Scalable, reproducible, and cost-effective processing of large-scale medical imaging datasets
Michael E. Kim, Karthik Ramadass, Chenyu Gao, Praitayini Kanakaraj, Nancy R. Newlin, Gaurav Rudravaram, Kurt G. Schilling, Blake E. Dewey, Derek Archer, Timothy J. Hohman, Zhiyuan Li, Shunxing Bao, Bennett A. Landman, and Nazirah Mohd Khairi. Scalable, reproducible, and cost-effective processing of large-scale medical imaging datasets. SPIE Medical Imaging: Imaging Informatics, 2025, February, San Diego, California.
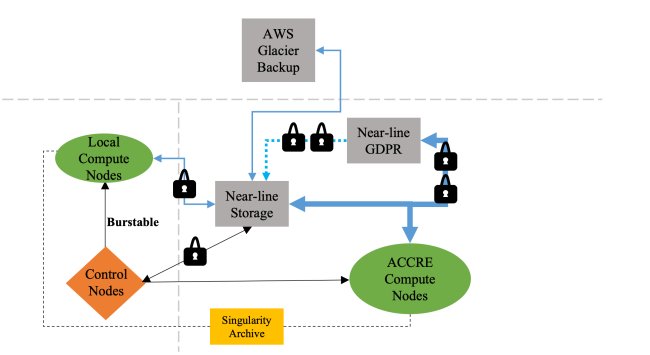
Figure 3. We propose an adaptive solution for scalable managing of large neuroimaging datasets. The storage is split between two separate servers: one for datasets that require additional security compliance measures (such as GDPR), and another for all other datasets. The data on the high-security server is symbolically linked to the general-purpose storage only for users with authorized access. Both the storage servers and all HPC compute nodes share a high bandwidth network are located at the ACCRE cluster at Vanderbilt University, which uses a SLURM scheduler for managing computation jobs. For running processing pipelines on a dataset, we design an automated data query to create processing scripts for all data that can be run through a pipeline and have not already been run successfully. A SLURM job array script is also generated that can easily be submitted for processing. All scripts run containerized code located in a Singularity archive. For instances when quick processing bursts are required or ACCRE is unavailable, our data query and script generator is compatible with local servers as well. To further reduce overhead while maintaining data integrity, data are backed up to an Amazon Glacier storage instance, which is comparatively cheaper than backup under ACCRE. All querying and job submission is done from local control nodes. Blue lines represent data transfer, single arrow black lines represent job submission, double arrow black lines represent query, and dotted lines represent symbolic links. Line thickness represents transfer speed.
Purpose: Curating, processing, and combining large-scale medical imaging datasets from national studies is a non-trivial task due to the intense computation and data throughput required, variability of acquired data, and associated financial overhead. Existing platforms or tools for large-scale data curation, processing, and storage have difficulty achieving a viable cost- to-scale ratio of computation speed for research purposes, either being too slow or too expensive. Additionally, management and consistency of processing large data in a team-driven manner is a non-trivial task.
Approach: We design a BIDS- compliant method for an efficient and robust data processing pipeline of large-scale diffusion-weighted and T1-weighted MRI data compatible with low-cost, high-efficiency computing systems.
Results: Our method accomplishes automated querying of data available for processing and process running in a consistent and reproducible manner that has long-term stability, while using heterogenous low-cost computational resources and storage systems for efficient processing and data transfer. We demonstrate how our organizational structure permits efficiency in a semi-automated data processing pipeline and show how our method is comparable in processing time to cloud-based computation while being almost 20 times more cost-effective. Our design allows for fast data throughput speeds and low latency to reduce the time for data transfer between storage servers and computation servers, achieving an average of 0.60 Gb/s compared to 0.33 Gb/s for using cloud-based processing methods.
Conclusion: The design of our workflow engine permits quick process running while maintaining flexibility to adapt to newly acquired data.