Resources : Software
Distributed Automation for XNAT
 The Vanderbilt University Institute for Imaging Science (VUIIS) Center for Computational Imaging (CCI) has developed a database built on XNAT housing over a quarter of a million scans. The database provides framework for (1) rapid prototyping, (2) large scale batch processing of images and (3) scalable project management. The system uses the web-based interfaces of XNAT and REDCap to allow for graphical interaction. A python middleware layer, the Distributed Automation for XNAT (DAX) package, distributes computation across the Vanderbilt Advanced Computing Center for Research and Education high performance computing center. All software are made available in open source for use in combining portable batch scripting (PBS) grids and XNAT servers.
The Vanderbilt University Institute for Imaging Science (VUIIS) Center for Computational Imaging (CCI) has developed a database built on XNAT housing over a quarter of a million scans. The database provides framework for (1) rapid prototyping, (2) large scale batch processing of images and (3) scalable project management. The system uses the web-based interfaces of XNAT and REDCap to allow for graphical interaction. A python middleware layer, the Distributed Automation for XNAT (DAX) package, distributes computation across the Vanderbilt Advanced Computing Center for Research and Education high performance computing center. All software are made available in open source for use in combining portable batch scripting (PBS) grids and XNAT servers.
- GitHub: https://github.com/VUIIS/dax
- Article: https://www.ncbi.nlm.nih.gov/pubmed/25988229
- Documentation / Tutorials: http://xnat.vanderbilt.edu/index.php/Main_Page
JIST: Java Image Science Toolkit
Java Image Science Toolkit (JIST) provides a native Java-based imaging processing environment similar to the ITK/VTK paradigm. Initially developed as an extension to MIPAV (CIT, NIH, Bethesda, MD), the JIST processing infrastructure provides automated GUI generation for application plug-ins, graphical layout tools, and command line interfaces.
This repository maintains the current multi-institutional JIST development tree and is recommended for public use and extension. JIST was originally developed at IACL and MedIC (Johns Hopkins University) and is now also supported by MASI (Vanderbilt University).
Covariate-Adjusted Restricted Cubic Spline Regression
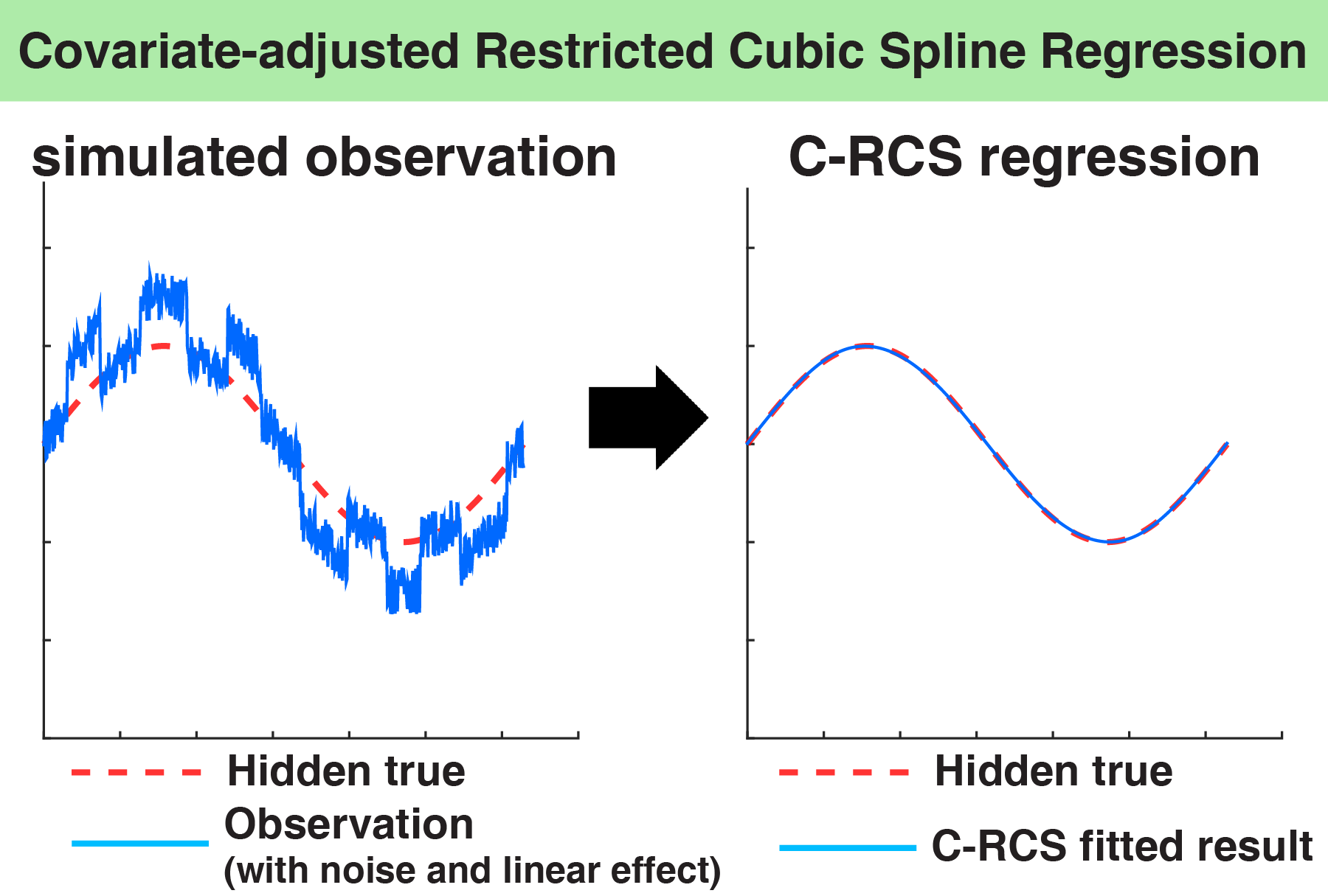 We propose to use covariate- adjusted restricted cubic spline (C-RCS) regression within a multi-site cross-sectional framework. This model allows for flexible consideration of nonlinear age-associated patterns while accounting for traditional covariates and interaction effects
We propose to use covariate- adjusted restricted cubic spline (C-RCS) regression within a multi-site cross-sectional framework. This model allows for flexible consideration of nonlinear age-associated patterns while accounting for traditional covariates and interaction effects
