Phenotype discovery of traumatic brain injury segmentations from heterogeneous multi-site data
Adam M. Saunders, Michael E. Kim, Gaurav Rudravaram, Lucas W. Remedios, Chloe Cho, Elyssa M. McMaster, Daniel R. Gillis, Yihao Liu, Lianrui Zuo, Bennett A. Landman, and Tonia S. Rex. Phenotype discovery of traumatic brain injury segmentations from heterogeneous multi-site data. Accepted to SPIE Medical Imaging: Image Processing, February 2026. https://arxiv.org/abs/2511.03767
Abstract
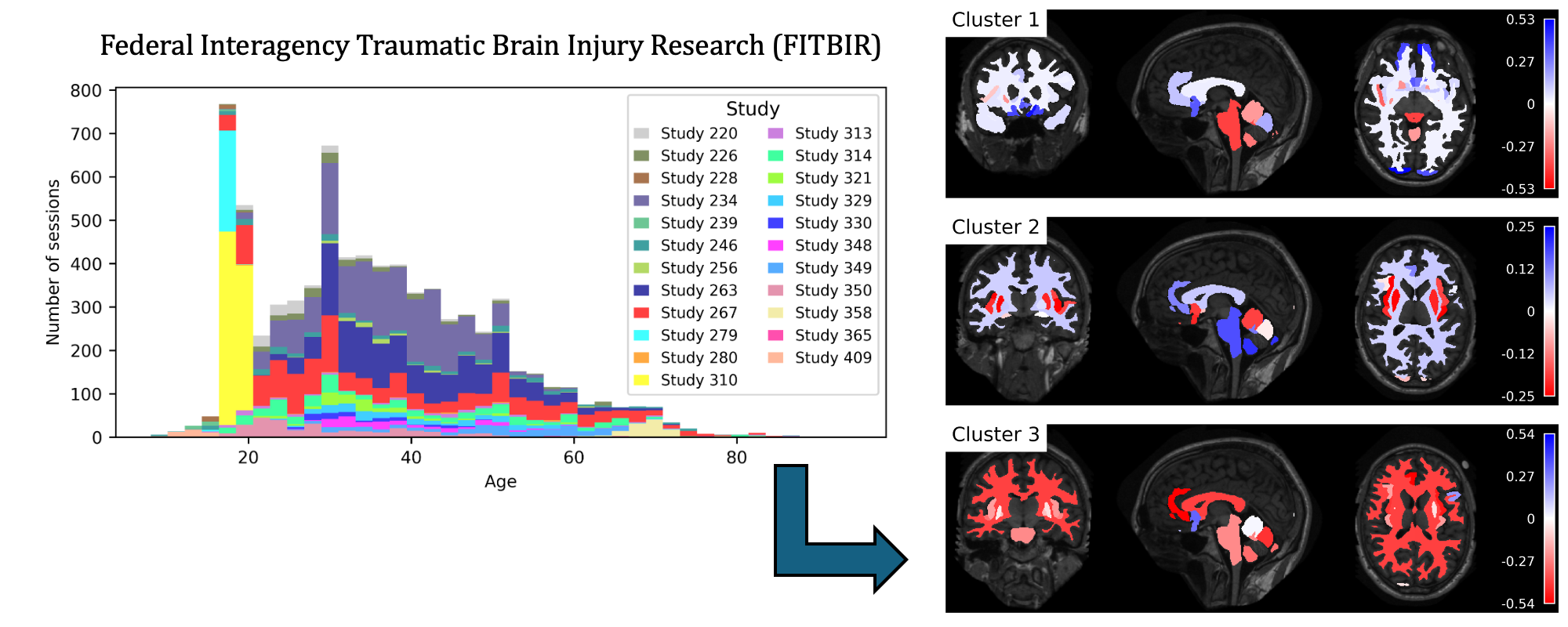
Traumatic brain injury (TBI) is intrinsically heterogeneous, and typical clinical outcome measures like the Glasgow Coma Scale complicate this diversity. The large variability in severity and patient outcomes render it difficult to link structural damage to functional deficits. The Federal Interagency Traumatic Brain Injury Research (FITBIR) repository contains large-scale multi-site magnetic resonance imaging data of varying resolutions and acquisition parameters (25 shared studies with 7,693 sessions that have age, sex and TBI status defined – 5,811 TBI and 1,882 controls). To reveal shared pathways of injury of TBI through imaging, we analyzed T1-weighted images from these sessions by first harmonizing to a local dataset and segmenting 132 regions of interest (ROIs) in the brain. After running quality assurance, calculating the volumes of the ROIs, and removing outliers, we calculated the z-scores of volumes for all participants relative to the mean and standard deviation of the controls. We regressed out sex, age, and total brain volume with a multivariate linear regression, and we found significant differences in 37 ROIs between subjects with TBI and controls (p < 0.05 with independent t-tests with false discovery rate correction). We found that differences originated in 1) the brainstem, occipital pole and structures posterior to the orbit, 2) subcortical gray matter and insular cortex, and 3) cerebral and cerebellar white matter using independent component analysis and clustering the component loadings of those with TBI.