Harmonization of white and gray matter features in diffusion microarchitecture for cross sectional studies
Prasanna Parvathaneni, Shunxing Bao , Allison Hainline , Yuankai Huo , Kurt G. Schilling , Hakmook Kang , Owen Williams , Neil D. Woodward , Susan M. Resnick , David H. Zald , Ilwoo Lyu , Bennett A. Landman “Harmonization of white and gray matter features in diffusion microarchitecture for cross sectional studies.” In International Conference on Clinical and Medical Image Analysis 2018 (ICCMIA’18) – Accepted
Abstract
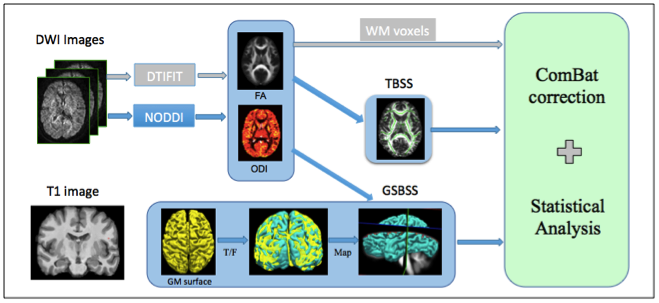
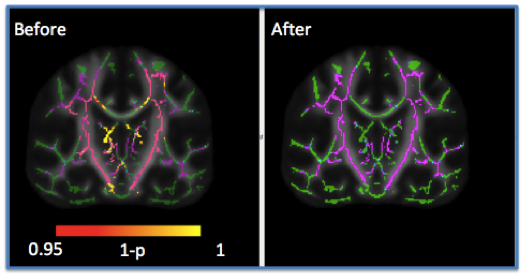
Understanding of the specific processes involved in development of brain microarchitecture and how these are altered by genetic, cognitive, or environmental factors is a key to more effective and efficient interventions. With the increasing number of publicly available neuroimaging databases, there is an opportunity to combine large-scale imaging studies to increase the power of statistical analyses to test common biological hypotheses. However, cross-study, cross-sectional analyses are confounded by inter-scanner variability that can cause both spatially and anatomically dependent signal aberrations. In particular, scanner related differences in the diffusion weighted magnetic resonance imaging (DWMRI) signal are substantially different in tissue types like cortical/subcortical gray matter and white matter. Recent studies have shown effective harmonization using the ComBat technique (adopted from genomics) to address inter-site variability in white matter using diffusion tensor imaging (DTI) microstructure indices like fractional anisotropy (FA) or mean diffusivity (MD). In this study, we propose 1) to apply the correction at voxel level using tract based spatial statistics (TBSS) in FA, 2) to correct variability across scanners with different gradient strengths in DTI and 3) to apply the ComBat technique to advanced DWMRI models, i.e., Neurite Orientation Dispersion and Density Imaging (NODDI), to correct for variability of orientation dispersion index (ODI) in gray matter using gray matter based spatial statistics tool (GSBSS). We show that the biological variability with age is retained or improved while correcting for variability across scanners.
Keywords: Harmonization, NODDI, brain microstructure, Gray matter surface based analysis