Citation: Yuankai Huo, Katherine Aboud, Hakmook Kang, Laurie E. Cutting, Bennett A. Landman, “Mapping Lifetime Brain Volumetry with Covariate-Adjusted Restricted Cubic Spline Regression from Cross-sectional Multi-site MRI”. In International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Athens, Greece, October 2016
- Download URL: https://www.nitrc.org/frs/?group_id=385
Augmented Reality Mirror
 The Augmented Reality Mirror is an open source rendering software to provide users with a virtual reality experience. With the provisions of a projection display, a laptop with multi-core GPU, and XBOX kinect, users can stand in front of the mirror and representative anatomical images will overlay the body. The images implemented are adapted to approximate the actual anatomy. The open source software will have an application programming interface (API) to allow users to develop additional content within the Augmented Reality Mirror.
The Augmented Reality Mirror is an open source rendering software to provide users with a virtual reality experience. With the provisions of a projection display, a laptop with multi-core GPU, and XBOX kinect, users can stand in front of the mirror and representative anatomical images will overlay the body. The images implemented are adapted to approximate the actual anatomy. The open source software will have an application programming interface (API) to allow users to develop additional content within the Augmented Reality Mirror.
Hadoop for Data Colocation
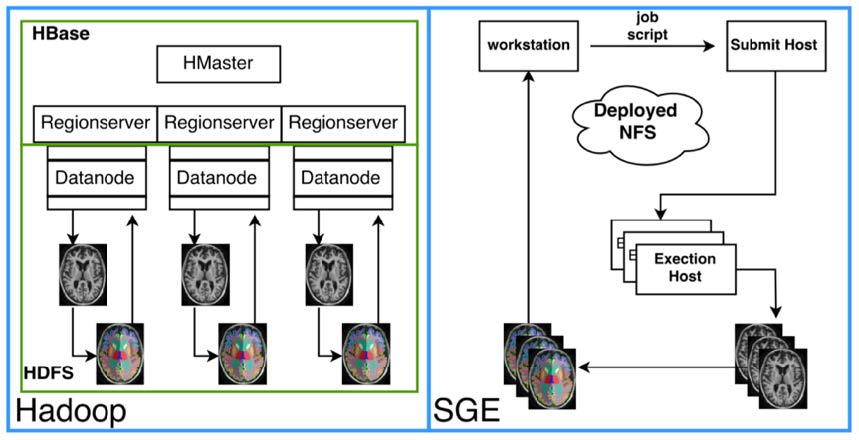 As imaging datasets and computing grid sizes grow larger, traditional computing’s separation of data and computational nodes create a problem. Moving data from where it is centrally stored to computational nodes can saturate a network with relatively few active processes. Under certain conditions, the bottleneck in the computing architecture becomes the network bandwidth. An inexpensive solution is to locate the data on the computational nodes to avoid the problem of saturating the network by copying data. This project provides our resources for performing medical image data colocation via Hadoop and Hbase.
As imaging datasets and computing grid sizes grow larger, traditional computing’s separation of data and computational nodes create a problem. Moving data from where it is centrally stored to computational nodes can saturate a network with relatively few active processes. Under certain conditions, the bottleneck in the computing architecture becomes the network bandwidth. An inexpensive solution is to locate the data on the computational nodes to avoid the problem of saturating the network by copying data. This project provides our resources for performing medical image data colocation via Hadoop and Hbase.
Robust Biological Parametric Mapping
Web Game for Collaborative Labeling
Continuous Integration Server
NeuroDebian
 The MASI lab has provided a mirror for the NeuroDebian project since 2010.
The MASI lab has provided a mirror for the NeuroDebian project since 2010.
