Automatically identifying white matter tracts using cortical labels
A. Bogovic, A. Carass, J. Wan, B. A. Landman, and J. L. Prince. “Automatically identifying white matter tracts using cortical labels,” In Proceedings of the 2008 IEEE International Symposium on Biomedical Imaging, Paris, France, May 2008 PMC2812932
Full Text: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2812932/
Abstract
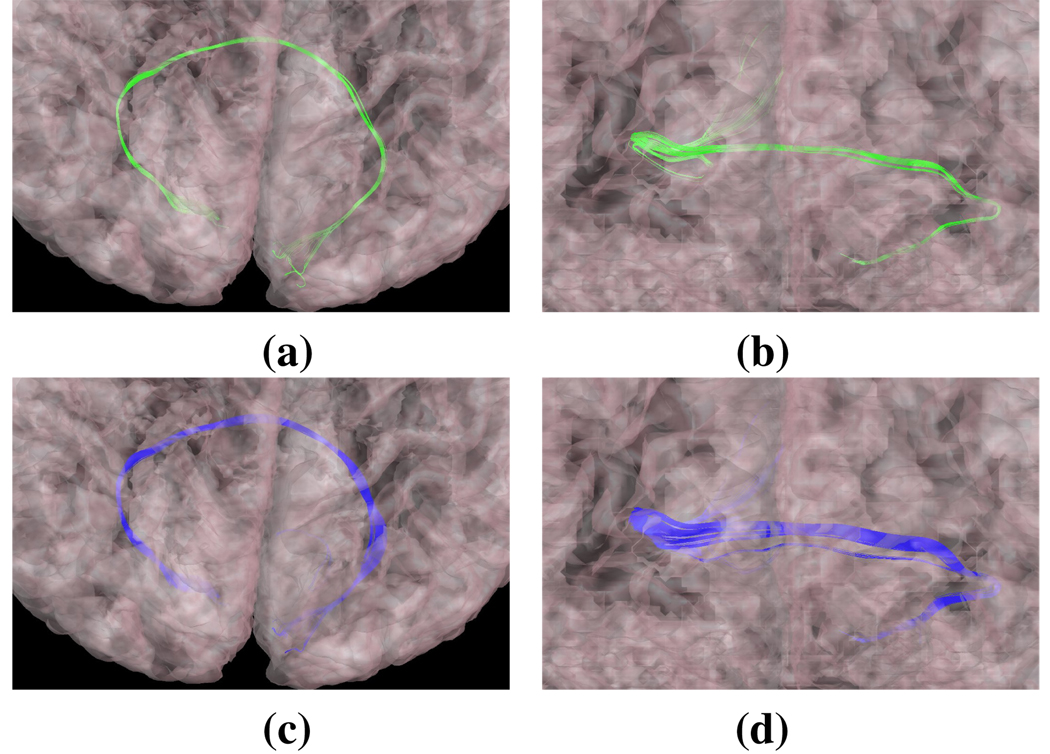
Diffusion tensor imaging (DTI) has become a standard clinical procedure in assessing the health of white matter in the brain. Tractography, the tracing of individual fibers in the brain using DTI data, has begun to play a more central role in neuroscience research, particularly in understanding the relationships between brain connectivity and behavior. The measuring of features related to bundles of fibers, i.e., tracts or fasciculi, is currently problematic because of the need for manual interaction. This article presents an algorithm for the automatic identification of selected white matter tracts. It extracts fibers using the FACT algorithm and finds cortical gyral labels using a multi-atlas deformable registration scheme. Tracts are identified as the fibers passing between selected cortical labels. The quality of automatic labels are compared both visually and numerically against a well-accepted manual approach. The automatic approach is shown to be more consistent with conventional definitions of tracts and more repeatable on separate scans of the same subject.