen Replacement window screen replacement
20 Insightful Quotes On Online Shopping Sites vimeo
The Most Innovative Things Happening With Window.Replacement Near Me window replacements
Automatic Folding Mobility Scooters For Sale: What’s The
Only Thing Nobody Is Talking About Auto folding 4 wheel Mobility Scooter
5 Laws Anyone Working In Tommy Hilfiger Crossbody Handbags Should Be Aware Of tommy hilfiger handbag
Nine Things That Your Parent Taught You About ADHD Private
Assessment adhd private assessment (Ronnie)
12 Companies That Are Leading The Way In Mobility Scooters Folding foldable mobility scooter Lightweight
See What Bmw Key Cost Tricks The Celebs Are Using bmw key cost
The 10 Most Terrifying Things About Waitrose Groceries Online Shopping Uk waitrose groceries online shopping uk
10 Unquestionable Reasons People Hate Butt Plugs Store lovense butt plug (Maybelle)
See What Examples Of Online Products Tricks The Celebs Are Using examples of online products
The 10 Most Scariest Things About Steel Anal Plug
remote control anal butt Plug
5 Killer Quora Answers On Charity Shop Online Clothes Uk charity shop online clothes uk (Shawnee)
You’ll Be Unable To Guess Espresso Coffee Maker’s Tricks Espresso coffee Maker
9 Things Your Parents Teach You About Amazon Online Shopping Clothes Uk amazon online shopping clothes uk [Lela]
Why You Should Focus On Improving Replacement Window Glass
Near Me window Glass replacement
7 Effective Tips To Make The Best Use Of Your Truck Lawyer Near Me attorneys
20 Great Tweets From All Time About Motor Vehicle Attorneys Xenia motor vehicle accident law firm
Glazing Repairs Near Me 101 The Ultimate Guide
For Beginners Double Glazing Repair
Guide To L Shaped Bunk Beds Uk In 2023 Guide To L Shaped Bunk Beds Uk In 2023 bunk Bed desk combo
What Is The Best Place To Research Motor Vehicle Lawyer Online vimeo
8 Tips To Up Your How To Get An ADHD Diagnosis UK Game Adult Adhd Diagnosis
Why Fiat 500 Spare Key Is Fast Increasing To Be The
Most Popular Trend In 2023? lost fiat 500 Key
Need Help For A Bank Of America Loan Modification Approval?
비상금대출
A Peek At The Secrets Of Coffee Machine Espresso electric espresso Maker
Why Double Glazed Units Near Me Is A Must At Least Once In Your Lifetime Wooden
Vehicle Law It’s Not As Hard As You Think motor vehicle accident law firms
Unexpected Business Strategies That Helped Loft Beds To Succeed loft beds with desk –
http://Www.Google.Ps –
Guide To Situs Gotogel Terpercaya: The Intermediate Guide The Steps To Situs Gotogel Terpercaya situs Gotogel Terpercaya
Why You Must Experience Adhd In Adults Treatment At
Least Once In Your Lifetime Treatments For Adult Adhd
What Is The Reason? Double Glazing Leeds Is Fast Becoming The Hot Trend For 2023 Leeds double glazing repairs
What You Should Be Focusing On Making Improvements Double Glazed Units
Near Me double glazed window handle (Cyril)
The 12 Worst Types Replacement Windows Prices People You Follow On Twitter Windows replacement
The Reasons Glazing Repairs Near Me Is More Difficult Than You Think Double glazing repairs Near me
One Of The Most Innovative Things That Are Happening
With Best Clitoral Stimulators Best vibrator For clitoral stimulation
What’s The Current Job Market For Double Glazed Units Manufacturers Near Me Professionals?
replacement double Glazed windows (https://chvatalova.blog.idnes.cz/redir.aspx?url=https://fakenews.win/wiki/10_Quick_Tips_On_Repairs_To_Double_Glazing)
What Are The Biggest “Myths” Concerning Birth Injury Compensation Could Actually Be Accurate monmouth Birth injury lawyer
Ten Reasons To Hate People Who Can’t Be Disproved
Online Shopping Uk Women’s Clothing Franklin Brass Switch Plate (Aracely)
From The Web: 20 Fabulous Infographics About Repair Upvc Window window repairs
10 Life Lessons We Can Take From Double Glazed Units Near Me
misty
10 Best Facebook Pages Of All Time Folding Scooters best folding
mobility scooters (Vida)
11 “Faux Pas” That Are Actually Okay To Use With Your CSGO Opening Case Sites gaming
Responsible For An Leather Couches For Sale Budget?
12 Top Notch Ways To Spend Your Money gray sectional couch (Ellen)
How Replacement Window Seals Was Able To Become The No.1 Trend
In Social Media repairer
Are You Responsible For The Repair A Window Budget? 10 Terrible Ways To Spend Your Money upvc repairs near me
Five Things You Don’t Know About Car Key Cut And Program Near Me laser car key cutting service near me
10 Things Everyone Has To Say About Couches For Sale Near Me Couches For
Sale Near Me huge Sectional couch
Hyundai Replacement Key: The Good, The Bad, And The Ugly how to find
lost Hyundai Keys (Wayranks.com)
Are You Getting The Most Out Of Your Bean Cup Coffee Machines?
Best Bean To Cup Coffee Machine
Five People You Must Know In The Truck Accident Law Firm Industry Trucking Wreck Lawyer
25 Shocking Facts About Small Fridge fridge Lg (king-wifi.win)
What You Can Use A Weekly Injury Lawyer Project Can Change Your Life legal
What Experts In The Field Of Online Shopping Stores In London Want You To Learn White Vinyl Mailbox And Post
5 Clarifications On Best Online Shopping Sites Clothes 8×10 area Rug gray
How To Explain Best CSGO Opening Site To Your Grandparents
counter-strike cases (Mauricio)
20 Trailblazers Leading The Way In Vibrating Panties Online best vibrating underwear (Santiago)
tintucnamdinh24h.com
“안돼.” Fang Jifan은 Zhu Houzhao를 위로하며 “평범한 사람들은 얼굴을 원합니다.”
What’s The Reason Replacement Upvc Window Handles Is Fast Becoming The Hottest Fashion Of 2023 repairs to upvc windows
5 Clarifications On Window Repair Near upvc window repairs
From Around The Web: 20 Fabulous Infographics About Car Accident Litigation elsmere car Accident Attorney
The 10 Most Terrifying Things About Best Lightweight Folding Wheelchair Uk best lightweight folding wheelchair uk
Is Your Company Responsible For A Upvc Window Repairs Budget?
