Exploring shared memory architectures for end-to-end gigapixel deep learning
Lucas W. Remedios, Leon Y. Cai, Samuel W. Remedios, Karthik Ramadass, Aravind Krishnan, Ruining Deng, Can Cui, Shunxing Bao, Lori A. Coburn, Yuankai Huo, Bennett A. Landman (2023). Exploring shared memory architectures for end-to-end gigapixel deep learning. MIDL 2023 short paper track
Full text: NIHMSID
Abstract
Deep learning has made great strides in medical imaging, enabled by hardware advances in GPUs. One major constraint for the development of new models has been the saturation of GPU memory resources during training. This is especially true in computational pathology, where images regularly contain more than 1 billion pixels. These pathological images are traditionally divided into small patches to enable deep learning due to hardware limitations. In this work, we explore whether the shared GPU/CPU memory architecture on the M1 Ultra systems-on-a-chip (SoCs) recently released by Apple, Inc. may provide a solution. These affordable systems (less than $5000) provide access to 128 GB of unified memory (Mac Studio with M1 Ultra SoC). As a proof of concept for gigapixel deep learning, we identified tissue from background on gigapixel areas from whole slide images (WSIs). The model was a modified U-Net (4492 parameters) leveraging large kernels and high stride. The M1 Ultra SoC was able to train the model directly on gigapixel images (16000×64000 pixels, 1.024 billion pixels) with a batch size of 1 using over 100 GB of unified memory for the process at an average speed of 1 minute and 21 seconds per batch with Tensorflow 2/Keras. As expected, the model converged with a high Dice score of 0.989 ± 0.005. Training up until this point took 111 hours and 24 minutes over 4940 steps. Other high RAM GPUs like the NVIDIA A100 (largest commercially accessible at 80 GB, ∼$15000) are not yet widely available (in preview for select regions on Amazon Web Services at $40.96/hour as a group of 8). This study is a promising step towards WSI-wise end-to-end deep learning with prevalent network architectures.
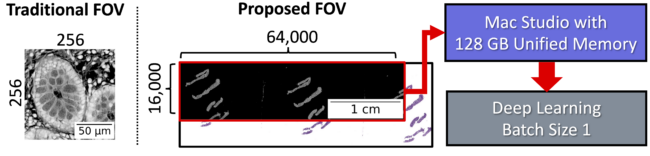
Usually, small patches (eg. 256×256 pixels) are used in computational pathology. Enabled by the unified memory architecture, we instead use a gigapixel area (16000×64000 pixels) from preprocessed images, which is larger than a traditional single tissue field of view.