Nucleus subtype classification using inter-modality learning
Lucas W. Remedios, Shunxing Bao, Samuel W. Remedios, Ho Hin Lee, Leon Y. Cai, Thomas Li, Ruining Deng, Can Cui, Jia Li, Qi Liu, Ken S. Lau, Joseph T. Roland, Mary K. Washington, Lori A. Coburn, Keith T. Wilson, Yuankai Huo, Bennett A. Landman (2024). Nucleus subtype classification using inter-modality learning. SPIE Medical Imaging 2024 : Digital and Computational Pathology
Full text: NIHMSID
Abstract
Understanding the way cells communicate, co-locate, and interrelate is essential to understanding human physiology. Hematoxylin and eosin (H&E) staining is ubiquitously available both for clinical studies and research. The Colon Nucleus Identification and Classification (CoNIC) Challenge has recently innovated on robust artificial intelligence labeling of six cell types on H&E stains of the colon. However, this is a very small fraction of the number of real cell types. In this paper, we propose to use inter-modality learning to label previously un-labelable cell types on virtual H&E. We leveraged multiplexed immunofluorescence (MxIF) histology imaging to identify 14 subclasses of cell types. We performed style transfer to synthesize virtual H&E from MxIF and transferred the higher density labels from MxIF to these virtual H&E images. We then evaluated the efficacy of learning in this approach. We identified helper T and progenitor nuclei with positive predictive values of 0.34 ± 0.15 (prevalence 0.03 ± 0.01) and 0.47 ± 0.1 (prevalence 0.07 ± 0.02) respectively on virtual H&E. This is a promising step towards automating annotation in digital pathology.
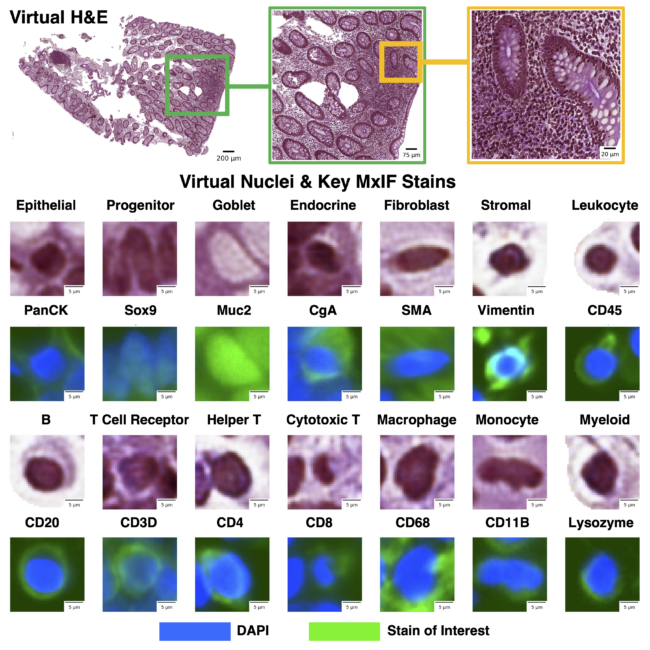
We leveraged inter-modality learning to investigate identification of cells on virtual H&E staining that are traditionally viewed with specialized staining. The realistic quality of our virtual H&E holds at multiple scales (top row). Representative nuclei from each of our 14 classes in both virtual H&E and MxIF illustrate intensity and morphological variation across cell types (lower section). Green is used to denote the MxIF stain of interest, which is a different stain for each of the 14 cell types in this figure. While the signal to identify these classes of nuclei is present in MxIF, the nucleus classes are more difficult to distinguish on virtual H&E.