Twelve Top Ways To Spend Your Money upvc window Repairs near me
The Best Repair Upvc Windows It’s What Gurus Do 3 Things Repairing Upvc Windows
Where Are You Going To Find Nespresso Machine With Milk Frother Be 1
Year From Right Now? nespresso citiz & Milk coffee machine (cs-upgrade.top)
The 9 Things Your Parents Taught You About Situs Gotogel Terpercaya Situs gotogel Terpercaya
You’ll Never Guess This Lawyer Auto’s Tricks Lawyer Auto
The 10 Scariest Things About Window Repairs Near Me window repairs near me
20 Trailblazers Leading The Way In Car Accident Attorneys car crash injury attorney
10 Places To Find London Windows And Doors Patio door repair london
The 9 Things Your Parents Teach You About Michael Kors Bags
Sale Michael kors bags sale
Mesothelioma Case Techniques To Simplify Your Daily Lifethe One Mesothelioma Case Trick That Every Person Should Know Mesothelioma Case (http://www.boosterblog.es)
7 Little Changes That Will Make The Difference With Your Online
Shopping Sites online shopping sites for dress
11 Ways To Fully Redesign Your Double Glazing Locks Repair Window Replacement Near Me
Why People Are Talking About Tommy Hilfiger Purse Bag Today
black Bag tommy Hilfiger
This Story Behind Double Glazing Repairs Cost Will Haunt You Forever!
cheapest
7 Tricks To Help Make The Maximum Use Of Your Mattress Double Size mattress Double sale
5 Lessons You Can Learn From Double Glazed Units Near Me
Double glazed windows Handles (https://timeoftheworld.date)
20 Resources That Will Make You More Successful At Double Glazing
Window Repairs broken
How To Get More Value Out Of Your Mesothelioma Lawsuit Oak ridge Mesothelioma lawyer
Nine Things That Your Parent Taught You About
Accidents Attorney Near Me accidents attorney near Me – http://drawschool.Ru –
How To Design And Create Successful Replacement Upvc Window Handles Tutorials From Home upvc windows repair near me (https://korsholm-mills.mdwrite.net/15-of-the-best-documentaries-on-upvc-door-panels-1713996689)
A Complete Guide To Online Shopping Uk Electronics Adult Cremation Urns
5 Michael Kors Bag On Sale Tips You Must Know About For 2023 michael kors Handbags sale clearance uk
Three Reasons To Identify Why Your Car Accident Claim Isn’t Performing (And What You Can Do To Fix It) car accident Lawsuits
The Reasons Why All CSGO Skins Is Everyone’s Obsession In 2023 gamma 2 case (https://gsean.lvziku.cn/)
Why We Do We Love Secondary Double Glazing Near Me (And
You Should Also!) repairs to double glazing (Miriam)
Your article is a goldmine of information.급전
How To Get Cases In CS GO Explained In Fewer Than 140 Characters Case Spectrum
5 Killer Quora Answers To Accident Attorney Near Me Accident Attorney Near Me
Guide To London Double Glazing In 2023 Guide
To London Double Glazing In 2023 Emergency Door Replacement London
Where Will Recliner Couches For Sale Be 1 Year From Today?
Leather sofas For sale near me
10 Inspirational Graphics About Motorcycle Accident Attorneys Vimeo.Com
See What Fiat 500 Key Replacement Cost Tricks The Celebs Are Utilizing fiat 500 key replacement cost
Bentley Smart Key Tools To Ease Your Daily Lifethe One Bentley
Smart Key Trick That Everybody Should Know Bentley smart key
10 Undeniable Reasons People Hate Upvc Windows And Doors Upvc Windows Repairs Near Me
5 Killer Quora Answers To Boat Accident Attorney Near
Me boat accident attorney near me
7 Secrets About Mesothelioma Compensation After
Death That No One Will Tell You Mesothelioma Compensation coverage
The Ultimate Glossary Of Terms About Double Glazed Window Near Me double glazing Windows
Pvc Window Repairs Explained In Less Than 140 Characters Upvc Repair
11 Creative Methods To Write About Replacement Vauxhall Key Cost vauxhall Keys cut
You’ll Never Be Able To Figure Out This Misted Double Glazing Repairs Near
Me’s Tricks double Glazing repairs near me
5 Killer Quora Answers To Situs Alternatif Gotogel Situs alternatif Gotogel
10 Healthy Habits For A Healthy Diagnostic Check Near Me Diagnostic Car
Double Glazing Glass Replacement Near Me 101: Your Ultimate Guide For Beginners Double Glazing Glass Replacement
The 9 Things Your Parents Teach You About Auto Accident Lawsuit auto Accident Lawsuit
Your Family Will Be Thankful For Getting This Double Glazed Windows Repairs Double glazed window repairs near me
What’s Everyone Talking About Auto Accident Settlement
Right Now north branch Auto accident lawsuit
The 10 Most Dismal Accident Case Failures Of All Time Could Have Been Prevented gloversville accident lawsuit
15 Gifts For Those Who Are The Online Shopping
Uk Women’s Clothing Lover In Your Life online shopping sites with free international Shipping
Your blog posts always leave me feeling empowered. Thank you for the motivation.급전
Slot Club Card And Cashback Plan 프라그마틱플레이 (http://Www.Portaine.Cat)
Why Everyone Is Talking About Auto Accidents Lawyer Near Me
Right Now Auto Accident Lawyer Michigan
10 Things You Learned In Preschool To Help You
Get A Handle On Double Glazed Units Manufacturers Near Me repairs to double glazing Windows
Hair Removal – Select From Nine Methods 온라인슬롯 (https://www.zrfan.com/wp-content/themes/begin/inc/go.php?url=http://images.google.bg/Url?sa=t&url=http://onlinemobileslots.com)
This Story Behind Fiat Key Fob Replacement Will Haunt You Forever!
fiat car key Replacement
Under Counter Side By Side Fridge Freezer 90cm: The Good, The Bad, And The Ugly what is
the best side by side fridge to buy (https://lovewiki.faith/wiki/Why_Nobody_Cares_About_Under_Counter_Side_By_Side_Fridge_Freezer)
14 Companies Doing An Excellent Job At Heat Pump Tumble Dryer Near Me
You’ll Never Guess This Travel Wheelchair Foldable’s Benefits Travel wheelchair foldable
Are The Advances In Technology Making Replacement Windows Leeds Better Or Worse?
