Neuroimaging
Motivation
Medical imaging has provided powerful insights into understanding the structural and functional architecture of human anatomy and is widely used for the diagnosis, intervention, and management of clinical disorders. As we explore ever more subtle anatomical correlations in health and disease, we must look towards efficiently acquiring data on diverse subjects and making best use of this information. Large-scale imaging studies could be commissioned to map population variability; however, these studies face theoretical, technical, and practical challenges. Impediments include stability of imaging protocols, subject recruitment, data storage, consistent analysis, and, of course, excessive cost. Under the federal data sharing mandate, a plethora of investigators are releasing diverse, but internally well-controlled, data on scores of subjects. With the vast array of potential datasets available, it can be a severe challenge to carry out appropriate processing steps despite publicly available tools that might already exist for many of the procedures.
Research objectives
- Develop new analysis techniques to model tissue architecture from quantitative and multi-modal MRI imaging data.
- Exploit high performance computing to perform very large-scale image analysis.
- Design robust system architectures to capture algorithmic failure in image analysis algorithms.
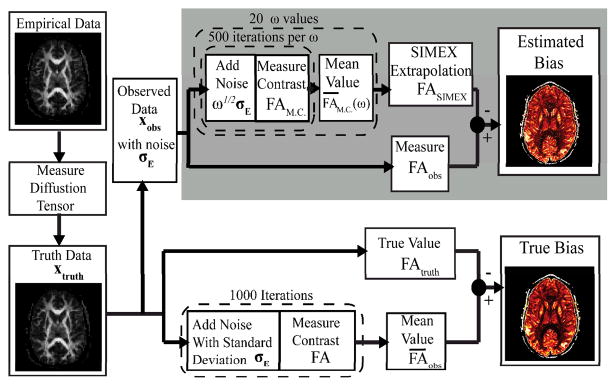
- Investigate techniques to characterize uncertainty in registration, segmentation and model fitting.
Progress
We developed and released the largest multi-modal resource of 3T MRI data to date (downloaded over 18,771 times) and have characterized large-scale pipelines for “Connectome”-like methods. We have continued developing and releasing image-processing tools; our primary platform – the Java Image Science Toolkit (JIST) has been downloaded over 13,477 times. We have created systems to characterize and predict algorithm runtime on heterogeneous imaging data and deploy image-processing software to multi-processor, traditional grid, and cloud computing systems using a common, modular code-base. We have created a novel Python-RedCAP-XNAT software package that allows for ready reuse of pipeline across projects and high throughput quality control of imaging data, which now serves as the primary analysis framework for more than a dozen funded NIH research grants.