Enabling Multi-shell b-Value Generalizability of Data-Driven Diffusion Models with Deep SHORE
Nath V, Lyu I, Schilling KG, Parvathaneni P, Hansen CB, Huo Y, Janve VA, Gao Y, Stepniewska I, Anderson AW, Landman BA. Enabling Multi-shell b-Value Generalizability of Data-Driven Diffusion Models with Deep SHORE. In International Conference on Medical Image Computing and Computer-Assisted Intervention 2019 Oct 13 (pp. 573-581). Springer, Cham.
Full text: https://arxiv.org/ftp/arxiv/papers/1907/1907.06319.pdf
Abstract
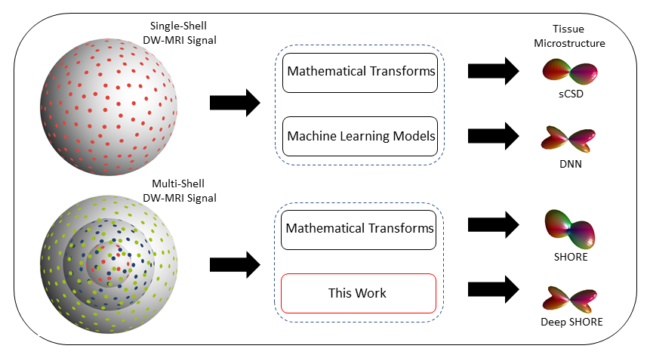
Intra-voxel models of the diffusion signal are essential for interpreting the organization of the tissue environment at the micrometer level with data at millimeter resolution. Recent advances in data-driven methods have enabled direct comparison and optimization of methods for in-vivo data with externally validated histological sections with both 2-D and 3-D histology. Yet, all existing methods
make limiting assumptions of either (1) model-based linkages between b-values or (2) limited associations with single shell data. We generalize prior deep learning models that used single shell spherical harmonic transforms to integrate the recently developed simple harmonic oscillator reconstruction (SHORE) basis. To enable learning on the SHORE manifold, we present an alternative formulation
of the fiber orientation distribution (FOD) object using the SHORE basis while representing the observed diffusion-weighted data in the SHORE basis. To ensure consistency of hyper-parameter optimization for SHORE, we present our DeepSHORE approach to learn on a data-optimized manifold. Deep SHORE is evaluated with eight-fold cross-validation of a preclinical MRI-histology data with four b-values. Generalizability of in-vivo human data is evaluated on two separate 3T MRI scanners. Specificity in terms of angular correlation (ACC) with the preclinical data improved on the single shell: 0.78 relative to 0.73 and 0.73, multi-shell: 0.80 relative to 0.74 (p < 0.001). In the in-vivo human data, Deep SHORE was more consistent across scanners with 0.63 relative to other multi-shell methods 0.39, 0.52 and 0.57 in terms of ACC. In conclusion, Deep SHORE is a promising method to enable data-driven learning with DW-MRI under conditions with varying b-values, number of diffusion shells, and gradient directions per shell.