Distributed deep learning for robust multi-site segmentation of CT imaging after traumatic brain injury
Remedios, Samuel, et al. “Distributed deep learning for robust multi-site segmentation of CT imaging after traumatic brain injury.” Medical Imaging 2019: Image Processing. Vol. 10949. International Society for Optics and Photonics, 2019.
Full text: PubMed Link
Abstract
Machine learning models are becoming commonplace in the domain of medical imaging, and with these methods comes an ever-increasing need for more data. However, to preserve patient anonymity it is frequently impractical or prohibited to transfer protected health information (PHI) between institutions. Additionally, due to the nature of some studies, there may not be a large public dataset available on which to train models. To address this conundrum, we analyze the efficacy of transferring the model itself in lieu of data between different sites. By doing so we accomplish two goals: 1) the model gains access to training on a larger dataset that it could not normally obtain and 2) the model better generalizes, having trained on data from separate locations. In this paper, we implement multi-site learning with disparate datasets from the National Institutes of Health (NIH) and Vanderbilt University Medical Center (VUMC) without compromising PHI. Three neural networks are trained to convergence on a computed tomography (CT) brain hematoma segmentation task: one only with NIH data, one only with VUMC data, and one multi-site model alternating between NIH and VUMC data. Resultant lesion masks with the multi-site model attain an average Dice similarity coefficient of 0.64 and the automatically segmented hematoma volumes correlate to those done manually with a Pearson correlation coefficient of 0.87, corresponding to an 8% and 5% improvement, respectively, over the single-site model counterparts.
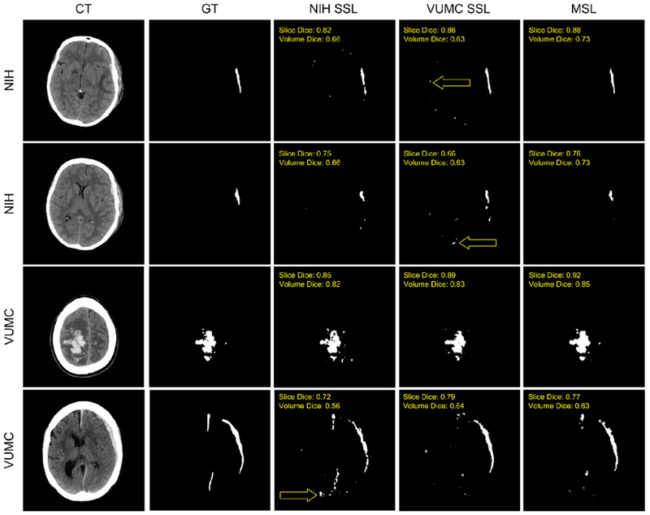
Examples of automatic segmentations are shown. From left to right, the images correspond to the CT, the manual “ground truth” (GT) segmentation, the NIF1 SSL, VUMC SSL, and MSL segmentations. Both the image volume and the specific image slice’s Dice coefficient are overlaid on that segmentation. Yellow arrows specify examples of false positives near the blood-brain barrier which were not present in the MSL segmentations.
Keywords: computed tomography (CT); deep learning; distributed; hematoma; lesion; multi-site; neural network; segmentation