Histological Validation of Diffusion MRI Fiber Orientation Distributions and Dispersion
Kurt G Schilling, Vaibhav Janve; Yurui Gao; Iwona Stepniewska; Bennett A Landman; Adam W Anderson. “Histological Validation of Diffusion MRI Fiber Orientation Distributions and Dispersion”. NeuroImage. 2018 Jan 15;165:200-221. doi: 10.1016/j.neuroimage.2017.10.046.
Full text: https://www.ncbi.nlm.nih.gov/pubmed/?term=Histological+Validation+of+Diffusion+MRI+Fiber+Orientation+Distributions+and+Dispersion
Abstract
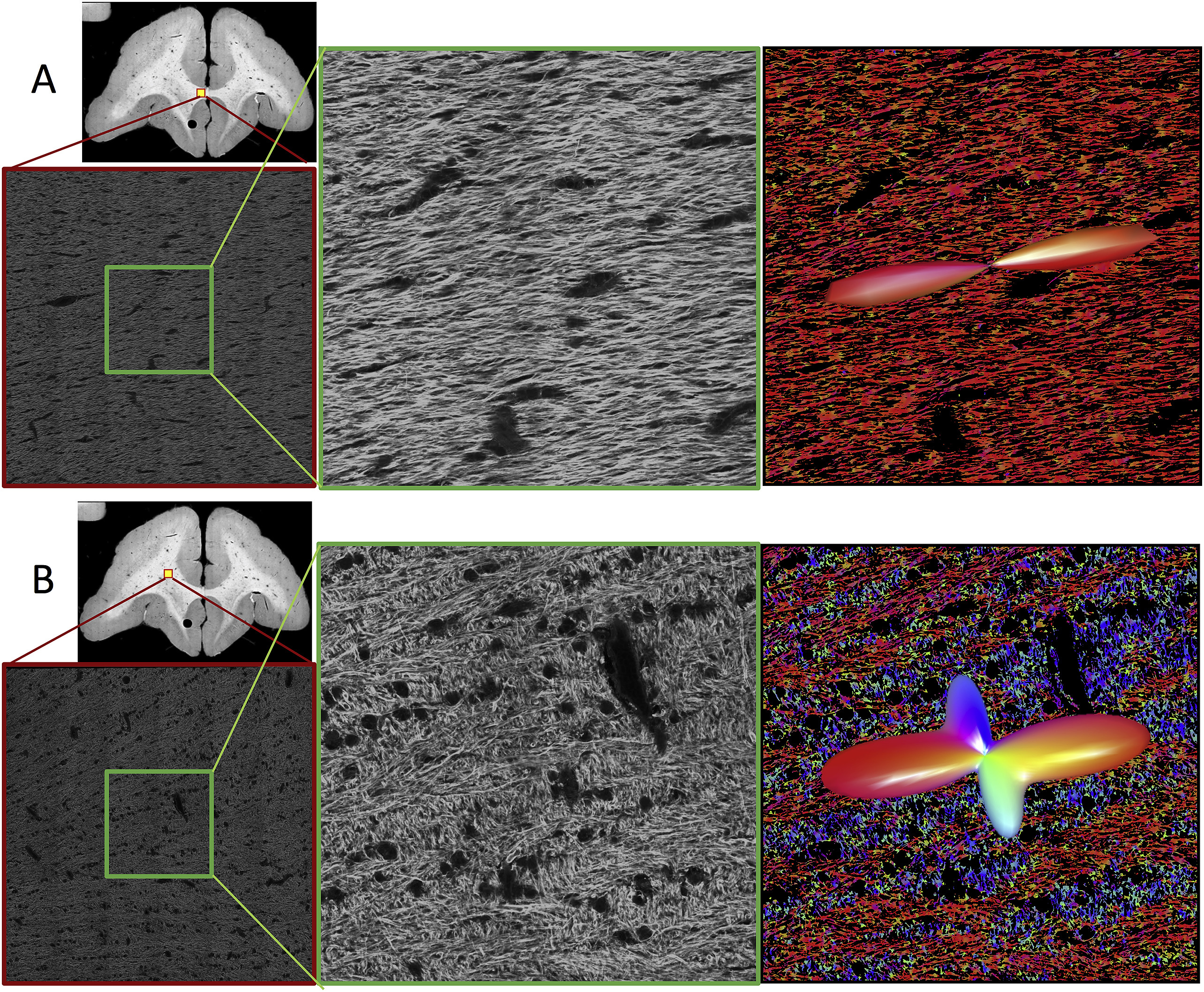
Diffusion magnetic resonance imaging (dMRI) is widely used to probe tissue microstructure, and is currently the only non-invasive way to measure the brain’s fiber architecture. While a large number of approaches to recover the intra-voxel fiber structure have been utilized in the scientific community, a direct, 3D, quantitative validation of these methods against relevant histological fiber geometries is lacking. In this study, we investigate how well different high angular resolution diffusion imaging (HARDI) models and reconstruction methods predict the ground-truth histologically defined fiber orientation distribution (FOD), as well as investigate their behavior over a range of physical and experimental conditions. The dMRI methods tested include constrained spherical deconvolution (CSD), Q-ball imaging (QBI), diffusion orientation transform (DOT), persistent angular structure (PAS), and neurite orientation dispersion and density imaging (NODDI) methods. Evaluation criteria focus on overall agreement in FOD shape, correct assessment of the number of fiber populations, and angular accuracy in orientation. In addition, we make comparisons of the histological orientation dispersion with the fiber spread determined from the dMRI methods. As a general result, no HARDI method outperformed others in all quality criteria, with many showing tradeoffs in reconstruction accuracy. All reconstruction techniques describe the overall continuous angular structure of the histologicalFOD quite well, with good to moderate correlation (median angular correlation coefficient > 0.70) in both single- and multiple-fibervoxels. However, no method is consistently successful at extracting discrete measures of the number and orientations of FOD peaks. The major inaccuracies of all techniques tend to be in extracting local maxima of the FOD, resulting in either false positive or false negative peaks. Median angular errors are ∼10° for the primary fiber direction and ∼20° for the secondary fiber, if present. For most methods, these results did not vary strongly over a wide range of acquisition parameters (number of diffusion weighting directions and b value). Regardless of acquisition parameters, all methods show improved successes at resolving multiple fiber compartments in a voxel when fiber populations cross at near-orthogonal angles, with no method adequately capturing low to moderate angle (<60°) crossing fibers. Finally, most methods are limited in their ability to capture orientation dispersion, resulting in low to moderate, yet statistically significant, correlation with histologically-derived dispersion with both HARDI and NODDI methodologies. Together, these results provide quantitative measures of the reliability and limitations of dMRI reconstruction methods and can be used to identify relative advantages of competing approaches as well as potential strategies for improving accuracy.
Keywords:
Diffusion magnetic resonance imaging; Dispersion; Fiber orientation distribution; HARDI; Histology; Reconstruction; Validation