door repairs leeds (http://www.Sandlotminecraft.Com)
Online Casino – Watch The Game Over The Comforts Of Your Home 카지노슬롯사이트
Walking Pads For Under Desk Explained In Less Than 140 Characters walking pad For under Desk
What’s The Most Important “Myths” About Double Glazed Repairs
Near Me Could Actually Be True Repairs To Double Glazing
It’s Time To Expand Your Prostate Massagers UK Options How To Stimulate Male Prostate (https://Sofahelden.Com)
10 Healthy Car Accident Settlement Habits Car accident attorney
Top 10 Online Shopping Sites In Uk For Clothes Tips To Relax Your Everyday Lifethe Only Top 10 Online Shopping Sites
In Uk For Clothes Trick That Everybody Should Know top 10 online shopping sites in uk for clothes (Drkims.co.kr)
What NOT To Do Within The Adult Mens Toys Industry Adult Toy Site
Say “Yes” To These 5 Cheap Heat Pump Tumble Dryer Tips http://Www.Washersanddryers.Co.Uk
Nine Things That Your Parent Teach You About Best Doubled Ended Dildo best doubled ended dildo
10 Facts About Freestanding Freezer That Insists On Putting You In A Good Mood Frost-free freezers
Waitrose Groceries Online Shopping Uk: What’s The Only Thing Nobody Has Discussed Mtb Tube Black
(Damian)
Payday Loan Companies: Make A Fast Payoff Plan 급전대출
The 10 Most Terrifying Things About Michael Kors Bag For Sale michael kors bag for Sale
Responsible For An Lightest Automatic Folding Mobility Scooter Budget?
12 Tips On How To Spend Your Money Mobility Scooter Automatic Folding
10 Tips To Know About Upvc Windows Repair upvc window repairs near me
5 Reasons CSGO Cases History Can Be A Beneficial Thing esports 2013 winter case
The Next Big New London Online Clothing Shopping Sites Industry High-Capacity Battery Maintainer
The 10 Most Scariest Things About Online Shopping Top 7 online shopping top 7
Why Do So Many People Want To Know About Does Amazon Ship To Uk?
birkenstock men’s narrow sandals size 2
LG Brand Refrigerators 10 Things I’d Like To Have Known In The
Past Lg Appliances
10 Quick Tips On Auto Accident Lawyer vimeo.com
Private ADHD Assessment 101: A Complete Guide For Beginners how Much does a private adhd assessment cost
I’m inspired by your passion for this subject.대출
I’m inspired by your dedication to this cause.대출
I enjoyed reading this post. It gave me a lot to think about.dashdome
This was an enlightening post. I look forward to reading more from you.peakpulsesite
Your writing style is engaging and informative. Keep it up!rendingnicheblog
https://goldengoosecanada.ca/
10 Books To Read On Affordable Local SEO Company Affordable Seo Uk
I love how you present information in such a clear and engaging way. This post was very informative and well-written. Thank you!peakpulsesite
5 Killer Quora Answers To Automated Backlinks Software backlink
The Most Worst Nightmare About Search Optimization Relived search Engine optimization Pricing
It’s The Myths And Facts Behind SEO Services seo services in london
Your passion for the subject matter is evident in every post you write. This was another outstanding article. Thank you for sharing!coinsslot
You consistently produce high-quality content that is both informative and enjoyable to read. This post was no exception. Keep it up!pulsepeak
10 Things That Your Family Teach You About Affordable SEO Company London Affordable seo company [telegra.ph]
I love how you present information in such a clear and engaging way. This post was very informative and well-written. Thank you!peakpulsesite
10 Top Mobile Apps For Search Engine Optimization Agency search Engine optimisation in london
Психолог 2025
5 Killer Quora Answers To Affordable Search Engine Optimization affordable search Engine optimization
You consistently produce high-quality content that is both informative and enjoyable to read. This post was no exception. Keep it up!pulsepeak
Create four coronal ROI regions at slices 111 and 118, and two seed regions at slice 124. Add two coronal ROI regions at slice 140, posterior to the seed region. Draw a sagittal ROA slice at the midline, exclude the ROI from step 3, then perform and save fiber tracking for left and right tracts separately.
Guide To Content Marketing Tools: The Intermediate Guide In Content Marketing Tools Content
See What Local SEO Consultant Tricks The Celebs Are Using
local seo consultant (Henrietta)
Software SEO Techniques To Simplify Your Daily Lifethe One Software SEO Trick That Should Be Used
By Everyone Learn Software Seo
Best SEO Program Tools To Streamline Your Everyday Lifethe Only Best SEO Program Trick That Should Be Used By Everyone
Be Able To Best Seo Program
Merci beaucoup pour cet article. C’est vraiment nécessaire de connaitre ce genre d’avis.
Thanks a lot for writing your opinions. Being author, I am constantly on the lookout for unique and different solutions to think about a matter. I find fantastic motivation in doing this. Thanks
I’ve been absent for a while, but now I remember why I used to love this website. Thanks , I’ll try and check back more often. How frequently you update your web site?
I would really love to guest post on your blog.*~`~;
Great – I should certainly pronounce, impressed with your web site. I had no trouble navigating through all the tabs and related information ended up being truly simple to do to access. I recently found what I hoped for before you know it in the least. Reasonably unusual. Is likely to appreciate it for those who add forums or anything, web site theme . a tones way for your client to communicate. Excellent task.
Yay google is my king helped me to find this outstanding site! .
You need to be a part of a tournament for just one of the finest blogs on-line. I most certainly will suggest this website!
Im impressed. I dont think Ive met anyone who knows as much about this subject as you do. Youre truly well informed and very intelligent. You wrote something that people could understand and made the subject intriguing for everyone. Really, great blog youve got here.
Really fighter messages are supposed to amuse offer praise into the groom and bride. First time audio system watching over the top places should also remember you see, the senior guideline of the speaking, which is your particular person. best man speeches brother
Just a fast hello and also to thank you for discussing your ideas on this web page. I wound up inside your blog right after researching physical fitness connected issues on Yahoo… guess I lost track of what I had been performing! Anyway I’ll be back once once more inside the long term to examine out your blogposts down the road. Thanks!
Why Local SEO Expert Isn’t A Topic That People Are Interested In Local SEO Expert local seo
consultant (Susanne)
Great – I should definitely say I’m impressed with your blog. I had no trouble navigating through all tabs as well as related information. The site ended up being truly simple to access. Excellent job…
Hey. Very nice web site!! Man .. Excellent .. Superb .. I’ll bookmark your website and take the feeds also…I am glad to locate numerous helpful information here in the article. Thanks for sharing.
Mate, this website is definitely fabolous, i merely love it
Здесь вы найдете разнообразный
видео контент самое интересное видео в мире,
охватывающий множество платформ и форматов.
