EMR Category
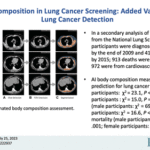
AI Body Composition in Lung Cancer Screening: Added Value Beyond Lung Cancer Detection
Aug. 31, 2023—Kaiwen Xu, Mirza S. Khan, Thomas Z. Li, Riqiang Gao, James G. Terry, Yuankai Huo, Thomas A. Lasko, John Jeffrey Carr, Fabien Maldonado, Bennett A. Landman, Kim L. Sandler Paper: https://pubs.rsna.org/doi/epdf/10.1148/radiol.222937 Abstract Background An artificial intelligence (AI) algorithm has been developed for fully automated body composition assessment of lung cancer screening noncontrast low-dose CT of the...
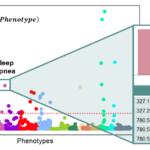
pyPheWAS Explorer: a visualization tool for exploratory analysis of phenome-disease associations
Aug. 31, 2023—Cailey I. Kerley, Karthik Ramadass, Tin Q. Nguyen, Laurie E. Cutting, Bennett A. Landman, Matthew Berger Paper: https://pubmed.ncbi.nlm.nih.gov/37021295/ Code: https://github.com/MASILab/pyPheWAS Abstract Objective To enable interactive visualization of phenome-wide association studies (PheWAS) on electronic health records (EHR). Materials and Methods Current PheWAS technologies require familiarity with command-line interfaces and lack end-to-end data visualizations. pyPheWAS Explorer allows users to...
Batch size: go big or go home? Counterintuitive improvement in medical autoencoders with smaller batch size
Nov. 13, 2022—Cailey I. Kerley*, Leon Y. Cai*, Yucheng Tang, Lori L. Beason-Held, Susan M. Resnick, Laurie E. Cutting, and Bennett A. Landman. *Equal first authorship Abstract Batch size is a key hyperparameter in training deep learning models. Conventional wisdom suggests larger batches produce improved model performance. Here we present evidence to the contrary, particularly when using autoencoders...
pyPheWAS: A Phenome-Disease Association Tool for Electronic Medical Record Analysis
Jan. 12, 2022—Kerley, C.I., Chaganti, S., Nguyen, T.Q. et al. pyPheWAS: A Phenome-Disease Association Tool for Electronic Medical Record Analysis. Neuroinform (2022). https://doi.org/10.1007/s12021-021-09553-4 Full text: NIHMSID, Springer Abstract Along with the increasing availability of electronic medical record (EMR) data, phenome-wide association studies (PheWAS) and phenome-disease association studies (PheDAS) have become a prominent, first-line method of analysis for uncovering...
Pancreas CT Segmentation by Predictive Phenotyping
Dec. 14, 2021—Y. Tang, R.Gao, H.H.Lee, Q.Yang, X.Yu,Y.Zhou, S.Bao, Y.Huo, J.Spraggins, J.Virostko, Z.Xu, B.A. Landman. “Pancreas CTSegmentation by Predictive Phenotyping”. International Conference on MedicalImage Computing and Computer Assisted Intervention(MICCAI), 2021 Full Text: https://link.springer.com/chapter/10.1007/978-3-030-87193-2_3 Abstract Pancreas CT segmentation offers promise at understanding the structural manifestation of metabolic conditions. To date, the medical primary record of conditions that impact...
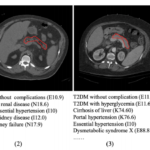
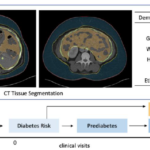
Prediction of Type II Diabetes Onset with Computed Tomography and Electronic Medical Records
Dec. 7, 2020—Yucheng Tang, Riqiang Gao, Ho Hin Lee, Quinn Stanton Wells, Ashley Spann, James Gregory Terry, Jeff Carr, Yuankai Huo, Shunxing Bao and Bennett A. Landman, “Prediction of Type II Diabetes Onset with Computed Tomography and Electronic Medical Records”, MICCAI CLIP, 2020. Full Text: Abstract Type II diabetes mellitus (T2DM) is a significant public health concern...
A cross-platform informatics system for the Gut Cell Atlas: integrating from clinical, anatomical and histological data
Dec. 6, 2020—Shunxing Bao, Sophie Chiron, Yucheng Tang, Cody N. Heiser, Austin N. Southard-Smith, Ho Hin Lee, Marisol A. Ramirez, Yuankai Huo, Mary K. Washington, Elizabeth A. Scoville, Joseph T. Roland, Qi Liu, Ken S. Lau, Keith T. Wilson, Lori A. Coburn, Bennett A. Landman, A cross-platform informatics system for the Gut Cell Atlas: integrating from clinical,...