Can increased spatial resolution solve the crossing fiber problem for diffusion MRI?
Kurt G Schilling, Yurui Gao, Vaibhav Janve, Iwona Stepniewska, Bennett A Landman, Adam W Anderson. “Can increased spatial resolution solve the crossing fiber problem for diffusion MRI?”. NMR in Biomedicine. (2017) 30(12),e3787. https://doi.org/10.1002/nbm.3787.
Full text: https://www.ncbi.nlm.nih.gov/pubmed/?term=Can+increased+spatial+resolution+solve+the+crossing+fiber+problem+for+diffusion+MRI%3F
Abstract
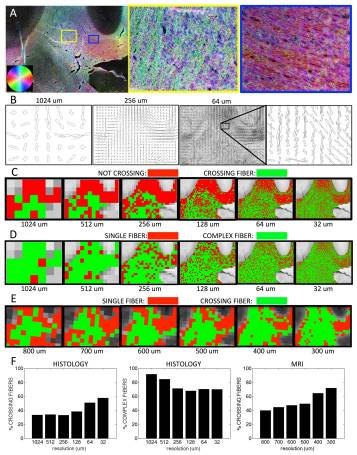
It is now widely recognized that voxels with crossing fibers or complex geometrical configurations present a challenge for diffusion MRI(dMRI) reconstruction and fiber tracking, as well as microstructural modeling of brain tissues. This “crossing fiber” problem has been estimated to affect anywhere from 30% to as many as 90% of white matter voxels, and it is often assumed that increasing spatialresolution will decrease the prevalence of voxels containing multiple fiber populations. The aim of this study is to estimate the extent of the crossing fiber problem as we progressively increase the spatial resolution, with the goal of determining whether it is possible to mitigate this problem with higher resolution spatial sampling. This is accomplished using ex vivo MRI data of the macaque brain, followed by histological analysis of the same specimen to validate these measurements, as well as to extend this analysis to resolutions not yet achievable in practice with MRI. In both dMRI and histology, we find unexpected results: the prevalence of crossing fibers increases as we increase spatial resolution. The problem of crossing fibers appears to be a fundamental limitation of dMRI associated with the complexity of brain tissue, rather than a technical problem that can be overcome with advances such as higher fields and stronger gradients.
Keywords:
crossing fibers; diffusion MRI; histology; macaque; validation; white matter