A cross-platform informatics system for the Gut Cell Atlas: integrating from clinical, anatomical and histological data
Shunxing Bao, Sophie Chiron, Yucheng Tang, Cody N. Heiser, Austin N. Southard-Smith, Ho Hin Lee, Marisol A. Ramirez, Yuankai Huo, Mary K. Washington, Elizabeth A. Scoville, Joseph T. Roland, Qi Liu, Ken S. Lau, Keith T. Wilson, Lori A. Coburn, Bennett A. Landman, A cross-platform informatics system for the Gut Cell Atlas: integrating from clinical, anatomical and histological data, SPIE 2021 Medical Imaging (accepted)
Full Text: NIHMSID
Contacts: shunxing dot bao@vanderbilt dot edu
Abstract
The Gut Cell Atlas (GCA), an initiative funded by the Helmsley Charitable Trust, seeks to create a reference platform to understand the human gut, with a specific focus on Crohn’s disease. Although a primary focus of the GCA is on focusing on single-cell profiling, we seek to provide a framework to integrate other analyses on multi-modality data such as electronic health record data, radiological images, and histology tissues/images. Herein, we use the research electronic data capture (REDCap) system as the central tool for a secure web application that supports protected health information (PHI) restricted access. Our innovations focus on addressing the challenges with tracking all specimens and biopsies, validating manual data entry at scale, and sharing organizational data across the group. We present a scalable, cross-platform barcode printing/record system that integrates with REDCap. The central informatics infrastructure to support our design is a tuple table to track longitudinal data entry and sample tracking. The current data collection (by June 2020) is illustrated with types and formats of the data that the system collects. We estimate that 1 terabyte is needed for data storage per patient study. Our proposed data sharing informatics system addresses the challenges with integrating physical sample tracking, large files, and manual data entry with REDCap.
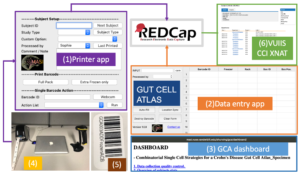
Figure 2. The data entry workflow of the GCA data collection. (1) According to patient types (i.e., healthy control vs Crohn’s disease), the printer app generates the barcodes). The barcodes are recorded to REDCap after printing. The barcode is human-readable, and scanner scannable for any further sample operations (i.e., distribute to other labs, store to a rack & box position, destroy a barcode). (2) The Location/Data entry app is a user-friendly software to help users enter data into REDCap to reduce manual input errors. (3) The GCA online dashboard is designed to show sample stats that users may care about, and it also allows for quality control and identification of potential inconsistencies with data entry in REDCap. (4) All apps are installed on a laptop workstation. The workstation connects to a barcode label printer and wireless barcode scanner. (5) A sample barcode for a frozen specimen. (6) The VUIIS CCI is a system based on XNAT that is used to store any large data in the GCA project.