Scalable quality control on processing of large diffusion-weighted and structural magnetic resonance imaging datasets
Michael E. Kim, Chenyu Gao, Nancy R. Newlin, Gaurav Rudravaram, Aravind R. Krishnan, Karthik Ramadass, Praitayini Kanakaraj, Kurt G. Schilling, Blake E. Dewey, David A. Bennett, Sid O’Bryant, Robert C. Barber, Derek Archer, Timothy J. Hohman, Shunxing Bao, Zhiyuan Li, Bennett A. Landman, Nazirah Mohd Khairi, The Alzheimer’s Disease Neuroimaging Initiative, The HABS-HD Study Team. “Scalable quality control on processing of large diffusion-weighted and structural magnetic resonance imaging datasets.” PLoS One 2025;20:e0327388. https://doi.org/10.1371/JOURNAL.PONE.0327388.
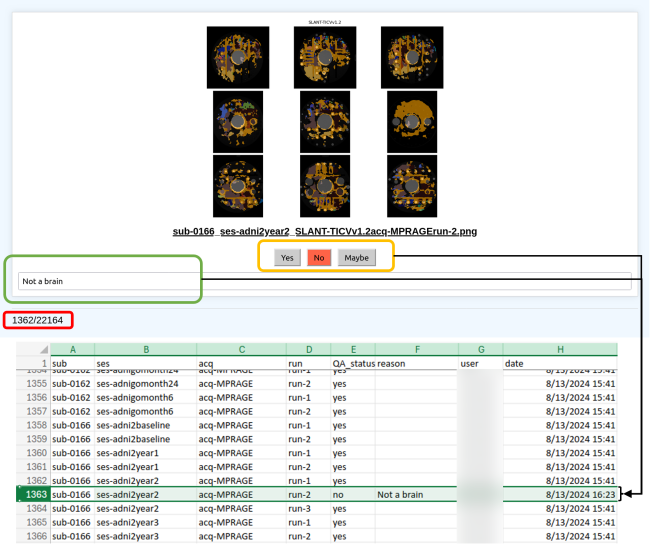
Purpose: Thorough quality control (QC) can be time consuming when working with large-scale medical imaging datasets, yet necessary, as poor-quality data can lead to erroneous conclusions or poorly trained machine learning models. Most efforts to reduce data QC time rely on quantitative outlier detection, which cannot capture every instance of algorithm failure. Thus, there is a need to visually inspect every output of data processing pipelines in a scalable manner.
Approach: We design a QC pipeline that allows for low time cost and effort across a team setting for a large database of diffusion-weighted and structural magnetic resonance images. Our proposed method satisfies the following design criteria: 1.) a consistent way to perform and manage quality control across a team of researchers, 2.) quick visualization of preprocessed data that minimizes the effort and time spent on the QC process without compromising the condition/caliber of the QC, and 3.) a way to aggregate QC results across pipelines and datasets that can be easily shared.In addition to meeting these design criteria, we also provide a comparison experiment of our method to an automated QC method for a T1-weighted dataset of N=1560 images and an inter-rater variability experiment for several processing pipelines.
Results: The experiments show mostly high agreement among raters and slight differences with the automated QC method.
Conclusion: While researchers must spend time on robust visual QC of data, there are mechanisms by which the process can be streamlined and efficient.