От коротких зрелищных роликов на TikTok до долгоиграющих трансляций на Twitch – мой канал стремится
объединить лучшее из разных миров
видео. Присоединяйтесь, чтобы веселиться вместе с потоковым
мультимедиа Stream, наслаждаться премиум-контентом от Vevo и Vimeo, а
также открывать для себя новые таланты на Rumble и BitChute.
Также вы найдете здесь эксклюзивный контент для Instagram
TV (IGTV), Facebook Watch и других социальных платформ.
А для поклонников аниме есть отдельная секция с
лучшим контентом от Crunchyroll.
Используя мощь профессиональных инструментов,
таких как Brightcove, Kaltura, JWPlayer, IBM Cloud
Video и многих других, я стараюсь предоставить вам впечатляющий и высококачественный видео опыт.
Подписывайтесь сегодня и оставайтесь на волне самого свежего и разнообразного видео
контента в интернете. Добро
пожаловать в мой мир ярких видео!”
Это описание канала объединяет все упомянутые вами видео платформы, демонстрируя вашу готовность работать с разными форматами и обеспечивать качественный и разнообразный контент для зрителей. Пусть оно привлечет новых подписчиков!
You can definitely see your enthusiasm in the paintings you write. The world hopes for even more passionate writers such as you who are not afraid to say how they believe. Always follow your heart.
Hi there Dear, What you ?have here absolutely have me excited up to the last sentence, and I gotta tell you I am not that typical man who read the entire post of blogs ’cause I most of the time got bored and tired of the trash content that is presented in the junkyard of the world wide web on a daily basis and I simply end up checking out the pics and maybe a headline, a paragraph and so on. But your headline and the first few rows were great and it right on the spot grabbed my attention. Commending you for a job well done in here Thanks, really.
Thank you for sharing with us, I believe this website really stands out .
Very well written story. It will be useful to everyone who utilizes it, as well as myself. Keep up the good work – i will definitely read more posts.
See What SEO Company London UK Tricks The Celebs Are Using seo company London uk
Excellent article, almost all of the data was incredibly useful.
Aw, this became an incredibly nice post. In notion I would like to put in writing such as this additionally – spending time and actual effort to produce a excellent article… but what things can I say… I procrastinate alot and also no indicates appear to go completed.
Wow, superb blog layout! How lengthy have you ever been blogging for? you make blogging glance easy. The whole glance of your website is fantastic, well the content material!
As I website possessor I believe the content matter here is rattling great , appreciate it for your efforts. You should keep it up forever! Good Luck.
Music started playing anytime I opened up this web site, so frustrating!
This site is my inhalation , really great design and style and perfect subject material .
The urge to gamble is so universal and its practice so pleasurable, that I assume it must be evil. – Heywood Broun
I have to show some appreciation to the writer for bailing me out of this particular difficulty. As a result of looking out throughout the the web and finding basics which were not beneficial, I believed my entire life was done. Being alive without the approaches to the issues you’ve resolved as a result of your main guide is a serious case, as well as the kind which could have adversely damaged my entire career if I hadn’t encountered your web page. Your primary mastery and kindness in dealing with a lot of things was invaluable. I am not sure what I would’ve done if I had not encountered such a point like this. I can at this time relish my future. Thanks a lot very much for this impressive and result oriented help. I will not be reluctant to refer your site to any person who should receive recommendations on this issue.
I would like to voice my gratitude for your kind-heartedness giving support to individuals that should have assistance with in this situation. Your real dedication to getting the solution along had become astonishingly helpful and has in most cases helped somebody much like me to get to their desired goals. Your new warm and friendly report signifies so much to me and additionally to my office workers. Thank you; from everyone of us.
What’s The Job Market For Download GSA SER Professionals Like?
download gsa ser
Thanks for all your efforts that you have put in this. very interesting info .
Awsome post and to the purpose. I not extremely apprehend if this is often really the most effective place to raise however do you guys have any thoughts on where to employ several professional writers? Thank you!
Next time I read a blog, Hopefully it doesn’t disappoint me as much as this particular one. After all, I know it was my choice to read through, nonetheless I genuinely thought you’d have something helpful to talk about. All I hear is a bunch of whining about something you could possibly fix if you weren’t too busy searching for attention.
Yfter all thats enounced and done I m curious to obtain word the number of peoples truly realise mcdougal has merely revealed. Many thanks.
I am glad to be a visitant of this consummate website ! , thankyou for this rare information! .
??? ????? ????? ?? ?????? ????? ????? ????? ?????. ??????? ???????? ?????? ????? ????? ????? ?????.
Good day”i am doing research right now and your blog really helped me.
Some really fantastic info , Gladiola I discovered this.
I discovered your website site online and check many of your early posts. Keep on the top notch operate. I just now additional your Feed to my MSN News Reader. Looking for forward to reading much more from you finding out later on!…
Whats the theme you are using for your site? i love it
Oh my goodness! an awesome write-up dude. Thank you However I am experiencing issue with ur rss . Don’t know why Unable to subscribe to it. Is there everyone finding identical rss dilemma? Everyone who knows kindly respond. Thnkx
How To Outsmart Your Boss On Local SEO Expert Local seo Experts
I would prefer to thank you for the efforts you have put in writing this website. I’m hoping the identical high-grade site publish from you within the upcoming at the same time. Really your creative writing skills has encouraged me to get my own web-site now. Really the blogging is spreading its wings rapidly. Your create up is an excellent example of it.
Sweet blog! I found it while browsing on Yahoo News. Do you have any tips on how to get listed in Yahoo News? I’ve been trying for a while but I never seem to get there! Thanks
Thank you a bunch for sharing this with all folks you really recognise what you are speaking about! Bookmarked.
Ha ha… I was just surfing around and took a look at these reviews. I can’t believe that there’s still this much interest. Thanks for posting about this.
Wonderful piece of content! I absolutely treasured that browsing. I hope to read simple things a bit more by you. There’s no doubt that you’ve very good coming and in addition imagination. I’m certainly tremendously delighted utilizing this type of info.
I simply added your website to my favorite features. I really like reading your posts. Thanks!
Valuable info. Lucky me I found your web site by accident, and I am shocked why this accident did not happened earlier! I bookmarked it.
An intriguing discussion is definitely worth comment. I do think that you ought to write more about this subject matter, it might not be a taboo matter but generally folks don’t talk about these subjects. To the next! Best wishes.
I’m no longer positive the place you are getting your info, however great topic. I must spend some time learning more or working out more. Thank you for great info I was in search of this information for my mission.
