Biological Parametric Mapping Accounting for Random Regressors with Regression Calibration and Model II Regression
Xue Yang, Carolyn B. Lauzon, Ciprian Crainiceanu, Brian Caffo, Susan M. Resnick, Bennett A. Landman. “Biological Parametric Mapping Accounting for Random Regressors with Regression Calibration and Model II Regression.” NeuroImage. 2012 Sep;62(3):1761-8. PMC22609453
Full text: https://www.ncbi.nlm.nih.gov/pubmed/22609453
Abstract
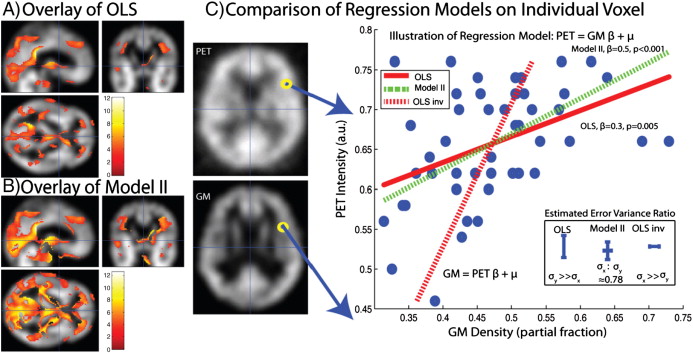
Massively univariate regression and inference in the form of statistical parametric mapping have transformed the way in which multi-dimensional imaging data are studied. In functional and structural neuroimaging, the de facto standard “design matrix”-based general linear regression model and its multi-level cousins have enabled investigation of the biological basis of the human brain. With modern study designs, it is possible to acquire multi-modal three-dimensional assessments of the same individuals — e.g., structural, functional and quantitative magnetic resonance imaging, alongside functional and ligand binding maps with positron emission tomography. Largely, current statistical methods in the imaging community assume that the regressors are non-random. For more realistic multi-parametric assessment (e.g., voxel-wise modeling), distributional consideration of all observations is appropriate. Herein, we discuss two unified regression and inference approaches, model II regression and regression calibration, for use in massively univariate inference with imaging data. These methods use the design matrix paradigm and account for both random and non-random imaging regressors. We characterize these methods in simulation and illustrate their use on an empirical dataset. Both methods have been made readily available as a toolbox plug-in for the SPM software