Efficient Abdominal Segmentation on Clinically Acquired CT with SIMPLE Context Learning
Zhoubing Xu, Ryan P. Burke, Christopher P. Lee, Rebeccah B. Baucom, Benjamin K. Poulose, Richard G. Abramson, Bennett A. Landman. “Efficient Abdominal Segmentation on Clinically Acquired CT with SIMPLE Context Learning.” In Proceedings of the SPIE Medical Imaging Conference. Orlando, Florida, February 2015. †
Full text: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4405802/
Abstract
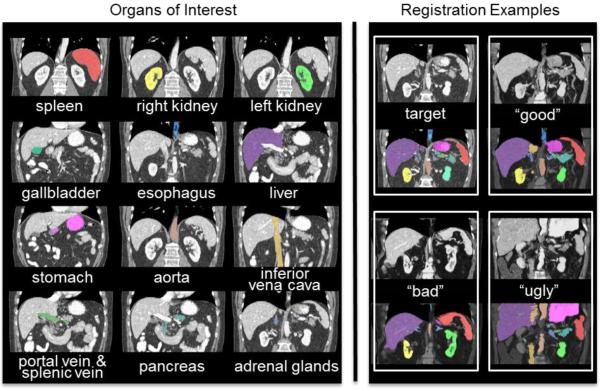
Abdominal segmentation on clinically acquired computed tomography (CT) has been a challenging problem given the inter-subject variance of human abdomens and complex 3-D relationships among organs. Multi-atlas segmentation (MAS) provides a potentially robust solution by leveraging label atlases via image registration and statistical fusion. We posit that the efficiency of atlas selection requires further exploration in the context of substantial registration errors. The selective and iterative method for performance level estimation (SIMPLE) method is a MAS technique integrating atlas selection and label fusion that has proven effective for prostate radiotherapy planning. Herein, we revisit atlas selection and fusion techniques for segmenting 12 abdominal structures using clinically acquired CT. Using a re-derived SIMPLE algorithm, we show that performance on multi-organ classification can be improved by accounting for exogenous information through Bayesian priors (so called context learning). These innovations are integrated with the joint label fusion (JLF) approach to reduce the impact of correlated errors among selected atlases for each organ, and a graph cut technique is used to regularize the combined segmentation. In a study of 100 subjects, the proposed method outperformed other comparable MAS approaches, including majority vote, SIMPLE, JLF, and the Wolz locally weighted vote technique. The proposed technique provides consistent improvement over state-of-the-art approaches (median improvement of 7.0% and 16.2% in DSC over JLF and Wolz, respectively) and moves toward efficient segmentation of large-scale clinically acquired CT data for biomarker screening, surgical navigation, and data mining.