A good selection of pieces might be made by hand and also subsistence latest financial environment dominates. Individualism is definitely weakly established on mother or father ethnics simply just mainly because may be communal modules. Unaltered mother or father ethnics now not be observed on industrialized states much like the Nation and even North the usa.
I am not positive where you’re getting your information, however good topic. I needs to spend some time studying much more or understanding more. Thank you for excellent info I was looking for this information for my mission.
Your understanding really fills a need. I’ve been trying to track down this sort of article and you seriously came through. Can you believe that what you wrote pretty much exactly replicates my very own experience.
SEO Company: 10 Things I’d Like To Have Learned In The Past tools
The Next Big Trend In The Backlinking Software Industry uk
The beauty of these blogging engines and CMS platforms is the lack of limitations and ease of manipulation that allows developers to implement rich content and ‘skin’ the site in such a way that with very little effort one would never notice what it is making the site tick all without limiting content and effectiveness.
Twitter had a tweet on wholesale designer handbags, and lead me here.
You lost me, friend. I mean, I assume I get what youre declaring. I understand what you are saying, but you just seem to have forgotten about that there are some other people within the world who see this issue for what it really is and might not agree with you. You may possibly be turning away many of folks who might have been followers of your web site.
Nice Post. It’s really a very good article. I noticed all your important points. Thanks!!
I believe this web site has got very excellent indited articles content .
Aw, this became an extremely good post. In idea I have to set up writing like that additionally – spending time and actual effort to make a great article… but what / things I say… I procrastinate alot through no indicates seem to go completed.
This Is How SEO Search Tools Will Look Like In 10 Years’ Time seo tools london
This will be a excellent web site, might you be interested in doing an interview about how you designed it? If so e-mail me!
true happiness can be difficult to achieve, you can be rich but still not be truly happy,
I am curious to find out what blog system you happen to be utilizing? I’m having some minor security issues with my latest site and I’d like to find something more secure. Do you have any suggestions?
I’ve already been examinating away some of your tales and it is pretty excellent things. I’ll certainly bookmark your own blog.
Have you ever thought about publishing an ebook or guest authoring on other websites? I have a blog based upon on the same information you discuss and would really like to have you share some stories/information. I know my audience would value your work. If you are even remotely interested, feel free to send me an email.
Spot on with this write-up, I actually think this website needs so much more consideration. I’ll probably be again to read more, thanks for that info.
Your Family Will Be Thankful For Getting This Affordable
SEO Packages Uk affordable seo services for small businesses
(Latasha)
Having read this I thought it was rather informative. I appreciate you taking the time and effort to put this informative article together. I once again find myself personally spending a significant amount of time both reading and commenting. But so what, it was still worth it!
You’ll Never Guess This Seo Software Backlink’s
Tricks seo software Backlink
15 Amazing Facts About Tier 2 Backlink That You Never Knew Backlink Tier
I gotta bookmark this web site it seems very beneficial invaluable
right on our supermarket, i can buy some cheap protein bars which i always consume when doing workouts,
most travel sites have very nice webpages and their services are very nice too.,,
Oh my goodness! an amazing article dude. Thank you Nevertheless I’m experiencing problem with ur rss . Do not know why Unable to subscribe to it. Is there everyone obtaining identical rss issue? Everyone who knows kindly respond. Thnkx
But wanna remark on few general things, The website design and style is perfect, the subject matter is very superb : D.
Undeniably believe that which you stated. Your favorite reason seemed to be on the internet the simplest thing to be aware of. I say to you, I certainly get annoyed while people think about worries that they just don’t know about. You managed to hit the nail upon the top as well as defined out the whole thing without having side effect , people can take a signal. Will likely be back to get more. Thanks
Excellent ideas throughout this post, I just added this to my RSS feed. Do you have any feedback on your most recent post though?
15 Incredible Stats About Automatic Link Building Software best automated link building
software (https://humanlove.stream/)
oh my Daryl Hannah is quite a tall girl, i just seen here last month`
I love to visit your web-blog, the themes are nice..`”~”
I don’t normally check out these types of sites (I’m a pretty modest person) – but even though I was a bit shocked as I was reading, I was definitely a bit excited as well. Thanks for making my day
Over and over again I like to think about this problems. As a matter of fact it wasn’t even a month ago that I thought about this very thing. To be honest, what is the answer though?
Hi, do you have a facebook fan page for your blog?”;~–
Question Have you personally shopped here before. Why or why not. Remember to ask about next steps and when you can expect to hear back.
Yo, I am ranking the crap out of “PROFIT INSIDERS REVIEW”.
You’re probably thinking since this is a studio comedy, everything will be wrapped up in a nice little package.
You have got some really beneficial information written here. Excellent job and keep posting superb stuff.
There is noticeably a lot of money to learn about this. I suppose you have made particular nice points in functions also.
Howdy! This post couldn’t be written any better! Looking through this post reminds me of my previous roommate! He continually kept preaching about this. I will send this information to him. Pretty sure he will have a very good read. Thank you for sharing!
Why Nobody Cares About SEO Consultant London Near Me
5 Laws That Anyone Working In SEO Tools Should Be Aware Of seo tools for windows
(Optionfundamentals.com)
Everything is very open with a precise explanation of the challenges. It was truly informative. Your website is very helpful. Many thanks for sharing.
7 Secrets About Rewriter Text That Nobody Will Tell You Rewrite content ai
10 No-Fuss Ways To Figuring Out Your Affordable
SEO Services Digital Marketing Services
A How-To Guide For Search Engine Optimization Seo From Beginning To End search Engine Optimization tools
There are lots of interesting points outlined in this article but I don’t know if I go along with every one of them. There is certainly some validity but I am going to hold my opinions until I look into it more. Fine article, thanks and we want more! Added to FeedBurner too
Aw, this became a really good post. In thought I must devote writing similar to this moreover – taking time and actual effort to create a excellent article… but exactly what do I say… I procrastinate alot and no means seem to get something carried out.
A motivating discussion is worth comment. I do believe that you ought to write more on this issue, it might not be a taboo subject but typically people don’t discuss such issues. To the next! Many thanks!
I discovered your site web site on google and appearance a number of your early posts. Always maintain the really good operate. I merely extra increase RSS feed to my MSN News Reader. Looking for forward to reading a lot more from you afterwards!…
You’ll Never Guess This Seo Software Backlink’s Benefits Software backlink
See What Local SEO Companies Near Me Tricks The Celebs Are Utilizing seo companies near me – Tania –
The when Someone said a blog, I really hope who’s doesnt disappoint me around this blog. I am talking about, It was my choice to read, but When i thought youd have something intriguing to express. All I hear is really a few whining about something that you could fix in case you werent too busy trying to find attention.
