Confirmation of a Gyral Bias in Diffusion MRI Fiber Tractography
Kurt G Schilling, Yurui Gao, Iwona Stepniewska, Bennett A. Landman, and Adam W Anderson. “Confirmation of a Gyral Bias in Diffusion MRI Fiber Tractography”. Human Brain Mapping. 2018 Mar;39(3):1449-1466. doi: 10.1002/hbm.23936.
Full text: https://www.ncbi.nlm.nih.gov/pubmed/?term=Confirmation+of+a+Gyral+Bias+in+Diffusion+MRI+Fiber+Tractography
Abstract
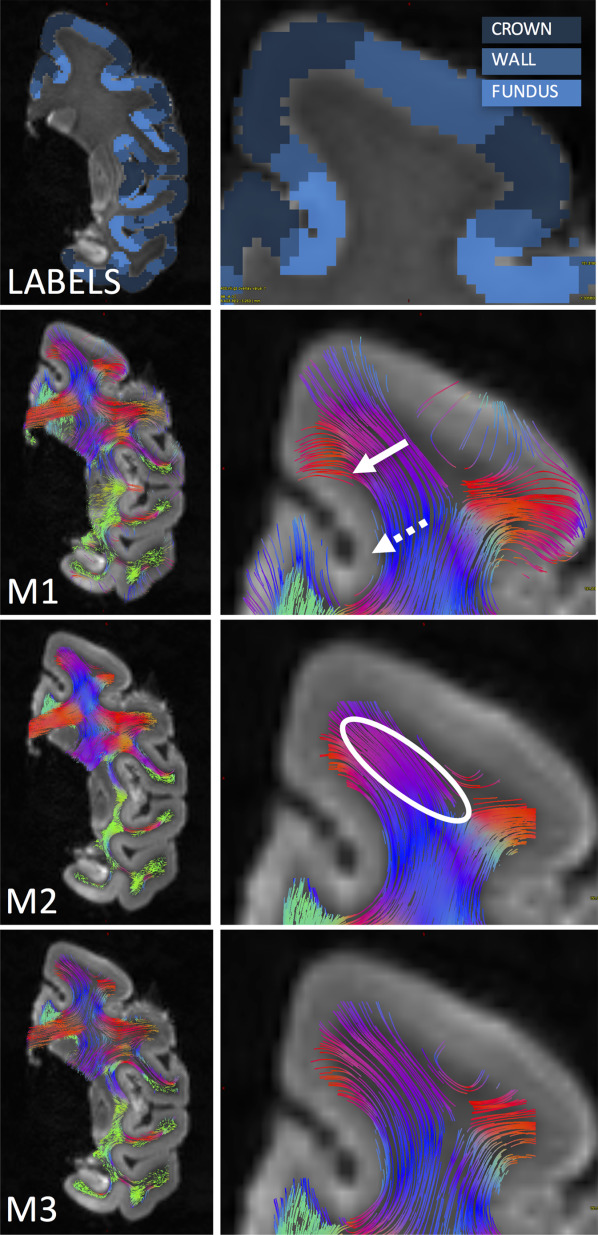
Diffusion MRI fiber tractography has been increasingly used to map the structural connectivity of the human brain. However, this technique is not without limitations; for example, there is a growing concern over anatomically correlated bias in tractography findings. In this study, we demonstrate that there is a bias for fiber tracking algorithms to terminate preferentially on gyral crowns, rather than the banks of sulci. We investigate this issue by comparing diffusion MRI (dMRI) tractography with equivalent measures made on myelin-stained histological sections. We begin by investigating the orientation and trajectories of axons near the white matter/gray matter boundary, and the density of axons entering the cortex at different locations along gyral blades. These results are compared with dMRI orientations and tract densities at the same locations, where we find a significant gyral bias in many gyral blades across the brain. This effect is shown for a range of tracking algorithms, both deterministic and probabilistic, and multiple diffusion models, including the diffusion tensor and a high angular resolution diffusion imaging technique. Additionally, the gyral bias occurs for a range of diffusionweightings, and even for very high-resolution datasets. The bias could significantly affect connectivity results using the current generation of tracking algorithms.
Keywords:
brain; connectivity; diffusion MRI; gyral bias; histology; tractography; validation