Automatic Labeling of Cortical Sulci using Convolutional Neural Networks in a Developmental Cohort
Lingyan Hao, Shunxing Bao, Yucheng Tang, Riqiang Gao, Prasanna Parvathaneni, Jacob Miller, Willa Voorhies, Jewelia Yao, Silvia Bunge, Kevin Weiner, Bennett Landman, Ilwoo Lyu. “Automatic Labeling of Cortical Sulci using Convolutional Neural Networks in a Developmental Cohort”. IEEE International Symposium on Biomedical Imaging (ISBI) 2020, IEEE, 412-415, Iowa City, Iowa, USA, 2020.
[Full text][Code]
Abstract
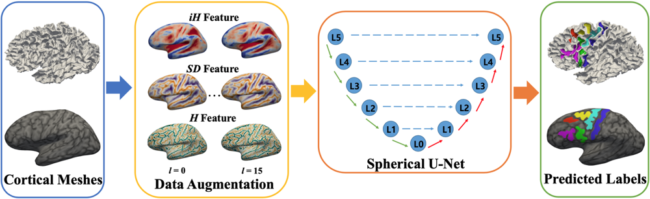
In this paper, we present the automatic labeling framework for sulci in the human lateral prefrontal cortex (PFC). We adapt an existing spherical U -Net architecture with our recent surface data augmentation technique to improve the sulcal labeling accuracy in a developmental cohort. Specifically, our framework consists of the following key components: (1) augmented geometrical features being generated during cortical surface registration, (2) spherical U -Net architecture to efficiently fit the augmented features, and (3) post-refinement of sulcal labeling by optimizing spatial coherence via a graph cut technique. We validate our method on 30 healthy subjects with manual labeling of sulcal regions within PFC. In the experiments, we demonstrate significantly improved labeling performance (0.7749) in mean Dice overlap compared to that of multi-atlas (0.6410) and standard spherical U-Net (0.7011) approaches, respectively (p <; 0.05). Additionally, the proposed method achieves a full set of sulcal labels in 20 seconds in this developmental cohort.