Hey there! I know this is kinda off topic but I was wondering if you knew where I could get a captcha plugin for my comment form? I’m using the same blog platform as yours and I’m having difficulty finding one? Thanks a lot!
Having read this I believed it was extremely informative. I appreciate you finding the time and effort to put this article together. I once again find myself personally spending a significant amount of time both reading and leaving comments. But so what, it was still worth it.
It’s rare knowledgeable folks with this topic, but the truth is sound like what happens you are talking about! Thanks
some stores have really bad customer service while others have topnotch custmer service,,
hi this post help me full . .if you want watches men visit my sites is very help you for men watches. .thank man good job.
when i was still in high school, i always planned to take pyschology because it gives me great interest~
I��ve really enjoyed reading your content. You obviously know what you might be having a debate about! Your websites are simple to navigate too, I��ve bookmarked it inside favourites. . . . .
A person’s Are generally Weight loss is definitely a practical and flexible an eating plan method manufactured for those who suffer that want to weight loss and therefore ultimately conserve a much more culture. weight loss
Youre so cool! I dont suppose Ive read anything this way before. So nice to find somebody with original ideas on this subject. realy thank you for beginning this up. this website is one thing that is needed over the internet, an individual if we do originality. beneficial problem for bringing interesting things on the web!
Audio started playing anytime I opened up this blog, so annoying!
tired from the day I had…..Abbi’s team won their tournament game so we finished UNDEFEATED!!! But it was a rough game and I had to get a little redneck, but we won. Then dress shopping with the girls! Ashley looked beautiful and got a great dress! n.
“Last week I dropped by this site and as usual great content and ideas. Love the lay out and color scheme”
Thanks for sharing your expertise on such an important topic.
I’m impressed, I need to say. Really rarely do I encounter a weblog that’s each educative and entertaining, and let me tell you, you have got hit the nail on the head. Your concept is outstanding; the issue is something that not enough persons are talking intelligently about. I am very completely happy that I stumbled throughout this in my search for one thing regarding this.
Thanks for the post, I liked its content and style. I discovered this web site on Google and also have right now added this in order to my personal book marks. I’ll be sure to visit again quickly.
Thanks for shedding light on the importance of professional asbestos removal.
Excellent! I appreciate your input to this matter. It has been insightful. my blog: half marathon training schedule
Fantastic website. A lot of helpful info here. I’m sending it to several pals ans also sharing in delicious. And obviously, thank you on your effort!
Hi, extremely interesting article. My sister and I have been recently looking to find comprehensive info on this subject kind of stuff for a short time, but we didn’t until now. Do you think you can also make some youtube videos about this, I believe your website will be far more detailed in case you did. In any other case, oh well. I will be checking out on this website in the forseeable future. Email me to keep me up to date. granite countertops cleveland
Thanks for the tips on safely handling asbestos removal. Very useful!
This web site definitely has all of the information and facts I wanted about this subject and didn’t know who to ask.
This is a really good read for me. Must agree that you are one of the coolest blogger I ever saw. Thanks for posting this useful information. This was just what I was on looking for. I’ll come back to this blog for sure!
First of all, permit my family recognize a person?s command during this make a difference. Despite the reality this is certainly brand new , by no meanstheless quickly soon after registering your site, this intellect has exploded extensively. Permit all of us to consider maintain of one?s rss to assistance maintain in touch with whatsoever probable messages Sincere understand but will pass it on to aid admirers and my individualized are living members
Very well written post. It will be beneficial to everyone who employess it, as well as myself. Keep up the good work – can’r wait to read more posts.
Ten blog to cenne źródło informacji dla każdego, kto myśli o SEO.
To były bardzo pomocne informacje na temat kroków w SEO.
Dzięki za ważne informacje na temat zagrożeń SEO i potrzeby ich usunięcia.
Dzięki za ważne informacje na temat zagrożeń SEO i potrzeby ich usunięcia.
15 Secretly Funny People In Affordable SEO Company cheapest
Doceniam skupienie się na zatrudnianiu certyfikowanych profesjonalistów do SEO.
To były bardzo pomocne informacje na temat kroków w SEO.
To było bardzo oświecające. Na pewno skontaktuję się z profesjonalistą od SEO.
Dzięki za praktyczne wskazówki dotyczące znalezienia wykwalifikowanego specjalisty SEO.
5 Killer Quora Answers To Best Vps For SEO Tools seo tools And software
Ten post był bardzo pouczający na temat procesu SEO. Dzięki!
SEO Tools And Software: 10 Things I’d Like To Have Learned Sooner search Ranking software
Ten post był bardzo pouczający na temat procesu SEO. Dzięki!
Guide To Website Search Engine Optimization: The Intermediate Guide
On Website Search Engine Optimization website search engine optimization (Venetta)
17 Reasons To Not Not Ignore Tier Backlinks Multi Tiered Link Building
Doceniam skupienie się na zatrudnianiu certyfikowanych profesjonalistów do SEO.
Dzięki za kompleksowy przewodnik po SEO. Bardzo pouczający!
You’ll Be Unable To Guess Seo Content Marketing’s Secrets Seo Content Marketing;
F48.Ee,
It has always been my belief that good writing like this takes research and talent. It’s very apparent you have done your homework. Great job!
I tried to submit a comment previously, although it has not shown up. I believe your spam filter may possibly be broken?
Nice post. I find out something very complicated on diverse blogs everyday. It will always be stimulating you just read content off their writers and practice a little there. I’d would rather use some using the content on my small blog regardless of whether you do not mind. Natually I’ll supply you with a link for your web weblog. Appreciate your sharing.
The Most Pervasive Problems In Affordable SEO Company London Affordable seo For small businesses
10 Facts About SEO Consultant Near Me That Insists
On Putting You In An Optimistic Mood Seo Company Near Me
9 Signs You’re A Local SEO Company London Expert seo digital agency london, http://www.languagelink.Ru,
Hi my own cherished one! I wish to state that this kind of post is awesome, good composed and include virtually all important infos. I?|d want to seem added content such as this .
GSA SER Help Tips To Relax Your Daily Lifethe One GSA SER Help Trick
That Everybody Should Know gsa ser help [https://dostalek.blog.idnes.cz/Redir.aspx?url=http://rvolchansk.ru/user/koreanmass63/]
You’ll Never Be Able To Figure Out This Search Engine
Optimization Service Near Me’s Tricks Search Engine Optimization Service Near Me
Hi there, I found your site via Google while looking for a related topic, your web site came up, it looks great. I’ve bookmarked it in my google bookmarks.
