pyPheWAS Explorer: a visualization tool for exploratory analysis of phenome-disease associations
Cailey I. Kerley, Karthik Ramadass, Tin Q. Nguyen, Laurie E. Cutting, Bennett A. Landman, Matthew Berger
Paper: https://pubmed.ncbi.nlm.nih.gov/37021295/
Code: https://github.com/MASILab/pyPheWAS
Abstract
Objective
To enable interactive visualization of phenome-wide association studies (PheWAS) on electronic health records (EHR).
Materials and Methods
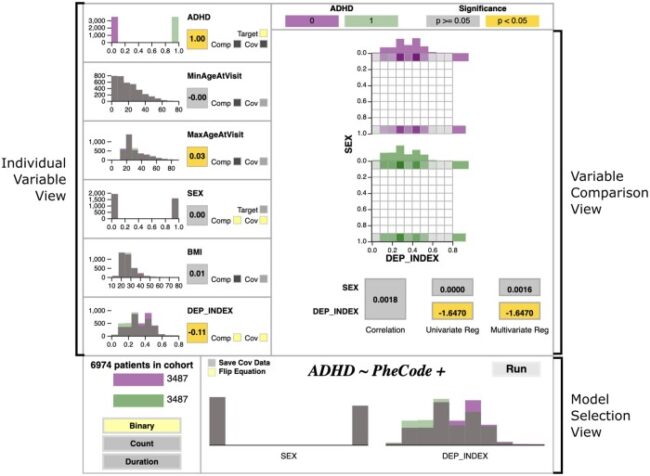
Current PheWAS technologies require familiarity with command-line interfaces and lack end-to-end data visualizations. pyPheWAS Explorer allows users to examine group variables, test assumptions, design PheWAS models, and evaluate results in a streamlined graphical interface.
Results
A cohort of attention deficit hyperactivity disorder (ADHD) subjects and matched non-ADHD controls is examined. pyPheWAS Explorer is used to build a PheWAS model including sex and deprivation index as covariates, and the Explorer’s result visualization for this model reveals known ADHD comorbidities.
Discussion
pyPheWAS Explorer may be used to rapidly investigate potentially novel EHR associations. Broader applications include deployment for clinical experts and preliminary exploration tools for institutional EHR repositories.
Conclusion
pyPheWAS Explorer provides a seamless graphical interface for designing, executing, and analyzing PheWAS experiments, emphasizing exploratory analysis of regression types and covariate selection.