Generalizing Deep Whole Brain Segmentation for Pediatric and Post-Contrast MRI with Augmented Transfer Learning
Bermudez, C., Blaber, J., Remedios, S.W., Reynolds, J.E., Lebel, C., McHugo, M., Heckers, S., Huo, Y., Landman, B.A. Generalizing Deep Whole Brain Segmentation for Pediatric and Post-Constrast MRI with Augmented Transfer Learning. SPIE Medical Imaging: Image Processing 2020. Houston, TX.
Abstract
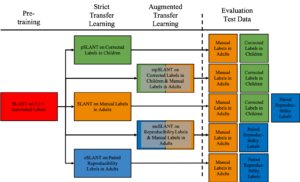
Generalizability is an important problem in deep neural networks, especially in the context of the variability of data acquisition in clinical magnetic resonance imaging (MRI). Recently, the Spatially Localized Atlas Network Tiles (SLANT) approach has been shown to effectively segment whole brain non-contrast T1w MRI with 132 volumetric labels. Enhancing generalizability of SLANT would enable broader application of volumetric assessment in multi-site studies. Transfer learning (TL) is commonly to update neural network weights for local factors; yet, it is commonly recognized to risk degradation of performance on the original validation/test cohorts. Here, we explore TL by data augmentation to address these concerns in the context of adapting SLANT to anatomical variation (e.g., adults versus children) and scanning protocol (e.g., non-contrast research T1w MRI versus contrast-enhanced clinical T1w MRI). We consider two datasets: First, 30 T1w MRI of young children with manually corrected volumetric labels, and accuracy of automated segmentation defined relative to the manually provided truth. Second, 36 paired datasets of pre- and post-contrast clinically acquired T1w MRI, and accuracy of the post-contrast segmentations assessed relative to the pre-contrast automated assessment. For both studies, we augment the original TL step of SLANT with either only the new data or with both original and new data. Over baseline SLANT, both approaches yielded significantly improved performance (pediatric: 0.89 vs. 0.82 DSC, p<0.001; contrast: 0.80 vs 0.76, p<0.001 ). The performance on the original test set decreased with the new-data only transfer learning approach, so data augmentation was superior to strict transfer learning.