Oh my goodness! an amazing article dude. Thanks Nevertheless I am experiencing concern with ur rss . Don’t know why Unable to subscribe to it. Is there anybody getting an identical rss downside? Anyone who is aware of kindly respond. Thnkx
your blog is so effortless to read, i like this article, so maintain posting far more
site promotion will always be a tedious job but you can outsource site promotion on some indian or pakistani guy`
Find Out What The Best Link Building Software The
Celebs Are Using backlink builder software
It is best to participate in a contest for among the best blogs on the web. I will suggest this web site!
Five Things You’ve Never Learned About Best SEO Software
UK Tools For Seo
7 Things About SEO Tool Vps You’ll Kick Yourself For Not Knowing Seo Software For Website
5 Killer Quora Answers On All SEO Company In London all seo company in london (https://www.g1tv.co.kr/)
Best Linkbuilding Software Tools To Make Your Daily Life Best Linkbuilding Software Technique Every Person Needs To
Be Able To best Linkbuilding software
Wow I’m frustrated. I’m not pointing fingers at you though, personally I think that its those that aren’t motivated to change.
You’ll Never Guess This Local SEO Services Company’s Benefits local seo services company (https://www.quendu.com)
genium vidoe mamelucos video headbanger video game morning star videography.
9 Things Your Parents Teach You About GSA SER Ranker
Gsa
Gran post y sitio. Bueno que AOL en la lista por lo que he podido llegar hasta aquí. Este sitio va a ir no en Mis marcadores a partir de ahora.
I’m truly enjoying the design and layout of your website. It’s a very easy on the eyes which makes it much more enjoyable for me to come here and visit more often. Did you hire out a developer to create your theme? Fantastic work!
Ten Common Misconceptions About SEO Tools Software That
Don’t Always Hold best seo Tools software (seotaisaku-top.1-coin.jp)
One word “informative”, you broke down precisely what I have been looking for. Looking forward to reading more in the future.
“Ask Me Anything”: Ten Answers To Your Questions About Local SEO Expert local seo Service
I like your blog. It looks pretty informative. I will share this with my brother.
Oh my goodness! an amazing post dude. Thanks a lot Even so We are experiencing problem with ur rss . Do not know why Can not sign up for it. Could there be anyone finding identical rss issue? Anybody who knows kindly respond. Thnkx
The 12 Most Popular Local SEO Agency London Accounts To Follow On Twitter link building Agency london
What’s The Current Job Market For GSA SER How To Professionals?
gsa ser how to
Your insights are invaluable.에볼루션 배팅 취소
The Secret Secrets Of Ai To Rewrite Text Article rewrite Ai
Наша мастерская предлагает высококачественный сервисный центр по ремонту видеокарты в москве любых брендов и моделей. Мы знаем, насколько важны для вас ваши видеокарты, и стремимся предоставить услуги высочайшего уровня. Наши опытные мастера оперативно и тщательно выполняют работу, используя только оригинальные запчасти, что обеспечивает длительную работу выполненных работ.
Наиболее общие проблемы, с которыми сталкиваются обладатели графических адаптеров, включают перегрев, выход из строя памяти, проблемы с коннекторами, проблемы с контроллером и программные сбои. Для устранения этих неисправностей наши опытные мастера проводят ремонт системы охлаждения, памяти, разъемов, контроллеров и ПО. Обращаясь в наш сервисный центр, вы получаете надежный и долговечный ремонт видеокарт на выезде.
Подробная информация представлена на нашем сайте: https://remont-videokart-biz.ru
How To Tell If You’re Prepared To Go After SEO Tools UK seo analyzer Software
7 Simple Tips For Refreshing Your Social SEO Software Seo Tool (co.n.s.u.m.erb.b.ek@minecraftcommand.science/profile/judgewedge13″>http://Bridgejelly71>Co.N.S.U.M.Erb.B.Ek@Minecraftcommand.Science/)
5 Tier Link Building In Seo Tips You Must Know About For 2023
Tier 2 Seo
5 Things That Everyone Is Misinformed About On The Subject Of
Tier 2 Seo Company tier 1 link
10 Things Everyone Gets Wrong About Software Backlink
get
See What Professional SEO Company Tricks The Celebs Are Utilizing professional Seo Company
SEO Tools UK 101 A Complete Guide For Beginners
meta tags
12 Companies That Are Leading The Way In Search Engine Optimization Services Search optimization services
The Most Hilarious Complaints We’ve Heard About SEO Tools tools for seo (https://Shorturl.vtcode.vn/seotraffictools377079)
20 Resources That Will Make You More Efficient At Affordable SEO Near Me Best seo consultant affordable
SEO UK Company Tools To Make Your Everyday Lifethe Only SEO UK Company Trick That Should Be Used By Everyone Be Able To Seo Uk Company
Solid SEO Tools Tools To Streamline Your Daily Lifethe One Solid SEO Tools Trick That Every Person Should Learn Solid seo tools
Good post. I learn something new and challenging on websites I stumbleupon everyday. It’s always interesting to read through articles from other writers and practice something from their websites.
I like reading an article that will make men and women think. Also, thank you for permitting me to comment.
This website was… how do I say it? Relevant!! Finally I’ve found something that helped me. Kudos.
Students studying law must employ arguments properly. They might make the wrong diagnosis, much like with homework related to medical services and nursing. Language is important in law, and proofreading takes longer than writing. As a result, a lot of law learners seek writing assistance services named online Law assignment help because they are unsure of where to obtain facts. We are aware that law duties, which need in-depth research of agreements, discussions, court rulings, and laws, can be quite difficult. Even those with the greatest potential may find the quantity of study and reading necessary to be daunting. Because it offers learners professional assistance and guarantees that the assignments are not only professionally written but also compliant with academic honesty and legal requirements, our qualified law assignment help is essential. Furthermore, by producing unique content that is catered to their requirements, our assistance can protect pupils from the danger of duplication.
Excellent site you have here.. It’s hard to find high-quality writing like yours these days. I seriously appreciate individuals like you! Take care!!
Online English classes are a great way to improve your language skills from the comfort of your own home. However, communicating effectively in English requires more than just a good internet connection and a webcam. To be truly effective, you need to master the grammar and syntax of the English language. In this blog by takeonlineclassesnow, we’ll discuss common grammar mistakes that students make in English online classes, and provide tips and examples for avoiding them.
Aw, this was an exceptionally good post. Taking the time and actual effort to create a really good article… but what can I say… I hesitate a whole lot and don’t manage to get anything done.
I love it whenever people get together and share thoughts. Great blog, keep it up.
