Cloud Engineering Principles and Technology Enablers for Medical Image Processing-as-a-Service
Shunxing Bao, Andrew Plassard, Bennett Landman and Aniruddha Gokhale. “Cloud Engineering Principles and Technology Enablers for Medical Image Processing-as-a-Service.” IEEE International Conference on Cloud Engineering (IC2E), Vancouver, Canada, April 2017.
Full text: NIHMSID
Abstract
Traditional in-house, laboratory-based medical imaging studies use hierarchical data structures (e.g., NFS file stores) or databases (e.g., COINS, XNAT) for storage and retrieval. The resulting performance from these approaches is, however, impeded by standard network switches since they can saturate network bandwidth during transfer from storage to processing nodes for even moderate-sized studies. To that end, a cloud-based “medical image processing-as-a-service” offers promise in utilizing the ecosystem of Apache Hadoop, which is a flexible framework providing distributed, scalable, fault tolerant storage and parallel computational modules, and HBase, which is a NoSQL database built atop Hadoop’s distributed file system. Despite this promise, HBase’s load distribution strategy of region split and merge is detrimental to the hierarchical organization of imaging data (e.g., project, subject, session, scan, slice).
This paper makes two contributions to address these concerns by describing key cloud engineering principles and technology enhancements we made to the Apache Hadoop ecosystem for medical imaging applications. First, we propose a row-key design for HBase, which is a necessary step that is driven by the hierarchical organization of imaging data. Second, we propose a novel data allocation policy within HBase to strongly enforce collocation of hierarchically related imaging data. The proposed enhancements accelerate data processing by minimizing network usage and localizing processing to machines where the data already exist. Moreover, our approach is amenable to the traditional scan, subject, and project-level analysis procedures, and is compatible with standard command line/scriptable image processing software. Experimental results for an illustrative sample of imaging data reveals that our new HBase policy results in a three-fold time improvement in conversion of classic DICOM to NiFTI file formats when compared with the default HBase region split policy, and nearly a nine-fold improvement over a commonly available network file system (NFS) approach even for relatively small file sets. Moreover, file access latency is lower than network attached storage.
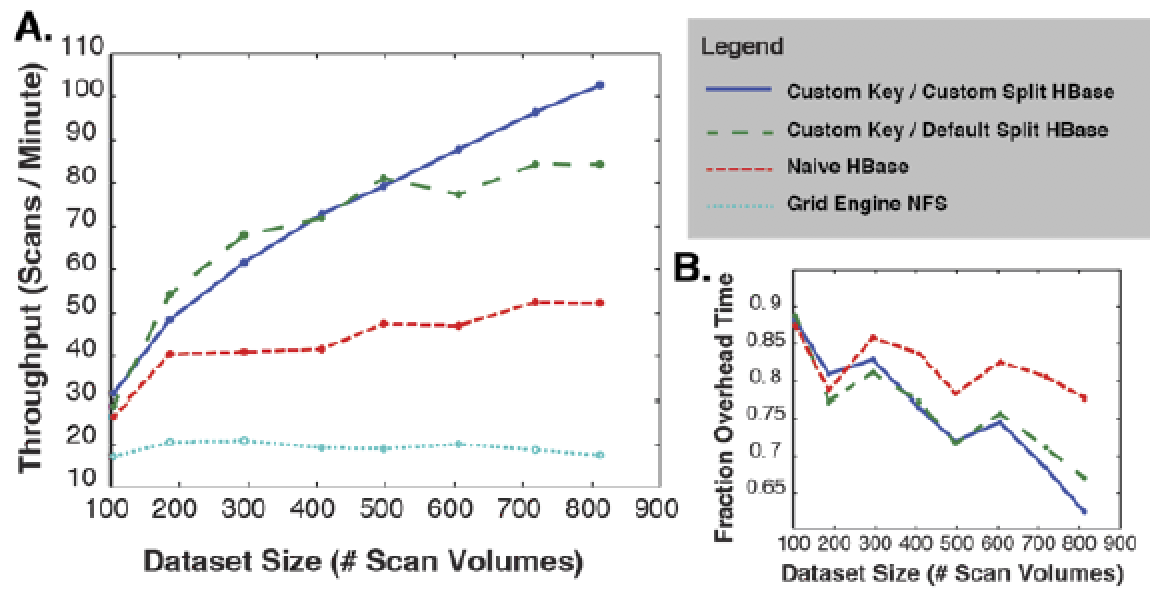
number of datasets processed per minute by each of the scenarios as a function
of the number of datasets selected for processing. (B) shows the fraction of
time spent on overhead relative to the number of datasets.