Using deep learning for a diffusion-based segmentation of the dentate nucleus and its benefits over atlas-based methods
Abstract
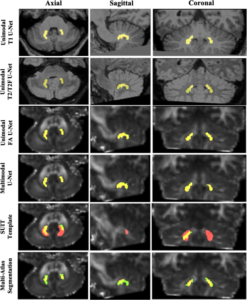
The dentate nucleus (DN) is a gray matter structure deep in the cerebellum involved in motor coordination, sensory input integration, executive planning, language, and visuospatial function. The DN is an emerging biomarker of disease, informing studies that advance pathophysiologic understanding of neurodegenerative and related disorders. The main challenge in defining the DN radiologically is that, like many deep gray matter structures, it has poor contrast in T1-weighted magnetic resonance (MR) images and therefore requires specialized MR acquisitions for visualization. Manual tracing of the DN across multiple acquisitions is resource-intensive and does not scale well to large datasets. We describe a technique that automatically segments the DN using deep learning (DL) on common imaging sequences, such as T1-weighted, T2-weighted, and diffusion MR imaging. We trained a DL algorithm that can automatically delineate the DN and provide an estimate of its volume. The automatic segmentation achieved higher agreement to the manual labels compared to template registration, which is the current common practice in DN segmentation or multiatlas segmentation of manual labels. Across all sequences, the FA maps achieved the highest mean Dice similarity coefficient (DSC) of 0.83 compared to T1 imaging (DSC = 0.76), T2 imaging (DSC = 0.79), or a multisequence approach (DSC = 0.80). A single atlas registration approach using the spatially unbiased atlas template of the cerebellum and brainstem template achieved a DSC of 0.23, and multi-atlas segmentation achieved a DSC of 0.33. Overall, we propose a method of delineating the DN on clinical imaging that can reproduce manual labels with higher accuracy than current atlas-based tools.