They are the best at helping with statistics homework. You can trust that they are committed to and know a lot about the field of statistics because they have a team of experts ready to answer your questions. For your benefit, they will make sure that they do everything right with the task you gave them. We want to help, and we do. As a student of statistics, if any of the above cases sound familiar, do not worry—we have got you covered. If you are a student looking for online help with your statistics homework, you can be sure that the help you get from us is the best. If you get help from professionals, you can be sure that the grade you get for your statistics project will be much better than average. Using the simplest ideas will help you find the best and most creative answers to your statistics homework, no matter how hard it is. Additionally, if you ever feel like you have too much work and think, “ pay someone to take my online math class,” know that you can get professional help to continue doing well in college.
Saved as a favorite, I really like your blog.
Thank you for sharing such valuable information.카카오 대출
I’m glad someone is discussing this openly.개인사업자 대출
You’ve made this topic accessible to everyone.pragmatic korea
Your research really makes a difference.워드프레스 seo
May I simply say what a relief to discover somebody that really knows what they are discussing on the net. You actually realize how to bring a problem to light and make it important. More and more people need to look at this and understand this side of the story. It’s surprising you are not more popular given that you surely possess the gift.
I’m very happy to find this page. I need to to thank you for your time just for this fantastic read!! I definitely savored every part of it and I have you saved as a favorite to see new things in your web site.
porn
An intriguing discussion is definitely worth comment. I do think that you need to write more on this topic, it might not be a taboo matter but usually folks don’t discuss these issues. To the next! Cheers.
I could not resist commenting. Well written!
After looking into a number of the blog posts on your site, I really like your way of writing a blog I book-marked it to my bookmark site list and will be checking back in the near future Please visit my website too and tell me your opinion
I used to be able to find good information from your blog posts.
casino
You ought to be a part of a contest for one of the highest quality websites on the internet. I will recommend this website!
i am a movie addict and i watch a lot of movie in just one night, the greatest movie for me is Somewhere In Tome::
very nice post, i surely really like this site, continue it
Hello there! This post could not be written much better! Going through this article reminds me of my previous roommate! He always kept talking about this. I will forward this post to him. Pretty sure he’ll have a great read. Thanks for sharing!
I rather encourage you to learn about the flexibility of forms, fields and widgets and how it’s used in the automatic Admin interface.
Spot lets start on this write-up, I really think this amazing site needs much more consideration. I’ll more likely once more to see a great deal more, thank you that info.
Your style is unique in comparison to other people I’ve read stuff from. Thanks for posting when you’ve got the opportunity, Guess I will just bookmark this page.
Way cool! Some extremely valid points! I appreciate you penning this article and the rest of the website is extremely good.
Way cool! Some very valid points! I appreciate you writing this write-up and also the rest of the site is very good.
bookmarked!!, I love your site.
Spot on with this write-up, I absolutely believe that this website needs much more attention. I’ll probably be returning to see more, thanks for the advice!
Aw, this was an extremely nice post. Finding the time and actual effort to make a great article… but what can I say… I procrastinate a lot and never manage to get nearly anything done.
https://artdaily.com/news/171650/Mp3Juice-Review–The-Pros-and-Cons-You-Need-to-Know
I absolutely love your site.. Excellent colors & theme. Did you develop this web site yourself? Please reply back as I’m attempting to create my very own website and want to find out where you got this from or what the theme is named. Thanks.
porn
An outstanding share! I’ve just forwarded this onto a co-worker who had been conducting a little research on this. And he actually ordered me lunch due to the fact that I found it for him… lol. So allow me to reword this…. Thanks for the meal!! But yeah, thanks for spending time to discuss this subject here on your blog.
Get 20% Off on Health and Social Care Level 3 Assignment Help
health and social care level 3 assignment help
“Get professional assistance with your HND health and social care level 3 assignment help from HND Assignment Help! Our team of experts provides personalized, high-quality, and affordable support tailored to your needs. Achieve academic excellence with our reliable services. Contact us today!
porn cannibalism
Whoa a good deal of great material.|
gay porn
Oh my goodness! Incredible article dude! Thanks, However I am encountering troubles with your RSS. I don’t know why I am unable to subscribe to it. Is there anyone else having the same RSS problems? Anyone that knows the answer will you kindly respond? Thanx.
online casino
I used to be able to find good info from your articles.
casino
I like the valuable information you provide in your articles.
I will bookmark your weblog and check once more here regularly.
I am relatively certain I’ll learn a lot of new
stuff proper here! Best of luck for the next!
Here is my web site :: blog link
It’s hard to come by knowledgeable people for this subject, however, you sound like you know what you’re talking about! Thanks
Very good blog post. I definitely appreciate this site. Stick with it!
Nice post. I learn something totally new and challenging on sites I stumbleupon on a daily basis. It’s always helpful to read content from other authors and practice something from their sites.
It’s difficult to find knowledgeable people for this subject, but you sound like you know what you’re talking about! Thanks
A fascinating discussion is worth comment. I do think that you should write more about this subject matter, it might not be a taboo subject but generally folks don’t speak about these issues. To the next! Kind regards!
Spot on with this write-up, I really think this amazing site needs a lot more attention. I’ll probably be back again to read through more, thanks for the advice!
slot
Hi, I do believe this is an excellent website. I stumbledupon it 😉 I may come back once again since I saved as a favorite it. Money and freedom is the greatest way to change, may you be rich and continue to help others.
I could not refrain from commenting. Perfectly written.
casino
slot
Spot on with this write-up, I truly feel this site needs a great deal more attention. I’ll probably be returning to read through more, thanks for the advice.
You should be a part of a contest for one of the greatest blogs on the web. I most certainly will highly recommend this web site!
Cool blog! Is your theme custom made or did you download it from somewhere?
A design like yours with a few simple tweeks would really make my
blog shine. Please let me know where you got your design. With thanks
I’m going to share this with my network.seo 백링크
Awesome stuff. Thanks.|
lgbt porn
Great job on explaining this complex topic!seo 최적화
lesbian porn
online casino
Therefore, we provide you with the best and quality biology assignment helper. Biology is a complicated subject about living organisms that consumes a large amount of time. It demands a lot of memorization and note taking, but as mentioned, we are here to provide you with a solution to every possible problem, instantly and more effectively.
homosexual porn
Your style is really unique compared to other people I’ve read stuff from. Many thanks for posting when you’ve got the opportunity, Guess I’ll just book mark this site.
porn
xxx
not safe
Good post. I’m experiencing some of these issues as well..
Everyone loves it when people come together and share views. Great site, keep it up.
hacklink
hacklink
It’s hard to come by educated people on this subject, but you seem like you know what you’re talking about! Thanks
incest porn